Abstract
Because of the association of β-sheet formation with the initiation and propagation of amyloid diseases, model systems have been sought to further our understanding of this process. WW domains have been proposed as one such model system. Whereas the folding of the WW domains from human Yes-associated protein (YAP) and Pin have been shown to obey single-exponential kinetics, the folding of the WW domain from formin-binding protein (FBP) 28 has been shown to proceed via biphasic kinetics. From an analysis of free-energy landscapes from atomic-level molecular dynamics simulations, the biphasic folding kinetics observed in the FBP WW domain may be traced to the ability of this WW domain to adopt two slightly different forms of packing in its hydrophobic core. This conformational change is propagated along the peptide backbone and affects the position of a tryptophan residue shown in other WW domains to play a key role in binding. The WW domains of Pin and YAP do not support more than one type of packing each, leading to monophasic folding kinetics. The ability of the FBP WW domain to assume two different types of packing may, in turn, explain the capacity of this WW domain to bind two classes of ligand, a property that is not shared by other WW domains. These findings lead to the hypothesis that lability with respect to conformations separated by an observable barrier as a requirement for function is incompatible with the ability of a protein to fold via single-exponential kinetics.
Keywords: protein folding, β-sheet, β-strand, packing
Many proteins are comprised of a series of modular domains that carry out diverse functions ranging from localization to catalysis to signaling. WW domains are a class of signaling modules that, similar to Src homology 3 (SH3) domains, carry out signal transduction via binding to proline-rich peptides in a type II polyproline helix conformation (1). Although the position of the ligand is maintained in all complexes solved to date, the bound ligand may adopt one of two orientations relative to the WW domain, which differ in the direction of the peptide backbone (2, 3). This feature arises from the approximate twofold symmetry of the type II polyproline helix and is shared by peptides that bind SH3 domains (1). Signaling processes mediated by WW domains have been implicated in a variety of human disorders including Alzheimer's disease, muscular dystrophy, Huntington's disease, and cancer (4). The WW domain, the name of which derives from a pair of conserved tryptophan residues, is comprised of a curved three-stranded antiparallel β-sheet; the concave face of the sheet contains the ligand-binding site, and the convex face contributes to a hydrophobic core involving the termini of the WW domain.
The study of helical peptides has led to simple models for their formation (5–7), but the greater sequence separation of interacting residues (8) and the dependence of the folding mechanism on the relative importance of hydrogen bonds and hydrophobic interactions (9, 10) has made the extension of such models to β-sheets difficult. The formation of nonnative β-sheet structures associated with several human diseases (11, 12) has brought to the forefront the search for a model β-sheet in which to study folding.
This quest has led to an interest in WW domains. After initial studies using the WW domain from human Yes-associated protein (YAP) (13, 14), focus shifted to the prolyl-isomerase Pin1 WW domain (15). Subsequent examination of the WW domain from formin-binding protein (FBP) 28 revealed a surprise; unlike the folding kinetics of the YAP and Pin WW domains, which were well described by a single exponential (14, 15), the folding kinetics of the FBP WW domain proved to be biphasic (16). Monophasic kinetics are indicative of a single barrier-crossing event, whereas the biphasic kinetics in the FBP WW domain suggest a more complex mechanism. Although a subsequent study has proposed aggregation as the origin of these biphasic kinetics (17), it is unclear how aggregation events on a time scale of hours is reflected in biphasic kinetics on the microsecond time scale.
Many small proteins (or protein domains) fold to a single distinct “native state” from which they are active (18). Evolutionary pressure is thought to have sculpted a free-energy surface that maximizes the population of molecules residing in this active state via two tactics. The first is the destabilization of nonnative states relative to the native state, which results in a funnel-shaped free-energy surface (19, 20). The second involves the design of a free-energy barrier separating the native state from nonnative states, which allows little conformational diversity within the native basin at physiological temperatures by ensuring that the population of partially folded molecules is small (21–23). These features common to the free-energy landscapes of many small proteins lead to other shared properties, which themselves are not a result of direct evolutionary pressure, such as folding by means of single-exponential kinetics on relatively fast time scales (24).
To understand the structural basis for the biphasic folding kinetics in the FBP WW domain, two studies were launched in collaboration. The experimental arm of this partnership identified several perturbations by which folding may be switched between monophasic and biphasic kinetics; these means include C-terminal truncation, variation of temperature, and mutation at particular sites (16). Meanwhile, theoretical studies of folding kinetics using sequence-dependent Cα-based Gō-like models were carried out and found to reproduce the respective folding kinetics of both the Pin and FBP WW domains (25). The biphasic kinetics observed in this simple model were traced to the mobility of the third β-strand within the context of otherwise folded conformations (25).
To extend the level of detail at which the origin of the biphasic folding kinetics of the FBP WW domain is understood, we have complemented our earlier analysis of the Cα-based Gō-like models by free-energy landscape calculations by using molecular dynamics with an atomic-resolution force field and an implicit representation of the solvent. After describing the structural origin of the biphasic folding kinetics, we rationalize the functional reasons that may have led to a free-energy landscape that differs from those of other small proteins.
