Stu2p/XMAP215/Dis1 family proteins are evolutionarily conserved regulatory factors that use αβ-tubulin-interacting TOG (tumor overexpressed gene) domains to catalyze fast microtubule growth. Catalysis requires that these polymerases discriminate between unpolymerized and polymerized forms of αβ-tubulin, but how they do so has remained unclear. We report the structure of the TOG1 domain from Stu2p bound to yeast αβ-tubulin. TOG1 binds αβ-tubulin in a way that excludes equivalent binding of a second TOG domain. Furthermore, TOG1 preferentially binds a “curved” conformation of αβ-tubulin that cannot be incorporated into microtubules, contacting α- and β-tubulin surfaces that do not participate in microtubule assembly. Conformation-selective interactions with αβ-tubulin explain how TOG-containing polymerases discriminate between unpolymerized and polymerized forms of αβ-tubulin, and how they selectively recognize the growing end of the microtubule.
Summary: A regulatory protein controls microtubule polymerization through conformation-selective interactions with αβ-tubulin
Microtubules are highly regulated, dynamic polymers of αβ-tubulin that have essential roles in intracellular organization and chromosome segregation. Microtubules grow faster in vivo than they do in vitro (reviewed in (1)). Proteins in the Stu2p/XMAP215/Dis1 family (2–4) are the major cellular factors that promote fast microtubule elongation. These proteins contain multiple TOG (TOG: tumor overexpressed gene) domains that function as αβ-tubulin binding modules(5) and that are required for the elongation-promoting activity of this family (5, 6). The molecular mechanisms underlying the activity of these microtubule polymerases are poorly understood. Recent studies of XMAP215 suggest that it functions as a catalyst, using at least two TOG domains (6, 7).
The ability to function as a catalyst for microtubule elongation suggests that XMAP215/Stu2p family proteins use their tubulin-binding TOG domains to discriminate between different forms of αβ-tubulin: unpolymerized, in the body of the microtubule, and at the growing end of the microtubule. How they do so has remained unclear. To establish the structural basis of TOG:tubulin recognition, we determined the crystal structure of the TOG1 domain from Stu2p bound to αβ-tubulin (Fig. 1A). We obtained crystals using a polymerization-blocked mutant of yeast αβ-tubulin (8, 9). The structure was determined by molecular replacement from crystals that diffracted anisotropically to 2.88 Å (minimum Bragg spacing 3.44 Å in the weakly diffracting direction, overall completeness 74.6%) (Tables S1, S2). The structure contains GTP (guanosine triphosphate) at the exchangeable nucleotide-binding site of αβ-tubulin (Fig. 1A, inset).
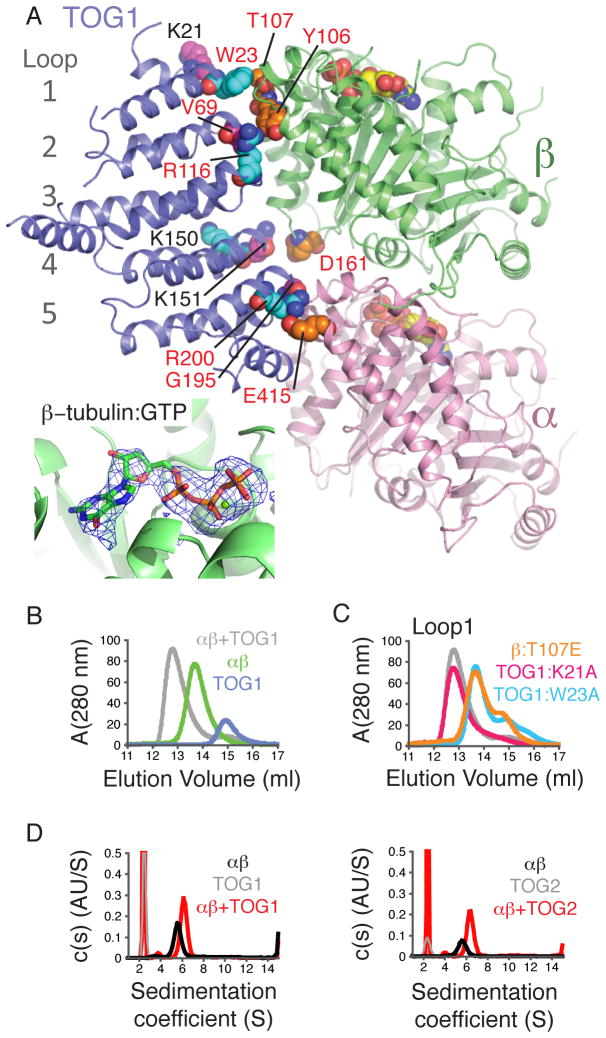
Figure 1. Structure of a TOG1:αβ-tubulin complex, revealing significant contacts with α- and β-tubulin.
