Abstract
Alignments of the sequences of the all members of the archaeal histone and Alba1 families of chromatin proteins identified isoleucine residues, I19 in HMtB and I39 in MtAlba, in Methanothermobacter thermautotrophicus, at locations predicted to be directly involved in DNA binding. In all other HMfB family members, residue 19 is an arginine (R19), and either arginine or lysine is present in almost all other Alba1 family members at the structural site equivalent to I39 in MtAlba. Electrophoretic mobility shift assays revealed that recombinant HMtB and MtAlba do not bind DNA, but variants constructed with R19 and R39, respectively, bound DNA; and whereas MtAlba(I19) did not bind RNA, MtAlba(R19) bound both single stranded RNA and tRNA. Amplification and sequencing of MT0254 (encodes HMtB) and MT1483 (encodes MtAlba) from several Methanothermobacter thermautotrophicus lineages has revealed that HMtB and MtAlba had arginine residues at positions 19 and 39, respectively, in the original isolate and that spontaneous mutations must have occurred, and been fixed, in some laboratory lineages that now have HMtB(I19) and MtAlba(I39). The retention of these variants suggests some continuing functions and fusion of the HMtB(I19) sequence to HMtA2 resulted in a protein that folds to form a histone fold heterodimer that binds and compacts DNA. The loss of DNA binding by HMtB(I19) does not therefore prevent HMtB from participating in DNA interactions as one partner of an archaeal histone heterodimer.
Keywords: Archaea, Chromatin proteins, Spontaneous mutations, Nucleic acid binding
Introduction
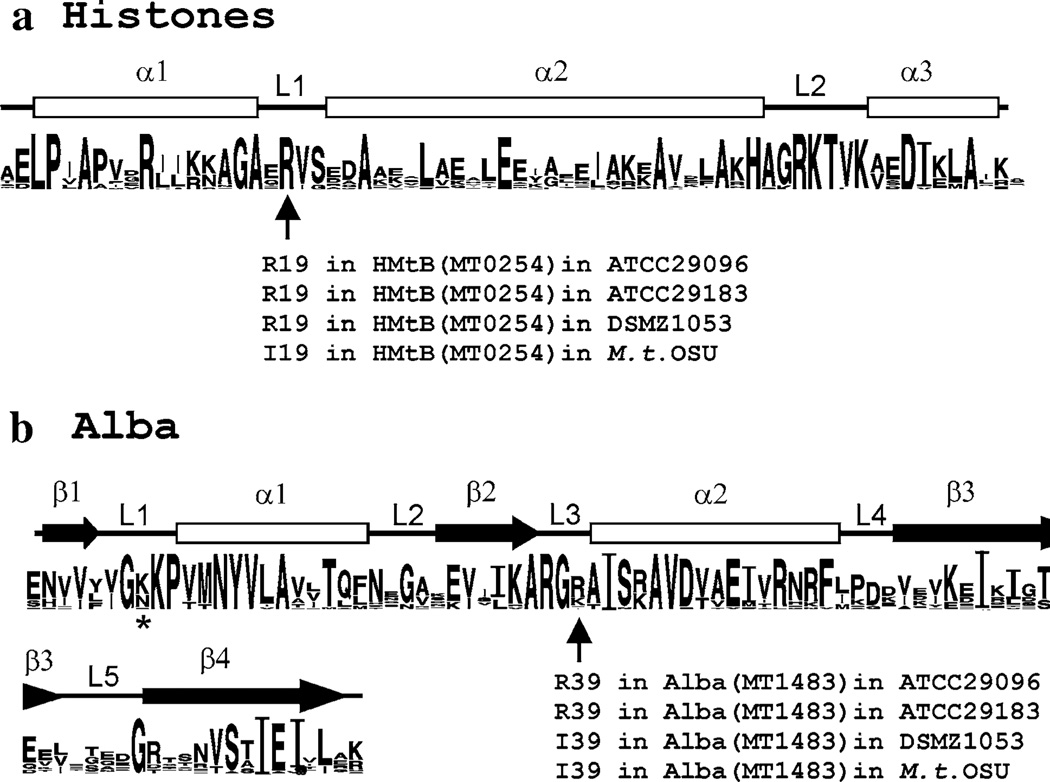
Several families of chromatin proteins have evolved in the Archaea that share the generic features of small size, positive charge and sequence-independent DNA binding, and likely have overlapping functions, but otherwise they are unrelated. Most Archaea contain representatives of more than one family of chromatin proteins, and many investigations have focused on members of the Sul7d, Sul10b (Alba), archaeal histone or MC1 families (Sandman and Reeve 2005; Samson and Reeve 2007). With ~50 archaeal genome sequences available, plus many partial sequences, consensus sequences can be established for protein families, and universally conserved and so likely essential residues can be identified. The genome sequence of Methanothermobacter thermautotrophicus [previously designated Methanobacterium thermoautotrophicum strain ΔH; (Smith et al. 1997)] encodes three archaeal histones, HMtA1, HMtA2 and HMtB, and one member of the Sul10b/Alba1 family, designated MtAlba. The consensus obtained by aligning all archaeal histone sequences currently available in GenBank (Crooks et al. 2004) confirmed that residues, identified by mutagenesis of HMfB from Methanothermus fervidus as essential for structure and function (Soares et al. 2000), are highly conserved but there was one striking anomaly (Fig. 1a). All members of the HMfB-family of archaeal histones have an arginine residue (R19 in the HMfB sequence) in the loop 1 region of the histone fold, positioned for DNA binding, except HMtB (encoded by MT0254) in Methanothermobacter thermautotrophicus. In HMtB, an isoleucine (I19) occupies this position and, with this noted, a similar anomaly was recognized in the consensus sequence established for the Alba family of proteins (Fig. 1b). A positively charged residue is almost universally conserved in loop 3, at a location predicted to contact DNA, but this is again an isoleucine (I39) in MtAlba (encoded by MT1483) from M. thermautotrophicus (Smith et al. 1997). The experiments reported were undertaken to investigate these anomalous isoleucine residues, and the results obtained document that the presence of I19 in HMtB and I39 in MtAlba result in loss of DNA binding ability. The sequences of the HMtB (MT0254) and MtAlba (MT1483) encoding genes amplified from M. thermautotrophicus lineages separated for >25 years from the lineage maintained at the Ohio State University (M. thermautotrophicus OSU) argue that AGA to ATA mis-sense mutations have occurred that inactivate nucleic acid binding by HMtB and Alba in some M. thermautotrophicus lineages. Their continued presence argues that they may still retain a positive function, and archaeal histone heterodimers constructed with HMtB(I19) as one partner, have been shown to retain the ability to bind and compact DNA.
