Abstract
G protein-coupled receptors are cell-surface seven-helical membrane proteins that undergo conformational changes on activation. The mammalian photoreceptor, rhodopsin, is the best-studied member of this superfamily. Here, we provide the first evidence that activation in rhodopsin may involve differential dynamic properties of side-chain versus backbone atoms. High-resolution NMR studies of α-15N-labeled receptor revealed large backbone motions in the inactive dark state. In contrast, indole side-chain 15N groups of tryptophans showed well resolved, equally intense NMR signals, suggesting restriction to a single specific conformation.
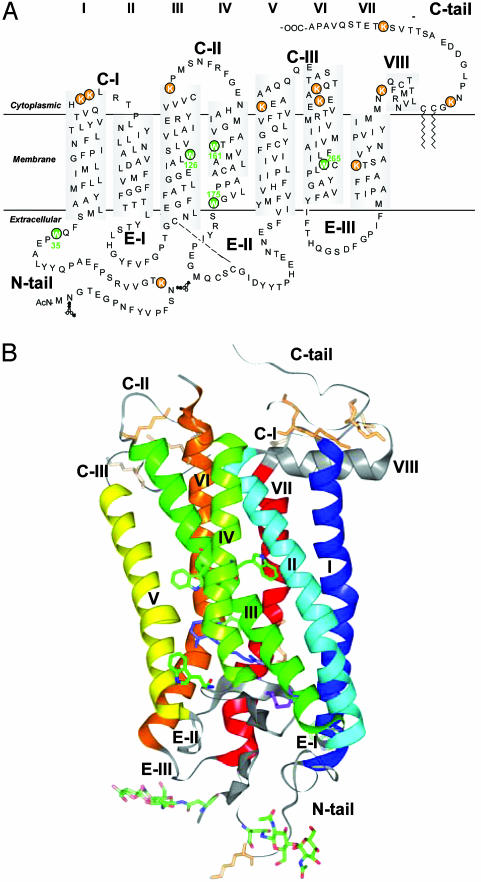
Rhodopsin, the vertebrate dim-light photoreceptor, is the prototypic member of the largest known superfamily of cell-surface receptors, the G protein-coupled receptors (GPCRs). These receptors perform extremely diverse functions that include responses to light, odorant molecules, neurotransmitters, hormones, and a variety of other signals. GPCRs all contain seven transmembrane (TM) helices (Fig. 1A) as shown by structural studies of rhodopsin by cryoelectron microscopy (1) and confirmed more recently in a 3D x-ray crystal structure in the inactive state (2–4) (Fig. 1B). The chromophore in rhodopsin, 11-cis-retinal, is covalently bound to the protein via a protonated Schiff base to the ε-amino group of Lys-296 located in TM7. Capture of a photon by rhodopsin results in isomerization of retinal to the all-trans form, which triggers a series of transient changes in the protein culminating in the active conformation. The application of NMR spectroscopy to the study of rhodopsin, the best studied member of GPCRs, has begun only recently (5). This has been made possible by the development of high level expression of a variety of rhodopsin mutants from the corresponding genes (6, 7). Previously, we reported on NMR spectroscopy of 19F-labeled rhodopsin mutants in detergent micelles. Distinct chemical shifts in the dark and changes in them on activation of rhodopsin were observed for 19F labels placed at different positions in the cytoplasmic domain (8). Further, nuclear Overhauser effects were observed between certain pairs of 19F labels (9). More recently, study of α-15N-lysine-labeled rhodopsin by high-resolution heteronuclear NMR spectroscopy (10) showed that whereas motions were detected on the nanosecond time scale in the single lysine located in the C terminus (Fig. 1 A), lysines present elsewhere in rhodopsin showed motions in micro- to millisecond time scales. In addition, more than the expected number of signals with variable intensities was detected. This finding indicated multiple backbone conformations. Here, we have studied α,ε-15N-tryptophan-labeled rhodopsin. In contrast with lysines in rhodopsin that are located mostly in the cytoplasmic domain (Fig. 1, orange), four of the five tryptophans are in the TM domain, and one is in the extracellular domain (Fig. 1, green). Here, we report that the indole side chains show the expected number of clearly defined signals with relatively homogeneous intensity; however, the backbone amide shows a larger than expected number of signals with varying intensities. These results indicate that whereas the indole side chains have one specific conformation, there are conformational fluctuations on the micro- to millisecond time scale in the backbone amide groups, as found previously for α-15N-lysine-labeled rhodopsin (10).