Methods
Molecular Dynamics. Simulations were carried out with the charmm molecular dynamics program (26). Each protein was represented by the all-hydrogen force field (param22) (27), with map-based backbone dihedral cross terms (28). Covalent bonds involving hydrogen atoms were constrained by using shake (29), and dynamics were propagated with a time step of 2 fs at a temperature of 285 K, selected to assure strongly native-promoting conditions. The generalized born molecular volume (GBMV) implementation (30, 31) of the generalized Born formalism (32) was used as an implicit representation of the solvent. Modified atomic radii were used to represent the solute–solvent interfacial region in the implicit solvent model (33). The hydrophobic effect was incorporated through the use of a term proportional to the molecular surface area, using a scaling factor of 0.015 kcal/mol·Å2. Ninety-six independent simulations of 6.4 ns in length were carried out for each protein, making the total simulation time 614 ns per protein. The first 0.8 ns of each simulation was not used in the analysis described below. Coordinates were saved for analysis every 2,000 steps of molecular dynamics.
Umbrella Sampling. To enhance exploration of the accessible conformational space, additional harmonic restraining potentials were applied to the Cα radius of gyration (34) and a continuous analog to the fraction of side-chain-native contacts formed (35). Native contacts of the FBP WW domain were defined as residues with nonhydrogen atoms within 4.5 Å in at least one model of the NMR ensemble (36) (PDB ID code 1E0L). The same definition was used to define native contacts for Pin WW domain using the crystal structure (37) (PDB ID code 1PIN). Umbrella potentials applied to the radius of gyration contained minima distributed between 9 and 12 Å in increments of 0.5 Å, and those applied to the fraction of native contacts formed contained minima distributed between 0.05 and 1.00 in increments of 0.05. Combination of each possible pair of these biasing potentials results in 140 unique “conditions.” This set was reduced to 96 conditions via exclusion of 44 pairs corresponding to unphysical extended states (i.e., large radius of gyration) in which most native contacts are formed. Initial studies verified that these indeed were regions of conformation space in which the free energy was much higher than the native state (results not shown). All force constants applied to the radius of gyration were set to 100 kcal/mol·Å2 for the first 0.4 ns to aid equilibration and then to 10 kcal/mol·Å2 for the remainder of the simulations. All force constants applied to the fraction of native contacts formed were set to 10,000 kcal/mol for the first 0.4 ns and then to 1,000 kcal/mol for the remainder of the simulations.
Replica Exchange. To enhance sampling further, an extension of the replica-exchange methodology (38) known as reus (39) was used, as implemented in the mmtsb tool set (40). This methodology involves carrying out independent simulations under different conditions and periodically swapping the conformations associated with a pair of these conditions, subject to the Metropolis criterion. In this case, exchanges took place between simulations in which the position of the minimum in the umbrella potentials differed. Exchanges were attempted only between neighboring replicas; neighbors were defined as those replicas in which one umbrella potential was identical and the other differed in the position of the minimum by only one increment. The overall exchange probability for each set of simulations was ≈20%. Exchanges were attempted every 2,000 steps of molecular dynamics to allow each conformation sufficient relaxation time after transferal to the energy surface associated with the new Hamiltonian.
Analysis. After completion of the simulations described above, data collected by using different umbrella potentials were combined by using the weighted histogram-analysis method (41–43); this method allows projection of the free energy onto any progress variable that is a function of the atomic coordinates.
Tryptophan burial was defined as inversely proportional to side-chain solvent-accessible surface area, in which the burial of the tryptophan in an AWA peptide was set to zero and the burial of the experimentally determined structure was set to unity. The solvent-accessible surface area was computed by the method of Lee and Richards (44).
Results and Discussion
Initial Thermodynamic Characterization. To gain an understanding of the general features of the free-energy landscapes for the Pin and FBP WW domains, the free energy was projected simultaneously onto several different pairs of global structural descriptors. If the free-energy landscape of the FBP WW domain includes one or more minima that are structurally distinct from the native state, these may act as kinetic traps and hence bring about biphasic folding kinetics.
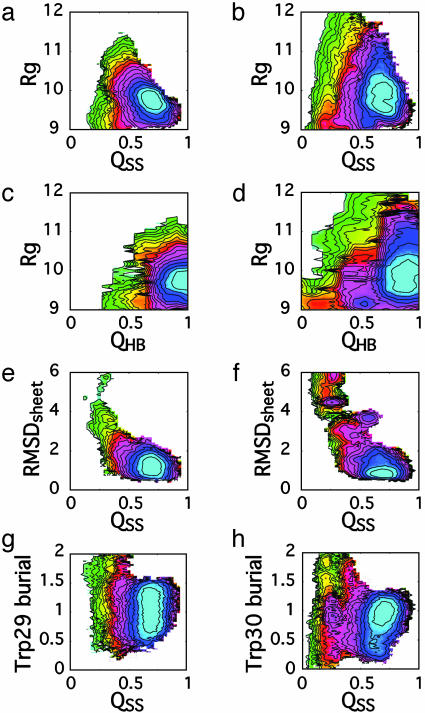
The first pair of variables selected were the radius of gyration (Rg) and the fraction of native side chain–side chain contacts formed (QSS) (Fig. 1 a and b). In both cases, the free-energy landscape appears funneled as these variables progress toward a single minimum, reflecting the strongly native-promoting conditions used (285 K). In both cases, this minimum lies at a native-like radius of gyration and near QSS ≈ 0.7. The breadth of this native basin arises from thermal fluctuations. Both free-energy landscapes are funneled similarly when projected onto Rg and the fraction of native hydrogen bonds formed (QHB) (Fig. 1 c and d), and again both contain a single minimum corresponding to the native state. The fact that the minimum in all cases contains all or most native contacts provides a strong indication that the free-energy minimum for each protein, given the set of potentials used, corresponds closely to the experimentally observed structure. The agreement in the β-sheets is underscored further by projection of the free energy onto the rms deviation (RMSD) from the published structures (36, 37) of Cα positions of atoms of the residues that comprise the β-sheets (RMSDsheet) and QSS (Fig. 1 e and f). RMSDsheet at the observed minimum is ≈1 Å from the experimentally derived structures, indicating that the conformations that comprise the minimum located at QSS = 0.7 contain a fully native-like β-sheet.
