Abstract
In eukaryotes, the Cdt1-bound replicative helicase core MCM2-7 is loaded onto DNA by the ORC-Cdc6 ATPase to form a pre-Replicative Complex (pre-RC) with a MCM2-7 double-hexamer encircling DNA. Using purified components in the presence of ATPγS, we have captured in vitro an intermediate in pre-RC assembly that contains a complex between the hetero-heptameric ORC-Cdc6 and the hetero-heptameric Cdt1-MCM2-7, called the OCCM complex. Cryo-EM studies of the 14-protein complex reveal that the two separate heptameric complexes are extensively engaged, with the ORC-Cdc6 N-terminal AAA+ domains latching onto the C-terminal AAA+ motor domains of the MCM2-7 hexamer. ORC-Cdc6 undergoes a concerted conformational change into a right-handed spiral with the helical symmetry identical to the DNA double helix. The results show a striking structural similarity between the ORC-Cdc6 helicase loader and the Replication Factor-C clamp loader and suggest a conserved mechanism of action.
The replication of eukaryotic chromosomes is a multi-step process that spans the G1 and the S phase of the cell division cycle1,2. The yeast Origin Recognition Complex (ORC) constitutively binds to the replication origin DNA3. Replication licensing occurs upon mitotic exit or during the G1 phase when Cdc6 and Cdt1 can interact with the ORC to load hexameric MCM2-7, the core of the DNA helicase complex4. Details of how MCM2-7 is loaded onto the DNA have not been biochemically defined so far. The loaded MCM2-7 is initially inactive and becomes activated during early S phase, when many other factors promote unwinding of the duplex DNA and the replisome is assembled4.
Low-resolution EM structures of ORC from S. cerevisiae and D. melanogaster have been reported5,6. Both ScORC and DmORC were found to have a slightly twisted half-ring structure with the same dimensions of ~160 Å long and ~120 Å wide7. The replication initiator Cdc6 was found to bind to one side of ScORC, thus completing an ORC-Cdc6 ring containing six proteins with predicted AAA+ architecture5,8-11. In vitro, the ScORC-Cdc6 complex was shown to load in a Cdt1 and ATP-hydrolysis dependent manner two MCM2-7 hexamers onto double stranded DNA to form an inactive but stable double-hexamer (DH) (Fig. 1a)12,13. During this reaction, Cdt1 was removed from the DH. Thus the ORC-Cdc6 ring acts as an ATP-hydrolysis dependent loading machine that enabled the ring-shaped MCM2-7 hexamers to encircle duplex DNA. The DNA-loaded MCM2-7 DH is subsequently activated by Cdc45 and GINS complex, forming the Cdc45-MCM2-7-GINS (CMG) complex that is shown to be the functional helicase14-17. EM structural studies revealed that Cdc45 and GINS together bridge a gap between Mcm2 and Mcm5 of the MCM2-7 ring18.
Figure 1. In vitro assembly of the OCCM complex.

(a) Model for Cdc6 recruitment to the replication origin readies the ORC for loading of MCM2-7. (b) Averaged cryo-EM images of the in vitro assembled OCCM. For scale, the box size is 27 nm. An enlarged view with the top area tentatively assigned to ORC-Cdc6 and the lower region to Cdt1-MCM2-7. (c) Mcm2 IP identifies the OCCM components. Using purified ORC, Cdc6, Cdt1, MCM2-7 (lanes 1-4) and origin DNA OCCM was assembled in the presence of ATPγS. OCCM was cleaved off from the plasmid DNA with DNase I and immunoprecipitated with an anti-Mcm2 antibody (lanes 5-7) or with anti-MBP control antibody (lanes 8-10). Asterisk marks nonspecific proteins from antibody-conjugated beads.
A cryo-EM derived structure of ORC-Cdc6 bound to origin DNA demonstrated that ORC assumed a different structure upon recruiting Cdc6 to the DNA and the length of DNA bound was almost double that bound by ORC alone5,11. Combined with the two-claw DNA-binding mode of the Archaeal Orc1/Cdc6 structure as revealed by X-ray crystallography19,20, we proposed that ScORC-Cdc6 bends and topologically wraps the origin DNA11 and it is this complex that is primed to recruit the MCM2-7 hexamers. Similar wrapping of DmORC has been proposed21.
Bioinformatic analyses of DNA replication licensing factors and crystallography of Archaeal homologues have shown that the Orc1-5, Cdc6 and MCM2-7 proteins belong to the AAA+ family of proteins5,20-24 (Supplementary Fig. 1). Orc1-5 and Cdc6 contain one or two C-terminal DNA-binding winged-helix domains (WHDs)5. Each Mcm protein is composed of an N-terminal domain (NTD) that complexes zinc and an ATP-binding C-terminal AAA+ domain (CTD)25,26. The NTDs and CTDs of the MCM2-7 hexamer form a two-tiered ring structure18,24,27. Orc6 and Cdt1 are structurally different, with Orc6 sharing partial similarity with the transcription factor TFIIB and Cdt1 containing two WHDs, but no ATP binding motif28,29.
A missing piece of crucial information is how ORC-Cdc6 recruits and interacts with Cdt1-MCM2-7 preceding the formation of the MCM2-7 DH (Fig. 1a). Using purified ScORC, Cdc6, Cdt1 and MCM2-7 proteins in the presence of origin DNA and ATPγS, we have captured a 1.1-MDa complex that contains all 14 proteins that are essential for replication origin licensing. Cryo-EM and subunit mapping have resulted in a model for the architecture of a key intermediate in the replication-licensing process. The structure provides insights into the molecular mechanism of MCM2-7 recruitment and helicase loading.
RESULTS
Capturing a pre-RC intermediate by cryo-EM
The fleeting nature of the initial encounter between ORC-Cdc6 and Cdt1-MCM2-7 has so far prevented capturing a pre-RC intermediate in vitro. We wondered if cryo-EM could visualize assembly intermediates prior to the formation of Mcm2-7 DH. To slow down the progression of MCM2-7 loading reaction, we initially used several crippling factors, including (1) a short origin DNA (the 86-bp ARS1-containing linear DNA), which was previously demonstrated to be too short to load the MCM2-7 DH; (2) ATPγS, which can be hydrolyzed only very slowly, instead of ATP; and (3) ORC containing an ATP-hydrolysis mutation in the Orc4 subunit (ORC-4R)5,30. Computational classification of the cryo-EM images of the heterogeneous reactants revealed 2D class averages that suggested the formation of a three-tiered complex structure (Fig. 1b). Based on the previous structural studies of ORC-Cdc6 and the MCM2-7 hexamer5,6,11,18,24,27,31, the top slanted tier is likely ORC-Cdc6, and the bottom two tiers belong to Cdt1-MCM2-7. Assembly under these conditions hindered MCM2-7 DH formation, and resulted in the pre-RC intermediate that we have called OCCM, for ORC-Cdc6-Cdt1-MCM2-7.