Fig. 1.
Secondary (A) and three-dimensional (B) structure model of bovine rhodopsin based on x-ray crystallographic analysis (2). The positions of the five tryptophan residues are indicated in green in comparison to the positions of lysine residues (orange).
Methods
Amino acids are numbered according to the general GPCR numbering scheme proposed by (11) and by their position in the rhodopsin sequence. All experimental methods have been described (10) with the following exceptions.
NMR Spectroscopy. NMR spectra were obtained at a spectrometer 1H frequency of ≈800 MHz by using a Bruker (Billerica, MA) spectrometer. Data were acquired and analyzed by using Bruker uxnmr v.2.1 software. Data processing and analysis was also carried out by using felix v.98.0.
Expression and Purification of Rhodopsin Containing Isotope-Labeled Amino Acids in HEK 293S Cell Lines. The preparation of HEK 293S stable cell lines containing the opsin gene for WT was described (6). Stable cell lines were grown in media with composition according to DMEM formulation, prepared from individual components. Thus, all solutions were prepared as 100× concentrated stock solutions, except glucose, NaCl, glutamine, and α,ε-15N-tryptophan, which were added as solids. The glutamine concentration was initially that of one-half of the DMEM formulation, and the same amount was then added on day 5 or 6, together with 6 ml of 20% (wt/vol) glucose and 4 ml of 8% (wt/vol) NaHCO3/5 mM sodium butyrate additions as described (7). To remove unlabeled amino acids from FBS, it was dialyzed three times against 10 liters of buffer A at 4°C with a tubing cut off of 1 kDa as described (12). Immunoaffinity purification of rhodopsin and preparation of samples for NMR spectroscopy was as described (10).
Prediction of Chemical Shift Values Based on the Rhodopsin Crystal Structure. 15N chemical shifts were calculated by using the programs shifts (13) and proshift (14). 1H chemical shifts were calculated by using the programs proshift (14) and shiftcalc (15). All programs were applied to both chains in the two available crystal structures with Protein Data Bank (PDB) ID codes 1F88 (resolution 2.8 Å) and 1L9H (resolution 2.6 Å), and the eight values were averaged for each of the five tryptophans in both structures (2, 4).
Results
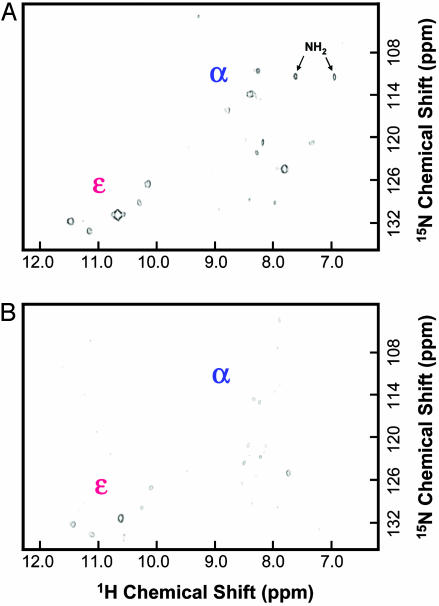
Conventional and Transverse Relaxation Optimized Spectroscopy (TROSY)-Type 2D Heteronuclear Single Quantum Correlation (HSQC) Spectra of α/ε-15N-Tryptophan-Labeled Rhodopsin. A 2D 1H,15N-HSQC spectrum of α- and ε-15N-tryptophan-labeled rhodopsin was recorded at 800 MHz (Fig. 2A). In the backbone region, proton chemical shifts from ≈7 ppm to 9 ppm, a total of 10 amide signals with variable intensities were observed instead of the five expected. This heterogeneity in the backbone region is unlikely to be due to deamination and scrambling of the 15N label into the α position of other amino acids as we have shown previously by using mass spectrometry (10). The amino signals indicated by arrow in Fig. 2 A presumably originated from the natural abundance signal from amino groups in rhodopsin because these were also observed in α-15N-lysine-labeled rhodopsin, and there was no evidence for scrambling of 15N to side-chain amino groups either (10). TROSY resulted in decreases in signal intensities overall, causing lowering of signal intensity below detection limit for some of the signals (Fig. 2B). On the other hand, analysis of the indole side-chain region, from ≈10 ppm to 12 ppm, showed the expected five signals. Their intensities were similar, except for a stronger intensity for the central signal at ≈10.5 ppm. This signal most likely corresponds to Trp-35(1.30) (Fig. 1), the single tryptophan in the extracellular domain in rhodopsin. The effect of TROSY in this region of the HSQC spectrum (Fig. 2B) was a smaller decrease in signal intensity than that observed for the backbone resonances.
