Abstract
Lipid transfer particle (LTP) is a high-molecular-weight, very high-density lipoprotein known to catalyze the transfer of lipids between a variety of lipoproteins, including both insects and vertebrates. Studying the biosynthesis and regulation pathways of LTP in detail has not been possible due to a lack of information regarding the apoproteins. Here, we sequenced the cDNA and deduced amino acid sequences for three apoproteins of LTP from the silkworm (Bombyx mori). The three subunit proteins of the LTP are coded by two genes, apoLTP-II/I and apoLTP-III. ApoLTP-I and apoLTP-II are predicted to be generated by posttranslational cleavage of the precursor protein, apoLTP-II/I. Clusters of amphipathic secondary structure within apoLTP-II/I are similar to Homo sapiens apolipoprotein B (apoB) and insect lipophorins. The apoLTP-II/I gene is a novel member of the apoB/large lipid transfer protein gene family. ApoLTP-III has a putative conserved juvenile hormone-binding protein superfamily domain. Expression of apoLTP-II/I and apoLTP-III genes was synchronized and both genes were primarily expressed in the fat body at the stage corresponding to increased lipid transport needs. We are now in a position to study in detail the physiological role of LTP and its biosynthesis and assembly.
Keywords: lipophorin, juvenile hormone-binding protein, lipid exchange and transport
Because lipids are water insoluble and do not exist at appreciable concentrations as individual molecules in an aqueous environment, their transport to various tissues via blood is intimately linked to lipoprotein metabolism. In insects, two major lipoproteins exist in the hemolymph, lipophorin and lipid transfer particle (LTP).
Lipophorin is a multifunctional lipid transport vehicle present during all life stages, in which it functions as a reusable shuttle to transport diacylglycerol (DAG), phospholipids, hydrocarbons, cholesterol, and carotenoids from sites of ingestion or synthesis to sites of utilization (1).
LTP is found in a variety of insects, including Manduca sexta (2), Locusta migratoria (3), Musca domestica (4), Periplaneta americana (5), Bombyx mori (6), and Rhodnius prolixus (7). LTP from B. mori is a very high density lipoprotein consisting of approximately 20% lipids and three apolipoproteins (apoLTP-I, apoLTP-II, and apoLTP-III) (6). In M. sexta, LTP is synthesized in the fat body and secreted into the hemolymph (8). LTP can catalyze the exchange and/or transfer of DAG between lipophorin particles with different densities (9), between lipophorins and human lipoproteins (10), or from the fat body to lipophorin, thus facilitating the formation of low-density lipophorin (LDLp) in vitro (11). The majority of stored lipids exist in the fat body as triacylglycerol (TAG) lipid droplets. TAG storage results from the transfer of dietary fat from the midgut to the fat body during the feeding stage (12). LTP may be involved in this process as well as in the transfer of lipids from the midgut to lipophorin (13). Lipid export from enterocytes does not involve de novo synthesis of lipophorin; rather, lipids are added directly to existing lipophorin particles in the hemolymph (14, 15). Taken together, these reports indicate that LTP plays an important role in facilitating lipophorin function and may mediate the transfer of many lipids, including hydrocarbons (5) and carotenoids (15), with its specificity being determined by the properties of putative lipid transfer factors in the target cell (16–18).
The mechanisms underlying LTP biosynthesis and assembly are virtually unknown. Due to its very large size, cloning and sequencing cDNA for LTP has been difficult, especially when apoLTP-I and apoLTP-II are both encoded on a single gene (apoLTP-II/I), as is the case with the precursor protein of lipophorin, apolipophorin-II/I (19, 20). Recently, however, a whole genome sequence database for silkworm (21, 22) has become available and genome annotation is ongoing. In this report, we present the cDNA and deduced amino acid sequences of apoLTP-I, apoLTP-II, and apoLTP-III from B. mori. Our results provide insights into the function of LTP as a novel member of the apoB/large lipid transfer protein (APO) family and represent an important step in the study of LTP biosynthesis and assembly.
MATERIALS AND METHODS
Isolation of LTP
The N4 strain of silkworm (B. mori) was maintained in a continuous laboratory colony and reared on an artificial diet. The larval LTP was isolated from the hemolymph of fifth instar larvae on day 4 according to the method described by Tsuchida et al. (6). The adult hemolymph was collected from the abdomen of adults at day 0 by cutting with a needle and dropped into an ice-cold bleeding solution (20 mM sodium phosphate pH 6.8, 150 mM NaCl, 5 mM EDTA, 1 mM glutathione, and 1 mM 4-2-aminoethyl benzenesulfonyl fluoride). The hemolymph was centrifuged at 800 g for 5 min to remove hemocytes. To the supernatant, 8.9 g potassium bromide (KBr) was added, and the volume was adjusted to 20 ml with bleeding solution. The solution was transferred to 36.2 ml OptiSealTM centrifuge tubes (Beckman Coulter, Brea, CA) and overlaid with bleeding solution. The tube was centrifuged at 4°C and 50,000 rpm for 4 h in a VTi 50 rotor (Beckman Coulter). After centrifugation, LDLp, high-density lipophorin (HDLp), and LTP formed three yellow bands in the ultracentrifuge tube. The fractions from the middle yellow band, which contained mainly HDLp and LTP, were pooled, and after desalting, were applied to DEAE Bio-Gel (Bio-Rad, Hercules, CA) and eluted with 20 mM sodium phosphate pH 7.5, containing a linear NaCl gradient (20–300 mM). The elutant was subjected to a Sephacryl S-300 column (GE Healthcare, Milwaukee, WI) and fractions were collected. Fractions containing LTP were examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (6).
SDS-PAGE and detection of protein glycosylation
SDS-PAGE was performed according to the method of Laemmli (23) in slab gels containing a 4–15% linear gradient of polyacrylamide. Gels were stained with Coomassie Brilliant Blue R-250 (Bio-Rad).
To detect glycoproteins in LTP subunit proteins, following SDS-PAGE of 5 μg of purified larval LTP, the protein was electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membrane was stained with fluorescein isothiocyanate conjugated to concanavalin A (FITC-Con A) (Calbiochem, San Diego, CA) according to Furlan, Perret, and Beck (24). Conjugates were visualized with ultraviolet light.
Estimation of LTP native molecular weight
Estimation of LTP native molecular weight was performed by blue-native PAGE (25, 26) and Western blot analysis (27). Samples of hemolymph (1 μl) from each fifth instar larva at day 4, pupal stage at day 3, and adult stage at day 0 were electrophoresed on a 3–10% blue-native acrylamide gradient gel at 4°C and a constant 150 V. A molecular weight marker set (Invitrogen, Carlsbad, CA) was used as the calibration standard. A rabbit polyclonal antibody was raised against purified apoLTP-I. After proteins were separated by blue-native PAGE and transferred to a PVDF membrane, the membrane was incubated with the anti-apoLTP-I antibody and an alkaline phosphatase-goat anti-rabbit IgG conjugate (Jackson ImmunoResearch Laboratory, West Grove, PA). The signals were detected using an alkaline phosphatase-conjugate substrate kit (Bio-Rad).
Preparation of LTP apoproteins and amino acid sequence determination
Three apoLTP subunits were separated by SDS-PAGE of purified LTP and transferred to a PVDF membrane. After Coomassie staining, membrane slices were excised. The N-terminal amino acid sequences of apoLTP-I and apoLTP-II were determined by Edman degradation (28), and the sliced membranes were incubated with lysyl endopeptidase. Digested peptides were separated by reverse-phase high-performance liquid chromatography (HPLC). Amino acid sequences of the peptides including the N-terminal amino acids were determined from six peptides of apoLTP-I, two peptides of apoLTP-II, and one peptide of apoLTP-III using a G1005A protein sequencing system (Hewlett-Packard, Palo Alto, CA).