To optimize OCCM preparation for cryo-EM 3D reconstruction, we found that OCCM formed readily on an ARS1-containing plasmid (~3000 bp) without the requirement for Orc4 arginine finger mutation as long as ATPγS was used in the reaction. The improved protocol involves first attaching a biotinylated ARS1 origin-containing DNA to streptavidin-coupled Dynabeads, adding the purified components (ORC, Cdc6, Cdt1 and MCM2-7) with ATPγS, washing the beads with low salt buffer (100 mM KGlu) to remove unbound proteins, then releasing OCCM from the beads by either DNase I or Alu I cleavage, and finally concentrating the sample for structural analyses. DNase I (29 kDa) binds ~10-bp DNA and leaves a ~4-bp overhang near the border, but cannot access DNA inside of protein/DNA complex32. The Alu I restriction enzyme introduces multiple cuts into the plasmid (Supplementary Fig. 2). The ARS1 replication origin is contained in a 238-bp Alu I fragment. We immunoprecipitated (IP) the DNase I-treated OCCM in ATPγS with anti-Mcm2 antibody, and observed co-precipitation of Mcm3-7, Cdt1 and ORC with Mcm2, but not with the anti-MBP control (Fig. 1c), This result demonstrates the integrity and composition of OCCM.
Cryo-EM and 3D reconstruction of the OCCM complex
Because of their large mass of 1.1 MDa, the OCCM particles embedded in vitreous ice have good contrast (Fig. 2a). Over 300 micrographs were recorded and over 80,000 particle images were selected. Fig. 2b shows seven reference-free 2D class averages in comparison with their approximately corresponding reprojections from the cryo-EM 3D map that is shown in Fig. 2c. Reference-based 2D averages and the corresponding reprojections are shown in Supplementary Fig. 3a. The Euler angle distribution of the raw particle images used for 3D reconstruction is shown in Supplementary Fig. 3b. The 3D map has an estimated resolution of 14 Å (Fig. 2d). The structure was further validated with the tilt pair technique33. We collected 142 pairs of untilted and 10° tilted particle images, determined their Euler angles by matching them with the 3D map-derived projections. The tilt geometry of each particle pair was then calculated and plotted as a dot in the polar coordinate system (Fig. 2e). Approximately 73% (47) of the plotted particles (64) were found to cluster around the experimental tilt angle 10° (average 9.65° with an RMSD of 4.52°) and the vertical tilt axis (average 85.22° with an RMSD of 14.02°). This result suggests that the cryo-EM structure and its associated handedness are correct.
Figure 2. Cryo-EM of the eukaryotic OCCM complex.

(a) A representative raw cryo-EM image of the purified OCCM complex embedded in vitreous ice. (b) Selected reference-free 2D class averages of the OCCM cryo-EM images (left panel) in comparison with their approximately corresponding reprojections from the 3D reconstruction (right panel). The box size is 34 nm. (c) Surface view of the cryo-EM 3D map of the OCCM complex rendered at the threshold that includes the expected molecular mass of 1.1 MDa. (d) Fourier shell correlation suggests that the 3D map has a resolution of 14 Å. (e) Tilt validation of the cryo-EM 3D map. The predicted tilt axis and tilt angle of each particle pair, based on the cryo-EM map, are plotted as a black dot. Most particle pairs cluster in a region demarcated by the red circle that is centered at the experimental tilt axis (90°) and tilt angle (10°).
Protein subunit and DNA mapping of the OCCM structure
A strategy of fusing the maltose binding protein (MBP) was initially developed for systematic subunit mapping within averaged 2D images of the yeast ORC34. The method has been applied successfully to several other protein complexes35,36. We systematically tested N-terminus, C-terminus or internal fusion of MBP to the OCCM proteins and examined their function in pre-RC assembly. We generated more than ten stable MBP-fused OCCM complexes and found MBP densities in 2D reference-free class averages or 3D reconstructions of six of these complexes (Fig. 3a-f). The six MBP-fused complexes were all functional: they formed a low-salt stable complex and supported the loading of a high-salt stable MCM2-7 DH (Supplementary Fig. 4a and 4b). MBP fused to Orc2 or Mcm2 is located to the right or at the bottom of OCCM, respectively, in the preferred and most informative side view; it is also visible in the 3D maps and their sections (Fig. 3a-b). However, MBP inserted in Mcm3, 5, 6, or Cdt1 is not visible in the 2D averages (Fig. 3c-f, left panels), indicating that the MBP is located in the front or back of OCCM in this view. Consistent with this assessment, we observed MBP in the front or back of the MCM region in 3D reconstructions of these OCCM (Fig. 3c-f, middle and right panels). We note that MBP densities appear rather small due to the combined effects of the relative small mass of MBP (38 kDa) as compared to the 1.1-MDa OCCM, and of the fact that fusion is flexible resulting in reduced densities in 3D reconstructions. However, the small density at the outer periphery of a fusion complex can be assigned to MBP because we have shown that the fusion complex retains structural and functional integrity18,37. We had better success in observing the tag when MBP was inserted internally into Mcm NTD or CTD. Insertion generates two linkages between MBP and Mcm, which likely results in a less flexible MBP; thus the improved detection rate.
Figure 3. Mapping protein and DNA components of the OCCM.

(a - f) 2D class averages and 3D reconstruction of OCCM with MBP fused to the C-terminus (CT) of Orc2 (Orc2-MBP) (a), the N-terminus (NT) of Mcm2 (MBP-Mcm2) (b), the NT of Mcm3 (MBP-Mcm3) (c), the CT of Mcm5 (MBP-Mcm5) (d), the NT of Cdt1 (MBP-Cdt1) (e), and the NT of Mcm6 (MBP-Mcm6) (f), respectively. In each left panel, the upper row shows two reference-free class averages, and the lower row shows the same images displayed at a higher contrast level (C=0.3). Each middle panel shows the surface rendered front, back, and bottom views of the 3D map of the MBP-fused OCCM complex. The peripheral MBP density is colored blue. The surface-rendering thresholds were lowered by ~20% to better visualize the small MBP density. Each right panel shows a vertical (a-b) or a horizontal section (c-f) of the 3D map of the MBP-fused OCCM (first column) in comparison to the corresponding section of that of the wild type OCCM (second column). The lower row is displayed at a higher contrast level than the upper row. The red arrows point to the MBP density at the peripheral of OCCM. All MBP fusion complexes were imaged by cryo-EM exception for MBP-Mcm3 (c) that was by negative stained EM. All fusion complexes were cleaved off the plasmid DNA by DNases I except for MBP-Mcm6 that was by Alu I (f). (g) Reference-free class averages of wtOCCM with their plasmid DNA digested either by DNase I (upper row) or by Alu I (lower row). Blue arrows point to dsDNA stub on the top ORC-Cdc6 region of OCCM. (h) MCM2-7 organization as mapped by the four MBP-fused Mcm subunits (red), viewed from the N-terminal end of MCM2-7. Box size is 37 nm in the left panels, 34 nm in the right panels in (a-f), and 31 nm in (g). 3D maps are on same scale.