Fig 1.
Consensus sequences for the HMfB family and Alba1 (Sul10b1) family of archaeal chromatin proteins. a The sequences of all ~50 members of the HMfB-family archaeal histones present in GenBank (in May 2008) were aligned, and from this the consensus sequence shown was generated by WebLogo (Crooks et al. 2004). Only amino acids that occurred at a site multiple times are noted, with font size indicating the relative frequency of occurrence of that amino acid at that location. Residues R52, K53, T54, and D59 were present in all sequences, and so comparison with their font size provides an estimate of percent conservation. The histone fold is formed by three alpha-helices (α1, α2, α3) separated by two β-strand looped regions (L1, L2) and, as indicated, the residue at position 19 in L1 (HMfB sequence numbers) is arginine in HMtB in M. thermautotrophicus ATCC29096, ATCC29183, DSMZ1053 but isoleucine in M. thermautotrophicus OSU. b The WebLogo consensus sequence based on alignment of the ~40 Sul10b/Alba1 sequences in GenBank. As illustrated, all Alba proteins fold to form four β-strands and two alpha-helices separated by five loop regions. The lysine residue (K16) in the L1 region of Sso10b that, when acetylated, reduces DNA affinity (Bell et al. 2002; Jelinska et al. 2005) is identified by an asterisk (*). As indicated, the residue at position 39 in L3 (MtAlba sequence numbers) is arginine in MtAlba in M. thermautotrophicus ATCC29096 and ATCC29183 but isoleucine in M. thermautotrophicus DSMZ1053 and OSU
Materials and methods
Methanothermobacter thermautotrophicus lineages
Methanothermobacter thermautotrophicus was isolated in 1972 (Zeikus and Wolfe 1972) and deposited with the American Type Culture Collection (ATCC) and is now catalogued as M. thermautotrophicus ATCC29096. The lineage maintained since 1988 at the Ohio State University (M. thermautotrophicus OSU) was provided then by collaborators (Reeve et al. 1989) who obtained ATCC29096 from the ATCC in 1981 (Jacobson et al. 1982). The M. thermautotrophicus genome sequence was determined from DNA samples purified from M. thermautotrophicus OSU in 1996 (Smith et al. 1997). Cultures of ATCC29096 were obtained again in 2005 from the ATCC, and from the German Microbiology Culture Collection (designated DSMZ strain 1053). A culture of an independent M. thermoautotrophicum isolate, separately deposited and now designated M. thermautotrophicus ATCC29183 was provided by Šmigán et al. (1984). This lineage was originally obtained from the ATCC in 1980 (Sauer et al. 1980).
Methanothermobacter thermautotrophicus growth and isolation of genomic DNA
Methanothermobacter thermautotrophicus cultures were grown as previously described (Morgan et al. 1997), the cells were harvested by centrifugation, resuspended in 10% sucrose, 20 mM Tris (pH 8), 5 mM EDTA, and lysed by incubation with pseudomurein endoisopeptidase (Pfister et al. 1998) for 1 h at 62°C and SDS addition to 0.2% (w/v) final concentration. Proteinase K (250 mg/ml) was added, incubation continued at 62°C for 1 h, NaCl added to 1 M final concentration and the mixture incubated on ice for 30 min. The resulting precipitate was removed, and nucleic acids precipitated from the supernatant by addition of isopropanol. RNA was removed by RNase digestion, and the genomic DNA remaining was purified by phenol–chloroform extraction and ethanol precipitation, and quantified by A260 measurements. Pfu-Turbo and the conditions recommended by the supplier (Stratagene) were used to PCR amplify genes from the genomic DNA preparations, and a sample of each PCR amplified DNA was sequenced directly.
