Abstract
Basonuclin is a zinc finger protein specific to basal keratinocytes and germ cells. In keratinocytes, basonuclin behaves as a stem cell marker and is thought to be a transcription factor that maintains proliferative capacity and prevents terminal differentiation. The human gene is located on chromosome 15. We have discovered in the chicken the existence of basonuclin 2, a basonuclin homolog. We also report the entire sequence of mouse and human basonuclin 2; the corresponding genes are located on mouse chromosome 4 and human chromosome 9. Although the amino acid sequence of basonuclin 2 differs extensively from that of basonuclin 1, the two proteins share essential features. Both contain three paired zinc fingers, a nuclear localization signal, and a serine stripe. The basonuclin 2 mRNA has a wider tissue distribution than the basonuclin 1 mRNA: it is particularly abundant in testis, kidney, uterus, and intestine. The extreme conservation of the basonuclin 2 amino acid sequence across vertebrates suggests that basonuclin 2 serves an important function, presumably as a regulatory protein of DNA transcription.
Basonuclin is a cell-type-specific zinc finger protein. The cDNA for basonuclin was originally isolated from cultured human keratinocytes (1), but the protein was later found in male and female germ cells, and in corneal and lens epithelia. No other cell types contain detectable amounts of basonuclin (1–4). The translated sequence of the cDNA showed that the protein contains three pairs of C2H2 zinc fingers (Zfs), a nuclear localization signal (NLS), and a serine-rich region (1). The human gene is located on chromosome 15 (5). Basonuclin is found in basal keratinocytes of epidermis and other stratified squamous epithelia but not in terminally differentiated cells. In rapidly growing keratinocytes basonuclin is mostly nuclear, whereas it becomes predominantly cytoplasmic in slowly growing cells (6). Basonuclin is thought to be a transcription factor that maintains the proliferative capacity of keratinocytes and prevents their terminal differentiation (7). In spermiogenesis, basonuclin moves from a nuclear to a cytoplasmic compartment (4). Nuclear import of basonuclin is thought to depend on an unphosphorylated state of a single serine residue (8). Basonuclin is quite conserved in evolution: mouse basonuclin is 88% identical in amino acid sequence to that of the human (9).
We have recently described the culture of chicken keratinocytes, a hitherto uncultivable cell type (A.V., T. Londero, N. Ghinea, and P.D., unpublished data). To study the differentiation of chicken keratinocytes, we decided to isolate the chicken orthologs of factors important for keratinocyte differentiation, such as basonuclin. In this process, we discovered that one EST present in a chick EST database encoded a hitherto undiscovered basonuclin homolog that we have designated basonuclin 2. We report the entire sequence of mouse and human basonuclin 2, as well as the partial sequences of the chicken and zebrafish proteins. Like basonuclin 1, basonuclin 2 is a zinc finger protein; it has a wider tissue distribution than basonuclin 1. The extreme conservation of basonuclin 2 among vertebrates strongly argues for an important function.
Materials and Methods
Cell Culture. Human epidermal keratinocytes derived from fore-skin of a newborn (strain YF23) were propagated on mitomycin-treated 3T3-J2F cells (2.3 × 104 3T3 cells per cm2) in a 1:1 mixture of Dulbecco–Vogt and F12 medium (Invitrogen) supplemented with 10% FCS (HyClone), 1.15 g/liter glucose (Sigma), 0.4 μg/ml hydrocortisone (Calbiochem), 5 μg/ml bovine insulin (Sigma), 2 × 10-9 M 3,3′,5-triiodo-l-thyronine (Sigma), 10-10 M cholera toxin (ICN), and 1.8 × 10-4 M adenine (Calbiochem). Recombinant human epidermal growth factor (Invitrogen) was added at the first medium change, at a concentration of 10 ng/ml (10–12). For experiments, cells were used in their seventh subculture.
Northern Analysis. Probes for human basonuclin 1 and 2 were generated by PCR amplification of genomic DNA (250 ng). The following primers were used: human basonuclin 2 (733 bp), 5′-CCA ATG GGT TTT ACC ACT CC (amino acids 471–477) and 5′-T GGG GTC TGT AAA TTC TTC C (amino acids 714–708) (see Fig. 5); and human basonuclin 1 (802 bp), 5′-TCT GAG AAC TAC AAG TGC CC (amino acids 434–440) and 5′-C TTT AGA ATC TTC AAG ACA AGG (amino acids 701–694) (1). The PCR product was the result of 30 cycles of amplification (95°C for 1 min, 57°C for 1 min, and 72°C for 1 min).