DNase I is a sequence non-specific enzyme that cleaves essentially all accessible DNA. In agreement with this property, we did not observe in the cryo-EM 2D class averages any densities protruding from OCCM treated with this enzyme (Fig. 3g, upper row). However, when DNA digestion was carried out using the DNA sequence-specific restriction enzyme Alu I, we consistently observed a thin density protruding from the top of the ORC-Cdc6 region of the OCCM structure in the averaged cryo-EM images, and therefore interpreted this linear and partially flexible density as DNA (Fig. 3g, lower row). Cryo-EM 3D reconstruction of the Alu I-treated OCCM with MBP insertion in Mcm6 NTD revealed both the MBP density at the bottom and the dsDNA stub at the top of the structure (Fig. 3f).
MBP inserted in the NTD of the Mcm subunits were located near the bottom, while insertion in the CTD located in the middle of the OCCM structure. This observation indicates that the MCM2-7 NTD faces outwards and the AAA+ domain-containing CTD of MCM2-7 interacts with ORC-Cdc6 at the top of OCCM. Such MCM2-7 orientation leaves the NTD free to interact with the NTD of the next MCM2-7 to eventually form a head-to-head MCM2-7 DH12,13. Our mapping results suggest that the subunits are arranged counterclockwise as Mcm5:Mcm3:Mcm7:Mcm4:Mcm6:Mcm2 when viewed from the bottom MCM2-7 NTD (Fig. 3h). This organization agrees with a large body of biochemical and structural data on MCM2-7 arrangement18,23,24,38.
The architecture of the OCCM complex
Fig. 4a-f show the overall architecture of OCCM in various views. To arrive at the subunit-assigned structure, we first subjected the cryo-EM 3D map to the semi-automatic segmentation procedure39. The subunit densities in the top ORC-Cdc6 region were well defined and the protein subunits could be segmented with little ambiguity. The location of Orc2 was directly mapped (Fig. 3a) and the remaining proteins were assigned based on the previously determined arrangement of Orc1:Orc4:Orc5:Orc2:Orc3, with Orc6 binding to Orc2, and Cdc6 bridging the gap between Orc1 and Orc311. This arrangement is best viewed in Fig. 4e. There is an additional density in the center of the top region that is colored gray. According to results shown in Fig. 3, this density belongs to the bound dsDNA that is protected by OCCM from DNase I digestion. In the bottom tier of the MCM2-7 region, the hexameric NTD crystal structure of an Archaeal MCM can be fitted40 (Fig. 4g). It is important to note that the very N-terminus of each of the known Archaeal Orc1/Cdc6 structures is located away from the N-terminal AAA+ domain and extends to and is part of the middle helical domain (HD)20,41 (Fig. 4h-i). The density identified as Cdc6 fits nicely to the rigid-body docked Archaeal homologue Orc1/Cdc6 crystal structure41, with the N-terminus in the middle of the structure (Fig. 4i). Based on the docking, Cdc6 N-terminus should be at a position marked by the blue asterisk in the OCCM structure (Fig. 4a-b). Interestingly, the Cdc6 C-terminal WHD is oriented upwards, in contact with the central DNA density, and the Cdc6 N-terminal AAA+ domain points down, reaching towards an AAA+ CTD of MCM2-7 (Fig. 4a-f). Given the similar architecture of the AAA+-containing ORC-Cdc6 subunits and their packing5,6, we expect that their N-termini are all located in the outer middle regions, and their C-terminal WHDs contact central DNA. We note that a centrally located Orc2 WHD is not in conflict with the outside/peripheral location of the MBP fused to the C-terminus of Orc2 (Fig. 3a), because there is a linker between the predicted WHD and the MBP.
Figure 4. Segmented cryo-EM structure of the OCCM.

(a – d) show four side views of OCCM obtained by consecutive 90°rotations around a vertical axis. (e and f) Top and bottom views by rotating +90° or -90° around the horizontal axis from the side view in (b). (g) Bottom and back side views of the OCCM map shown in semi-transparent surface, docked with homology crystal structure of the Archaeal MCM NTD hexamer (PDB ID: 1LTL)40. Surface-rendering threshold is set to enclose the expected 1.1-MDa mass of OCCM. (a – g) are on the same scale. Mcm2-7 are abbreviated as M2-7, and Orc1-6 as O1-6, respectively. (h) Ribbon presentation of the Archaeal Orc1/Cdc6 crystal structure, showing the N-terminal peptide meanders away from NT AAA+ domain and joins the middle helical domain (HD) (PDB ID: 2QBY). (i) The segmented Cdc6 density is shown as semi-transparent surface and the rigid-body docked Archaeal Orc1/Cdc6 structure shown as yellow ribbons (PDB ID: 1FNN)41. The N-terminal peptide shown in blue is at the middle HD region, away from the N-terminal AAA+ domain. The approximate location of N-terminal peptide of Cdc6 in the OCCM structure is labeled with a blue asterisk in (a) and (b), respectively. The C-terminal WHD of Cdc6 contacts the assigned DNA density visible in (e), whereas the N-terminal AAA+ domain reaches down and interacts with the CTD of Mcm3. (j) The Mcm3 density is shown as semi-transparent surface with the rigid-body docked Archaeal Mcm homology structure shown as salmon ribbons (PDB ID: 3F9V)27.