Fig. 1.
Initial thermodynamic characterization of the Pin (a, c, e, and g) and FBP (b, d, f, and h) WW domains, in which the free energy is projected simultaneously onto two structural descriptors. (a and b) Radius of gyration (Rg) and the fraction of native contacts formed (QSS). (c and d) Radius of gyration and the fraction of native hydrogen bonds formed (QHB). (e and f) The Cα RMSD of residues that comprise the β-sheets (RMSDsheet) and QSS. (g and h) The relative burial of the C-terminal tryptophan (zero is fully exposed, and unity is the burial in the experimentally determined structure) and QSS. Contour lines in all cases represent kBT (T = 285 K). The color scheme is for illustrative purposes only.
Given the presence of only one minimum when the free energy was projected onto these structural variables, we next selected a structural probe designed to mimic the experimental probe used to monitor the progress of folding (16). The experimental probe was tryptophan fluorescence, and through mutation the specific reporter of biphasic folding kinetics was shown to be Trp-30 (16). We use the relative burial of this tryptophan (Trp-29 in Pin) as a reflection of its environment, and simultaneously project the free energy onto this progress variable and QSS, for both the Pin and FBP WW domains (Fig. 1 g and h).
The use of this pair of structural descriptors demonstrates a qualitative difference between the free-energy landscapes of the Pin and FBP WW domains. The tryptophan burial of conformations of the Pin WW domain that occupy the native state basin, as distinguished by QSS = 0.7, ranges from 0.7 to 1.4 relative to the burial in the crystal structure (Fig. 1g). The free-energy surface of the FBP WW domain (Fig. 1h), by contrast, exhibits a minimum with native-like tryptophan burial (burial = 1.0) but also a small local minimum nearby. This additional minimum lies on the edge of the native state basin and differs from typical members of the native state basin by the presence of slightly fewer native contacts and an increase in the exposure of Trp-30 (burial = 0.4). The difference in the tryptophan environment of this minimum relative to the native state suggests that the rearrangement of the structure from this local minimum to that of the native basin may be responsible for the observed biphasic folding kinetics (16).
Structural Description of States Accessible to the FBP WW Domain. Having identified a measure that differentiates the native state basin from this “additional” minimum, we return to the ensemble of structures used to build these free-energy surfaces. By using Trp-30 burial and QSS, it is possible to extract many examples of conformations within each free-energy minimum and search for further distinguishing features.
Having noted earlier that the β-sheet is predominantly native-like in all these conformations (Fig. 1f), one may envision several scenarios involving the chain termini by which conformations from each of these two minima could differ. One could envision a series of conformations in which one or both of the chain termini move to the opposite side of the β-sheet (the concave side in the native state) or in which the termini have exchanged positions relative to those they occupy in the native state. One must also consider a vast array of other mispacked states including those that differ from the native packing in the relative position of two or more side chains. Because the barrier associated with protein folding is thought to generally correspond to the desolvation of side chains required to form a well packed hydrophobic core (35, 45–48), reorganization from a specific mispacked state to the native packing would be expected to proceed on a relatively slow time scale. To determine the structural features associated with each free-energy minimum, we examine representative conformations in detail.
Representative structures from the more-populated free-energy minimum, in which Trp-30 is more buried, show a significant level of agreement with the experimentally determined NMR ensemble of structures (36). Contacts not present in these structures generally involve the extreme N terminus, consistent with the high degree of flexibility in this region inferred from both the wide variation of conformations occupied by the N terminus in the ensemble of NMR structures (36) and the fact that N-terminal truncation has little effect on stability (16).
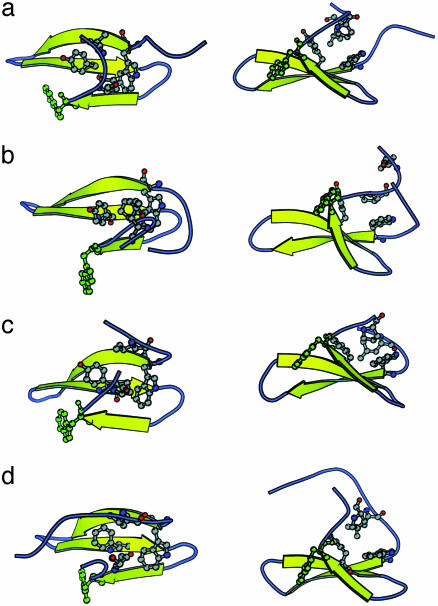
Examination of numerous examples of conformations from the less-populated free-energy minimum, in contrast, were often seen to incorporate a particular shift from the native state: a relatively subtle difference in the packing of the hydrophobic core. This alternate packing is seen most clearly through comparison with the packing found in the NMR structures (36) and the more-populated free-energy minimum. In the native FBP packing, a leucine (Leu-36) and a proline (Pro-33) lie equidistant from a tryptophan (Trp-8) and a tyrosine (Tyr-20) on the convex face of the β-sheet in an approximate tetrahedral arrangement (Fig. 2a). In the “alternate” packing, a representative of which is shown in Fig. 2b, Pro-33 moves to a position directly above Trp-8 and Tyr-20, forming a triangular arrangement that displaces Leu-36 into the solvent. Adoption of this alternate packing tilts the C-terminal end of the third β-strand toward the hydrophobic core, increasing the curvature of the β-sheet. Because Trp-30 lies across the β-sheet in the native packing (Fig. 2a), this increase in curvature induces a steric clash between Trp-30 and the second β-strand; this clash is resolved by reorientation of Trp-30 away from the β-sheet (Fig. 2b). A concerted conformational change such as that just described provides an explanation for the ability of Trp-30 burial to act as a reporter of the packing involving residues on the opposite face of the β-sheet. The native and alternate packing will, for the purposes of additional discussion, be referred to as N-packing and A-packing, respectively.