Identification and sequence analysis of apoLTP-I and apoLTP-II
We obtained candidate sequences containing apoLTP-I and apoLTP-II peptide sequences as described above by a TBLASTN search of the Silkworm Genome Database using the peptides as query sequences. A protein homology search using the candidate apolipoprotein sequences suggested that the B. mori apoLTP-I and apoLTP-II were encoded by one gene homologous to Drosophila melanogaster CG15828. To determine the full-length cDNA sequence of apoLTP-II/I, the 5′ and 3′ ends of the apoLTP-II/I sequence were obtained by rapid amplification of cDNA ends (RACE) using a SMART RACE cDNA amplification kit (Clontech, Mountain View, CA) with the following primers: 5′-AGGCTGGTGTCTTCTTGGCCCCGGACG-3′ for the first 5′-RACE product, 5′-GAGACGGCGCCTATGAATTTCTCCGCACG-3′ for the nested 5′-RACE product, 5′-CAGCTGGCAGGACTTCCTCAAGACCCCG-3′ for the first 3′-RACE product, and 5′-TGATCGGCGAGGCCTTGAACACGATCGG-3′ for the nested 3′-RACE product. Primers used for RACE were designed using the hypothetical partial cDNA sequences of the silkworm apoLTP-II/I gene, which are homologous to those of D. melanogaster CG15828. Total RNA for RACE was isolated from the fat body of fifth instar larvae of strain N4 on day 4 using TRIzol reagent (Invitrogen). Because a guanine-cytosine-rich region occurs from positions 12,203 to 12,342 bp in the B. mori apoLTP-II/I gene, full-length apoLTP-II/I cDNA was not amplified by polymerase chain reaction (PCR) with a primer pair generated from the 5′- and 3′-untranslated regions (UTRs) obtained by RACE. Instead, two partial apoLTP-II/I cDNA fragments from the fat body were amplified by reverse transcription (RT)-PCR using KOD FX DNA polymerase (Toyobo, Tokyo, Japan) with the following primer pairs: 5′-CGGTGGGCGAAACGTTTGGACATGGATAT-3′ (forward) and 5′-ACGATATTTCTATTGGGTCAGT-3′ (reverse), and 5′-CACTGACCCAATAGAAATATCG-3′ (forward) and 5′-AATTATCAACTAAGCGACGGTATGGTGGGG-3′ (reverse). These four primers were designed based on the partial sequences determined by 5′- and 3′-RACE. Sequences of the amplified fragments were determined by direct sequencing. The determined sequences were then combined to obtain a full-length cDNA sequence of apoLTP-II/I encoding all eight of the apoLTP-I and apoLTP-II peptides determined by amino acid sequence analyses. The apoLTP-II/I cDNA cloning and sequence methods are diagramed in supplementary Fig. I.
Identification and sequence analysis of apoLTP-III
The amino acid sequence of purified apoLTP-III was determined as described above. We obtained candidate protein sequences of apoLTP-III containing the peptide sequence by a TBLASTN search of the Silkworm Genome Database using the peptide as a query sequence. A protein homology search with the candidate protein sequences revealed that B. mori apoLTP-III was encoded by one gene homologous to Tribolium castaneum (XP972731). To determine the full-length cDNA sequence of apoLTP-III, the 5′ and 3′ ends of the apoLTP-III sequence were obtained by RACE using a SMART RACE cDNA amplification kit (Clontech) with primer 5′-CCAATTTGTCGAGCTCCGACTGAAC-3′ for the 5′-RACE product and primer 5′-TACCCGAGGAGGTGTCGAGTGAAG-3′ for the 3′-RACE product. Both primers were designed based on the silkworm genomic sequence encoding the putative apoLTP-III protein. Total RNA was prepared using methods described previously. The determined sequences were combined to obtain the full-length cDNA sequence of apoLTP-III encoding the apoLTP-III peptide.
Phylogenetic analysis of deduced amino acid sequences
Amino acid sequences of 33 large lipid transfer proteins (LLTPs) were collected from the National Center for Biotechnology Information (NCBI) protein database. The deduced amino acid sequence of apoLTP-II/I was aligned with the 33 LLTP sequences using CLUSTALX2 (29). Alignments were edited and corrected manually with Genetyx version 9.0.1 software (Genetyx Corporation, Tokyo, Japan). Accession numbers of the LLTP sequences collected from the database are listed in supplementary Table I. Twenty-two N-terminal conserved motifs of the large lipid transfer (LLT) modules (N1–N22) were extracted and aligned according to previous reports (30, 31), and the conjugated sequences (supplementary Fig. II) were employed for subsequent phylogenetic analysis. A maximum-likelihood tree was constructed using the MEGA 5 (version 5.05) program (32). The RtREV (F+I+G) model was selected as the best fitted amino acid substitution model, and the number of discrete γ categories was defined as five. All positions containing gaps were eliminated. The bootstrap tests were replicated 1,000 times for each node.
The deduced amino acid sequence of apoLTP-III was used as the query in a BLASTP search against the NCBI protein database. Thirty-five amino acid sequences corresponding to the best hits were aligned using CLUSTALX2, and the alignments were edited and corrected manually with Genetyx version 9.0.1 (supplementary Fig. III). The maximum-likelihood tree was constructed using the same parameters as used for the apoLTP-II/I tree, except that the WAG (F+G) model was adopted as the best-fitted amino acid substitution model.
Prediction of the amphipathic secondary structure in apoLTP-II/I
Amphipathic α-helixes and β-strand regions with high lipid affinity were predicted using the computer program LOCATE, developed by Segrest et al. at the University of Alabama, Birmingham (33, 34). All amino acid sequences, except that of B. mori apoLTP II/I, were obtained from the NCBI protein database. These included the Homo sapiens apolipoprotein B-100 precursor protein (AA35549.1), the D. melanogaster CG15828 protein (NP_995670), and the M. sexta apolipophorin precursor protein (AAB53254.1).
Northern blot analysis
The midgut, fat body, silk gland, Malpighian tube, testis, and ovary were dissected from fifth instar larvae at day 0, and total RNA was extracted from each tissue separately using TRIzol reagent. To identify the expression of apoLTP-II/I and apoLTP-III, 20 μg total RNA from each tissue was applied to a 0.7% agarose gel in 2% formaldehyde. After electrophoresis, RNA was blotted to a Hybond-N+ membrane (GE Healthcare) and hybridized in ULTRAhyb (Ambion, Austin, TX). Radiolabeled hybridization probes for apoLTP-I and apoLTP-II were generated from cDNA for each region (cloned into the pGem-T vector) using the Riboprobe system SP6 (Promega, Madison, WI) and [α-32P]CTP; therefore, the pGem-T vectors contained nucleotides from positions 702 to 1,749 and 9,613 to 10,543 [the ATG start codon of each open reading frame (ORF) was indicated to be at positions 1–3] of apoLTP-II and apoLTP-I, respectively. ApoLTP-III mRNA was detected using a [α-32P]CTP-labeled single-stranded RNA probe synthesized from nucleotide positions 35 to1,083 (the ATG was indicated to be at positions 1–3) of the apoLTP-III cDNA sequence using the method described above. Bound radioactivity was detected with a Typhoon FLA7000 image analyzer (GE Healthcare).
Developmental profiles of apoLTP-II/I and apoLTP-III mRNA expression in the fat body
Changes in apoLTP-II/I and apoLTP-III mRNA expression in the fat body were analyzed from the fourth instar larva to the adult stages. Total RNA was prepared from the fat body of fourth and fifth instar larvae, pupae, and adults, and used for quantitative real-time PCR (qPCR) analyses of apoLTP-II/I and apoLTP-III gene expression. Single-stranded cDNAs were synthesized from total RNA using Superscript III reverse transcriptase (Invitrogen) and an oligo-dT primer, and then treated with RNaseH (Takara, Otsu, Japan). Quantification of the transcripts was carried out by qPCR using the cDNAs as templates with LightCycler FastStartDNA MasterPLUS SYBR Green I reagent (Roche, Darmstadt, Germany) and a LightCycler DX400 thermocycler (Roche). The specific primer pairs used for apoLTP-II/I and apoLTP-III were 5′-CTGACTGTCGATATGTTTGGCGAGT-3′ and 5′-TTCATGTTCAAAGGCAAACCGCATCCG-3′, and 5′-TGTTCCAGTTTAGGAACTGCC-3′ and 5′-TGCATAGTTCCAAGAGTGAG-3′, respectively. Transcript levels of the genes were normalized to the level of the ribosomal protein L3 (rpL3) in the same samples (35) using the primer pair 5′-TTCCCGAAAGACGACCCTAG-3′ and 5′-CTCAATGTATCCAACAACACCGAC-3′. mRNA levels were expressed relative to that found in fourth instar larvae at day 0.