Fig. 2.
Two-dimensional HSQC spectra of α/ε-15N-tryptophan WT rhodopsin at 800 MHz. (A) Conventional HSQC spectra of 0.4 mM rhodopsin in dodecyl maltoside at 37°C. (B) TROSY-HSQC spectra. Tryptophan backbone regions of the spectra are labeled “α,” and side-chain regions are labeled “ε.” Background signals from amino groups are indicated by arrow and label “NH2.”
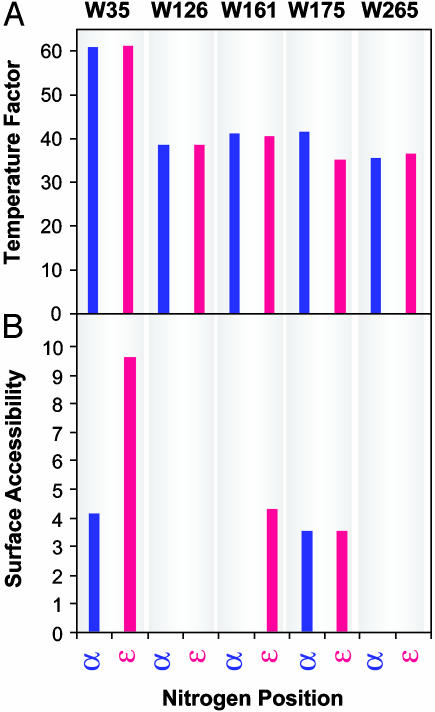
Comparison of Tryptophan Temperature Factors in the Rhodopsin Crystal Structure. The temperature factors of side-chain and backbone nitrogen atoms for all of the five tryptophan residues were extracted from the rhodopsin crystal structure [PDB ID code 1F88 (2)] and are shown in Fig. 3A. The four TM tryptophan residues show similar temperature factors, ≈35, for both nitrogen atoms in each residue. On the other hand, Trp-35(1.30) in the extracellular domain shows temperature factors significantly elevated with values for both nitrogens of ≈60. Thus, within any given tryptophan, the temperature factors of the two nitrogens are essentially identical.
Fig. 3.
Properties of backbone (α) and side-chain (ε) nitrogen atoms of tryptophan residues in the crystal structure. (A) Temperature factors. The factors were obtained from the published PDB ID code 1F88 (2). (B) Surface-accessible area. The surface-accessible area was determined by using Connolly surface calculations by insight ii software.
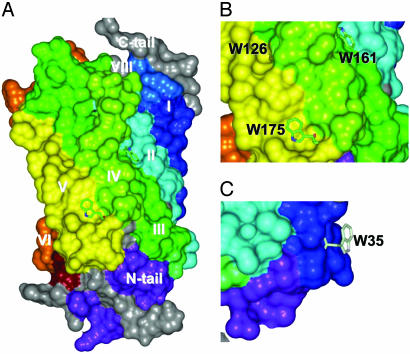
Solvent Accessibilities and Steric Restrictions of Nitrogens Vary With the Position of Tryptophan Residues but Not Within α or ε-Positions in the Same Tryptophan. The conformational restriction of indole side chains relative to the fluctuations in the amide groups is opposite to that commonly observed in proteins, especially on surfaces. Unless restricted by tertiary contacts, side chains can have more freedom than the backbone. Thus, protein NMR spectra usually indicate higher degrees of conformational averaging in the side chains than in the backbone. Therefore, we inspected the local environment of the tryptophan residues in the crystal structure model for differences in the immediate steric packing of the indole side chains and the backbone. Connolly surfaces were calculated for these residues and the surface accessibility is plotted in Fig. 3B. The surfaces of the rhodopsin molecule surrounding the tryptophans are shown in Fig. 4. Large differences in the degree of steric restrictions for each tryptophan are found. Trp-35(1.30) (Fig. 4C), and to a lesser extent Trp-175(4.65) (Fig. 4B), and the side-chain nitrogen of Trp-161(4.50) (Fig. 4B) are solvent-accessible. However, the nitrogen atoms of Trp-126(3.41) and all atoms of Trp-265(6.48) (Fig. 4 A and B) are completely buried and solvent-inaccessible. Despite the above differences between tryptophans, each individual tryptophan shows a very similar environment of the backbone amide group and the indole side chain.