Fig. 2.
A comparison of hydrophobic core packing in several WW domains, shown from two perspectives. (a) The N-packing of FBP, solved with NMR. (b) The A-packing of FBP, extracted from the molecular dynamics simulations. (c) Packing in the Pin WW domain, which resembles A-packing in FBP. (d) Packing in the YAP WW domain, which resembles N-packing in FBP. The C-terminal tryptophan is also shown explicitly in each case (green).
Using this insight from structural analysis, it now is possible to design additional structural descriptors to delineate these two minima. Given the assertion that the packing is described by the position of Pro-33 relative to Trp-8 and Tyr-20, the “nativeness” of the packing will be reflected by the all-atom RMSD of Pro-33 relative to its position in an experimentally determined structure (36) after alignment by minimization of the all-atom RMSD of the positions of Trp-8 and Tyr-20. We will refer to this measure as PrmsdN (Pro-33 RMSD from N-packing). Conversely, the degree to which the packing resembles the A-packing may be captured by using the same measure but computing the RMSD to an example of A-packing (the conformation shown in Fig. 2b was used). We will refer to this measure as PrmsdA (Pro-33 RMSD from A-packing). Each projection involving these variables was first “filtered” by using QSS, and only conformations with QSS > 0.5 were used in calculation of the free-energy surface; this filtering was used to reduce contributions from conformations distant from the native state, which would act as noise in PrmsdN and PrmsdA.
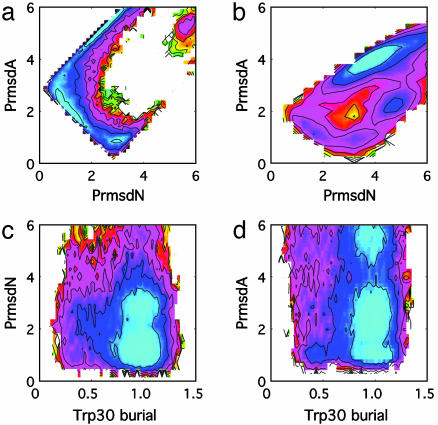
We demonstrate first that these measures differentiate the two forms of packing by simultaneously projecting the free energy onto both PrmsdN and PrmsdA (Fig. 3a). Two minima are apparent on this surface: one is located at PrmsdN = 1 Å, PrmsdA = 2 Å and corresponds to the native packing, whereas the other is located at PrmsdN = 3 Å, PrmsdA = 1 Å and corresponds to the alternate packing. As expected, the free-energy minimum corresponding to the native packing is more populated under these conditions.
Fig. 3.
Characterization of the FBP free-energy surface using PrmsdN and PrmsdA. The free energy is projected simultaneously onto PrmsdA and PrmsdN using the full-length FBP WW domain (a), PrmsdA and PrmsdN using the C-terminally truncated FBP WW domain (b), PrmsdN and Trp-30 burial using the full-length FBP WW domain (c), and PrmsdA and Trp-30 burial using the full-length FBP WW domain (d). Contour lines in all cases represent kBT. The color scheme is for illustrative purposes only.
The free energy is projected next onto Trp-30 burial as well as either PrmsdN or PrmsdA to demonstrate the ability of Trp-30 to act as a reporter for the packing of the hydrophobic core (Fig. 3 c and d). Based these projections, it is apparent that conformations in which Trp-30 is largely exposed (burial = 0.4) typically have PrmsdN > 2 Å and PrmsdA < 1 Å, indicative of the type of packing shown in Fig. 2b. It is important to note that many conformations that contain A-packing allow Trp-30 to be buried to an extent similar to that present in the native packing (Fig. 3 c and d); this form of packing does not require that Trp-30 become exposed but merely enhances the probability of Trp-30 exposure. Conversely, conformations in which Trp-30 is largely exposed almost always contain A-packing (Fig. 3 c and d). It is the sensitivity of fluorescence measurements toward this change in distribution of the Trp-30 environment that we anticipate allows an experimental probe of the type of packing and in turn allows observation of biphasic folding kinetics (16). Using additional free-energy projections, Trp-8 was found to be buried to a similar amount regardless of the type of packing (results not shown), consistent with the inability of this tryptophan to act as a reporter of biphasic folding kinetics in Trp-30 → Phe and Trp-30 → Ala mutants (16).
Based on our analysis of the free-energy landscape for this system, probed by contrasting multiple “cuts” via projection onto many different sets of coordinates, we propose a structural model consistent with the biphasic folding kinetics observed in the FBP WW domain. After initiation of folding, the chain rapidly adopts a conformation in which the β-sheet is fully native-like; the packing, however, may exist in one of two forms. The free energy is seen to be essentially downhill for this component of folding (Fig. 1 b, d, and f), supporting assignment of this step to the observed fast phase. The subset of conformations that have adopted A-packing subsequently undergo rearrangement to N-packing on a slower time scale, associated with a barrier-crossing event (Fig. 3a).