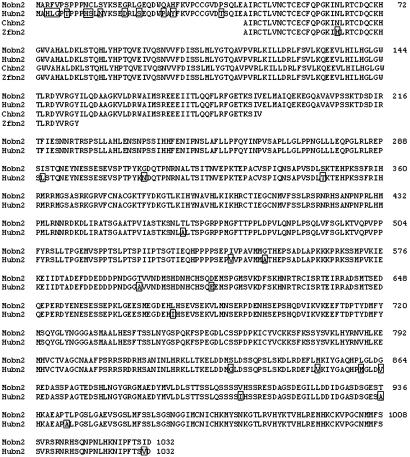
Fig. 5.
Alignment of the amino acid sequences of basonuclin 2 of various species. The deduced amino acid sequences of the entire human and mouse basonuclin 2 have been aligned with partial sequences of the chicken and zebrafish. Divergent residues are boxed. The protein is practically identical in all species examined. Mouse sequence (Mobn2) was from GenBank entry NM_172870. The human sequence (Hubn2) was assembled from exons found on BACs AL450105, AL450003, and AL449983 (see Fig. 7). The chicken sequence (Chbn2) was derived from EST 603508405F1 (http://chick.umis-t.ac.uk). The zebrafish sequence (Zfbn2) was from two exons on BAC BX000462.
RNA blots were purchased from Clontech. Hybridization, washes, and dehybridizations were carried out as described by the manufacturer.
RT-PCR. Tissues were resected from two male mice (C57 black) and immediately frozen by immersion in liquid nitrogen, except for the epidermis, which was separated from the dermis with thermolysine before freezing (13). The frozen tissue was ground with mortar and pestle under nitrogen. Total RNA was extracted by using the TriPure isolation reagent (Roche Applied Science). Human cultured keratinocytes (six confluent 100-mm plates) were directly scraped into the TriPure reagent.
RT-PCR was carried out by using the SuperScript First-Strand synthesis system for RT-PCR (Invitrogen Life Technologies). First-strand synthesis was initiated from 4 μg of total RNA with oligo(dT). For human keratinocytes, the PCR primers were those used to generate the basonuclin 1 and 2 probes described under Northern analysis. For mouse tissues, the following primers were used: mouse basonuclin 2 (894 bp), 5′-AGC AGT GAA TCT GAA GTA TCG (amino acids 299–305) and 5′-GCC GTC ATT AGG ATC GTC G (amino acids 596–591); and mouse basonuclin 1 (810 bp), 5′-AC CTG GCA AGC TCT GAG ACC (amino acids 399–405) and 5′-C TTT AGA CTC TTC CAG AAA AGG (amino acids 669–662). Cycling temperatures and number of cycles used to amplify the two mouse basonuclins were the same as those used to amplify the human basonuclins from genomic DNA (see Northern Analysis). This process generated enough PCR product to allow detection by ethidium bromide staining after agarose gel electrophoresis.
Bands on agarose gels were quantitated with an Agfa digital light-sensing scanner and the fotolook v.2.07 program; images were analyzed with the imagej program (nih image for PC). The relation of the intensity of the fluorescent band to the amount of basonuclin 1 or basonuclin 2 mRNA present in the sample was determined by serial dilutions of the cDNAs before PCR amplification. Under the conditions used, the amount of PCR product generated was proportional to the amount of basonuclin 1 or basonuclin 2 cDNA present in the sample and therefore presumably to the amount of mRNA.
Results
Identification of Chicken ESTs Encoding Basonuclin 1 and 2. The Biotechnology and Biological Sciences Research Council chick database (http://chick.umist.ac.uk) contains nearly 340,000 EST sequences derived from a variety of adult and embryonic tissues (14). When the database was searched with the keyword basonuclin, three ESTs were identified, all derived from early embryos (stages 20–22 corresponding to 3–4 days after fertilization). The accession numbers of the ESTs were 603796838F1, 603508405F1, and 603755772F1, but in this paper they will be designated as 38F1, 05F1, and 72F1. 38F1 and 72F1 partially overlapped and their sequences were identical in the overlapping part. 05F1 did not show any identity with the other two sequences.
For each EST, the database provides a report of a similarity search of GenBank by using blastx (six-frame translation of the query). The reports showed that translated 38F1 and 72F1 aligned with the central part of mouse and human basonuclin (GenBank proteins Q01954 and O035914, respectively), whereas 05F1 aligned with the N terminus of basonuclin. However, we were surprised to find that all three ESTs also gave highly significant alignments with an unknown putative 1,032-residue mouse protein (Swiss-Prot Q8BMQ3). In the case of 05F1, the alignment with Q8BMQ3 was better than with mouse or human basonuclin. We concluded that 05F1 encoded the chicken ortholog of Q8BMQ3, a protein related to but distinct from basonuclin, whereas 38F1 and 72F1 encoded fragments of the chicken ortholog of basonuclin. To distinguish it from the previously known basonuclin (1) we designate the sequence of Q8BMQ3 as basonuclin 2. We will thereafter designate the basonuclin originally described by Tseng and Green (1) as basonuclin 1.