Data deposition
The apoLTP-II/I and apoLTP-III cDNA sequences from the B. mori N4 strain were deposited in the DNA Data Bank of Japan with accession numbers AB700597 and AB700598, respectively.
RESULTS
cDNA sequence and deduced amino acid sequence of apoLTP-I and apoLTP-II
Three LTP subunits were separated by SDS-PAGE of purified LTPs, and the amino acid sequence of eight peptides, including the N-terminal amino acid sequences of apoLTP-I and apoLTP-II, were determined. Based on these peptide sequences, we determined the full-length cDNA of apoLTP-II/I by searching the Silkworm Genome Database, 5′- and 3′-RACE, and RT-PCR. Methods for sequence determination of apoLTP-II/I cDNA are illustrated in supplementary Fig. I.
The full-length cDNA for apoLTP-II/I has a single 12,363 bp ORF, beginning with ATG at nucleotide positions 1–3 and extending to a stop codon at positions 12,364–12,366. The ORF is predicted to encode a 4,121 amino acid protein. Lengths of the 5′- and 3′-UTR were 63 bp and 92 bp, respectively.
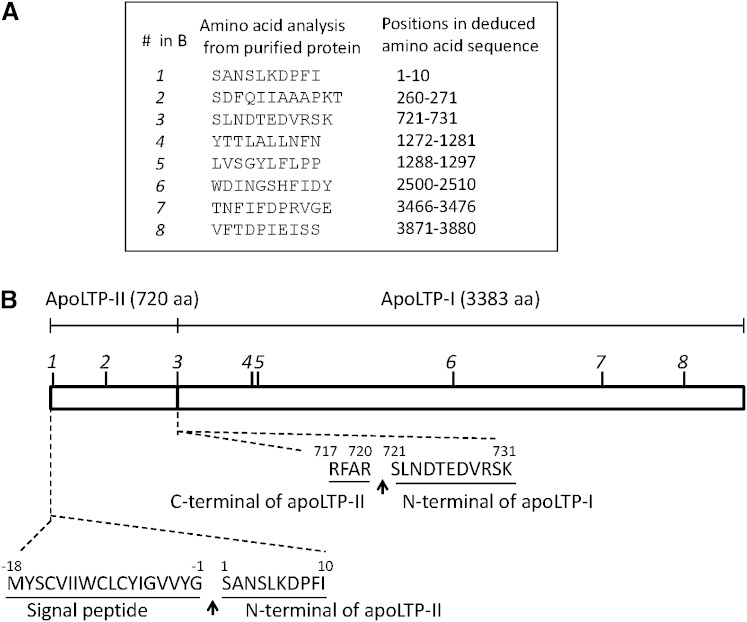
Six peptides from apoLTP-I, SLNDTEDVRSK, YTTLALLNFN, LVSGYLFLPP, WDINGSHFIDY, TNFIFDPRVGE, and VFTDPIEISS, and two peptides from apoLTP-II, SANSLKDPFI, and SDFQIIAAAPKT, which were determined by amino acid sequence analysis, were found in the apoLTP-I and apoLTP-II amino acid sequences deduced from cDNA (Fig. 1A). Because the N-terminal apoLTP-II (SANSLKDPFI) sequence was determined to begin at position 19 in the cDNA, the first 18 amino acids were assumed to be the signal peptide, which was consistent with the prediction by the SignalP 4.0 program (36). Signal peptide cleavage may occur after residue 18 to produce the 4,103 amino acid apoLTP-II/I protein (Fig. 1B).
Fig. 1.
Characterization of apoLTP-II/I from B. mori. A: Amino acid sequence of the eight peptides from apoLTP-I and apoLTP-II. Positions of the eight peptides in the deduced amino acid sequence are indicated by their position number in (A); their positions are also shown in (B) as the peptide initialized number. B: Signal peptide and cleavage site of apoLTP-II and apoLTP-I. Arrows show cleavage sites of the signal peptide and precursor protein (apoLTP-II/I). Numbers on the amino acid represent the residue number of each amino acid sequence (the N terminus of apoLTP-II was indicated to be at position 1).
These results indicate that the apoLTP-II/I protein is arranged with apoLTP-II at the N-terminal end and apoLTP-I at the C-terminal end. The cleavage site was found between positions 720 and 721 in the precursor protein (the N terminus of apoLTP-II was indicated to be at position 1). The N-terminal amino acid sequence (SLNDTEDVRSK) from the purified apoLTP-I was found at positions 721–731 in the precursor protein following the RFAR amino acid sequence at the C terminus of apoLTP-II (Fig. 1B). The calculated molecular masses of apoLTP-I and apoLTP-II based on the total of the deduced amino acid sequences were 385,826 and 82,303 Da, respectively. Using NetNGlyc 1.0 software, apoLTP-I and apoLTP-II were predicted to contain 52 and 3 potential N-glycosylation sites, respectively (NXT/S, where X is any residue other than P).
The apoLTP-II/I gene for B. mori (strain p50), constructed from the Silkworm Genome Database, consisted of 68 exons separated by 67 introns and spanned more than 87 kb. The gene is located at position 3.2 Mb on chromosome 28 of the B. mori genome.
Homology search of apoLTP-II/I, phylogenetic analysis of the LLTP superfamily, and structural organization of apoLTP-II/I
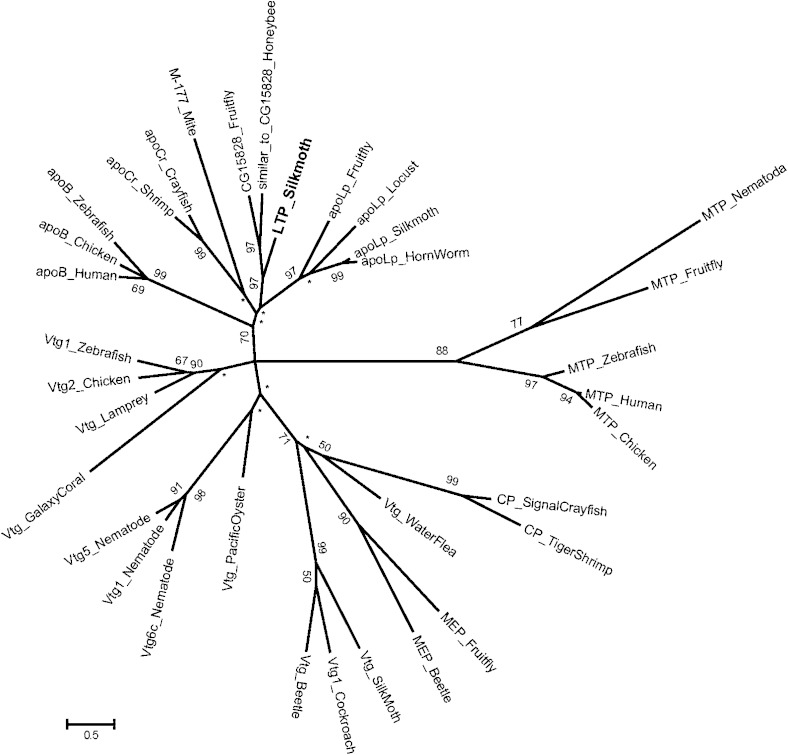
The similarities between the deduced amino acid sequences of the B. mori apoLTP-II/I and H. sapiens apoB or L. migratoria apolipophorin-II/I were analyzed using the NCBI BLAST search, and the results showed that the first 1,000 amino acid residues with the highest similarity were 43% similar to H. sapiens apoB and 44% similar to L. migratoria apolipophorin-II/I. Alignment of apoLTP-II/I with human and insect apolipoproteins showed that apoLTP-II/I belongs to the LLTP superfamily (supplementary Fig. II) (37, 38). The LLTP superfamily contained three distinguishable groups: the APO family, which includes apoB, apolipophorin-II/I, and apolipocrustacein (apoCr); the Vtg/CP family, including vitellogenin (Vtg), melanine-engaging protein (MEP) and the crustacean clotting protein (CP); and the microsomal triglyceride transfer protein (MTP) family. The B. mori apoLTP-II/I fell within the APO family, although the bootstrap values were not high. The APO family also contains D. melanogaster protein CG15828 and Apis mellifera protein similar to CG15828, and the apoLTP-II/I subfamily consisting of these three proteins was separated from the apoB, apoCr, and apolipophorin-II/I subfamilies in the APO family (Fig. 2).