Other Folding Studies of the FBP WW Domain. The wealth of variants of the FBP WW domain examined experimentally (16) allow ample opportunity for comparison to both validate the model presented here and rationalize the experimental observations.
The variant most directly related to the model for folding that we propose above involves truncation of the four C-terminal residues. In this variant, termed mini-FBP, Leu-36 is removed but Pro-33 is maintained; one therefore would expect such a construct to adopt A-packing exclusively. To test this hypothesis, the simulations described above were repeated by using this C-terminally truncated construct. Projection of the free energy onto the same progress variables used in Fig. 1 showed a drastic destabilization relative to the full-length FBP WW domain (results not shown), which also was observed experimentally (16). Associated with this destabilization, projection of the free energy onto PrmsdN and PrmsdA for mini-FBP shows the presence of an additional minimum at PrmsdN = 4 Å, PrmsdA = 4 Å in which the hydrophobic core is not formed. The minimum associated with A-packing remains at PrmsdN = 3 Å, PrmsdA = 1 Å, whereas the minimum associated with N-packing moves toward the A-packed state (i.e., higher PrmsdN and lower PrmsdA). The decreased structural separation and lower barrier between these two minima are suggestive additionally of fluidity in the packing of the hydrophobic core in mini-FBP. Experimental characterization has shown that the folding of this variant follows single-exponential kinetics with a rate similar to the faster phase observed in the full-length FBP WW domain (16). The structural model presented here suggests an explanation for this observation, arising from the absence of competition between the two forms of packing in this construct. Experimental truncation beyond Pro-33, meanwhile, resulted in nonnative conformers and aggregates (16), which is consistent with the inability of such a construct to form a buried hydrophobic core with either type of packing.
Additional experiments probing the effect of temperature on the observed biphasic folding kinetics of the full-length FBP WW domain showed that the slower phase was eliminated during refolding at higher temperature (16). Given that the importance of the hydrophobic effect increases with temperature (49), one would expect that the penalty in free energy associated with adopting the A-packing instead of the N-packing would increase at higher temperature because of the solvent exposure of Leu-36 that accompanies A-packing. By destabilizing A-packing relative to N-packing, increasing temperature eliminates this kinetic trap.
Mutation of residues Tyr-11 and Tyr-19, which reside on the first two β-strands and face away from the hydrophobic core, showed little effect on the experimentally observed kinetics (16), which also is consistent with the structural model we propose.
As described in the Introduction, results from a simple model implicated mobility of the third β-strand in the biphasic folding kinetics (25). The importance of this mobility was verified experimentally via demonstration that mutation of Leu-26 (located on the N-terminal portion of the third β-strand) to alanine affected the slower of the two phases but not the faster (16). The results from this simple model are in striking agreement with the present observation that the effect of the alternate packing is propagated along the peptide backbone to the third β-strand (Fig. 2 a and b). The existence of the alternate packing, meanwhile, could not have been observed in the simple model because of the lack of favorable nonnative interactions in the model (25).
Comparison with Other WW Domains. Although the WW domain from FBP28 shows biphasic folding kinetics, the Pin WW domain has been shown to fold via monophasic kinetics (15). Given that the biphasic folding kinetics in the FBP WW domain arise from the availability of two alternate forms of packing in the hydrophobic core, does this suggest that only one type of packing is available to the Pin WW domain?
Examination of the packing present in the Pin WW domain (2, 37) (Fig. 2c) confirms that this indeed is the case. The packing in the Pin WW domain resembles the A-packed form of the FBP WW domain; Pin contains a proline at the position analogous to Pro-33 in FBP, which lies directly over the analogous tryptophan and tyrosine residues on the face of the β-sheet. Additional stabilization in the Pin WW domain derives from an inward-facing leucine residue on the N-terminal tail. The Pin WW domain harbors an asparagine at the position analogous to Leu-36 of FBP, which explains why Pin cannot assume packing more similar to the N-packing of FBP. The observation of packing analogous to the A-packed state of the FBP WW domain in an unrelated WW domain provides additional support for the applicability of such a state to the FBP WW domain.
Monophasic folding kinetics also have been observed by using the WW domain from YAP (14). Unlike the Pin WW domain, however, the YAP WW domain adopts packing similar to the N-packed state of the FBP WW domain (50) (Fig. 2d). Although the positions of the participating side chains are very similar, their location in sequence differs slightly; the position occupied by Leu-36 in the FBP WW domain is occupied instead by Leu-4 in YAP. The monophasic folding kinetics of the YAP WW domain suggests that the free-energy difference between the N- and A-packed states must be larger in the YAP WW domain than in the FBP WW domain. One would anticipate a large free-energy difference, given that the A-packing in FBP results in exposure of Leu-36 to solvent. The question therefore may be posed: Why is the free-energy difference between the two types of packing so small in the FBP WW domain but not in that of YAP? A reasonable hypothesis is that strain is placed on the intervening sequence between Pro-33 and Leu-36 in FBP when these residues participate in N-packing. Examination of the 10 members of the ensemble of NMR structures, which all exhibit N-packing, supports this hypothesis: in each of the 10 members of the ensemble, one of the two intervening residues (Gln-34 and Glu-35) lies in a region of the Ramachandran plot described as “generously allowed” (51, 52). This provides evidence for strain in the N-packed state of the FBP WW domain, which is not present in the N-packed state of the YAP WW domain, in turn accounting for the monophasic folding kinetics of the YAP WW domain.