Plasmid constructions
MT0254 was PCR amplified from M. thermautotrophicus OSU genomic DNA using primers that added flanking NdeI and BamHI sites. An aliquot of the amplified DNA was digested with NdeI and BamHI and ligated with NdeI and BamHI digested pT7-7 (Novagen) resulting in pKS643. The QuikChange procedure (Stratagene) was used to change codon 19 of MT0254 in pKS643 from ATA to AGA to obtain a plasmid (pKS724) that encodes HMtB (R19). The genes encoding HMtA1 and HMtA2, MT0821 and MT1696, respectively, were similarly PCR amplified from genomic DNA with flanking sequences that facilitated ligation and cloning into NdeI and BamHI digested pKS691, a derivative of pET30a that also expresses the E. coli methionine aminopeptidase (MAP) encoding (map) gene. The presence of additional MAP helps ensure complete N-terminal processing of recombinant proteins synthesized in E. coli (Sandman et al. 1995). These plasmids were transformed into E. coli NovaBlue(DE3) (Novagen) for isopropyl-β-d-thiogalactopyranoside (IPTG) induced over-expression of the archaeal histone encoding genes (Sandman et al. 2001).
Plasmid pKS731 was constructed from pT7-7 to facilitate the in-frame covalent linkage of two archaeal histones via a peptide (TAPDRRG) that naturally connects two archaeal histone folds in a single protein (VNG0134G) in Halobacterium NRC1 (Ng et al. 2000; Bailey et al. 2002). Either MT0254 (I19) or MT0254 (R19) was cloned into pKS731 as the N-terminal histone, and MT1696 was cloned as the C-terminal histone. The resulting plasmids, pKS735 and pKS736, respectively, were transformed into E. coli BL21(DE3) for over-expression of the encoded HMtB(I19)-HMtA2 or HMtB(R19)-HMtA2 fusion proteins.
MT1483, encoding MtAlba (I39), was PCR amplified from M. thermautotrophicus OSU genomic DNA using primers that added flanking restriction sites and eliminated an internal NdeI site without changing the encoded amino acid sequence. The amplified DNA was digested with NdeI and BamHI, and ligated with NdeI and BamHI digested pKS773 to construct plasmid pKS780. In pKS780, the MT1483 sequence is immediately followed in-frame by six histidine codons and so encodes MtAlba (I19) with a C-terminal his6 tag. QuikChange was used to change codon 39 of MT1483 in pKS780 from ATA to AGA, resulting in plasmid pKS781 that encodes MtAlba (R39) with a C-terminal his6 tag. pKS780 and pKS781 were transformed into E. coli BL21(DE3) for over-expression of the MtAlba-encoding genes.
Expression and purification of recombinant proteins
E. coli cultures (1 l) were grown at 37°C in Luria-Bertani medium to an OD600 of ~0.7, IPTG was added (final concentration of 1 mM) and incubation continued at 37°C for 3 h. Cells were then harvested by centrifugation, lysed and recombinant histones were purified and quantified exactly as previously described (Sandman et al. 2001), except for the following protein-specific modifications. For HMtB(I19) and HMtB(R19), the clarified E. coli lysates were passed through a Sephacryl S-100 HR gel filtration column (GE Healthcare), and aliquots of the fractions collected were screened by SDS-PAGE and Coomassie blue staining for the presence of the recombinant histone. Fractions containing the histone were pooled and this was then purified by heparin-affinity chromatography, as previously described (Sandman et al. 2001). For HMtB(I19)-HMtA2 and HMtB(R19)-HMtA2, after heat-treatment, the cell lysates were dialyzed against 200 mM NaCl, 50 mM Tris–HCl (pH 8), loaded onto a SP-FF cation exchange column (GE Healthcare) and bound proteins were eluted using a 0.2–1 M NaCl gradient dissolved in 50 mM Tris–HCl (pH 8). Fractions that contained the purified fusion protein were identified by SDS-PAGE and Coomassie blue staining, and combined.
E. coli cells containing MtAlba(I39) or MtAlba(R39) were harvested, resuspended in 5 ml of 100 mM NaCl, 50 mM Tris–HCl (pH 8) per gram of wet weight, and lysed by two passages through a French pressure cell at 20,000 psi. The lysate was clarified by centrifugation for 90 min at 114,000 × g at 4°C, and the resulting supernatant loaded on a column with a Co2+-charged affinity matrix. Proteins that bound were washed with 100 mM NaCl, 50 mM Tris–HCl (pH 8) and then eluted using a 0–150 mM imidazole gradient in 100 mM NaCl, 50 mM Tris–HCl (pH 8). An aliquot of each fraction was screened by SDS-PAGE and staining for the presence of an abundant ~11 kDa polypeptide. Fractions containing this polypeptide were pooled, loaded on a Q-XL anion exchange column (GE Healthcare), and bound proteins were eluted using a 100–500 mM NaCl gradient dissolved in 50 mM Tris–HCl (pH 8). Fractions containing the desired protein, identified by SDS-PAGE and staining, were pooled, the protein concentrated and quantified by amino acid measurements (Molecular Structure Facility, UC Davis, CA).