Fig. 2.
Phylogenetic tree of the LLTP superfamily. The maximum-likelihood tree of the LLTP superfamily created on the N-terminal conserved motifs in the LLT modules of the APO, Vtg/CP, and MTP families are shown. Numbers indicate the percentage of bootstrap tests replicated 1,000 times for each node. Bootstrap values under 50% were replaced with asterisks. Multiple alignments of the 33 amino acid sequences of conserved motifs in the LLT module of the LLTP superfamily are shown in supplementary Fig. II.
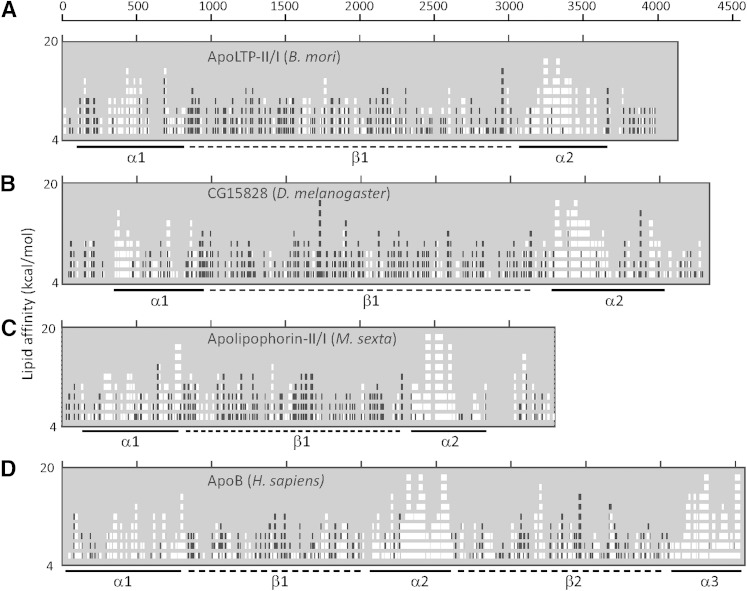
The LLT domain comprises a large N-terminal domain of approximately 1,000 amino acids that is proposed to bind lipids or transfer lipids to apolipoproteins (30). An amphipathic α-helix/β-strand region with high lipid affinity was predicted using the LOCATE computer program (33, 34). We analyzed the amphipathic clusters of apoLTP-II/I and found that it had a very similar arrangement as the lipophorin precursor protein (30), which contains three regions enriched in amphipathic α-helical and amphipathic β-strands organized as N-α1-β-α2-C (Fig. 3A). In apoLTP-II/I, the α1, β, and α2 domains were located between residues 1 to 800, 800 to 2,900, and 3,100 to 3,600, respectively. Predicted clusters of amphipathic secondary structure within the apoLTP-II/I protein were found to share some similarity with H. sapiens apoB and M. sexta apolipophorin-II/I (Fig. 3).
Fig. 3.
Predictions of amphipathic clusters within apoLTP-II/I. Individual panels show the results of LOCATE analyses of (A) B. mori apoLTP II/I, (B) D. melanogaster CG15828 protein, (C) M. sexta precursor protein of lipophorin, and (D) H. sapiens apoB. Numbers on the x-axis indicate the residue number of each amino acid sequence. The y-axis indicates the lipid affinity value, which varied from 4.0 to 20 kcal/mol. Regions of the predicted amphipathic α-helix with high lipid affinity are indicated with white boxes in the graph and solid bars below the graph (α), and the regions of predicted amphipathic β-strands with high lipid affinity are indicated with black boxes in the graph and dotted bars below the graph (β).
cDNA sequence and deduced amino acid sequence of apoLTP-III
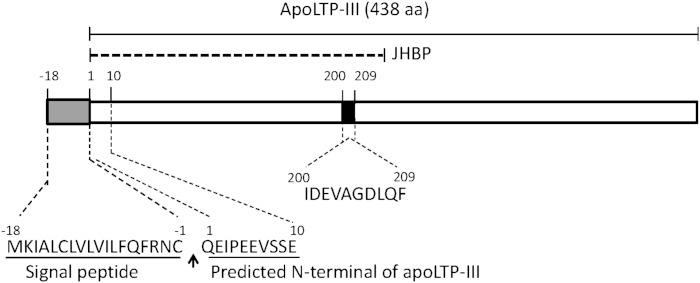
The cDNA for apoLTP-III has a 1,368 bp ORF (Fig. 4) and a deduced amino acid sequence with 456 amino acid residues and a calculated molecular mass of 51,074 Da. The 5′- and 3′-UTR lengths were 31 and 123 bp, respectively. The sequence included an 18 amino acid signal sequence predicted by the SignalP 4.0 program (36). Mature apoLTP-III is composed of 438 amino acids, corresponding to a calculated molecular mass of 48,969 Da. The existence of four potential N-glycosylation sites suggests the potential for an increase in the molecular mass of the protein. The amino acid sequence obtained following digestion of the purified apoLTP-III (IDEVAGDLQF) was identical to a region of the translated deduced cDNA sequence (Fig. 4). The B. mori apoLTP-III gene in the Silkworm Genome Database was located at position 14.2 Mb on chromosome 15 and consisted of three exons spanning more than 3 kb. The coding sequence of the apoLTP-III gene in strain p50 was identical to the N4 strain sequence.
Fig. 4.
Cloning of cDNA encoding apoLTP-III from B. mori. The 18 amino acid signal peptide of apoLTP-III (amino acid positions −18 to −1 shown in the gray box) was predicted by SignalP 4.0. The IDEVAGDLQF peptide was obtained following digestion of purified apoLTP-III and is shown in the black box. The JHBP superfamily conserved domain is indicated by the dashed bold line.
Homology search and phylogenetic analysis of apoLTP-III
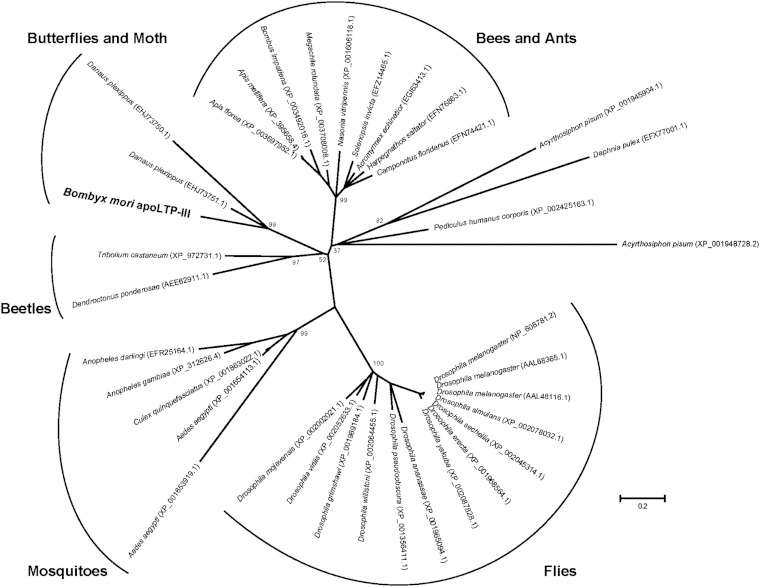
The deduced amino acid sequence of B. mori apoLTP-III shared 62% identity with a predicted protein of unknown function from Danaus plexippus (EHJ73751.1) identified in a NCBI BLAST database search. In addition, 34 similar proteins from insects and one protein from the water flea (Daphnia pulex; EFX77001.1), all of which are of unknown function, were found from a sequence homology search using NCBI BLAST. All of these proteins, including the B. mori apoLTP-III, had a putative conserved juvenile hormone-binding protein (JHBP) superfamily domain (Fig. 4). A phylogenetic tree of the insect proteins was constructed based on their primary amino acid sequences (Fig. 5). B. mori apoLTP-III, D. plexippus (EHJ73750.1), and D. plexippus (EHJ73751.1) proteins were clustered in a Lepidoptera-specific group.