Implications for Binding Specificity. To summarize, the FBP WW domain may access two alternate packing modes, whereas the Pin and YAP WW domains only allow one each. The conformational change associated with movement from one type of packing to the other is propagated along the peptide backbone, altering the tilt angle of the third β-strand and thus the curvature of the β-sheet. This in turn affects the conformational preference of the tryptophan located on the third β-strand.
This tryptophan residue is notable in that it plays a key functional role in all WW domain structures solved in complex with peptides. Complexes of the YAP and Pin WW domains with peptide substrates show that this tryptophan intercalates into the groove formed by the type II polyproline helix, stacking against a proline in the ligand (2, 50). The indole NH of this tryptophan additionally forms a hydrogen bond to either a carbonyl (50) or a hydroxyl (2) group in the ligand. Mutation of this tryptophan leads to a loss of binding affinity in both cases (2, 53).
Another important difference between the FBP WW domain and those of YAP and Pin is their ligand predilection. The YAP WW domain belongs to the set of group I WW domains, which bind polypeptides containing a consensus (L/P)PXY sequence (54). The Pin WW domain, meanwhile, binds the consensus (phospho-S/T)P sequence characteristic of group IV WW domains (55). The FBP28 WW domain binds both ligands that contain a PPPXXQ motif (56) as well as ligands that do not (the binding region for such ligands has not been established yet) (57). Unlike the YAP and Pin WW domains, which each bind a single consensus sequence, the FBP WW domain therefore binds at least two distinct classes of ligand. Given the ability of the FBP WW domain to assume an alternate packing that affects both the β-sheet curvature as well as the position of a residue likely to be important for binding (Trp-30), it is tempting to associate the specificity of the FBP WW domain toward each type of ligand with a different type of packing.
A straightforward experimental test of this hypothesis could be performed by measuring the effect of particular mutations on the affinity of the FBP WW domain toward each type of ligand. The N-packed state could be stabilized by relief of the strain in the region between Pro-33 and Leu-36; in particular, the strain reflected by the positive φ dihedral angle of Gln-34 found in several members of the NMR ensemble (36) could be eased via mutation to glycine. The A-packed state could be stabilized by either C-terminal truncation or mutation of Leu-36 to a polar residue. If each type of packing is associated with binding a specific class of ligand, modifications to the WW domain that shift the balance of these two types of packing in favor of a particular type of packing will enhance binding to the correlate class of ligand at the expense of the other class. Additional experiments will be required to validate this hypothesis as well as to match each type of packing with the mode of substrate binding it promotes.
Conclusions
Signal transduction through WW domains, as with SH3 domains, occurs via binding events of high specificity and low affinity; the former ensures fidelity in the flow of information, whereas the latter allows for dynamic modulation of the signal (58). Comparison of WW domain structures from proteins other than FBP28 with and without bound substrates show relatively minor rearrangements after ligand binding (2, 3, 50), which suggests that these WW domains have been designed to occupy a single conformation each: the one required for binding. The optimal free-energy landscape for a protein that must carry out this simple functional requirement would contain a single barrier separating the active conformation from nonactive conformations and be devoid of other competing local minima. Accordingly, folding on such a free-energy landscape would proceed by means of two-state kinetics, as seen in the WW domains from Pin and YAP. Similarly, the function of a variety of other small proteins often used as examples of two-state folding is a single binding event; examples include chymotrypsin inhibitor 2, SH3 domains, and the IgG-binding domains. These proteins are called on to carry out the simplest possible function and accordingly have evolved the simplest possible free-energy landscape on which to do so. It is also interesting to note that the rate-limiting step in the folding of the Pin WW domain involves formation of loop 1, which contains key residues for ligand binding (15).
The WW domain from FBP28 carries out at least two distinct functions: binding peptides that harbor one of at least two distinct motifs. This requires a more complex free-energy landscape that contains two distinct “active” states, which in turn leads to biphasic folding kinetics. The manifestation of functional requirements on the free-energy landscape is seen even more dramatically in the case of proteins that remain unstructured under physiological conditions (59, 60).
Although protein domains have been long classified on the basis of monophasic and biphasic folding kinetics, it has not been possible to predict apriori the class to which a given protein will belong; the only insight to date has been the observation that protein domains <110 aa often fold via monophasic kinetics, whereas larger proteins often exhibit biphasic kinetics (18). On the basis of the discussion above, we assert that biphasic folding kinetics should be expected for any protein domain that must perform a conformational change to carry out its complete function, provided that the spectroscopic probe is able to discern this conformational change (i.e., the observable quantity changes by a significant amount on a measurable time scale). Although the conformational change that must be supported by the FBP WW domain involves rearrangement of packing, other types of changes are possible. Another type of conformational change involves reorientation of two parts of a protein with respect to each other; such motions are common in the active sites of enzymes. One would expect that the functional advantage of allowing such a motion is paid for with a loss of cooperativity in the formation of these parts, and hence folding kinetics will not be monophasic. This prediction seems to hold in enzymes such as acylphosphatase (61), hen lysozyme (62), and dihydrofolate reductase (63) through the use of spectroscopic probes associated with ligand binding. The kinetic mechanism by which a protein folds thus offers insight into the nature of the underlying free-energy surface, which is the direct product of evolutionary optimization for function.