Fig. 4.
Views of surface-accessible area of the surroundings of the tryptophan residues. Helices are colored as in Fig. 1B. (A) Overall structure. (B) Expansion of region around Trp-126(3.41), Trp-161(4.60), and Trp-175(4.65). (C) Molecule turned around z axis and expanded to show region around Trp-35(1.30).
Prediction of Chemical Shifts Based on the Rhodopsin Crystal Structures. Finally, we tested whether a subset of the larger than expected number of signals of backbone amide groups in HSQC spectra could be attributed to a conformation similar to the crystal structure model. The chemical shift values for both nitrogen and hydrogen for backbone amide groups were calculated by using the programs shifts, proshifts, and shiftcalc (see Methods), both for the two chains with PDB ID codes 1F88 and 1L9H. The averages, SDs, and minima/maxima of predicted chemical shifts at each of the five tryptophan positions are listed in Table 1. Very little overlap between predicted and experimentally observed chemical shifts was observed.
Table 1. Chemical shift predictions based on the rhodopsin crystal structure models with PDB ID codes 1F88 and 1L9H.
| Amino acid position | Average 1H chemical shift, ppm | SD 1H chemical shift, ppm | Minimum 1H chemical shift, ppm | Maximum 1H chemical shift, ppm | Average 15N chemical shift, ppm | SD 15N chemical shift, ppm | Minimum 15N chemical shift, ppm | Maximum 15N chemical shift, ppm |
|---|---|---|---|---|---|---|---|---|
| Trp35 | 7.93 | 0.26 | 7.59 | 8.25 | 119.1 | 2.0 | 117.1 | 122.6 |
| Trp126 | 7.95 | 0.27 | 7.41 | 8.22 | 118.6 | 0.3 | 118.2 | 119.1 |
| Trp161 | 7.83 | 0.17 | 7.52 | 8.03 | 121.4 | 2.7 | 117.2 | 124.7 |
| Trp175 | 7.90 | 0.08 | 7.78 | 8.03 | 118.9 | 1.8 | 116.1 | 120.8 |
| Trp265 | 8.03 | 0.08 | 7.94 | 8.21 | 117.5 | 1.5 | 115.9 | 120.9 |
Several prediction methods were applied to chain A and chain B in both models (see Methods), and the average, SD, and minima/maxima of the predicted chemical shift values were calculated.
Discussion
The motions in the backbone amide groups of the polypeptide chain indicated by the HSQC spectra of α,ε-15N-tryptophan-labeled rhodopsin at 37°C in detergent solution is in agreement with the previous results with α-15N-lysine-labeled rhodopsin based on two observations (1). More than the expected number of signals was observed, indicating the presence of multiple conformations. We tested whether a subset of the signals reflects the rhodopsin crystal structure conformation by predicting the chemical shifts by using the crystal structure as a template. However, there was little overlap between predicted and experimentally observed chemical shift values. This may be due to changes in experimental chemical shifts as a result of the micelle environment that is lacking in the reference chemical shifts calculated by using soluble proteins. Furthermore, large variation in predicted chemical shifts were observed for the two different crystal structure models, and for each of the chains within the rhodopsin dimers, suggesting that the low resolution of the crystal structures may further limit the accuracy of chemical shift predictions (2). Exchange broadening causes low intensity of the majority of signals in conventional HSQC spectra, and using the TROSY scheme resulted in a decrease of sensitivity. This observation would be expected for a system with exchange contributions to the T2 relaxation. It would be desirable to quantify these exchange contributions both experimentally and through modeling. However, the low sensitivity of the signals due to the combined effects of high-molecular-weight exchange contributions and signal intensity distribution over multiple signals prevents application of NMR spectroscopic approaches to measure relaxation time directly. Estimation of the exchange contributions through modeling of the relaxation sources in the local environments of the tryptophans based on the crystal structure is unreliable because of the above described differences between the crystal structure and the aqueous micellar system. Future perdeuteration will likely enhance the quality of TROSY spectra (16) and may thus make the quantification of the exchange contributions experimentally measurable, but perdeuteration is currently not feasible by using our mammalian expression system. The conformational exchange observed here for rhodopsin may prove general for TM proteins, as evidence for microsecond–millisecond exchanges was also observed for OmpA (17, 18), bacteriorhodopsin (19, 20), and diacylglycerol kinase (21) in micelles.