Relatedness of Basonuclin 1 and 2 of the Mouse. Protein Q8BMQ3 is the translated sequence of a single large ORF found in a 3,385-bp mouse mRNA, whose sequence has been deposited in the GenBank database under accession number NM_172870. This sequence is likely to encode the entire basonuclin 2 protein. The first ATG codon at nucleotide 110 is predicted to be a strong initiator because it is followed by a G and preceded by a purine, 3 bases upstream (15). A termination codon is present ≈200 nt before the end of the mRNA sequence. The deduced amino acid sequence of basonuclin 2 is shown in Fig. 1 aligned with that of mouse basonuclin 1. The predicted protein contains 1,032 residues; its molecular weight is 115 kDa. Like basonuclin 1, basonuclin 2 contains six zinc finger motifs in separated pairs, a NLS, and a serine stripe (1).
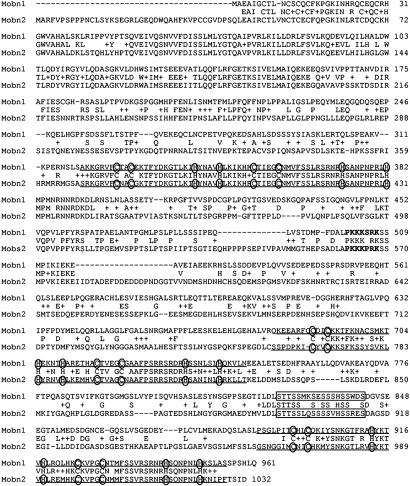
Fig. 1.
Alignment of the amino acid sequences of mouse basonuclin 1 and 2. The main features of the basonuclin 2 sequence (Mobn2) are the six zinc fingers (underlined) with their cysteines and histidines (circled), the NLS (bold type), and the serine stripe (framed). These features are also found in basonuclin 1 (Mobn1). The basonuclin 1 sequence is from Matsuzaki et al. (9). The middle sequence corresponds to residues shared by basonuclin 1 and 2. +, Conservative amino acid replacement.
The overall identity of mouse basonuclin 1 and 2 is 43.4% at the amino acid level, but identities are not evenly distributed. A segment of 158 aa (residues 43–200 in basonuclin 2) toward the N terminus is highly conserved between the two proteins (79.1% identity), although it does not contain any obvious structural elements that could be related to function (Fig. 1).
In the rest of the molecule, the similarity is strongest between the zinc fingers. In basonuclin 2, as in basonuclin 1, zinc fingers 1, 3, and 5 contain the characteristic sequence C-X2-C-X12-H-X4-H, whereas zinc fingers 2, 4, and 6 contain the sequence C-X4-C-X12-H-X6–8-H (1). The spacing between the zinc-binding cysteine and histidine residues is identical in each corresponding zinc finger of the two basonuclins. Zinc fingers 1 and 2 are particularly conserved (92.1% identity), zinc finger 2 being virtually identical in the two proteins. Zinc fingers 4 and 6 are also well conserved, but zinc fingers 3 and 5 (particularly 3) show numerous divergences. The X12 region between the second cysteine and the first histidine contains the amino acids that establish contact with DNA (16). In finger 3, numerous divergences lie in this region. However all of the lysine residues are conserved as well as one serine and the only phenylalanine residue. Lysine and serine are common base-contacting amino acids in other zinc finger proteins (17). It had been noted by Tseng and Green (1) that all of the zinc fingers of basonuclin had an aromatic residue at the fourth position of the X12 region.
Like basonuclin 1, basonuclin 2 possesses an NLS, located between the second and third zinc fingers. The NLS of basonuclin 2 (PKKKPRK) differs from that of basonuclin 1 (PKKKSRK) by a serine to proline replacement (Fig. 1) but is still in good agreement with the class A NLS consensus sequence (XXK-R/K-X-R/KX) (18).
A serine-rich region, the serine stripe, is found between the fourth and fifth zinc fingers of basonuclin 1. Basonuclin 2 contains a serine stripe in a similar position, and all of the serine residues but one are conserved between basonuclin 1 and 2. A similar stripe is present in the zinc finger protein PRDII BFI (19). The serine stripe may have an important function in all of these proteins.