Fig. 5.
Phylogenetic tree of B. mori apoLTP-III and similar proteins. The maximum-likelihood tree was constructed with B. mori apoLTP-III and 34 similar insect proteins and one water flea collected from the NCBI protein database. Five distinguished groups, bees and ants group, butterflies and moths group, beetles group, mosquitoes group, and flies group, are denoted by arcs. The taxon name represents the scientific name of each species with the protein accession number. Numbers at each node indicate the percentage of bootstrap tests replicated 1,000 times. Bootstrap values inside each group have been omitted. Multiple alignment of the 36 amino acid sequences is shown in supplementary Fig. III.
Detection of apoLTP protein glycosylation
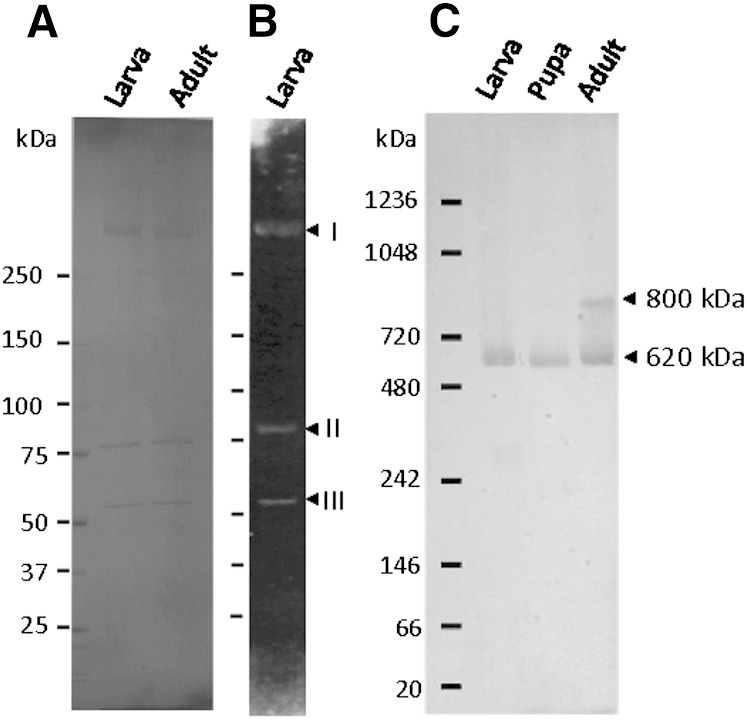
According to SDS-PAGE, both LTPs purified from the hemolymph of larvae and adults contained three protein subunits; no protein other than the three subunits was observed (Fig. 6A). Additionally, the molecular masses of apoLTP-I, apoLTP-II, and apoLTP-III were estimated to be approximately 350, 85, and 60 kDa, respectively. Because apoLTP-I is extremely large and the largest protein used for a SDS-PAGE molecular mass standard was approximately 250 kDa, the molecular mass of apoLTP-I was not estimated precisely by SDS-PAGE.
Fig. 6.
Estimation of the molecular mass of native LTP. A: Purified LTP from the hemolymph of fifth instar larvae and adults was subjected to SDS-PAGE and stained with Coomassie Brilliant Blue R-250. B: Purified LTP from the hemolymph of fifth instar larvae was subjected to SDS-PAGE and transferred to a PVDF membrane. The blot was incubated with FITC-Con A solution. Detection of glycoproteins bound to FITC-Con A was carried out under ultraviolet light. Arrows indicate apoLTP-I (I), apoLTP-II (II), and apoLTP-III (III) from the top, respectively. C: One microliter each of the hemolymph from fifth instar larva, pupae, and adults was electrophoresed by 3–10% blue-native PAGE. Separated proteins were transferred to a PVDF membrane. Native LTP was detected by Western blot analysis using the anti-apoLTP-I antibody. Arrows indicate the 620 kDa and 800 kDa LTP. Numbers on the left of each panel represent the molecular masses for protein standard.
Larval LTP was subjected to SDS-PAGE and transferred to a PVDF membrane. As shown in Fig. 6B, the three subunits of LTP showed reactivity with FITC-Con A; therefore, the three subunits appear to contain carbohydrate chains.
Molecular mass of intact LTP
The molecular mass of intact LTP from B. mori was estimated by a combination of blue-native PAGE and Western blot analysis using anti-apoLTP-I rabbit serum. The molecular masses of the B. mori intact LTP from the hemolymph of the larval, pupal, and adult stages were estimated at approximately 620 kDa (Fig. 6C). In the hemolymph of adults at day 0, both the 620 kDa LTP and larger molecular mass LTP of about 800 kDa were found. The larger intact LTP was not found in the hemolymph of fifth instar larvae at day 4 and pupae at day 3 (Fig. 6C).
Tissue-specific expression of apoLTP-II/I and apoLTP-III in fifth instar larvae
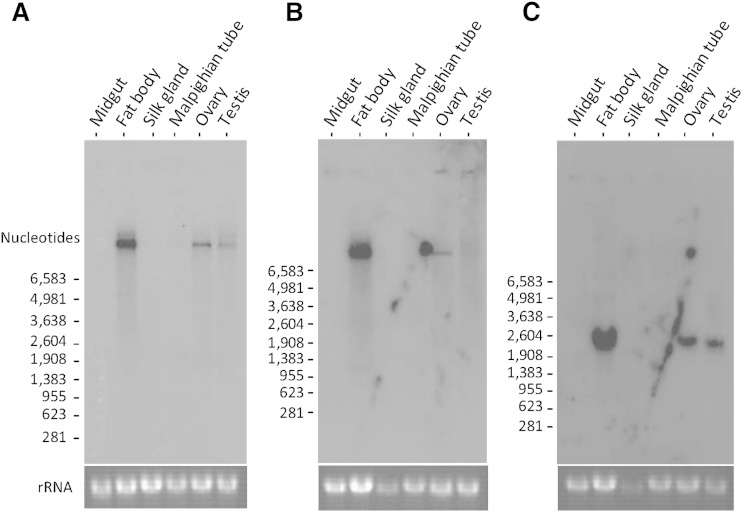
Northern hybridization experiments were performed to identify tissue expression patterns of apoLTP-II/I and apoLTP-III in fifth instar larvae at day 0 (Fig. 7). Both 32P-labeled apoLTP-I-specific (Fig. 7A) and apoLTP-II-specific probes (Fig. 7B) detected a single product of over 12,000 nucleotides in the Northern blot analysis, whereby the size was estimated by comparison with RNA standards. As shown in Fig. 7A, B, the fat body, testis, and ovary accumulated the apoLTP-II/I transcript, whereas no expression was observed in the midgut, silk gland, or Malphigian tube. The highest levels of apoLTP-II/I mRNA were observed in the fat body.
Fig. 7.
Tissue-specific gene expression of apoLTP-II/I and apoLTP-III from B. mori. Northern hybridization with apoLTP-I-specific probes (A) and apoLTP-II-specific probes (B) revealed transcripts for apoLTP-II/I in the fat body, ovary, and testis of fifth instar larvae at day 0, which were larger than the 6,583 nucleotide RNA marker. ApoLTP-III-specific probes detected the approximately 2,300 nucleotide transcript in the fat body, ovary, and testis (C). No transcripts for apoLTP-II/I (A, B) or apoLTP-III (C) were detected in other tissues, including the midgut, silk gland, and Malpighian tube of fifth instar larvae at day 0. The rRNA bands stained with ethidium bromide were used to monitor equal loading of the sample. Size of the RNA markers is shown on the left.