In the lower Cdt1-MCM2-7 region of the OCCM structure, segmentation of subunit densities for Mcm3, Mcm4, Mcm5, and Mcm7 were nearly automatic and unambiguous (Fig. 4a-f). However, the densities belonging to Cdt1, Mcm2, and Mcm6 were intertwined. Therefore, their boundaries are less certain and subjected to interpretation, although the relative locations are defined. After assigning all seven proteins in the Cdt1-MCM2-7 region, there was one segmented density remained unaccounted for. This density, colored light gray, is in the central region of MCM2-7, and best viewed in the bottom view (Fig. 4f). The identity of the density has not been established (See the question mark in Fig. 4f), but we speculate that it is from the eukaryotic specific N-terminal extensions of some Mcm subunits and perhaps contains DNA emerging from the bottom end of the OCCM structure (see below). In the well-defined Mcm3 density (Fig. 4b, 4j), the crystal structure of the near full-length Archaeal MCM can be fitted by rigid-body docking with the NTD pointing to the bottom and the AAA+ domain-containing CTD facing ORC-Cdc6 above27 (Fig. 4j).
The physical proximity between the individual subunits in the heptameric ORC-Cdc6 and subunits of the heptameric Cdt1-MCM2-7 can be summarized as following: Orc1:(Mcm4 + Mcm7), Orc2:(Mcm2+Mcm6), Orc3:(Mcm2+Mcm5), Orc4:Mcm4, Orc5:(Mcm4+Mcm6), Orc6:Mcm2, and Cdc6:(Mcm3+Mcm7) (Fig. 4a-f). Although the putative density assigned to Orc6 is on the same side as Cdt1 in the OCCM, they are not touching. Earlier studies suggested that interaction between Orc6 and Cdt1 was critical for MCM2-7 loading onto origin DNA by ORC-Cdc642, but recent studies suggested otherwise43,44
We previously found that N- and C-terminal MBP-fusions to Orc1-5 support ORC-Cdc6 complex formation34. Here, we observed efficient OCCM and DH formation with all N-terminal ORC-MBP-fusions (Supplementary Fig. 5a). This is in agreement with the deduced OCCM model predicting the surface localization of the N-termini of the ORC subunits. However, MBP fused to the C-terminus of Orc1 (CO1) or Orc4 (CO4) reduced recruitment of MCM2-7 (low salt) and blocked DH formation (high salt), although they did not affect ORC association with DNA. It is possible that MBP fused to the Orc1 and Orc4 C-termini interferes with the establishment of the correct DNA path, which in turn blocks MCM2-7 recruitment. On the other hand, MBP attached to Orc2 C-terminus did not interfere with the loading function, indicating that not all Orc C-termini function in the same way.
Cdc6 is central for pre-RC formation9,10,45. The OCCM model predicts the proximity of Cdc6 and Mcm3. We examined their potential interaction in an independent IP approach. Mcm3 interacted efficiently with MBP-Cdc6, but not with MBP, while all other Mcm subunits bound only weakly to Cdc6 (Supplementary Fig. 5b). In the case of Mcm4 we observed non-specific interaction with MBP rendering the result for Mcm4 inconclusive. Furthermore, the data indicated that the Mcm3 C-terminus is orientated to ORC-Cdc6. An analysis of the MCM2-7 C-termini shows that Mcm3 contains a long C-terminal extension that is not present in other MCM subunits (Supplementary Fig. 1b). We reasoned that this extension could be important for the interaction between Cdc6 and MCM2-7. For this reason a MCM2-7-ΔC3 mutant, missing aa740-971 was generated. Interestingly, we observed that MCM2-7-ΔC3 interacted weaker than wtMCM2-7 with MBP-Cdc6 (Supplementary Fig. 5c). Although it was able to interact with Cdt1 (Supplementary Fig. 5d), MCM2-7-ΔC3 did not associate with ORC-Cdc6 in low salt buffer and failed to load high salt resistant MCM2-7 onto DNA (Supplementary Fig. 5e). Our result is consistent with the recent report on the importance of Mcm3 C-terminal domain on MCM2-7 loading43. The IP results confirm the model-predicted proximity of Cdc6 and Mcm3 and indicate the functional importance of their interaction.
Yeast Cdt1-MCM2-7 in comparison with Drosophila MCM2-7
Fig. 5a-c show Cdt1-MCM2-7 structure extracted from the OCCM structure in comparison with Drosophila MCM2-7 structure18 (Fig. 5b). The top view is in stereo that better shows the likely DNA density inside the MCM2-7 hexamer chamber (Fig. 5c). In this structure, the helicase-loading factor Cdt1 binds primarily to the N-terminal half of Mcm2, but extends further to Mcm5 near its N-terminus and to Mcm6 at the C-terminus. Cdt1 interaction with Mcm2 and Mcm6 was reported13,46. The N-terminal region of Cdt1 is in a position that it could keep the NTD of Mcm2 away from the NTD of Mcm5, resulting in a gap between Mcm2 and Mcm5 in the bottom N-terminal side of the MCM2-7 structure. On the contrary, the C-terminal side of MCM2-7 better resembles a ring structure (Fig. 5c), but with two caveats: the Mcm2 CTD is notably higher than that of the other Mcm subunits; and there appears to be a gap between Mcm4 and Mcm6. The gap or weak association between Mcm4 and Mcm6 was also present in the negative stain EM structure of the DmMCM2-7 structure. The significance of the potential Mcm4-Mcm6 gap in eukaryotic MCM2-7 is unclear but it may function as an alternative DNA gate in the Mcm467 hexamer that has in vitro helicase activity47,48.
Figure 5. Cryo-EM structure of the yeast Cdt1-MCM2-7 in the context of the OCCM complex in comparison with the Drosophila MCM2-7 structure.

(a) The Cdt1-MCM2-7 density is isolated from the OCCM complex and shown in three surface views. Individual subunits are labeled. (b) The negatively stained EM structure of the Drosophila MCM2-7 in the bottom N-terminal view (EMDB ID: 1134)18. (c) The ORC-facing C-terminal stereo view of the yeast Cdt1-MCM2-7. Note that the light gray density inside the hexamer chamber resembles DNA. Mcm2-7 are abbreviated as M2-7. (a – c) are on the same scale.