In contrast to the flexibility of backbone groups, the indole side chains in tryptophan residues appear to be restricted to one conformation. The difference does not appear to result from increased steric freedom of side chains that would lead to rapid averaging of conformations in contrast to the amide groups. This conclusion comes from the study of each tryptophan in the crystal structure model: both indole-side-chain and backbone groups show similar accessibility and steric environments for individual tryptophans. Larger differences are only observed between tryptophans. Even though NMR and x-ray crystallographic analyses measure different types of dynamics, steric properties of the crystal structure model suggest that the differences in side-chain and backbone groups observed in the HSQC spectra are not due to overall increased mobility of side chains because of lack of steric constraints.
At present, the reason for the observation of heterogeneity in signal number and intensity is not clear. One possibility is the presence of heterogeneous glycosylation causing fluctuations in the backbone conformations of rhodopsin, a possibility that can now be tested with the recent development of a rhodopsin expression system in which glycosylation can be suppressed (22). Backbone flexibility may also be modulated by the specific detergent micelle structure present. The dynamic properties of the dark state may have importance for the process of light-activation of rhodopsin from the inactive state. The first event in this process, after light-induced retinal isomerization, is a perturbation in the TM helical bundle that leads to tertiary structure changes in the cytoplasmic domain (reviewed in refs. 5, 23, and 24). The conformational flexibility in the backbone of the polypeptide chain demonstrated here for tryptophans and previously for lysines (10) may be important for allowing the light-induced conformational changes to occur at the microsecond time scale of the light-activated Meta II state formation. In the absence of light, or in the absence of ligand for GPCRs in general, specific contacts lock the receptors in an inactive conformation and the release of these constraints is a key event in the activation process of rhodopsin (25, 26), the β2-adrenergic (27–30), α1B-adrenergic (31, 32), muscarinic acetylcholine (33), gonadotropin releasing hormone receptor (34), μ opioid (35), dopamine D2 (36), cannabinoid (37), histamine H2 (38), serotonin 5HT2A, and C5a (39) receptors, and probably GPCRs in general (reviewed in ref. 40). These constraints cluster in conserved domains, including a cluster of aromatic amino acids surrounding the ligand binding pockets (25, 27, 41–44). In rhodopsin, the highly conserved Trp-265(6.48) is in direct contact with the retinal, as demonstrated by crosslinking (25) and x-ray crystallography (2). On light-activation, retinal no longer crosslinks to Trp-265(6.48) but instead to Ala-169(4.58) (45), a residue located >10 Ångstroms away from the ionone ring in the dark state crystal structure (2). This would enhance the conformational flexibility of Trp-265(6.48), allowing a rearrangement of its indole side chain, shown to be important for activation of rhodopsin (46, 47) and the β2-adrenergic receptor (27). Another highly conserved tryptophan, Trp-161(4.50), may contribute to this process (45, 46). The results described in this paper provide experimental evidence for differential dynamics on the amino acid level in a membrane protein. This suggests that only some strong constraints exist, whereas the majority of the molecule experiences conformational flexibility within the same domains, and even within the same amino acid. In particular, we showed that tryptophan side chains are more restricted in conformation than their backbone. Thus, it appears that the indole side-chain contacts in part contribute to restricting the conformation in a “locked” dark state, without fully restricting motional fluctuations in the overall molecule including the helical bundle itself. The inference is that it would not be necessary to break and form thousands of specific contacts within nanoseconds after retinal isomerization in rhodopsin (or ligand binding in other GPCRs). Rather, a few specific contacts restricting the inactive state need to break on activation, and these changes are transmitted through the entire membrane protein because of its dynamic plasticity.
Acknowledgments
This work was supported by the National Institutes of Health (H.G.K.), the National Science Foundation (H.G.K. and J.K.-S.), the Deutsche Forschungsgemeinschaft (H.S.), the Howard Hughes Medical Institute (J.K.-S.), and the Humboldt Foundation/Zukunftsinvestitionsprogramm der Bundesregierung Deutschland (J.K.-S.). This is paper no. 54 in the Structure and Function in Rhodopsin series.
Abbreviations: GPCR, G protein-coupled receptor; HSQC, heteronuclear single quantum correlation; TM, transmembrane; TROSY, transverse relaxation optimized spectroscopy.