Agarose gel shift and plasmid topology assays
The agarose gel shift assay used to measure archaeal histone binding and compaction of linear DNA molecules, and topology assay used to monitor binding and wrapping of relaxed circular DNAs, have been described previously (Musgrave et al. 1991; Sandman et al. 2001).
Acrylamide gel shift assays
Single stranded (ss) DNA (5′TAAAGCCCGTGGAAGAGCCATAAGCCGGGCTG and 5′CAGCCCGGCTTATGGCTCTTCCACGGGCTTTAA), and ss RNA (5′-UUAAAGCCCGUGGAAGAGCC) molecules were purchased from Integrated DNA Technologies (IDT; Coralville, IA), and 5′-[32P]-end labeled by incubation with T4 polynucleotide kinase (NEB) and γ-32P-ATP. The [32P]-labeled nucleic acids were separated from unincorporated 32P-ATP by passage through a Sephadex G50 spin column and purified by phenol–chloroform extraction and ethanol precipitation. Double-stranded (ds) DNA molecules (5′-ATCAATTAAAGCGTTCTACGGCGTTTTAGATCTGAT) were generated by EcoRV digestion of pKS584 DNA that contains 36 tandem copies of the sequence separated by EcoRV sites. The 36 bp ds DNA molecules that remained in solution after polyethylene glycol precipitation of the vector DNA were ethanol precipitated, and 5′-[32P]-end labeled using T4 polynucleotide kinase and γ-32P-ATP. RNA preparations with the E. coli tRNAPro sequence (5′-GGGCGAGUAGCGCAGCUUGGUAGCGCAACUGGUUUGGGACCAGUGGGUCGGAGGUUCGAAUCCU CUCUCGUCCACCA) were generated and [32P]-labeled by in vitro transcription from the tRNAPro gene cloned in pHR419, using T7 RNA polymerase (Invitrogen) and α-[32P]-UTP. Aliquots of each of the [32P]-labeled nucleic acids were incubated with increasing concentrations of MtAlba (I39) or MtAlba (R39) and the reaction products were subjected to polyacrylamide gel electrophoresis. The gels were dried and 32P-labeled molecules and complexes were visualized and quantified by phosphorimaging. The protein concentrations (apparent Kd) at which 50% of the nucleic was bound into a complex and so migrated more slowly than the protein-free nucleic acid were determined.
Results and discussion
HMtB and MtAlba from M. thermautotrophicus OSU lack consensus residues
The consensus sequences, generated by WebLogo (Crooks et al. 2004), for the HMfB-family of archaeal histones, and Alba1 (Sul10b) family are shown Fig. 1 with the α-helical, β-strand and loop regions identified (Samson and Reeve 2007). The genes encoding HMtB (MT0254) and MtAlba (MT1483) were PCR amplified and sequenced from M. thermautotrophicus ATCC29096, DSMZ1053, ATCC29183 and from a current culture of M. thermautotrophicus OSU. The MT0254 sequences were identical and all encoded an HMtB with arginine at position 19 [HMtB(R19)], except for MT0254 from M. thermautotrophicus OSU that had one nucleotide difference that changed codon 19 from AGA to ATA and so encoded HMtB(I19). The MT1483 sequences were similarly all identical except for the same single nucleotide difference that resulted in codon 39 being either AGA or ATA. In this case, MT1483 amplified from M. thermautotrophicus ATCC29096 and ATCC29183 encoded MtAlba(R39) whereas MT1483 from M. thermautotrophicus DSMZ1053 and OSU encoded MtAlba(I39).