ORC-Cdc6 transforms into a right-handed spiral in the OCCM
Our previous determination of the DNA-bound ORC-Cdc6 structure in the absence of Cdt1-MCM2-711 (Fig. 6a) and the current structure in the presence of Cdt1-MCM2-7 (Fig. 6b) provides an opportunity to examine how ORC-Cdc6 functions to recruit the MCM2-7 hexamer. Although both structures are ring shaped with similar sizes, ORC-Cdc6 has undergone profound conformational changes upon interaction with Cdt1-MCM2-7. First, ORC-Cdc6 alone is nearly flat but bends into a dome-like structure towards Cdt1-MCM2-7 when it contacts the latter (Fig. 6a-b). This transformation is largely accomplished by movement of the individual “C-shaped” subunits; specifically, by the N-terminal AAA+ domains reaching down to interact with the CTDs of the MCM2-7 subunits (Fig. 4a-d). Furthermore, at one edge of ORC-Cdc6, Orc6, Orc3, and Cdc6 have undergone a series of concerted movements (Fig. 6a-b, the middle panels). It appears that the mass tentatively assigned to Orc6 initiates the transition since it has to move out of the way to allow MCM2-7 interaction with ORC-Cdc6. This is so because, in the absence of Cdt1-MCM2-7, Orc6 occupies the region that MCM2-7 will bind ORC-Cdc6 (Fig. 6a middle panel). We suggest that upon encountering Cdt1-MCM2-7, Orc6 rotates ~60° downwards, which would push Orc3 upwards and away from Orc2. The upward movement of Orc3 narrows the gap between Orc1 and Orc3 that Cdc6 bridges, and this in turn forces Cdc6 to rotate ~ 45° in order for it to fit into the now much narrowed gap (Fig. 6a-b middle and right panels). Accompanying the Cdc6 rotation, the N-terminal domain of Orc1 (labeled 1N) moves out and away from the center (Fig. 6a-b, left panels, the black arrow). The movement of ORC-Cdc6 upon Cdt1-MCM2-7 binding also brings the Orc1 and Orc4 subunits into closer contact, potentially allowing Orc4 to activate the ATPase activity of Orc1, an activity necessary for pre-RC assembly30.
Figure 6. Upon recruitment of Cdt1-MCM2-7, ORC-Cdc6 undergoes concerted conformational changes into a right-handed spiral structure.
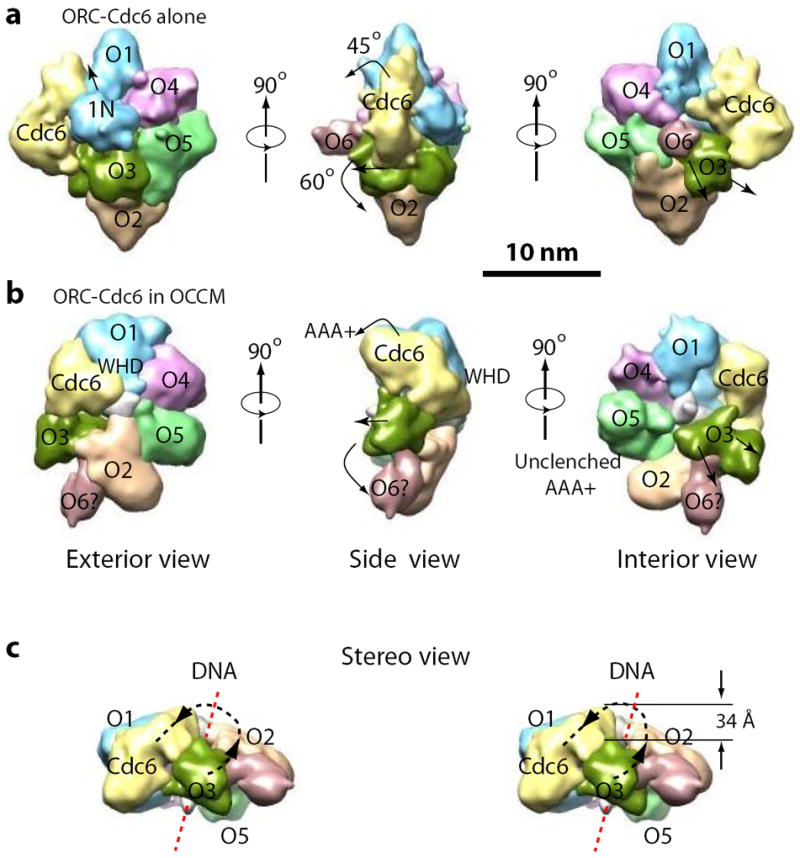
(a) The ORC-Cdc6-DNA structure alone11. (b) The ORC-Cdc6-DNA structure extracted from the OCCM structure. The question mark indicates the tentative assignment of Orc6. (c) Stereo side view of ORC-Cdc6 extracted from the OCCM structure. The dashed black curve traces the right-handed helical rise in the order of Orc3, Orc2, Orc5, Orc4, Orc1, and Cdc6. The vertical rise from the lowest Orc3 to the highest Cdc6 is 34 Å as labeled. The dashed red line illustrates the bound dsDNA. Orc1-6 are abbreviated as O1-6.
Importantly, in the presence of Cdt1-MCM2-7, ORC-Cdc6 forms a right-handed spiral structure (Fig. 6c; supplemental movies 1 and 2). Orc3 is at the lowest position in the spiral, followed by Orc2, Orc5, Orc4, Orc1, and finally Cdc6 at the highest position between Orc1 and Orc3. The total vertical rise from Orc3 to Cdc6 is 34 Å, which translates to ~ 5.6 Å axial rise per protein subunit (34 Å/6) (Fig. 6c). The 34-Å rise in the ORC-Cdc6 structure is remarkable because it is equal to the helical pitch of the B-form double stranded DNA. Therefore, the six AAA+ domain-containing subunits in the ORC-Cdc6 hetero-heptamer form a spiral structure with its helical symmetry exactly matching the dsDNA to which they bind. The symmetry match suggests that ORC-Cdc6 stably anchors DNA at the center of OCCM, which may be important for MCM2-7 loading.
MCM2-7 hexamer in the OCCM may be partially loaded
There is a linear, nearly continuous density that passes through OCCM from the outside into the ORC-Cdc6 region and further into the MCM2-7 central chamber (Fig. 7). The central density is better viewed in the eight consecutive sections by a horizontal plane moving from the top (position 0) to the bottom (position 7), as marked by blue arrows (Fig. 7a-b). This density is also visible in the 3D map when the front Orc3 and Mcm5 are removed (Fig. 7c). We have experimentally identified the top region of the linear density as dsDNA (Fig. 3f-g), and now suggest that the entire stretch of density may be from the dsDNA (Fig. 7c, the pair of red curves). Therefore, our cryo-EM structure suggests that the first MCM2-7 may be partially loaded and encircling the dsDNA in the absence of extensive ATP hydrolysis, before the second MCM2-7 hexamer is recruited. This is so because we used non- or weakly-hydrolyzable ATPγS in our preparation, and there is only one MCM2-7 hexamer in the OCCM. The loading is partial and must have not completed because OCCM can be washed off of DNA by a high salt buffer.
Figure 7. The DNA apparently passes through the middle of the OCCM complex.