One apoLTP-III transcript of approximately 2,300 nucleotides was detected in the fat body, ovary, and testis using a single-stranded RNA probe synthesized from a DNA fragment representing apoLTP-III. Northern blot analyses showed that the highest level of apoLTP-III mRNA was detected in the fat body with slightly lower, but still relatively high, levels in the ovary and testis (Fig. 7C).
Developmental changes in apoLTP-II/I and apoLTP-III expression
We analyzed changes in apoLTP-II/I and apoLTP-III mRNA expression in the fat body throughout the fourth instar larval to adult stages. qPCR analysis was performed to examine the expression levels of apoLTP-II/I and apoLTP-III mRNA. mRNA expression levels were expressed relative to that found in fourth instar larvae at day 0.
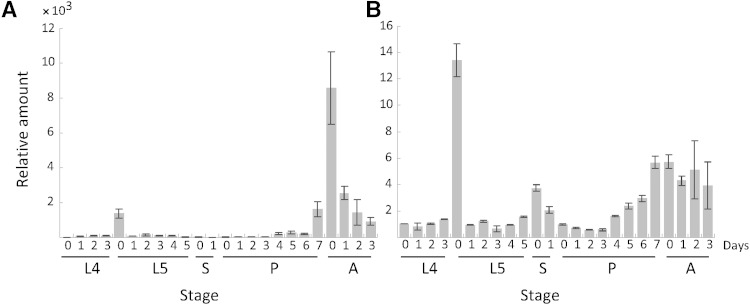
In the fat body, high levels of apoLTP-II/I mRNA were observed in fifth instar larvae at day 0, immediately after initiation of feeding. Later, during the larval feeding stage and the pupal stage, levels gradually declined and remained relatively low and constant. From the late pupal stage to emergence, the transcript level increased sharply, after which it stayed relatively high until death (Fig. 8A).
Fig. 8.
Developmental profile of apoLTP-II/I (A) and apoLTP-III (B) gene expression. Total RNA was prepared from the fat body of fourth and fifth instar larvae, pupae, and adults. Quantification of transcripts was carried out by qPCR. The y-axis (relative amount) indicates the fold-increase in mRNA expression compared with that of fourth instar larvae at day 0 (L4-0). The y-axis bars on L4-0 in both (A) and (B) are set to one. Results shown are the mean ± SEM (n = 3). Stages are shown as L4, fourth instar larval stage; L5, fifth instar larval stage; S, spinning cocoon stage; P, pupal stage; A, adult stage.
The expression pattern of apoLTP-III mRNA was similar to that of apoLTP-II/I mRNA. In the fourth instar larvae the level was low, but increased and became high at day 0 of the fifth instar. Following day 0 of the fifth instar, the levels decreased gradually until the spinning stage. At day 0 of the spinning stage, apoLTP-III expression increased and remained at a low level during the early pupal stage. ApoLTP-III expression increased again from day 4 of the pupal stage to day 0 of the adult stage and then stayed relatively high until death (Fig. 8B).
DISCUSSION
Until now, apoLTP-II/I and apoLTP-III cDNAs have not been isolated from any species, and the gene structures of apoLTP-II/I and apoLTP-III were unknown. This is the first study to report the LTP cDNA sequence, the gene structure of LTP, and the amino acid sequences of three apoproteins of LTP.
The three subunit proteins of B. mori LTP are coded by two genes, apoLTP-II/I and apoLTP-III. ApoLTP-II/I from the silkworm (B. mori) is a novel member of the APO family, which is similar to H. sapiens apoB and insect lipophorins. However, a major difference exists between apoB and apoLTP-II/I. ApoB is not cleaved during its association with lipids, but apoLTP-II/I is a proapoprotein that is cleaved to become two subunits (apoLTP-I and apoLTP-II) of LTP arranged with apoLTP-I at the C-terminal end and apoLTP-II at the N-terminal end. In H. sapiens apoB, the whole βα1 domain at the N-terminal end was proposed to be a lipid pocket for initiation of lipoprotein particle assembly (39, 40), and the N-terminal sequence of apoB is known to be critical for TAG-rich lipoprotein assembly and secretion (41). ApoLTP-II is 720 amino acids long; the α1 domain covers the entire apoLTP-II sequence. Blacklock and Ryan (42) examined the catalytically important LTP apoprotein using LTP apoprotein-specific antibodies in a lipid transfer inhibition and reported that apoLTP-II plays a key role in lipid transfer activity. Posttranslational cleavage of apoLTP-II/I for building of the two subunits is assumed to be important for formation of the lipid-binding and transfer region required to recruit and pack DAG instead of TAG. Indeed, the two subunits of insect DAG-rich lipophorin, apolipophorin-I and apolipophorin-II, were also shown to result from cleavage of apolipophorin-II/I and are arranged in the same manner as apoLTP-II/I (19, 20). In addition, the subunit cleavage enzyme for apoLTP-II/I and apolipophorin-II/I may be common because it is accordant to that of the limited endoproteolytic cleavage that occurs after a sequence containing two or more basic residues (K or R) (43–46). The amino acid sequence RFAR was found at the C-terminal end of the putative B. mori apoLTP-II, and RGRR was also found in both M. sexta and B. mori apolipophorin-II/I (20, 47). Our results presented here do not rule out a role for apoLTP-I and apoLTP-II. However, we completed sequencing cDNA for the 4,103 amino acid apoLTP-II/I and are now in a position to produce LTP protein with and without cleaved apoLTP-II/I to determine whether cleavage of apoLTP-II/I is required to produce a biologically active LTP.
Thirty-five apoLTP-III protein homologs were found in insects and water fleas from a sequence homology search using NCBI BLAST. All identified proteins were of unknown function but all shared a putative conserved JHBP superfamily domain (48). The JHs are acyclic sesquiterpenoids that regulate insect development and reproduction (49). The juvenile hormone (JH)-binding activity of apoLTP-III and the JH transport or JH transfer/exchange activity between lipophorin as the functional activity of LTP have not been studied.
Palm et al. (50) referred to the protein encoded by D. melanogaster CG15828 as apoLTP and observed its function using RNAi knockdown. Their results showed that the D. melanogaster CG15828-encoded protein facilitated lipid export from the gut to lipophorin, indicating a function similar to that of LTPs of other insects (13). Because CG15828 protein is equivalent to apoLTP-II/I, apoLTP-III may not function on lipid transfer in D. melanogaster. Palm et al. (50) suggested that if apoLTP-III exists in this fly, it is not likely to be derived from the apoLTP precursor protein. In D. melanogaster, three apoLTP-III homologs were found in NCBI BLAST database (accession numbers NP-608781.2, AAL68365.1, and AAL48116.1) that may be apoLTP-III. However, we did not address how apoLTP-III works within the LTP particle.
We determined that the molecular mass of intact B. mori LTP was approximately 620 kDa (Fig. 6C). Based on SDS-PAGE, LTP showed three subunits of molecular masses 350, 85, and 60 kDa, respectively (Fig. 6A), and all three apoproteins were glycosylated (Fig. 6B). However, based on the present results, the calculated molecular masses of apoLTP-I, apoLTP-II, and apoLTP-III proteins from their cDNA sequences were 385,826, 82,303, and 48,969 Da, respectively. Therefore, apoLTP-I was considered to be larger than 385,826 Da rather than 350,000 Da. Considering the total molecular mass of the three subunit proteins (>530 kDa) and the lipid composition of LTP (about 20%) together, we conjectured that intact B. mori LTP may consist of one apoLTP-I, one apoLTP-II, and one apoLTP-III molecule. Because we did not confirm the molecular masses of the three subunits of B. mori LTP, quantitative carbohydrate and other protein modification analyses of separated subunits of B. mori LTP will be required to confirm subunit composition. Ryan et al. (2, 9) have extensively studied the properties of M. sexta LTP and reported that the molecular mass of LTP is greater than 500 kDa or over 670 kDa, which was estimated by gel permeation chromatography or pore-limiting gradient PAGE of purified LTP, respectively, and that all three subunits were glycosylated proteins containing carbohydrate (5% by weight). Additionally, LTP becomes unstable and easy to aggregate when LTP purification is progressing. Particle aggregation might preclude an accurate molecular mass determination. To avoid this problem, we applied the hemolymph rather than purified LTP on blue-native PAGE and detected LTP by Western blot analysis using an anti-apoLTP-I antibody.