References
- 1.Unger, V. M. & Schertler, G. F. (1995) Biophys. J. 68, 1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 3.Teller, D. C., Okada, T., Behnke, C. A., Palczewski, K. & Stenkamp, R. E. (2001) Biochemistry 40, 7761-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okada, T., Fujiyoshi, Y., Silow, M., Navarro, J., Landau, E. M. & Shichida, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein-Seetharaman, J. (2002) Chembiochem 3, 981-986. [DOI] [PubMed] [Google Scholar]
- 6.Reeves, P. J., Thurmond, R. L. & Khorana, H. G. (1996) Proc. Natl. Acad. Sci. USA 93, 11487-11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves, P. J., Klein-Seetharaman, J., Getmanova, E. V., Eilers, M., Loewen, M. C., Smith, S. O. & Khorana, H. G. (1999) Biochem. Soc. Trans. 27, 950-955. [DOI] [PubMed] [Google Scholar]
- 8.Klein-Seetharaman, J., Getmanova, E. V., Loewen, M. C., Reeves, P. J. & Khorana, H. G. (1999) Proc. Natl. Acad. Sci. USA 96, 13744-13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewen, M. C., Klein-Seetharaman, J., Getmanova, E. V., Reeves, P. J., Schwalbe, H. & Khorana, H. G. (2001) Proc. Natl. Acad. Sci. USA 98, 4888-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein-Seetharaman, J., Reeves, P. J., Loewen, M. C., Getmanova, E. V., Chung, J., Schwalbe, H., Wright, P. E. & Khorana, H. G. (2002) Proc. Natl. Acad. Sci. USA 99, 3452-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballesteros, J. & Weinstein, H. (1995) Methods Neurosci. 25, 366-428. [Google Scholar]
- 12.Eilers, M., Reeves, P. J., Ying, W., Khorana, H. G. & Smith, S. O. (1999) Proc. Natl. Acad. Sci. USA 96, 487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, X. P. & Case, D. A. (2001) J. Biomol. NMR 21, 321-333. [DOI] [PubMed] [Google Scholar]
- 14.Meiler, J. (2003) J. Biomol. NMR 26, 25-37. [DOI] [PubMed] [Google Scholar]
- 15.Williamson, M. P. & Asakura, T. (1997) Methods Mol. Biol. 60, 53-69. [DOI] [PubMed] [Google Scholar]
- 16.Pervushin, K. (2000) Q. Rev. Biophys. 33, 161-197. [DOI] [PubMed] [Google Scholar]
- 17.Tamm, L. K., Abildgaard, F., Arora, A., Blad, H. & Bushweller, J. H. (2003) FEBS Lett. 555, 139-143. [DOI] [PubMed] [Google Scholar]
- 18.Arora, A., Abildgaard, F., Bushweller, J. H. & Tamm, L. K. (2001) Nat. Struct. Biol. 8, 334-338. [DOI] [PubMed] [Google Scholar]
- 19.Orekhov, V., Pervushin, K. V. & Arseniev, A. S. (1994) Eur. J. Biochem. 219, 887-896. [DOI] [PubMed] [Google Scholar]
- 20.Orekhov, V., Abdulaeva, G. V., Musina, L. & Arseniev, A. S. (1992) Eur. J. Biochem. 210, 223-229. [DOI] [PubMed] [Google Scholar]
- 21.Oxenoid, K., Sonnichsen, F. D. & Sanders, C. R. (2002) Biochemistry 41, 12876-12882. [DOI] [PubMed] [Google Scholar]
- 22.Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. (2002) Proc. Natl. Acad. Sci. USA 99, 13419-13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng, E. C. & Bourne, H. R. (2001) Trends Pharmacol. Sci. 22, 587-593. [DOI] [PubMed] [Google Scholar]
- 24.Hubbell, W. L., Altenbach, C., Hubbell, C. M. & Khorana, H. G. (2003) Adv. Protein Chem. 63, 243-290. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, T. A. & Khorana, H. G. (1990) J. Biol. Chem. 265, 15762-15769. [PubMed] [Google Scholar]