(a) 3D cryo-EM map of OCCM with the positions of horizontal sections labeled from the top 0 to the bottom 7. (b) 2D sections of OCCM with resolved DNA density labeled by the blue arrow. (c) A side view of the OCCM structure with the front Orc3 and Mcm5 densities removed to allow viewing of the interior elongated DNA densities extending from the top to the bottom, as outlined by a pair of dashed red lines. (d) The central axis of MCM2-7 hexamer is tilted 12° away from the helical axis of the spiral ORC-Cdc6 structure. (e) A ribbon representation of the crystal structure of the yeast RFC-PCNA complex (PDB ID: 1SXJ)51, showing the 9° mismatch between the central axis of the PCNA ring and the spiral axis of the RFC-A-RFC-E pentamer. Note that only the AAA+ motor domains in RFC are helically arranged. Mcm2-7 are abbreviated as M2-7, and Orc1-6 as O1-6, respectively. (a, c, d, e) are on the same scale. The box size in (b) is 22 nm.
The ORC-Cdc6 spiral resembles the PCNA-loading RFC spiral
There is clearly a mismatch, about 12°, between the central axis of the MCM2-7 hexameric ring and the helical axis of the ORC-Cdc6 (Fig. 7d). This mode of interaction between ORC-Cdc6 and Cdt1-MCM2-7 bears striking similarity with the eukaryotic PCNA (Proliferating Cell Nuclear Antigen) DNA polymerase clamp and its loading by the ATPase complex Replication Factor C (RFC) (Fig. 7e). RFC is an ATP-dependent machine that loads the DNA polymerase clamp PCNA onto primer-template DNA so that PCNA can then recruit and tether the DNA polymerase at the replication fork49,50. In the crystal structure of the RFC-PCNA complex, the five AAA+ motor domains of RFC-A to RFC-E form a spiral structure and the gap between RFC-A and RFC-E is bridged by the extra domain (domain IV) of RFC-A50,51. The helical axis of the RFC spiral tilts 9° away from the PCNA ring axis (Fig. 7e). Therefore, the six-AAA+ subunits of ORC-Cdc6 form a spiral in the OCCM that resembles the PCNA ring-loading five-subunit AAA+ RFC spiral50,52.
DISCUSSION
Using cryo-EM, an intermediate in the loading onto DNA of the eukaryotic DNA helicase core complex by ORC-Cdc6 and Cdt1 was captured and characterized, thereby establishing the architecture of the OCCM complex. The cryo-EM structure reveals that several AAA+-like domains of ORC-Cdc6 unclench and latch onto the CT AAA+ motor domains of the MCM2-7 hexamer, leading to the loading of MCM2-7 onto the dsDNA. During the loading process, ORC-Cdc6 transforms into a spiral structure.
The DNA gate in the MCM2-7 hexamer is widely believed to be located between Mcm2 and Mcm518,38. In the OCCM, a gap exists between the NTDs of Mcm2 and Mcm5, but the two CTDs are not separated (Fig. 4f, and Fig. 5). Interestingly, Orc3 makes contact with both CTDs of Mcm2 and Mcm5 (Fig. 4b). Thus there is a possibility that a slight descent of Orc3 would further separate Mcm2 and Mcm5 and fully open up the MCM2-7 ring.
The DNA in OCCM is nearly perpendicular to the spiral surface of ORC-Cdc6. In this DNA binding model, the length of the DNA that can be protected by ORC-Cdc6 is estimated to be shorter than 30 bp: the first AAA+ protein (Orc3) may protect up to 20 bp DNA19,20, and the following five subunits in the spiral would cover additional 10 bp in one DNA helical pitch (Fig. 6c). However, it has been well established that ORC alone has a DNase I footprint of 48 bp3, and when Cdc6 binds ORC, the DNase I footprint extends to nearly 80 bp53. Therefore, the DNA binding mode in ORC-Cdc6 alone before encountering Cdt1-MCM2-7 must be different from that seen in the OCCM structure, and the DNA in ORC-Cdc6 likely undergoes a profound transition when Cdt1-MCM2-7 is recruited. Indeed, the proposed DNA binding model in our previous ORC-Cdc6 alone structure is significantly different: the origin DNA is proposed to be bent and wrapped around in the interior of the ORC-Cdc6 structure, in accordance with the DNA binding model in the crystal structure of Archaeal Orc1 and in agreement with the extended DNase I footprint11. We suggest that the DNA transitions to the current vertical position, accompanying the subunit rearrangement in ORC-Cdc6 following the interaction with Cdt1-MCM2-7 (Fig. 6). Clearly, DNA rearrangements in ORC-Cdc6 during helicase loading are important and require further investigation.
In the T4 bacteriophage clamp loading system in the presence of DNA and ATP hydrolysis, the clamp ring opens up, and converts to a spiral shape whose helical symmetry matches that of the DNA as well as the clamp loader50. Furthermore, in the negative stain EM structure of an Archaeal PCNA-RFC complex with DNA and ATP, the clamp ring is also open and resembles a washer54. In the case of ORC-Cdc6, six AAA+-containing components form a spiral encircling the dsDNA, the ORC-Cdc6 helical axis is tilted 12° away from the MCM2-7 ring axis, and the MCM2-7 ring appears to be partially open. One important difference between these two ring-loading systems is that in RFC, the third and C-terminal helical domains form a tightly sealed collar that blocks DNA passage, whereas in ORC-Cdc6, the corresponding six WHDs from the six AAA+ proteins form an open collar through which dsDNA passes (Fig. 3f-g, Fig. 6c and Fig. 7c). Another obvious difference is that MCM2-7 is an ATPase ring whereas the PCNA ring is not, so the respectively ring opening mechanism could be different. Despite these differences, it is striking that two structurally related ATP-dependent protein machines, ORC-Cdc6 and RFC, respectively load ring-shaped proteins MCM2-7 and PCNA that are later involved in movement along DNA during replication fork progression.
A recent study also revealed an analogy between the bacterial DnaB helicase loading by DnaC and the PCNA loading by RFC55. The observed organization of the yeast OCCM may provide insights into the mechanism of helicase loading for higher eukaryotes and Archaea, as in these systems structural information on the helicase loader – helicase complex have not yet been obtained.
ONLINE METHODS
Pre-RC assay
The pre-RC was performed as described12. 40 nM ORC, 80 nM Cdc6, 40 nM Cdt1, 40 nM MCM2-7, 6 nM pUC19-ARS1 beads in 50 μl buffer A (50 mM Hepes-KOH pH 7.5, 100 mM KGlu, 10 mM MgAc, 50 μM ZnAc, 3 mM ATP, 5 mM DTT, 0.1% Triton X-100, and 5% glycerol) were incubated for 15 min at 24 °C. After 3 washes with buffer A or B (50 mM Hepes-KOH pH 7.5, 1 mM EDTA, 500 mM NaCl, 5% Glycerol, 0.1% Triton X-100, and 5 mM DTT) the complex was eluted with 1 U of DNase I in buffer A + 5 mM CaCl2.