We found a larger size of LTP (about 800 kDa) compared with the 620 kDa LTP in adult hemolymph (Fig. 6C). Although in the adult hemolymph, the larger size of LTP is assumed to have an increased amount of DAG, one cannot exclude that these molecules have another protein component other than the three subunits. Indeed, after KBr density gradient ultracentrifugation of adult hemolymph, the 800 kDa LTP coexists in the HDLp fraction, which has a density of 1.12–1.15 g/ml, but the larval LTP (620 kDa) was recovered from the higher density fractions (1.20–1.23 g/ml) than HDLp. Moreover, as indicated by SDS-PAGE, no protein other than the three subunits was observed in the LTP purified from these fractions (Fig. 6A). In insects, during the flight activity of an adult, the amount of lipids mobilized in the hemolymph increases and forms HDLp to LDLp. LTP exchange or DAG transfer between two different density lipophorins progresses LDLp formation. During this process, larger LTP (800 kDa), which has an increased amount of DAG, may be formed without association of additional exchangeable apoproteins. The amount of lipid in LTP may increase up to approximately 30% to form the 800 kDa lower density LTP in adult hemolymph.
Northern blot analysis suggests that the apoLTP-II/I and apoLTP-III genes are more actively transcribed in the B. mori fat body than in the ovary and testis (Fig. 7). Gene expression levels of both apoLTP-II/I and apoLTP-III were synchronized in the fat body (Fig. 8). Both the apoLTP-II/I and apoLTP-III genes were strongly expressed at day 0 of the fifth instar larval stage and at the early adult stage corresponding to increased DAG transport needs. ApoLTP-II/I and apoLTP-III expression might be enhanced by the onset of feeding. In adults, the high expression of apoLTP-II/I and apoLTP-III may correlate with LTP function during flight-related lipophorin conversion of HDLp to LDLp in response to adipokinetic hormone. These observations support the finding that the LTP concentration in the hemolymph is highest in adults (8, 15).
While LTP plays an essential role in promoting lipid transfer from cells to lipophorin, and exchanges lipids between lipophorins, the present results suggest that LTP is a novel member of the APO family, and that LTP may play a major role as a lipid carrier in the hemolymph, similar to that of lipophorin. Finally, the molecular characterization of LTP reported here may not only open a new field of research on the biosynthesis, lipid recruitment, and assembly of LTP, but may also allow its function in insect lipid metabolism to be clarified.
Supplementary Material
Footnotes
Abbreviations:
- APO
- apoB/large lipid transfer protein
- apoCr
- apolipocrustacein: apoLp, apolipophorin
- CP
- crustacean clotting protein
- DAG
- diacylglycerol
- FITC-Con A
- fluorescein isothiocyanate conjugated to concanavalin A
- HDLp
- high density lipophorin
- JH
- juvenile hormone
- JHBP
- juvenile hormone-binding protein
- LDLp
- low density lipophorin
- LLT
- large lipid transfer
- LLTP
- large lipid transfer protein
- LTP
- lipid transfer particle
- MEP
- melanine-engaging protein
- MTP
- microsomal triglyceride transfer protein
- NCBI
- National Center for Biotechnology Information
- ORF
- open reading frame
- PVDF
- polyvinylidene difluoride
- qPCR
- quantitative real-time PCR
- RACE
- rapid amplification of cDNA ends
- TAG
- triacylglycerol
- UTR
- untranslated region
- Vtg
- vitellogenin
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan and the Teimei Empress Memorial Foundation (Japan).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight figures.
REFERENCES
- 1.Chino H. 1985. Lipid transport: biochemistry of hemolymph lipophorin. Comprehensive Insect Physiology, Biochemistry and Pharmacology. Kerkut G.A., Gilbert L.I., Vol. 10, Pergamon Press, Oxford: 115–136. [Google Scholar]
- 2.Ryan R. O., Wells M. A., Law J. H. 1986. Lipid transfer protein from Manduca sexta hemolymph. Biochem. Biophys. Res. Commun. 136: 260–265. [DOI] [PubMed] [Google Scholar]
- 3.Hirayama Y., Chino H. 1990. Lipid transfer particle in locust hemolymph: purification and characterization. J. Lipid Res. 31: 793–799. [PubMed] [Google Scholar]
- 4.Capurro M. L., De Bianchi A. G. 1990. A lipid transfer particle in Musca domestica haemolymph. Comp. Biochem. Physiol. Part B. 97: 649–653. [Google Scholar]
- 5.Takeuchi N., Chino H. 1993. Lipid transfer particle in the hemolymph of the American cockroach: evidence for its capacity to transfer hydrocarbons between lipophorin particles. J. Lipid Res. 34: 543–551. [PubMed] [Google Scholar]
- 6.Tsuchida K., Soulages J. L., Moribayashi A., Suzuki K., Maekawa H., Wells M. A. 1997. Purification and properties of a lipid transfer particle from Bombyx mori. Comparison to the lipid transfer particle from Manduca sexta. Biochim. Biophys. Acta. 1337: 57–65. [DOI] [PubMed] [Google Scholar]
- 7.Golodne D. M., Van Heusden M. C., Gondim K. C., Masuda H., Atella G. C. 2001. Purification and characterization of a lipid transfer particle in Rhodnius prolixus: phospholipid transfer. Insect Biochem. Mol. Biol. 31: 563–571. [DOI] [PubMed] [Google Scholar]
- 8.Van Heusden M. C., Yepiz-Plascencia G. M., Walker A. M., Law J. H. 1996. Manduca sexta lipid transfer particle, synthesis by fat body and occurrence in hemolymph. Arch. Insect Biochem. Physiol. 31: 39–51. [DOI] [PubMed] [Google Scholar]
- 9.Ryan R. O., Senthilathipan K. R., Wells M. A., Law J. H. 1988. Facilitated diacylglycerol exchange between insect hemolymph lipophorins. Properties of Manduca sexta lipid transfer particle. J. Biol. Chem. 263: 14140–14145. [PubMed] [Google Scholar]
- 10.Ryan R. O., Wessler A. N., Price H. M., Ando S., Yokoyama S. 1990. Insect lipid transfer particle catalyzes bidirectional vectorial transfer of diacylglycerol from lipophorin to human low density lipoprotein. J. Biol. Chem. 265: 10551–10555. [PubMed] [Google Scholar]
- 11.Van Heusden M. C., Law J. H. 1989. An insect lipid transfer particle promotes lipid loading from fat body to lipoprotein. J. Biol. Chem. 264: 17287–17292. [PubMed] [Google Scholar]
- 12.Tsuchida K., Wells M. A. 1988. Digestion, absorption, transport and storage of fat during the larval stadium of Manduca sexta. Changes in the role of lipophorin in the delivery of dietary lipid to the fat body. Insect Biochem. 18: 263–268. [Google Scholar]
- 13.Canavoso L. E., Wells M. A. 2001. Role of lipid transfer particle in delivery of diacylglycerol from midgut to lipophorin in larval Manduca sexta. Insect Biochem. Mol. Biol. 31: 783–790. [DOI] [PubMed] [Google Scholar]
- 14.Prasad S. V., Fernando-Warnakulasuriya G. J. P., Sumida M., Law J. H., Wells M. A. 1986. Lipoprotein biosynthesis in the larvae of the tobacco hornworm, Manduca sexta. J. Biol. Chem. 261: 17174–17176. [PubMed] [Google Scholar]
- 15.Tsuchida K., Arai M., Tanaka Y., Ishihara R., Ryan R. O., Maekawa H. 1998. Lipid transfer particle catalyzes transfer of carotenoids between lipophorins of Bombyx mori. Insect Biochem. Mol. Biol. 28: 927–934. [DOI] [PubMed] [Google Scholar]