A Cartoon representation of the complex (pink: α-tubulin, lime: β-tubulin, slate: TOG1). Contacts probed by mutagenesis are represented as spheres (colored to match panel C and Fig. S3), as are GTPs. (inset) mF0-DFc omit electron density map contoured at 3.5 σ and computed from a model without nucleotides: β-tubulin is bound to GTP.
B Size-exclusion chromatography assay for TOG1:αβ-tubulin interactions.
C TOG1:αβ-tubulin binding assay using interface mutants on Loop 1 (see Fig. S3).
D TOG1 (left) and TOG2 (right) (grey) each form a 1:1 complex (red) with αβ-tubulin (black) as detected by analytical ultracentrifugation (Table S4). Curves are shown from S=1 to eliminate a slowly sedimenting contaminant in the TOG2 run.
TOG1 forms a flat, layered structure similar to that observed for other TOGs (10, 11)(Fig. 1A and Fig. S1). Almost the entire narrow, evolutionarily conserved face of TOG1 (10, 11) interacts with αβ-tubulin (Fig. 1A, Fig. S1), burying approximately 1600 Å 2 of surface area, 58% attributable to the partial interface with β-tubulin. The ‘asymmetric’ mode of TOG1 binding apparently excludes analogous binding of TOG2 to the same heterodimer. This is unexpected because it had been thought that multiple TOGs could simultaneously engage a single αβ-tubulin (5, 7, 10).
We probed the importance of TOG1:αβ-tubulin contacts using site-directed mutagenesis and a gel filtration binding assay(10, 11) (Fig. 1B,C, Fig. S2,S3). TOG1:αβ-tubulin interactions were affected by mutations on α- or β-tubulin contacting loops of TOG1, or on contacted surfaces of α- or β-tubulin (e.g. W23A or R200A on TOG1, T107E on β-tubulin, E415A on α-tubulin) (Fig. 1A,C). The importance of TOG1:W23 confirms earlier studies (10, 11). The salt-bridge between TOG1:R200 and α-tubulin:E415 (Fig. S4) that is required for robust TOG1:αβ-tubulin interactions (Fig. 1C) rationalizes the strong evolutionarily conservation of R200 in TOG domains(10). Analytical ultracentrifugation revealed that TOG1 and TOG2 each bind αβ-tubulin efficiently at low μM concentration (Fig. 1D,E). Thus, TOG1:αβ-tubulin interactions detected in solution require simultaneous engagement with both α- and β-tubulin as observed in the crystal, and TOG2 can interact with unpolymerized αβ-tubulin.
We used conditional depletion of Stu2p (12) and a rescue assay to investigate how mutations on the tubulin-interacting surfaces of TOG1 or TOG2 affect Stu2p function in vivo. Wild-type Stu2p and Stu2p with a TOG1 mutation (K21A) that did not interfere with αβ-tubulin binding completely rescued the growth defect arising from the depletion of endogenous Stu2p. By contrast, Stu2p with mutations in TOG1 (W23A, V69D, or R200A) that affect αβ-tubulin binding was compromised for rescue (Fig. 2A). Mutations on the presumptive αβ-tubulin-interacting surface of TOG2 (W341A and R519A) affected rescue similarly to their TOG1 equivalents (W23A and R200A) (Fig. 2A). The R200A mutant of Stu2p displayed a defect in mitotic spindle elongation (Fig. 2B), similar to the complete removal of the TOG1 domain(5).
Figure 2. Disruptive point mutations on the tubulin-binding interfaces of TOG1 or TOG2 affect Stu2p function in vivo.