Pre-RC assembly for EM analyses
Pre-RC assembly was performed as described above12, however ATPγS was used instead of ATP. The DNaseI or AluI elution was performed in 5 μl buffer C (50 mM Hepes-KOH pH7.5, 100 mM KAc, 5 mM MgAc, 5 mM CaCl2). For the initial experiment (Fig. 1b) an ARS1 origin DNA of 86 base pairs was used (TTTGTGCACTTGCCTGCAGGCCTTTTGAAAAGCAAGCATAAAAGATCTAAACATAAAATCTGTAAAATAACAAGATGTAAAGATAA) and proteins and DNA were mixed at equimolar ratios.
Immunoprecipitation of OCCM
The pre-RC complexes were prepared as described (pre-RC assay) and then immunoprecipitated with anti-Mcm256 and anti-MBP (E8032L; NEB) antibody coupled to protein G beads for 7.5 min at 24 °C, washed three times with buffer A and analyzed by Western blot with anti-Mcm256 antibody.
MBP-Cdc6 and MBP-Cdt1 immunoprecipitation
100 ng of MBP and MBP-Cdc6, MBP-Cdt1 were immobilized on 2 μl anti-MBP antibody beads (NEB) in 50 μl buffer D (buffer A with 300 mM KGlu, 0.25% Triton X-100, 0.5% NP-40, 0.1% BSA, and 10% glycerol). A bacterial extract containing equal amounts of overexpressed GST-Mcm2-7 in buffer D was incubated with MBP or MBP-Cdc6 beads for 10 min at 24 °C. Purified wtMCM2-7 or MCM2-7-ΔC3 in buffer D was incubated with MBP or MBP-Cdt1 beads for 10 min at 24 °C. Afterwards the beads were washed three times. The bound proteins were analyzed by western blotting with anti-GST antibody (3G10/1B3; Abcam) or an anti-Mcm2-7 antibody.
Cloning of GST-Mcm2-7
The individual MCM genes were amplified by PCR (primers available on request) and cloned via XmaI-NotI into pGEX6-P1 (GE Healthcare) resulting in pCS328 (Mcm2), pCS329 (Mcm3), pCS330 (Mcm4), pCS331 (Mcm5), pCS332 (Mcm6) and pCS333 (Mcm7).
Cloning of MBP-Mcm3, Mcm5-MBP, MBP-Mcm6 and Mcm3ΔC
Using site directed mutagenesis a restriction site was inserted after aa111 (Mcm3), aa591 (Mcm5) and aa124 (Mcm6). MBP was amplified from pMAL-c2X (NEB) with primers incorporating flexible linkers (sequences available on request) and inserted in the restriction sites generating pCS444 (pESC-URA-MBP-Mcm3/Mcm5), pCS470 (pESC-URA-Mcm3/Mcm5-MBP) and pCS473 (pESC-TRP-Mcm4/MBP-Mcm6). Mcm2-MBP pCS280 was described in45. aa740-971 of Mcm3 was deleted generating pCS493 (pESC-URA-MCM3ΔC740-971-MCM5).
Protein purification
The untagged ORC complex and the MBP-tagged ORC complexes34 were expressed and purified as described57. Cdc6 and Cdt1 were expressed in bacteria and purified as described5,12. MCM2-7 wild type or mutants were expressed in yeast and purified as described12.
Cryo-EM
To prepare cryo-EM grids, we first evaporated a thin layer of carbon film (~2 nm) on a freshly-cleaved mica in a Edwards vacuum evaporator, then floated the carbon film off the mica surface in deionized water, and deposited film onto the lacey carbon-coated EM grids. The dried EM grids were glowed discharged in the 100 mTorr Argon atmosphere for 40 seconds. Sample vitrification was carried out in an FEI Vitrobot plunge-freezing device, which was set to the operating temperature of 11°C, 70% relative humidity, -1.0 mm blotting pad height offset. Three μl of the OCCM sample was pipetted onto the freshly glow-discharged EM grid, after 30 s, the grid was blotted for 5 s, and plunged into the liquid ethane. The cryo-EM grids were transferred in liquid nitrogen into a Gatan 626 cryo specimen holder. The specimen was maintained at below -170°C during data collection. Cryo-EM was performed in the JEM-2010F operated at the accelerating voltage of 200 kV. Cryo-EM images were recorded at 50,000X mag on a Gatan 4K×4K UltraScan 4000 CCD camera with an electron dose of 15 e-/Å2, corresponding to 2.12 Å/pixel sampling at the specimen level.
2D image analyses
Computational image analyses and 3D reconstruction of the OCCM images followed procedures as outlined58. Briefly, we used the software package EMAN for most of our image processing needs59. Raw particle images were selected semi-automatically with program e2boxer.py in EMAN259. The particle images were manually inspected and “bad” particles were rejected at this stage, leaving ~90,000 particles in the final wild type OCCM dataset. The contrast transfer function (CTF) was determined on the whole micrographs, and its effects corrected in program ctfit. The raw particles were pooled, phase flipped, edge normalized, and high-pass filtered (hp=1). Reference-free 2D classification and averaging of the raw dataset were carried out by the program refine2d.py. A large number of class averages (up to 500) were produced by running the program for 9 cycles with at least 20 particles in each class. Careful inspection of the averaged images lead us to conclude that the OCCM structure was homogeneous at the 1-2 nm resolution concerned here.
3D reconstruction
We used the 2D class averages as input into the program e2initialmodel.py to produce 10 starting models. We then carefully inspected the consistency between the model reprojections and the original reference-free class averages, and selected three models for the full-scale refinement. We used their low-pass filtered versions (99-Å resolution) as the starting models to minimize initial model bias. Refinement was carried out in the EMAN1.8 with options dfilt and refine turned on. Refinement was carried out in a 144-CPU Dell Linux cluster. The resolution of the cryo-EM 3D map was estimated from Fourier shell correlation at the threshold of 0.5. Refinement with the three models resulted in essentially the same final 3D map at the stated 14-Å resolution. This result is likely due to the good contrast of the 1.1-MDa particle images and the distinct shape of the OCCM structure. 3D reconstruction of the MBP-fusion OCCM complexes followed essentially the same procedure, with the 40-Å low-pass filtered wild type OCCM 3D map as the starting model for refinement. The number of particles used and the 3D map resolution for each fusion complex were 7036 and 20 Å for OCCM_Orc2-MBP, 4029 and 28 Å for OCCM_MBP-Cdt1, 20793 and 20 Å for OCCM_MBP-Mcm2, 1654 and 30 Å for OCCM_MBP-Mcm3 (negatively-stained images), 6491 and 26 Å and for OCCM_Mcm5-MBP, and 6036 and 22 Å for OCCM_MBP-Mcm6, respectively.