DNA binding and compaction by HMtB(R19) but not by HMtB(I19)
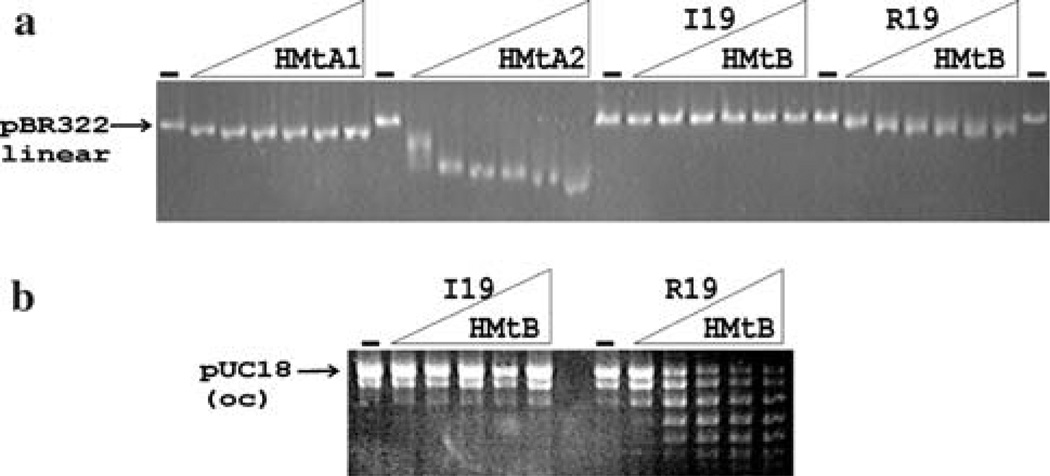
Previous studies with HMfB, an archaeal histone from Methanothermus fervidus, documented the importance of residues in loop 1 and 2 of the histone fold for DNA binding. HMfB variants constructed with R19I, R19Q and R19S substitutions did not bind and compact DNA (Soares et al. 2000). The DNA binding and compaction abilities of the recombinant archaeal histones encoded from M. thermautotrophicus OSU were assayed by incubation with linear pBR322 DNA and subsequent separation of the reaction products by agarose gel electrophoresis. Incubation with HMtA1 (encoded by MT0821) and HMtA2 (encoded by MT1696) resulted in histone–DNA complexes that migrated faster during agarose gel electrophoresis than the histone-free DNA (Fig. 2a). This gel-acceleration result is typical and diagnostic of DNA binding and compaction by archaeal histones (Sandman et al. 2001). Incubation of pBR322 DNA with HMtB(I19), however, had no effect on the mobility of the DNA (Fig. 2a). In contrast, incubation with HMtB(R19), generated by site-directed mutagenesis of MT0254 from M. thermautotrophicus OSU, expression and purification from E. coli, resulted in complexes with increased mobility relative to histone-free DNA (Fig.2a). To confirm the participation of R19 in DNA wrapping, relaxed circular DNA molecules were incubated with HMtB(I19) or HMtB(R19). Incubation with HMtB(I19) had no effect on the topology of the plasmid DNA whereas incubation with HMtB(R19) introduced supercoils, consistent with binding and wrapping of the DNA into archaeal nucleosomes [Fig. 2b; (Musgrave et al. 1991; Sandman et al. 2001)].
Fig 2.
Electrophoretic separation of archaeal histone–DNA complexes, and of the topoisomers formed by histone binding to relaxed circular DNAs. a Aliquots (50 ng) of EcoRI linearized pBR322 were incubated without (dash) and with 10, 20, 40, 60, 80 or 100 ng of recombinant HMtA1, HMtA2, HMtB(I19) or HMtB(R19). The reaction products were subjected to agarose gel electrophoresis, and visualized by ethidium bromide staining. b Aliquots (500 ng) of relaxed, circular pUC18 DNA were incubated without (dash) or with 100, 200, 300, 400 or 500 ng of HMtB(I19) or HMtB(R19). Plectonemic supercoils were removed by incubation with topoisomerase I, proteins were removed by phenol–chloroform extraction, and the ethanol-precipitated pUC18 DNA topoisomers generated were separated by agarose gel electrophoresis
Nucleic acid binding by MtAlba(R39) but not by MtAlba(I39)
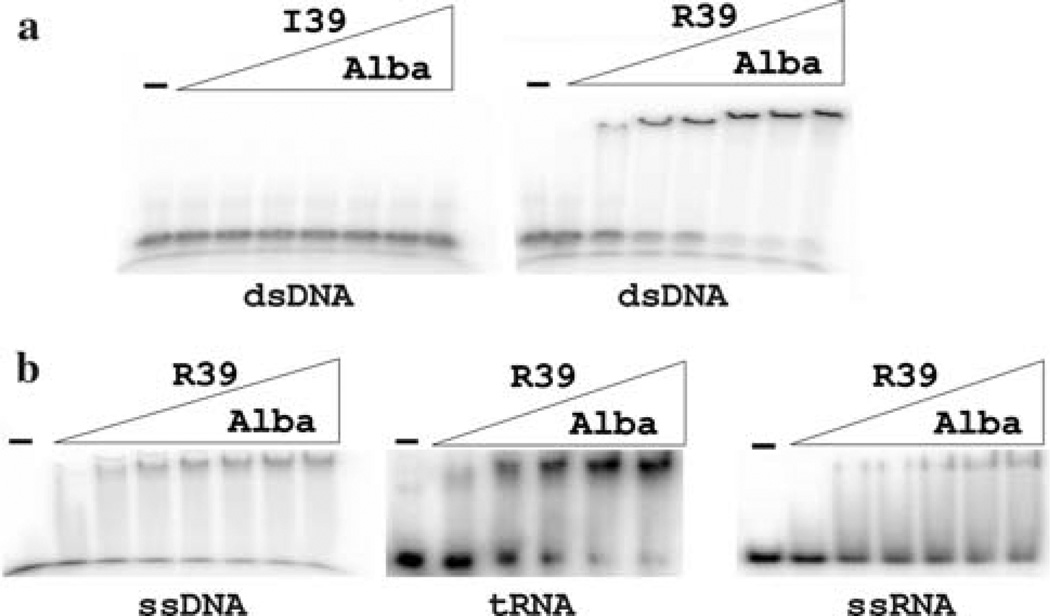
Research with members of the Alba family has primarily focused on DNA binding, although it is well documented that Alba family members also bind RNA (Guo et al. 2003; Biyani et al. 2005; Marsh et al. 2005; Wang et al. 2003; Wardleworth et al. 2002; Hada et al. 2008). Electrophoretic mobility shift assays (EMSA) were therefore undertaken to determine the abilities of MtAlba(I39) and MtAlba(R39) to bind ds DNA, ss DNA, ss RNA and tRNA (Fig. 3). Purified MtAlba(R39) bound to all of these nucleic acids, whereas MtAlba(I19) had no detectable effects on the mobility of any of the nucleic acids (Fig. 3). From the concentrations of MtAlba(R39) required to bind 50% of the nucleic acid into complexes, determined in at least three independent experiments, the apparent Kds for MtAlba(R39) binding to dsDNA, ssDNA, ssRNA and tRNA were ~0.2, 3.5, 1.5, and 1 µM, respectively, values similar to those reported for other members of the Alba family (Xue et al. 2000; Bell et al. 2002; Guo et al. 2003; Biyani et al. 2005; Marsh et al. 2005; Kumarevel et al. 2008). To date, there is no experimentally established structure for an Alba-nucleic acid complex, but several structures have been modeled in which residues in loops 1, 3 and 5 participate directly in DNA binding (Wardleworth et al. 2002; Chou et al. 2003; Zhao et al. 2003; Wang et al. 2003; Kumarevel et al. 2008). Changes in the NMR spectra of Alba in solution on nucleic acid addition (Cui et al. 2003; Biyani