- 16.Sakudoh T., Sezutsu H., Nakashima T., Kobayashi I., Fujimoto H., Uchino K., Banno Y., Iwano H., Maekawa H., Tamura T., et al. 2007. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc. Natl. Acad. Sci. USA. 104: 8941–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakudoh T., Iizuka T., Narukawa J., Sezutsu H., Kobayashi I., Kuwazaki S., Banno Y., Kitamura A., Takada N., Fujimoto H., et al. 2010. A CD36-related transmembrane protein is coordinated with an intercellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J. Biol. Chem. 285: 7739–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakudoh T., Kuwazaki S., Iizuka T., Narukawa J., Yamamoto K., Uchino K., Sezutsu H., Banno Y., Tsuchida K. 2013. CD36 homolog divergence is responsible to the selectivity carotenoid species migration to the silk gland of the silkworm, Bombyx mori. J. Lipid Res. 54: 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weers P. M. M., Van Marrewijk W. J. A., Beenakkers A. M. T., Van der Horst D. J. 1993. Biosynthesis of locust lipophorin. Apolipophorins I and II originate from a common precursor. J. Biol. Chem. 268: 4300–4303. [PubMed] [Google Scholar]
- 20.Sundermeyer K., Hendricks J. K., Prasad S. V., Wells M. A. 1996. The precursor protein of the structural apolipoproteins of lipophorin: cDNA and deduced amino acid sequence. Insect Biochem. Mol. Biol. 26: 735–738. [DOI] [PubMed] [Google Scholar]
- 21.The International Silkworm Genome Consortium. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38: 1036–1045. [DOI] [PubMed] [Google Scholar]
- 22.Xia Q., Guo Y., Zhang Z., Li D., Xuan Z., Li Z., Dai F., Li Y., Cheng D., Li R., et al. 2009. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science. 326: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 24.Furlan M., Perret B. A., Beck E. A. 1979. Staining of glycoproteins in polyacrylamide and agarose gels with fluorescent lectins. Anal. Biochem. 96: 208–214. [DOI] [PubMed] [Google Scholar]
- 25.Schägger H., Cramer W. A., Vonjagow G. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230. [DOI] [PubMed] [Google Scholar]
- 26.Wittig I., Braun H. P., Schagger H. 2006. Blue native PAGE. Nat. Protoc. 1: 418–428. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of protein from polyacrylamide gel to nitrocellulose sheet: procedure and some applications. Proc. Natl. Acad. Sci. USA. 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niall H. D. 1973. Automated Edman degradation: the protein sequenator. Methods Enzymol. 27: 942–1010. [DOI] [PubMed] [Google Scholar]
- 29.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babin P. J., Bogerd J., Kooiman F. P., Van Marrewijk W. J., Van der Horst D. J. 1999. Apolipophorin II/I, apolipoprotein B, vitellogenin, and microsomal triglyceride transfer protein genes are derived from a common ancestor. J. Mol. Evol. 49: 150–160. [DOI] [PubMed] [Google Scholar]
- 31.Van der Horst D. J., Roosendaal S. D., Rodenburg K. W. 2009. Circulatory lipid transport: lipoprotein assembly and function from an evolutionary perspective. Mol. Cell. Biochem. 326: 105–119. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segrest J. P., Jones M. K., Mishra V. K., Anantharamaiah G. M., Garber D. W. 1994. ApoB-100 has a pentapartite structure composed of three amphipathic alpha-helical domains alternating with two amphipathic beta-strand domains. Detection by the computer program LOCATE. Arterioscler. Thromb. 14: 1674–1685. [DOI] [PubMed] [Google Scholar]
- 34.Segrest J. P., Jones M. K., Mishra V. K., Pierotti V., Young S. H., Boren J., Innerarity T. L., Dashti N. 1998. Apolipoprotein B-100: conservation of lipid-associating amphipathic secondary structural motifs in nine species of vertebrates. J. Lipid Res. 39: 85–102. [PubMed] [Google Scholar]
- 35.Niwa R., Sakudoh T., Namiki T., Saida K., Fujimoto Y., Kataoka H. 2005. The ecdysteroidgenic P450 Cryp302al/disembodied from the silkworm, Bombyx mori, is transcriptionally regulated by prothoracicotropic hormone. Insect Mol. Biol. 14: 563–571. [DOI] [PubMed] [Google Scholar]
- 36.Petersen T. N., Brunak S., von Heijne G., Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 37.Avarre J. C., Lubzens E., Babin P. J. 2007. Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. BMC Evol. Biol. 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smolenaars M. M. W., Madsen O., Rodenburg K. W., Van der Horst D. J. 2007. Molecular diversity and evolution of the large lipid transfer protein superfamily. J. Lipid Res. 48: 489–502. [DOI] [PubMed] [Google Scholar]
- 39.Segrest J. P., Jones M. K., Dashti N. 1999. N-terminal domain of apolipoprotein B has structural homology to lipovitellin and microsomal triglyceride transfer protein: a “lipid pocket” model for self-assembly of apoB-containing lipoprotein particles. J. Lipid Res. 40: 1401–1416. [PubMed] [Google Scholar]
- 40.Manchekar M., Richardson P. E., Forte T. M., Datta G., Segrest J. P., Dashti N. 2004. Apolipoprotein B-containing lipoprotein particle assembly: lipid capacity of the nascent lipoprotein particle. J. Biol. Chem. 279: 39757–39766. [DOI] [PubMed] [Google Scholar]
- 41.Wang L., Jiang Z. G., McKnight C. J., Small D. M. 2010. The interfacial properties of apolipoprotein B292-593 (B6.4-13) and B611-782 (B13-17). Insights into the structure of the lipovitellin homology region in apoB. Biochemistry. 49: 3898–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blacklock B. J., Ryan R. O. 1995. Structural studies of Manduca sexta lipid transfer particle with apolipoprotein-specific antibodies. J. Lipid Res. 36: 108–116. [PubMed] [Google Scholar]
- 43.Wise R. J., Barr P. J., Wong P. A., Kiefer M. C., Brake A. J., Kaufman R. J. 1990. Expression of a human proprotein processing enzyme: correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc. Natl. Acad. Sci. USA. 87: 9378–9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N. G., Israel A. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA. 95: 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou A., Webb G., Zhu X., Steiner D. F. 1999. Proteolytic processing in the secretory pathway. J. Biol. Chem. 274: 20745–20748. [DOI] [PubMed] [Google Scholar]
- 46.Smolenaars M. M. W., Kasperatis M. A. M., Richardson P. E., Rodenburg K. W., Van der Horst D. J. 2005. Biosynthesis and secretion of insect lipoprotein: involvement of furin in cleavage of the apoB homolog, apolipophorin-II/I. J. Lipid Res. 46: 412–421. [DOI] [PubMed] [Google Scholar]
- 47.Hanada Y., Sekimizu K., Kaito C. 2011. Silkworm apolipophorin protein inhibits Staphylococcus aureus virulence. J. Biol. Chem. 286: 39360–39369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kołodziejczyk R., Kochman M., Bujacz G., Dobryazycki P., Ozyhar A., Jaskolski M. 2003. Crystallization and preliminary crystallographic studies of juvenile hormone-binding protein from Galleria mellonella haemolymph. Acta Crystallogr. D Biol. Crystallogr. 59: 519–521. [DOI] [PubMed] [Google Scholar]
- 49.Riddiford L. M. 1993. Hormone receptors and the regulation of insect metamorphosis. Receptor. 3: 203–209. [PubMed] [Google Scholar]
- 50.Palm W., Sampaio J. L., Brankatschk M., Carvalho M., Mahmoud A., Scevchenko A., Eaton S. 2012. Lipoproteins in Drosophila melanogaster- assembly, function, and influence on tissue lipid composition. PLoS Genet. 8: e1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.