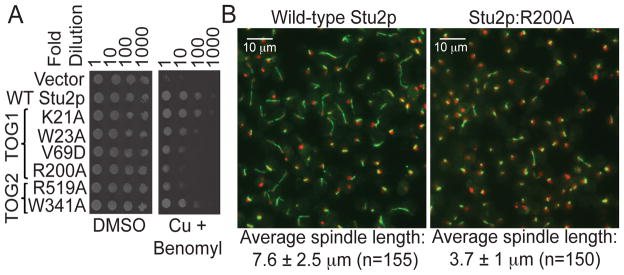
A Yeast carrying plasmid-based rescue constructs of Stu2p were plated at serial dilutions on media containing DMSO (control) or 500 μM CuSO4 (to deplete endogenous Stu2p) plus 20 μg/ml benomyl (to cause microtubule stress). TOG1 or TOG2 impaired for αβ-tubulin interactions only partially compensate for the depletion of endogenous Stu2p.
B Fluorescence images one hour after release from hydroxyurea arrest (green: αβ-tubulin; red: DNA) of yeast depleted of endogenous Stu2p and rescued with wild-type (left) or R200A (right) Stu2p. Spindle elongation is compromised when TOG1:αβ-tubulin interactions are impaired (R200A).
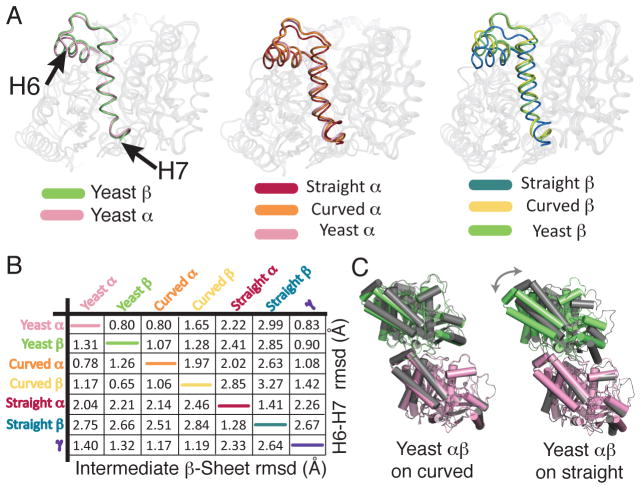
The conformations of α- and β-tubulin in the TOG1 complex are remarkably similar to each other (Fig. 3A,B, Fig. S5), to “curved” α- and β-tubulin monomer conformations previously described(13), and to γ-tubulin (14) (Fig. 3B, Figs. S5, S6). A 13° rotation is required to superimpose the α- and β-tubulin chains in the TOG1 complex. This quaternary arrangement also closely resembles that of a “curved” heterodimer (13) (12° rotation), characteristically distinct from the “straight” heterodimer (15)(~1° rotation) (Fig. 3C,D). This “curved” conformation could thus instead represent a conserved ground state of αβ-tubulin (see below). Together, these observations add further support to a model in which the role of GTP is to promote assembly by tuning the strength of polymerization contacts (16) and/or by decreasing the free energy difference between straight and curved conformations(17).
Figure 3. αβ-tubulin-GTP adopts a curved conformation.
A Superposition of yeast α (pink) and β-tubulin (lime) shows similar positioning of the H6-H7 segment (represented with darker colors).
B Superposition of yeast α- and β-tubulin onto curved (α: orange, β: yellow) and straight (α: maroon, β: dark blue) structures shows the H6-H7 segments of yeast tubulins arranged as in prior curved structures.
C Pairwise Cα r.m.s. coordinate deviations between yeast α- and β-tubulin and prior structures, computed for the subdomains indicated (Table S3).
D The quaternary structure of yeast αβ-tubulin (pink and green) resembles that of the “curved” form (grey, left), and differs from the straight form (grey, right).