Cryo-EM 3D map validation
We followed the Henderson-Rosenthal tilt pair technique as implemented in EMAN2 version 2.0633. The critical part of the technique was to identify cryo-EM grids that produced high particle contrast. Tilt image pairs were recorded in CCD camera, with the 10° tilted images recorded first and followed by the untilted images, at the accumulative dose of 30 e-/Å2. The particle pairs were selected in program e2RCTboxer.py. Then, in program e2projectmanager.py, the CTF effects were corrected for the tilted and untilted particles separately, and the program e2tiltvalidate.py was used to find the Euler anglers for all particle images by using the default parameters as suggested in the EMAN2 documentation. The relative tilt axis and angle of each tilt pair was calculated and plotted as a dot in the polar coordinate system. We limited the out-of-plane tilt angle to 0.3°, which removed ~ 50% of the particle pairs. Clustering of the plotted dots around the experimental tilt geometry indicates the correctness of the 3D map and its associated handedness. We found that the ORC-Cdc6 structure in the OCCM was consistent with the mirrored version of the previous ORC-Cdc6 map11. Therefore, the ORC-Cdc6 map shown in Fig. 7a (EMDB ID: 5381) has been mirrored.
Density segmentation and crystal structure docking
The 3D density map was segmented with the program Segger39. The homolog crystal structures were docked into 3D maps with the fit function in Chimera. Surface-rendered figures were also prepared in Chimera60. Although crystal structures of some Archaeal replication initiators are known, homology modeling and molecular dynamics flexible fitting are not carried out, because eukaryotic replication initiators are larger and often contained several additional domains that have no known homolog structures, and therefore cannot be modeled confidently, and also because of the medium resolution of the OCCM 3D map.
Supplementary Material
Acknowledgments
We thank Marie Smulczeski and Shuanglu Zhang for helping to manually select a large number of particles from raw cryo-EM micrographs and Emanuela Gardenal and Christian Winkler for the MCM2-7-Cdc6 interaction analysis. This work was supported by National Institutes of Health grant nos. GM45436 (to B.S.) and GM74985 (to H.L.) and the United Kingdom Medical Research Council (to C.S.). H.K. was supported by Postdoctoral Fellowships for Research Abroad from the Japan Society for the Promotion of Science and the Uehara Memorial Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
J.S., C.E., S.S, A.F., A.R., and H.K. performed the specimen preparation and biochemistry. J.S. collected the cryo-EM data, performed the cryo-EM reconstructions. J.S., B.S., C.S., and H.L. designed experiments and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
ACCESSION NUMBER.
The cryo-EM 3D density map of S. cerevisiae OCCM complex (ORC-Cdc6-Cdt1-MCM2-7 on dsDNA) has been deposited in the Electron Microscopy Data Bank under accession number EMD-5625.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–84. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–34. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–7. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–71. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarey MG, et al. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol. 2006;13:684–90. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Stillman B. The origin recognition complex: a biochemical and structural view. Subcell Biochem. 2012;62:37–58. doi: 10.1007/978-94-007-4572-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. Embo J. 1996;15:6671–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A. 1997;94:5611–6. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, et al. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20:534–44. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–5. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–30. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–58. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A. 2012;109:6042–7. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–7. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–6. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 20.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–3. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 21.Clarey MG, Botchan M, Nogales E. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol. 2008;164:241–9. doi: 10.1016/j.jsb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, et al. Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res. 2012;40:3208–17. doi: 10.1093/nar/gkr1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci U S A. 2012;109:11999–2004. doi: 10.1073/pnas.1209406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onesti S, Macneill SA. Structure and evolutionary origins of the CMG complex. Chromosoma. 2013 doi: 10.1007/s00412-013-0397-x. [DOI] [PubMed] [Google Scholar]
- 25.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–31. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–83. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brewster AS, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci U S A. 2008;105:20191–6. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci U S A. 2003;100:9150–5. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khayrutdinov BI, et al. Structure of the Cdt1 C-terminal domain: conservation of the winged helix fold in replication licensing factors. Protein Sci. 2009;18:2252–64. doi: 10.1002/pro.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–78. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 31.Pape T, et al. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–83. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suck D, Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986;321:620–5. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 33.Henderson R, et al. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J Mol Biol. 2011;413:1028–46. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, et al. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2008;105:10326–31. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Chavan M, Schindelin H, Lennarz WJ, Li H. Structure of the oligosaccharyl transferase complex at 12 A resolution. Structure. 2008;16:432–40. doi: 10.1016/j.str.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–91. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau PW, Potter CS, Carragher B, MacRae IJ. DOLORS: versatile strategy for internal labeling and domain localization in electron microscopy. Structure. 2012;20:1995–2002. doi: 10.1016/j.str.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–73. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–38. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fletcher RJ, et al. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10:160–7. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, et al. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol Cell. 2000;6:637–48. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007;21:2897–907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–43. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez-Cid A, et al. An ORC/Cdc6/MCM2-7 Complex Is Formed in a Multistep Reaction to Serve as a Platform for MCM Double-Hexamer Assembly. Mol Cell. 2013;50:577–588. doi: 10.1016/j.molcel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 45.Evrin C, et al. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawasaki Y, Kim HD, Kojima A, Seki T, Sugino A. Reconstitution of Saccharomyces cerevisiae prereplicative complex assembly in vitro. Genes Cells. 2006;11:745–56. doi: 10.1111/j.1365-2443.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- 47.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–13. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 48.Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J Biol Chem. 2007;282:33795–804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- 49.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–51. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 50.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–80. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 52.O’Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Speck C, Stillman B. Cdc6 ATPase activity regulates ORC × Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem. 2007;282:11705–14. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyata T, et al. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci U S A. 2005;102:13795–800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arias-Palomo E, O’Shea VL, Hood IV, Berger JM. The Bacterial DnaC Helicase Loader Is a DnaB Ring Breaker. Cell. 2013;153:438–48. doi: 10.1016/j.cell.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–96. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 58.Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat Protoc. 2010;5:1697–708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