et al. 2005), and directed mutagenesis of loop 1 residues (Bell et al. 2002; Zhao et al. 2003; Marsh et al. 2005) support these predicted interactions. Surprisingly, the lack of nucleic acid binding by MtAlba(I39) is the first direct experimental evidence for a loop 3 residue being essential for nucleic acid binding. In terms of the models for Alba-nucleic acid complexes, it seems noteworthy that this residue is apparently essential for binding DNA and RNA, in either single or double-stranded configuration. The residue at this position is either arginine or lysine in all Alba sequences, except for the isoleucine in MtAlba(I39), alanine in four Alba sequences from different Pyrobaculum species and a serine in an Alba sequence from Caldivirga maquilingensis (Fig. 1b).
Fig 3.
Electrophoretic mobility shift assays (EMSA) of MtAlba binding to nucleic acids. a Aliquots of 32P-labeled 36 bp double stranded (ds) DNA were incubated without (dash) or with 100, 200, 300, 400, 500, 600 or 700 ng of MtAlba(I39) or MtAlba(R39). The reaction products were separated by electrophoresis through 8% polyacrylamide gels, visualized and quantified by phosphor-imaging. b Aliquots of 32P-labeled 36 bp single-stranded (ss) DNA, tRNAPro or ss RNA were incubated without (dash) or with 50, 100, 150, 200 or 250 ng of MtAlba(R39). The reaction products were separated by electrophoresis through 6% polyacrylamide gels, visualized and quantified by phosphor-imaging. MtAlba(I39) binding to these nucleic acids was also determined by EMSA but, as shown in (a) for ds DNA, incubation of ssDNA, tRNA or ssRNA with MtAlba(I19) at these concentrations did not result in gel shifts
Histone heterodimers containing HMtB(I19) bind and compact DNA
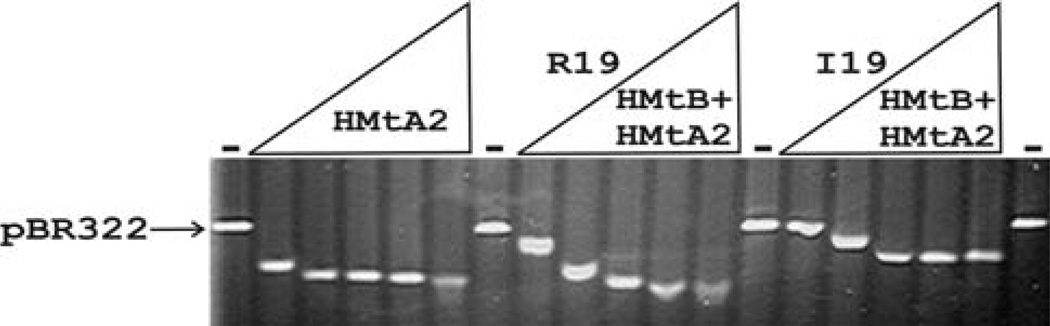
Based on the genomic sequences, mutations have apparently occurred in the M. thermautotrophicus OSU lineage during laboratory domestication that eliminate DNA binding by HMtB, and DNA and RNA binding by Alba. This might provide an advantage under laboratory growth conditions but, if so, this advantage is not apparently provided by the complete loss of these proteins. There are ten codons in MT0254, and four in MT1483 where a G to T mutation would result in a nonsense codon and so prevent protein production (Smith et al. 1997), but the only G to T mutations that have been fixed in the lineage are missense mutations that change AGA to ATA and so maintain the synthesis of full length proteins but with arginine to isoleucine substitutions. Native archaeal histone preparations purified from M. thermautotrophicus OSU have been confirmed by mass spectrometry to contain HMtB(I19), consistent with retaining an in vivo function. The histone fold is only stable in a dimer configuration, and as solutions of HMtB(I19) and HMtB(R19) have very similar circular dichroism spectra (result not shown), the R19I substitution does not prevent dimer formation or cause a major distortion of the folded structure of a (HMtB)2 homodimer. Archaeal histones assemble to form both homodimers and heterodimers (Sandman and Reeve 2005; Samson and Reeve 2007) and so HMtB(I19) monomers could form heterodimers in vivo with HMtA1 and/or HMtA2 monomers that retain DNA binding ability. To determine if a HMtB(I19)-HMtA2 heterodimer can bind DNA, MT0254 [encoding either HMtB(I19) or HMtB(R19)] and MT1696 (encoding HMtA2) were fused in frame to encode a single protein that would fold to form an archaeal histone-fold heterodimer (Bailey et al. 2002). Incubation of either HMtB(I19)-HMtA2 or HMtB(R19)-HMtA2 with DNA resulted in binding and the assembly of complexes that migrated faster during agarose gel electrophoresis than the histone-free DNA (Fig. 4). The complexes formed by HMtB(I19)-HMtA2 migrated slower than those formed by HMtB(R19)-HMtA2, consistent with less compact structures, but HMtB(I19)-HMtA2 heterodimers did clearly have DNA binding and compaction ability.
Fig 4.