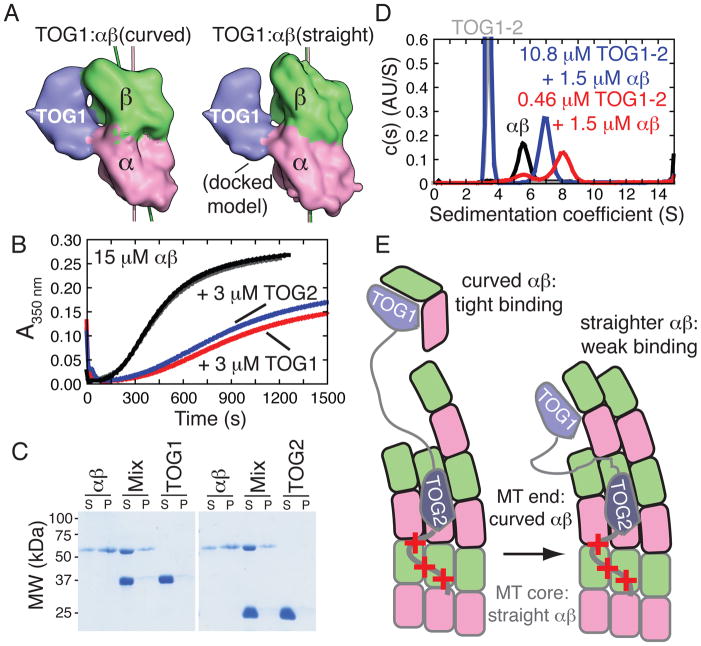
The regions of curved αβ-tubulin that engage TOG1 move relative to each other in the transition to the straight conformation (Fig. 4A). TOG1:αβ-tubulin interactions might thus be sensitive to αβ-tubulin quaternary structure, with a preference for curved αβ-tubulin. If TOG1 binds preferentially to curved αβ-tubulin, it should inhibit in vitro microtubule formation by stabilizing a microtubule-incompatible conformation of αβ-tubulin. We tested this counterintuitive prediction using microtubule assembly reactions, observing strong inhibition when TOG1 was present (Fig. 4B, Fig. S8), extending prior observations (11). Inhibition is not observed for TOG1 mutants (e.g. W23A or R200A, Fig. S9) that affect αβ-tubulin binding. TOG1 does not bind appreciably to “straight” αβ-tubulin in pre-formed microtubules (Fig. 4C) despite the apparent accessibility of the TOG1-interacting epitopes on the outside of the microtubule (Fig. S7). Thus, TOG1 binds preferentially to “curved” αβ-tubulin. We obtained similar results using TOG2 (Fig. 4B,C) indicating that it also binds preferentially to an αβ-tubulin conformation that cannot exist in the body of the microtubule.
Figure 4. TOG1:αβ-tubulin interactions are conformation-selective.
A The structure of the TOG1:αβ-tubulin complex (left) and a docked model with straight αβ-tubulin (right) illustrates how TOG1-contacting epitopes on α- and β-tubulin move relative to each other in the two conformations.
B Microtubule assembly reactions (15 μM animal αβ-tubulin) containing 3 μM TOG1 (red) or TOG2 (blue) are inhibited relative to control reactions (black and grey) that received only buffer.
C Microtubule co-sedimentation showing that TOG1 or TOG2 do not appreciably bind microtubules, even though the TOG-interacting epitopes are accessible on the outside of the microtubule (Fig. S7). S: supernatant, P: pellet.
D Size distributions showing that substoichiometric concentrations of TOG1-TOG2 mixed with αβ-tubulin (red) behave as a complex that sediments faster than αβ-tubulin alone (black) and the TOG1-TOG2:(αβ)1 complex that results when TOGs are in molar excess over αβ-tubulin (blue).
E Minimal cartoon model illustrating how conformation-selective TOG:αβ-tubulin interactions contribute to function. ‘+++’ denotes a basic region that provides microtubule affinity. TOG1 can efficiently capture unpolymerized αβ-tubulin in its naturally curved state (left). The straight/straighter conformation of αβ-tubulin in the MT has lower affinity interactions with TOG1 (right) and may be recognized by TOG2.
TOG2 binds to GTP- or GDP-bound αβ-tubulin with approximately equal affinity (200–300 nM) (Fig. S10), supporting a model in which the curvature of unpolymerized αβ-tubulin does not change appreciably as a function of the bound nucleotide. For hand-off to the microtubule to be efficient, the affinity of αβ-tubulin:microtubule interactions must at least be comparable to that of TOG:αβ-tubulin interactions. We also used analytical ultracentrifugation to demonstrate that TOG1-TOG2 and αβ-tubulin interact in a manner most consistent with a fast interchange between 1:1 and 1:2 TOG1-TOG2:αβ-tubulin complexes (Fig. 4D, red trace). The observation of a TOG1-TOG2:(αβ-tubulin)2 complex is surprising because prior studies(5, 7) suggested multiple TOG domains could simultaneously engage the same αβ-tubulin. Some of these prior studies were conducted using a gel filtration binding assay similar to the one we used, so it is possible that complexes with multiple αβ-tubulins were overlooked (we initially overlooked TOG2:αβ-tubulin interactions for the same reason (Fig. S2)). Our data also show that the complex formed depends on the relative stoichiometry of TOG domains to αβ-tubulin.