Note Added in Proof. The dual binding specificity of the FBP28 WW domain has recently been confirmed in a high-throughput proteomic stuty (64).
Acknowledgments
We thank Professor M. Sudol and M. Bedford for their helpful correspondence. This work was supported by the National Institutes of Health (Grant GM48807) and a predoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada and the La Jolla Interfaces in Science Interdisciplinary Program (to J.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SH3, Src homology 3; YAP, Yes-associated protein; FBP, formin-binding protein; RMSD, rms deviation; PrmsdN, Pro-33 RMSD from N-packing; PrmsdA, Pro-33 RMSD from A-packing.
References
- 1.Zarrinpar, A. & Lim, W. A. (2000) Nat. Struct. Biol. 7, 611-613. [DOI] [PubMed] [Google Scholar]
- 2.Verdecia, M. A., Bowman, M. E., Lu, K. P., Hunter, T. & Noel, J. P. (2000) Nat. Struct. Biol. 7, 639-643. [DOI] [PubMed] [Google Scholar]
- 3.Huang, X., Poy, F., Zhang, R., Joachimiak, A., Sudol, M. & Eck, M. J. (2000) Nat. Struct. Biol. 7, 634-638. [DOI] [PubMed] [Google Scholar]
- 4.Sudol, M. & Hunter, T. (2000) Cell 103, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 5.Zimm, B. H. & Bragg, J. K. (1959) J. Chem. Phys. 31, 526-535. [Google Scholar]
- 6.Lifson, S. & Roig, A. (1961) J. Chem. Phys. 34, 1963-1974. [Google Scholar]
- 7.Qian, H. & Schellman, J. A. (1992) J. Phys. Chem. 96, 3987-3994. [Google Scholar]
- 8.Smith, C. K. & Regan, L. (1997) Acc. Chem. Res. 30, 153-161. [Google Scholar]
- 9.Zhou, R., Berne, B. J. & Germain, R. (2001) Proc. Natl. Acad. Sci. USA 98, 14931-14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinosa, J. F., Munoz, V. & Gellman, S. H. (2001) J. Mol. Biol. 306, 397-402. [DOI] [PubMed] [Google Scholar]
- 11.Serpell, L. C. (2000) Biochim. Biophys. Acta. 1502, 16-30. [DOI] [PubMed] [Google Scholar]
- 12.Pruisner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koepf, E. K., Petrassi, H. M., Sudol, M. & Kelly, J. W. (1999) Protein Sci. 8, 841-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crane, J. C., Koepf, E. K., Kelly, J. W. & Gruebele, M. (2000) J. Mol. Biol. 298, 283-292. [DOI] [PubMed] [Google Scholar]
- 15.Jager, M., Nguyen, H., Crane, J. C., Kelly, J. W. & Gruebele, M. (2001) J. Mol. Biol. 311, 373-393. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen, H., Jager, M., Moretto, A., Gruebele, M. & Kelly, J. W. (2003) Proc. Natl. Acad. Sci. USA 100, 3948-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, N., Berriman, J., Petrovich, M., Sharpe, T. D., Finch, J. T. & Fersht, A. R. (2003) Proc. Natl. Acad. Sci. USA 100, 9814-9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, S. E. (1998) Folding Des. 3, R81-R91. [DOI] [PubMed] [Google Scholar]
- 19.Wolynes, P. G., Onuchic, J. N. & Thirumalai, D. (1995) Science 267, 1619-1620. [DOI] [PubMed] [Google Scholar]
- 20.Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. (1997) Annu. Rev. Phys. Chem. 48, 545-600. [DOI] [PubMed] [Google Scholar]
- 21.Luque, I., Leavitt, S. A. & Freire, E. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 235-256. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Mira, M. M., Sadqi, M., Fischer, N., Sanchez-Ruiz, J. M. & Munoz, V. (2002) Science 298, 2191-2195. [DOI] [PubMed] [Google Scholar]
- 23.Hao, M. H. & Scheraga, H. A. (1998) J. Mol. Biol. 277, 973-983. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D. E., Gu, H. & Baker, D. (1998) Proc. Natl. Acad. Sci. USA 95, 4982-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karanicolas, J. & Brooks, C. L., III (2003) Proc. Natl. Acad. Sci. USA 100, 3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks, B. R., Bruccoleri, R. E., Olafson, B., States, D., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem. 4, 187-217. [Google Scholar]
- 27.MacKerell, A. D., Jr., Bashford, D., Bellott, M., Dunbrack, J. D., Evanseck, M. J., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Phys. Chem. B 102, 3586-3616. [DOI] [PubMed] [Google Scholar]
- 28.Feig, M., MacKerell, A. D., Jr., & Brooks, C. L., III. (2003) J. Phys. Chem. B 107, 2831-2836. [Google Scholar]
- 29.Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. (1977) J. Comput. Phys. 23, 327-341. [Google Scholar]
- 30.Lee, M. S., Salsbury, F. R., Jr., & Brooks, C. L., III (2002) J. Chem. Phys. 116, 10606-10614. [Google Scholar]
- 31.Lee, M. S., Feig, M., Salsbury, F. R., Jr., & Brooks, C. L., III (2003) J. Comput. Chem. 24, 1348-1356. [DOI] [PubMed] [Google Scholar]
- 32.Bashford, D. & Case, D. A. (2000) Annu. Rev. Phys. Chem. 51, 129-152. [DOI] [PubMed] [Google Scholar]
- 33.Nina, M., Beglov, D. & Roux, B. (1997) J. Phys. Chem. 101, 5239-5248. [Google Scholar]
- 34.Boczko, E. M. & Brooks, C. L., III (1995) Science 269, 393-396. [DOI] [PubMed] [Google Scholar]
- 35.Sheinerman, F. B. & Brooks, C. L., III (1998) Proc. Natl. Acad. Sci. USA 95, 1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macias, M. J., Gervais, V., Civera, C. & Oschkinat, H. (2000) Nat. Struct. Biol. 7, 375-379. [DOI] [PubMed] [Google Scholar]
- 37.Ranganathan, R., Lu, K. P., Hunter, T. & Noel, J. P. (1997) Cell 89, 875-886. [DOI] [PubMed] [Google Scholar]
- 38.Sugita, Y. & Okamoto, Y. (1999) Chem. Phys. Lett. 314, 141-151. [Google Scholar]
- 39.Sugita, Y. & Okamoto, Y. (2002) in Lecture Notes in Computational Science and Engineering, eds. Gan, H. H. & Schlick, T. (Springer, Berlin), pp. 303-331.