The Genomic Sequence of Basonuclin 2. A blastn (nucleotide/nucleotide) similarity search of GenBank by using the entire sequence of the mRNA for mouse basonuclin 2 revealed that the gene was spread over four nonoverlapping bacterial artificial chromosomes (BAC). All four BACs were derived from chromosome 4, on which the mouse basonuclin 2 gene must therefore reside. This search also identified the corresponding human exons spread over three BACs derived from chromosome 9. In the zebrafish genome, sequences similar to human and mouse exons 3 and 4 were also found on a single BAC (see Fig. 7, which is published as supporting information on the PNAS web site). We conclude that basonuclin 2 is present in fishes. The nucleotide sequences of the BACs had been submitted directly to GenBank by the Wellcome Trust Sanger Institute (Hinston, U.K.).
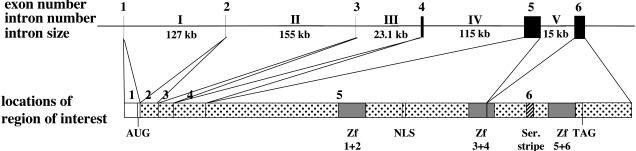
The alignment of the basonuclin 2 mRNA sequence with the four mouse BACs showed the presence of six exons. The size of the mouse gene could not be measured precisely because some consecutive exons lay on nonoverlapping BACs and the sizes of the introns separating them (introns I, II, and IV) could therefore not be determined. However, the sizes of introns I, II, and IV could be exactly determined in the human. Assuming that the sizes of introns I, II, and IV are conserved between the mouse and the human, the size of the mouse basonuclin 2 gene would be 435 kb. The positions of the exons and introns are shown in Fig. 2. Exons 1–4 are short and do not contain any of the regions of interest recognized in the mRNA sequence. Exon 5 is 1,857 bp in length and contains the first three zinc fingers and the NLS. This exon ends within the fourth zinc finger, one nucleotide before the histidine codon. Exon 6 encodes the rest of the fourth finger, as well as zinc fingers 5 and 6 and the serine stripe.
Fig. 2.
Map of the mouse basonuclin 2 gene. Exons are represented by black boxes; they are designated by Arabic numerals. Introns are indicated by Roman numerals. Introns are not drawn to scale. The regions of interest of each of the six exons are assigned.
The transcription start site could not be precisely identified. The genomic region upstream of the initiating methionine is extremely GC rich and contains numerous Sp1 sites, but no TATA-box could be identified. The promoter of basonuclin 2, like that of basonuclin 1, is likely to be a TATA-less promoter.
The overall structure of the basonuclin 2 gene is very similar to that of basonuclin 1. The basonuclin 2 gene possesses one additional exon, at its 5′ end, whose main purpose seems to provide an initiating methionine codon. Otherwise, the five other exons of basonuclin 2 appear to be homologous to the five basonuclin 1 exons: their sizes are in good agreement, and they are interrupted by introns at exactly the same positions (Fig. 3). The two basonuclin genes must have been generated by duplication of an ancestral gene. We found in GenBank a truncated cDNA that is likely to encode zinc fingers 5 and 6 of zebrafish basonuclin 1 (accession no. AI877747). Like basonuclin 2, basonuclin 1 is present in fish, and the duplication that generated the two genes must have occurred in the ancestral fish lineage. The gene for basonuclin 1 has been mapped to human chromosome 15 (5), whereas the basonuclin 2 gene is located on human chromosome 9; the considerable length of time during which the two genes have coexisted probably explains why they are located on different human chromosomes.
Fig. 3.
Comparison of intron boundaries of basonuclin 1 and 2. Numbers in parentheses indicate the amino acid position of mouse basonuclin 2 (see Fig. 1) at which intron interrupt exons.
Additional Exons and Alternate Splicing. A blastn similarity search of GenBank by using NM_172870 revealed a second mouse basonuclin 2 mRNA sequence (accession no. AK031440) and five partial human basonuclin 2 mRNA sequences. Some of these mRNAs differ from NM_172870 and from each other in that they contain additional exons or use different poly(A) addition sites. The various splicing isoforms are shown in Fig. 4.
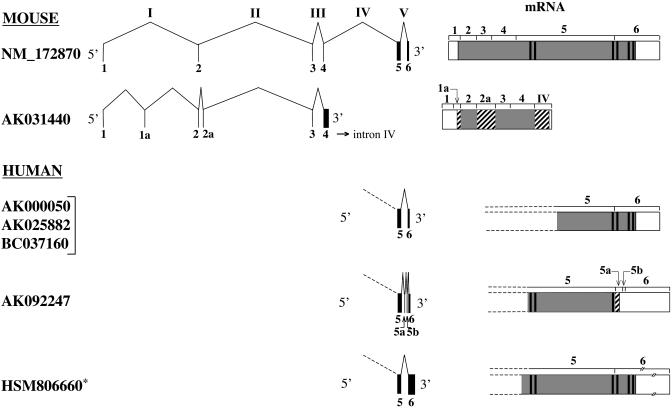
Fig. 4.