We hypothesize that the structure we determined provides a model for substrate recognition in which TOG1 (which is dispensable for plus-end binding (5)) of microtubule-bound Stu2p would capture unpolymerized subunits and/or stabilize a collision complex through its relatively strong interactions with naturally curved αβ-tubulin (Fig. 4E). Selective microtubule-end association is presumably the combined effect of a basic region in Stu2p providing microtubule lattice affinity (5) and TOG2 preferentially recognizing an end-specific conformation of αβ-tubulin. We speculate based on the polarity of TOG:αβ-tubulin engagement that the ordering of TOGs and the basic region dictates plus-end specificity. Indeed, if TOG2 and the C-terminal basic domain together mediate plus-end recognition, only at the plus end would TOG2 be able to engage non-straight αβ-tubulins nearest the microtubule plus end while the basic region engages surfaces more distant from the end. The conformational straightening in αβ-tubulin that accompanies lattice incorporation will result in lower affinity TOG1 interactions (Fig. 4E). In this ‘hand-off’ mechanism, polymer incorporation and release of TOG1 for a subsequent round of capture would be intrinsically coupled by virtue of the conformational preferences of TOG1. Hand-off will only become efficient when TOG1 is tethered to free αβ-tubulin binding sites at the end of the microtubule; this explains the requirement for at least two TOGs (6), and why isolated TOG1 or TOG2 inhibit microtubule assembly despite functioning to promote assembly when part of Stu2p.
Collectively, our observations indicate that Stu2p/XMAP215 family proteins discriminate between unpolymerized and polymerized forms of αβ-tubulin using conformation-selective TOG:αβ-tubulin interactions. This suggests by extension that assembly-dependent conformational change in αβ-tubulin plays an important role in dictating microtubule polymerization dynamics.
Supplementary Material
Acknowledgments
We thank: D. Borek for assistance with diffraction data processing, D. Tomchick in the UT Southwestern Structural Biology Core Facility for advice and assistance, M. Rosen, Y. Jiang and M. Moritz for sharing equipment/reagents, and S. Padrick for help with fluorescence anisotropy. H. Yu, X. Zhang, and M. Rosen gave critical comments on the manuscript. LMR is the Thomas O. Hicks Scholar in Medical Research. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, beamline 19-ID. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported by grants I-1692 from the Robert A. Welch Foundation, and GM-098543 from the NIH. Coordinates have been deposited in the PDB, accession code 4FFB.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohkura H, et al. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 1988;7:1465. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang PJ, Huffaker TC. Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol. 1997;139:1271. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widlund PO, et al. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. P Natl Acad Sci Usa. 2011;108:2741. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson V, Ayaz P, Huddleston P, Rice LM. Design, overexpression, and purification of polymerization-blocked yeast alphabeta-tubulin mutants. Biochemistry. 2011;50:8636. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- 9.Materials and methods are available as supplementary material on Science Online.
- 10.Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosco KA, et al. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravelli RBG, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 14.Aldaz H, Rice LM, Stearns T, Agard DA. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature. 2005;435:523. doi: 10.1038/nature03586. [DOI] [PubMed] [Google Scholar]
- 15.Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 16.Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: Structural studies of alpha beta- and gamma-tubulin clarify the role of GTP in microtubule assembly. P Natl Acad Sci Usa. 2008;105:5378. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buey RM, Díaz JF, Andreu JM. The nucleotide switch of tubulin and microtubule assembly: a polymerization-driven structural change. Biochemistry. 2006;45:5933. doi: 10.1021/bi060334m. [DOI] [PubMed] [Google Scholar]
- 18.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 19.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nogales E, Wang HW. Structural mechanisms underlying nucleotide-dependent self-assembly of tubulin and its relatives. Curr Opin Struct Biol. 2006;16:221. doi: 10.1016/j.sbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Schrodinger, LLC. 2010 [Google Scholar]
- 23.des Georges A, et al. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laue TM, Shah BD, Ridgeway RM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe AJ, Horton JC, editors. The Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 27.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys J. 2005;89:651. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.