Electrophoretic separation of archaeal histone–DNA complexes. Aliquots (50 ng) of linear pBR322 DNA were incubated without (dash) and with 20, 50, 100, 150 or 200 ng of HMtA2, or HMtB(R19)-HMtA2, or HMtB(I19)-HMtA2. The reaction products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining
Conclusion
When isolated, M. thermautotrophicus ATCC29096 and ATCC29183, had the capacity to synthesize three members of the archaeal histone family (HMtA1, HMtA2, and HMtB) and one member of the Alba family (MtAlba). All four proteins had nucleic acid binding ability. During laboratory passage, two spontaneous mutations have been fixed that inactivate the DNA binding ability of HMtB and DNA and RNA binding ability of Alba. Both mutations have been present in M. thermautotrophicus OSU for over a decade (Smith et al. 1997). Only the MtAlba inactivating mutation is present in M. thermautotrophicus DSMZ1053. The viability of M. thermautotrophicus OSU and DSMZ1053 is consistent with the viability of methanococcal and methanosarcina mutants that have archaeal histone- or Alba-encoding genes specifically inactivated but these mutants do have reduced growth rates and distorted patterns of global gene expression (Heinicke et al. 2004; Weidenbach et al. 2008). In contrast, the loss of nucleic acid binding by HMtB and Alba in M. thermautotrophicus OSU has no detrimental effect on growth rate when compared with the growth rates of the M thermautotrophicus lineages that contain nucleic acid-binding HMtB and Alba. The loss of nucleic acid binding by HMtB and Alba could still have a significant effect on overall genome expression but, if so, the resulting changes have no obvious negative effects under laboratory growth conditions.
Acknowledgments
This research was supported by grant GM53185 from the US National Institutes of Health. We thank Drs. P. Šmigáň and H. Roy for providing the culture of M. thermautotrophicus ATCC29183, and pHR419, respectively.
References
- Bailey KA, Marc F, Sandman K, Reeve JN. Both DNA and histone fold sequences contribute to archaeal nucleosome stability. J Biol Chem. 2002;277:9293–9301. doi: 10.1074/jbc.M110029200. [DOI] [PubMed] [Google Scholar]
- Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- Biyani K, Kahsai MA, Clark AT, Armstrong TL, Edmondson SP, Shriver JW. Solution structure, stability, and nucleic acid binding of the hyperthermophile protein Sso10b2. Biochem. 2005;44:14217–14230. doi: 10.1021/bi051266r. [DOI] [PubMed] [Google Scholar]
- Chou C-C, Lin T-W, Chen C-Y, Wang AH-J. Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 angstroms. J Bacteriol. 2003;185:4066–4073. doi: 10.1128/JB.185.14.4066-4073.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Tong Y, Xue H, Huang L, Feng Y, Wang J. Two conformations of archaeal Ssh10b. The origin of its temperature dependent interaction with DNA. J Biol Chem. 2003;278:51015–51022. doi: 10.1074/jbc.M308510200. [DOI] [PubMed] [Google Scholar]
- Guo R, Xue H, Huang L. Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol Microbiol. 2003;50:1605–1615. doi: 10.1046/j.1365-2958.2003.03793.x. [DOI] [PubMed] [Google Scholar]
- Hada K, Nakashima T, Osawa T, Shimada H, Kakuta Y, Kimura M. Crystal structure and functional analysis of an archaeal chromatin protein Alba from the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci Biotechnol Biochem. 2008;72:749–758. doi: 10.1271/bbb.70639. [DOI] [PubMed] [Google Scholar]
- Heinicke I, Muller J, Pittelkow M, Klein A. Mutational analysis of genes encoding chromatin proteins in the archaeon Methanococcus voltae indicates their involvement in the regulation of gene expression. Mol Gen Genom. 2004;272:76–87. doi: 10.1007/s00438-004-1033-5. [DOI] [PubMed] [Google Scholar]
- Jacobson FS, Daniel L, Fox JA, Walsh CT, Orme-Johnson WH. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1982;257:3385–3388. [PubMed] [Google Scholar]
- Jelinska C, Conroy MJ, Craven CJ, Hounslow AM, Bullough PA, Waltho JP, Taylor GL, White MF. Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure. 2005;13:963–971. doi: 10.1016/j.str.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Kumarevel T, Sakamoto K, Gopinath SCB, Shinkai A, Kumar PKR, Yokoyama S. Crystal structure of an archaeal specific DNA-binding protein (Ape10b2) from Aeropyrum pernix K1. Proteins. 2008;71:1156–1162. doi: 10.1002/prot.21807. [DOI] [PubMed] [Google Scholar]