Alternative splicing of basonuclin 2 gene. The accession numbers of the various mouse and human mRNAs found in GenBank are given. The asterisk indicates an error in the sequence of HSM806660 reported in GenBank. An adenine between nucleotides 1254 and 1255 is missing; this adenine is found on BAC AL449983. The primary transcript can be spliced in different ways (introns are designated with Roman numerals) and translated into different proteins, some of which lack all or some of the zinc fingers (thick vertical bars in the protein). Regions of proteins shared with basonuclin 2 (NM_172870) are shaded, whereas regions that differ are cross-hatched. UTRs are empty. Dashed lines show that some protein sequences are incomplete because they are deduced from partial-length cDNAs. AK031440 contains two additional exons (1a and 2a). Exon 1a encodes the putative initiating methionine; exon 2a is in frame with exons 2 and 4. The AK031440 coding region terminates within intron IV. AK000050, AK025882, and BC037160 are similar to NM_172870. AK092247 contains two additional small exons (5a and 5b) at its 3′ end and terminates within exon 5a. It therefore lacks zinc fingers 4–6. HSM806660 uses a poly(A) addition site located 2 kb downstream from that used by the other human transcripts. It encodes the usual protein. Slashes indicate that exon 6 of HSM806660 is not drawn to scale.
AK031440, which is derived from mouse embryonic testis, shares with basonuclin 2 exons 1–4; it possesses two additional exons in the 5′ region (exons 1a and 2a) but entirely lacks exons 5 and 6 and is therefore predicted to encode a short protein. Its sequence is shown aligned with the corresponding part of mouse basonuclin 2 in Fig. 8, which is published as supporting information on the PNAS web site. An ORF encoding 288 aa starts in exon 1a and extends to the 5′ end of intron IV, where it terminates after ≈180 nt. Because exon 1a is out of frame with exon 1, the first ATG is located in exon 1a and not in exon 1 as in basonuclin 2. The first ATG codon of AK031440 conforms to the Kozak consensus sequence (15). The extra exons 1a and 2a are in the same reading frame used by basonuclin 2 in exons 2–4. As a result, AK031440 and basonuclin 2 share the same sequence in the parts encoded by exons 2–4. Because it lacks exons 5 and 6, AK031440 does not contain either zinc fingers or a NLS. Although the human orthologs of mouse exons 1a and 2a could not be found among the human basonuclin 2 mRNA sequences deposited in GenBank, they were identified on the human BACs (Fig. 7). The nucleotide sequences of exons 1a and 2a show 97% and 95% identity, respectively, between mouse and human, whereas the surrounding intronic sequences show only ≈75% identity between the two species. Exon 1a was also found in the chicken basonuclin 2 cDNA 05F1.
The human basonuclin 2 mRNAs found in GenBank are all truncated at their 5′ end. Three classes can be distinguished. The first class that includes AK000050, AK025882, and BC037160 conforms to mouse basonuclin 2. A single mRNA is found in the second class (AK092247); it contains two small extra exons (5a and 5b) between exons 5 and 6. Because exon 5a places a stop codon immediately downstream of exon 5, the entire sequence encoded by exon 6 is lacking. As a result, the putative protein lacks zinc fingers 4–6 and the serine stripe, but retains the NLS. The third class mRNA (HSM806660) possesses a very long 3′ UTR (2582 bases) because it uses a poly(A) addition site located ≈2 kb downstream from the one used by the other human transcripts. The protein encoded by HSM806660 conforms to mouse basonuclin 2.
Conservation of Basonuclins in Vertebrates. Although all human basonuclin 2 mRNA sequences found in GenBank were incomplete, we could assemble the entire human sequence by using the mouse exons as guides in the identification of the human exons present on the three BACs shown in Fig. 7. The nucleotide sequences and the deduced amino acid sequences of human and mouse basonuclin 2 were aligned with those of the chicken (translation of EST 05F1), and with a partial sequence of zebrafish basonuclin 2 (exons 3 and 4) found on BAC BX000462 (Fig. 5). The protein is extremely conserved over its all length, more so than basonuclin 1. A comparison of mouse and human basonuclin 2 coding regions showed 91% identity at the nucleotide level and 97.2% identity at the amino acid level. The corresponding values for basonuclin 1 are 87% and 88%, respectively (9). Silent substitutions represent 89% of total nucleotide divergences between the human and mouse basonuclin 2 coding regions; the corresponding value for basonuclin 1 is 55% (see table 2 in ref. 2). In the parts of the coding region that were compared between chicken, zebrafish, and mouse, virtually all nucleotide substitutions were silent (Table 1). The large number of silent nucleotide substitutions in the basonuclin 2 coding regions of the species examined suggests that the protein is under strong selective pressure.