- 40.Feig, M., Karanicolas, J. & Brooks, C. L., III (2004) J. Mol. Graph. Model, in press. [DOI] [PubMed]
- 41.Ferrenberg, A. M. & Swendsen, R. H. (1989) Phys. Rev. Lett. 63, 1195-1198. [DOI] [PubMed] [Google Scholar]
- 42.Boczko, E. M. & Brooks, C. L., III (1993) J. Phys. Chem. 97, 4509-4513. [Google Scholar]
- 43.Kumar, S., Bouzida, D., Swendsen, R. H., Kollman, P. A. & Rosenberg, J. (1992) J. Comput. Chem. 13, 1011-1021. [Google Scholar]
- 44.Lee, B. & Richards, F. M. (1971) J. Mol. Biol. 55, 379-400. [DOI] [PubMed] [Google Scholar]
- 45.Jernigan, R. L. & Bahar, I. (1996) Curr. Opin. Struct. Biol. 6, 195-209. [DOI] [PubMed] [Google Scholar]
- 46.Bilsel, O. & Matthews, C. R. (2000) Adv. Protein Chem. 53, 153-207. [DOI] [PubMed] [Google Scholar]
- 47.Shea, J.-E., Onuchic, J. N. & Brooks, C. L., III (2002) Proc. Natl. Acad. Sci. USA 99, 16064-16068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khorasanizadeh, S., Peters, I. D. & Roder, H. (1996) Nat. Struct. Biol. 3, 193-205. [DOI] [PubMed] [Google Scholar]
- 49.Baldwin, R. L. (1986) Proc. Natl. Acad. Sci. USA 83, 8069-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pires, J. R., Taha-Nejad, F., Toepert, F., Ast, T., Hoffmuller, U., Schneider-Mergener, J., Kuhne, R., Macias, M. J. & Oschkinat, H. (2001) J. Mol. Biol. 314, 1147-1156. [DOI] [PubMed] [Google Scholar]
- 51.Morris, A. L., MacArthur, M. W., Hutchinson, E. G. & Thornton, J. M. (1992) Proteins 12, 345-364. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 53.Toepert, F., Pires, J. R., Landgraf, C., Oschkinat, H. & Schneider-Mergener, J. (2001) Angew. Chem. Int. Ed. Engl. 40, 897-900. [DOI] [PubMed] [Google Scholar]
- 54.Kasanov, J., Pirozzi, G., Uveges, A. J. & Kay, B. K. (2001) Chem. Biol. 8, 231-241. [DOI] [PubMed] [Google Scholar]
- 55.Lu, P. J., Zhou, X. Z., Shen, M. & Lu, K. P. (1999) Science 283, 1325-1328. [DOI] [PubMed] [Google Scholar]
- 56.Goldstrohm, A. C., Albrecht, T. R., Sune, C., Bedford, M. T. & Garcia-Blanco, M. A. (2001) Mol. Cell. Biol. 21, 7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bedford, M. T., Chan, D. C. & Leder, P. (1997) EMBO J. 16, 2376-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen, J. T., Turck, C. W., Cohen, F. E., Zuckermann, R. N. & Lim, W. A. (1998) Science 282, 2088-2092. [DOI] [PubMed] [Google Scholar]
- 59.Lee, C., Park, S. H., Lee, M. Y. & Yu, M. H. (2000) Proc. Natl. Acad. Sci. USA 97, 7727-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright, P. E. & Dyson, H. J. (1999) J. Mol. Biol. 293, 321-331. [DOI] [PubMed] [Google Scholar]
- 61.Chiti, F., Taddei, N., Giannoni, E., van Nuland, N. A., Ramponi, G. & Dobson, C. M. (1999) J. Biol. Chem. 274, 20151-20158. [DOI] [PubMed] [Google Scholar]
- 62.Matagne, A., Radford, S. E. & Dobson, C. M. (1997) J. Mol. Biol. 267, 1068-1074. [DOI] [PubMed] [Google Scholar]
- 63.Heidary, D. K., O'Neill, J. C., Roy, M. & Jennings, P. A. (2000) Proc. Natl. Acad. Sci. USA 97, 5866-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu, H., Columbus, J., Zhang, Y., Wu, D., Lian, L., Yang, S., Goodwin, J., Luczak, C., Carter, M., Chen, L., et al. (2004) Proteomics, in press. [DOI] [PubMed]