- Marsh VL, Peak-Chew SY, Bell SD. Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J Biol Chem. 2005;280:21122–21128. doi: 10.1074/jbc.M501280200. [DOI] [PubMed] [Google Scholar]
- Morgan RM, Pihl TD, Nölling J, Reeve JN. Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum deltaH. J Bacteriol. 1997;179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrave DR, Sandman KM, Reeve JN. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc Natl Acad Sci USA. 1991;88:10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschroder M, Spudich JL, Jung KW, Alam M, Freitas T, Hou S, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang P, Riley M, Hood L, DasSarma S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacterium phage psiM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- Reeve JN, Beckler GS, Cram DS, Hamilton PT, Brown JW, Krzycki JA, Kolodziej AF, Alex L, Orme-Johnson WH, Walsh CT. A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain delta H encodes a polyferredoxin. Proc Natl Acad Sci USA. 1989;86:3031–3035. doi: 10.1073/pnas.86.9.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Reeve JN. DNA binding proteins and chromatin Archaea: In: Cavicchioli R, editor. molecular and cellular biology. Washington DC: ASM Press; 2007. pp. 110–119. [Google Scholar]
- Sandman K, Bailey KA, Pereira SL, Soares D, Li W-T, Reeve JN. Archaeal histones and nucleosomes. Methods Enzymol. 2001;334:116–129. doi: 10.1016/s0076-6879(01)34462-2. [DOI] [PubMed] [Google Scholar]
- Sandman K, Grayling RA, Reeve JN. Improved N-terminal processing of recombinant proteins synthesized in Escherichia coli. Biotechnology (NY) 1995;13:504–506. doi: 10.1038/nbt0595-504. [DOI] [PubMed] [Google Scholar]
- Sandman K, Reeve JN. Archaeal chromatin proteins: different structures but common functions? Curr Opin Microbiol. 2005;8:656–661. doi: 10.1016/j.mib.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sauer FD, Erfle JD, Mahadevan S. Methane production by the membranous fraction of Methanobacterium thermoautotrophicum. Biochem J. 1980;190:177–182. doi: 10.1042/bj1900177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmigáň P, Friederová A, Rusňák P, Greksák M. Effect of 2,4-dinitrophenol and ionophores on growth and methanogenesis in Methanobacterium thermoautotrophicum. Folia Microbiol. 1984;29:353–358. [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski AS, Church G, Daniels CJ, Mao J, Rice P, Nölling J, Reeve JN. Complete genome sequence of Methanobacterium thermoautotrophicum delta H: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares DJ, Sandman K, Reeve JN. Mutational analysis of archaeal histone-DNA interactions. J Mol Biol. 2000;297:39–47. doi: 10.1006/jmbi.2000.3546. [DOI] [PubMed] [Google Scholar]
- Wang G, Guo R, Bartlam M, Yang H, Xue H, Liu Y, Rao Z. Crystal structure of a DNA binding protein from the hyper-thermophilic euryarchaeon Methanococcus jannaschii. Protein Sci. 2003;12:2815–2822. doi: 10.1110/ps.03325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardleworth BN, Russell RJM, Bell SD, Taylor GL, White MF. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenbach K, Glöer J, Ehlers C, Sandman K, Reeve JN, Schmitz RA. Deletion of the archaeal histone in Methanosarcina mazei Gö1 results in reduced growth and genomic transcription. Mol Microbiol. 2008;67:662–671. doi: 10.1111/j.1365-2958.2007.06076.x. [DOI] [PubMed] [Google Scholar]
- Xue H, Guo R, Wen Y, Liu D, Huang L. An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J Bacteriol. 2000;182:3929–3933. doi: 10.1128/jb.182.14.3929-3933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J Biol Chem. 2003;278:26071–26077. doi: 10.1074/jbc.M303666200. [DOI] [PubMed] [Google Scholar]
- Zeikus JG, Wolfe RS. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J Bacteriol. 1972;109:707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]