Table 1. Divergence of mouse basonuclin 2 coding region from that of other species.
| Sequence examined, residue position | Amino acid replacements | Nucleotide substitutions | Silent nucleotide substitutions | |
|---|---|---|---|---|
| Human | 1-1032 | 29/1,032 (2.8%) | 280/3,096 (9%) | 249/280 (88.9%) |
| Chicken | 44-191 | 0/148 (0%) | 57/444 (12.8%) | 57/57 (100%) |
| Zebrafish | 44-155 | 1/112 (0.9%) | 61/336 (18.1%) | 60/61 (98.4%) |
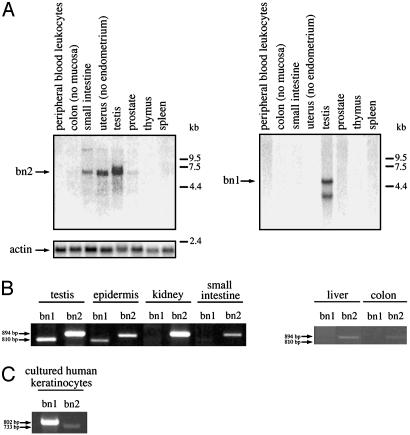
Presence of Basonuclin 2 mRNA in Various Tissues. To avoid any possible cross-hybridization of the basonuclin 2 probe with the basonuclin 1 mRNA, we used as basonuclin 2 probe a 733-bp PCR fragment corresponding to the region located between zinc fingers 2 and 3. This region differs extensively between basonuclin 1 and 2 (Fig. 1). When a collection of human tissue samples was examined by Northern analysis, a single transcript at ≈6 kb was detected with the probe used. It was found that testis, uterus, and small intestine were the tissues that gave the strongest signals. Colon and prostate were weakly positive. No mRNA was detected in either blood, thymus, or spleen. When the same blot was rehybridized with a basonuclin 1-specific probe, testis was the only tissue of eight examined that gave a detectable signal.
To quantify more precisely the relative amounts of the basonuclin 1 and 2 mRNAs in various tissues, we prepared total RNA from a number of mouse tissues and carried out RT-PCR reactions specific for each of the two basonuclin mRNAs. This analysis revealed the presence of basonuclin 2 mRNA in all of the tissues examined, but in very variable amounts. The basonuclin 2 mRNA was most abundant in testis and kidney, and less abundant in epidermis and small intestine. Small amounts were found in liver and colon. The amount in testis and kidney was estimated to be five times that of epidermis and eight times that of small intestine. As expected, the basonuclin 1 mRNA was detected only in testis and epidermis (7), where its amount was about one-third to one-half that of the basonuclin 2 transcript (Fig. 6B).
Fig. 6.
Presence of basonuclin 2 mRNA in various tissues. (A) A Northern blot containing 2 μg of poly(A)+ RNA prepared from various human tissues was purchased from Clontech. RNAs were successively hybridized to a 733-bp basonuclin 2-specific probe (Left), an 802-bp basonuclin 1 probe (Right), and a human actin probe. Testis, uterus, and intestine gave appreciable signals for basonuclin 2, prostate gave a weak signal, and colon gave a trace signal. All other tissues were negative. The human basonuclin 2 mRNA was estimated at 6 kb. Testis was the only tissue that gave a signal for basonuclin 1; two transcripts at 4.6 and 3.2 kb were present, as described in ref. 3. The sources of the RNAs are indicated. (B) RT-PCR analysis of total RNA prepared from various adult mouse tissues by using oligonucleotide primers designed to amplify specifically an 894-bp basonuclin 2 fragment and an 810-bp basonuclin 1 fragment. The PCR products were resolved on a 1% agarose gel and visualized by ethidium bromide staining. The mouse basonuclin 2 mRNA is found in all tissues examined. It is particularly abundant in testis and kidney and less abundant in epidermis and small intestine. Liver and colon contain trace amounts (the image is overexposed). The basonuclin 1 mRNA is only found in testis and epidermis, in which it is less abundant than the basonuclin 2 mRNA. (C) RT-PCR analysis of total RNA prepared from human cultured keratinocytes. The mRNA for basonuclin 1 (802-bp fragment) is ≈10 times more abundant than that for basonuclin 2 (733-bp fragment).