- 26.Cohen, G. B., Yang, T., Robinson, P. R. & Oprian, D. D. (1993) Biochemistry 32, 6111-6115. [DOI] [PubMed] [Google Scholar]
- 27.Shi, L., Liapakis, G., Xu, R., Guarnieri, F., Ballesteros, J. A. & Javitch, J. A. (2002) J. Biol. Chem. 277, 40989-40996. [DOI] [PubMed] [Google Scholar]
- 28.Samama, P., Cotecchia, S., Costa, T. & Lefkowitz, R. J. (1993) J. Biol. Chem. 268, 4625-4636. [PubMed] [Google Scholar]
- 29.Rasmussen, S. G., Jensen, A. D., Liapakis, G., Ghanouni, P., Javitch, J. A. & Gether, U. (1999) Mol. Pharmacol. 56, 175-184. [DOI] [PubMed] [Google Scholar]
- 30.Ballesteros, J. A., Jensen, A. D., Liapakis, G., Rasmussen, S. G., Shi, L., Gether, U. & Javitch, J. A. (2001) J. Biol. Chem. 276, 29171-29177. [DOI] [PubMed] [Google Scholar]
- 31.Scheer, A., Fanelli, F., Costa, T., De Benedetti, P. G. & Cotecchia, S. (1996) EMBO J. 15, 3566-3578. [PMC free article] [PubMed] [Google Scholar]
- 32.Greasley, P. J., Fanelli, F., Rossier, O., Abuin, L. & Cotecchia, S. (2002) Mol. Pharmacol. 61, 1025-1032. [DOI] [PubMed] [Google Scholar]
- 33.Hogger, P., Shockley, M. S., Lameh, J. & Sadee, W. (1995) J. Biol. Chem. 270, 7405-7410. [DOI] [PubMed] [Google Scholar]
- 34.Ballesteros, J., Kitanovic, S., Guarnieri, F., Davies, P., Fromme, B. J., Konvicka, K., Chi, L., Millar, R. P., Davidson, J. S., Weinstein, H. & Sealfon, S. C. (1998) J. Biol. Chem. 273, 10445-10453. [DOI] [PubMed] [Google Scholar]
- 35.Huang, P., Li, J., Chen, C., Visiers, I., Weinstein, H. & Liu-Chen, L. Y. (2001) Biochemistry 40, 13501-13509. [DOI] [PubMed] [Google Scholar]
- 36.Wilson, J., Lin, H., Fu, D., Javitch, J. A. & Strange, P. G. (2001) J. Neurochem. 77, 493-504. [DOI] [PubMed] [Google Scholar]
- 37.Abadji, V., Lucas-Lenard, J. M., Chin, C. & Kendall, D. A. (1999) J. Neurochem. 72, 2032-2038. [DOI] [PubMed] [Google Scholar]
- 38.Alewijnse, A. E., Timmerman, H., Jacobs, E. H., Smit, M. J., Roovers, E., Cotecchia, S. & Leurs, R. (2000) Mol. Pharmacol. 57, 890-898. [PubMed] [Google Scholar]
- 39.Baranski, T. J., Herzmark, P., Lichtarge, O., Gerber, B. O., Trueheart, J., Meng, E. C., Iiri, T., Sheikh, S. P. & Bourne, H. R. (1999) J. Biol. Chem. 274, 15757-15765. [DOI] [PubMed] [Google Scholar]
- 40.Gether, U. (2000) Endocr. Rev. 21, 90-113. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama, T. A. & Khorana, H. G. (1991) J. Biol. Chem. 266, 4269-4275. [PubMed] [Google Scholar]
- 42.Han, M., Lou, J., Nakanishi, K., Sakmar, T. P. & Smith, S. O. (1997) J. Biol. Chem. 272, 23081-23085. [DOI] [PubMed] [Google Scholar]
- 43.Ridge, K. D., Bhattacharya, S., Nakayama, T. A. & Khorana, H. G. (1992) J. Biol. Chem. 267, 6770-6775. [PubMed] [Google Scholar]
- 44.Visiers, I., Ballesteros, J. A. & Weinstein, H. (2002) Methods Enzymol. 343, 329-371. [DOI] [PubMed] [Google Scholar]
- 45.Borhan, B., Souto, M. L., Imai, H., Shichida, Y. & Nakanishi, K. (2000) Science 288, 2209-2212. [DOI] [PubMed] [Google Scholar]
- 46.Lin, S. W. & Sakmar, T. P. (1996) Biochemistry 35, 11149-11159. [DOI] [PubMed] [Google Scholar]
- 47.Chabre, M. & Breton, J. (1979) Photochem. Photobiol. 30, 295-299. [DOI] [PubMed] [Google Scholar]