We then measured by RT-PCR the relative amounts of the basonuclin 1 and 2 mRNAs in cultured human keratinocytes. In contrast to mouse epidermis, the basonuclin 1 mRNA predominated, and in three experiments, its amount was found to be ≈10 times that of the basonuclin 2 mRNA (Fig. 6C).
Discussion
A pair of zinc fingers appears to be the smallest unit conferring specific DNA binding to proteins with multiple separated paired zinc fingers (20, 21). Because basonuclin 1 and 2 each contain three different pairs of zinc fingers, they could each bind three different DNA motifs. It has been shown that the first pair, but not the second pair of basonuclin 1, binds to the rRNA promoter (22). As shown in Fig. 1, the first pair of zinc fingers is virtually identical in basonuclin 1 and 2. It is therefore likely that basonuclin 2 will also bind the rRNA promoter. Basonuclin 1 and 2 would therefore possess the common function of increasing transcription of the ribosomal RNA genes (23). The second and third pairs of zinc fingers, which differ significantly between basonuclin 1 and 2, may have functions related to different targets.
There is a remarkable conservation of the amino acid sequence of basonuclin 2 across species as distant as the zebrafish, the chicken, and the mammals. Interspecies comparisons at the nucleotide level show that the gene has undergone numerous silent nucleotide substitutions (Table 1), suggesting a strong selective pressure against amino acid replacements over the whole length of the molecule. In contrast, the basonuclin 1 gene is mostly conserved in functionally meaningful regions, such as the zinc fingers (9). When the basonuclin 1 and 2 amino acid sequences are compared, the zinc fingers are also the most conserved regions, but also conserved are the N-terminal 160 aa to which no function has yet been attributed.
Another extremely conserved regulatory protein of epithelial cells is p63, which shows 95% identity between the chicken and the human at the amino acid level (24). p63 is most abundant in the basal layer of a number of epithelia (25). It must serve an essential function in keratinocytes, because targeted disruption of the gene leads to absence of all stratified squamous epithelia (26). Basonuclin 2 is also likely to have an essential function in the cells in which it is found.
One could ask what the significance is of the numerous splicing variants that we have observed (Fig. 4), particularly when they encode shortened proteins lacking some or all of the zinc fingers, as does AK031440. It is worth noting that (i) the first methionine codon of the large ORF of AK031440 is predicted to be a strong initiator, (ii) the extra exons of AK031440 are in the same reading frame as the exons shared with basonuclin 2, and (iii) these extra exons are well conserved across species as distant as the human and the chicken. One hypothesis would be that AK031440 dimerizes with basonuclin 2 and regulates its activity by a dominant negative effect. Such a hypothesis would be consistent with the fact that both mRNAs are found in testis. Alternatively, AK031440 could serve a function totally unrelated to that of basonuclin 2.
Two antibodies have been extensively used in the study of basonuclin 1. The antibody prepared by Tseng and Green (7) was raised against residues 72–203, a region of basonuclin 1 that is substantially conserved with basonuclin 2 (Fig. 1). The second antibody, described by Iuchi and Green (8), was prepared by using as antigen nearly all of basonuclin 1. Because both antibodies were polyclonal, it is very likely that both would contain antibody specificities directed against epitopes shared by the two basonuclins. Both antibodies would therefore be expected to show some level of cross-reactivity with basonuclin 2. In sections of testis stained with the antibody of Tseng and Green, basonuclin 1 was found in the germ cells (4), and in sections of skin and hair follicles stained with the antibody of Iuchi and Green, basonuclin 1 was confined to keratinocytes (27). In neither testis nor skin was there any appreciable staining of the mesenchymal tissue. We conclude that basonuclin 2 is similarly absent from mesenchymal tissue and is therefore likely to be confined to germ cells in testis and to epithelial cells in tissues such as kidney, intestine, and uterus where its mRNA is abundant (Fig. 6). Antibodies specific to each of the two basonuclins will have to be generated to distinguish the two basonuclins in germ cells and keratinocytes.
In mouse epidermis, basonuclin 1 is mainly concentrated in the cytoplasm, whereas in growing cultured human keratinocytes it is strongly concentrated in the cell nuclei (6). In mouse epidermis, the mRNA for basonuclin 2 is more abundant than that for basonuclin 1, whereas in cultured human keratinocytes, the basonuclin 1 mRNA is far more abundant than the basonuclin 2 mRNA (Fig. 6). Because only nuclear basonuclin 1 and 2 are likely to be active, we conclude that in keratinocytes the action of basonuclin 1 is likely to be predominant over that of basonuclin 2.
Note Added in Proof. Basonuclin 2 has been independently discovered by H. Tseng (personal communication).
Supplementary Material
Acknowledgments
We thank Dr. Howard Green for his kind gifts of the 3T3-J2F line and the YF23 keratinocyte strain. This work was supported by the Centre National de la Recherche Scientifique.
Abbreviations: BAC, bacterial artificial chromosome; NLS, nuclear localization signal.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. NM_172870).
References
- 1.Tseng, H. & Green, H. (1992) Proc. Natl. Acad. Sci. USA 89, 10311-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tseng, H., Matsuzaki, K. & Lavker, R. M. (1999) Differentiation 65, 221-227. [DOI] [PubMed] [Google Scholar]
- 3.Yang, Z.-h., Ian-Gallicano, G., Yu, Q.-C. & Fuchs, E. (1997) J. Cell Biol. 137, 657-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahoney, M. G., Tang, W., Xiang, M. M., Moss, M. B., Gerton, G. L., Stanley, J. R. & Tseng, H. (1998) Biol. Reprod. 59, 388-394. [DOI] [PubMed] [Google Scholar]
- 5.Teumer, J., Tseng, H. & Green, H. (1997) Gene 188, 1-7. [DOI] [PubMed] [Google Scholar]
- 6.Iuchi, S., Easley, K., Matsuzaki, K., Weiner, L., O'Connor, N. & Green, H. (2000) Exp. Dermatol. 9, 178-184. [DOI] [PubMed] [Google Scholar]
- 7.Tseng, H. & Green, H. (1994) J. Cell Biol. 126, 495-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iuchi, S. & Green, H. (1997) Proc. Natl. Acad. Sci. USA 94, 7948-7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzaki, K., Iuchi, S. & Green, H. (1997) Gene 195, 87-92. [DOI] [PubMed] [Google Scholar]
- 10.Rheinwald, J. G. & Green, H. (1975) Cell 6, 331-343. [DOI] [PubMed] [Google Scholar]
- 11.Allen-Hoffmann, B. L. & Rheinwald, J. G. (1984) Proc. Natl. Acad. Sci. USA. 81, 7802-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon, M. & Green, H. (1984) Cell 36, 827-834. [DOI] [PubMed] [Google Scholar]
- 13.Walzer, C., Benathan, M. & Frenk, E. (1989) J. Invest. Dermatol. 92, 78-81. [DOI] [PubMed] [Google Scholar]
- 14.Boardman, P. E., Sanz-Ezquerro, J., Overton, A. M., Burt, D. W., Bosch, E., Fong, W. T., Tickle, C., Brown, W. R. A., Wilson, S. A. & Hubbard, S. J. (2002) Curr. Biol. 12, 1965-1969. [DOI] [PubMed] [Google Scholar]
- 15.Kozak, M. (1989) J. Cell Biol. 108, 229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavletich, N. P. & Pabo, C. O. (1991) Science 252, 809-817. [DOI] [PubMed] [Google Scholar]
- 17.Wolfe, S. A., Nekludova, L. & Pabo, C. O. (1999) Annu. Rev. Biophys. Biomol. Struct. 3, 183-212. [DOI] [PubMed] [Google Scholar]
- 18.Dang, C. V. & Lee, W. M. F. (1989) J. Biol. Chem. 264, 18019-18023. [PubMed] [Google Scholar]
- 19.Fan, C. M. & Maniatis, T. (1990) Genes Dev. 4, 29-42. [DOI] [PubMed] [Google Scholar]
- 20.Keller, A. D. & Maniatis, T. (1992) Mol. Cell. Biol. 12, 1940-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iuchi, S. (2001) Cell. Mol. Life Sci. 58, 625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iuchi, S. & Green, H. (1999) Proc. Natl. Acad. Sci. USA 96, 9628-9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian, Q., Kopf, G. S., Brown, R. S. & Tseng, H. (2001) Development (Cambridge, U.K.) 128, 407-416. [DOI] [PubMed] [Google Scholar]
- 24.Yasue, A., Tao, H., Nohno, T., Moriyama, K., Noji, S. & Ohuchi, H. (2001) Mech. Dev. 100, 105-108. [DOI] [PubMed] [Google Scholar]
- 25.Yang, A., Kaghad, M., Wang, Y., Gillett, E., Fleming, M. D., Dötsch, V., Andrews, N. C., Caput, D. & McKeon, F. (1998) Mol. Cell 2, 305-316. [DOI] [PubMed] [Google Scholar]
- 26.Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R. T., Tabin, C., Sharpe, A., Caput, D., Crum, C. & McKeon, F. (1999) Nature 398, 714-718. [DOI] [PubMed] [Google Scholar]
- 27.Weiner, L. & Green, H. (1998) Differentiation 63, 263-272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.