Abstract
RNA-binding proteins of the PUF family are well conserved post-transcriptional regulators that control a variety of developmental processes. The C. elegans protein PUF-8 is essential for several aspects of germ cell development including the maintenance of germline stem cells (GSCs). To explore the molecular mechanisms underlying its function, we have identified 160 germline-expressed mRNAs as potential targets of PUF-8. We generated GFP::H2B-3′ UTR fusions for 17 mRNAs to assay their post-transcriptional regulation in germ cells. Twelve transgenes were not expressed in the mitotic germ cells, and depletion of PUF-8 led to misexpression of six of them in these cells. In contrast, the expression of 3′ UTR fusion of hip-1, which encodes the HSP-70 interacting protein, was dependent on PUF-8. These results indicate that PUF-8 may regulate the expression of its targets both negatively as well as positively. We investigated the PUF-8-mediated post-transcriptional control of one mRNA, namely pal-1, which encodes a homeodomain transcription factor responsible for muscle development. Our results show that PUF-8 binds in vitro to specific sequences within pal-1 3′ UTR that are critical for post-transcriptional suppression in GSCs. Removal of PUF-8 resulted in PAL-1 misexpression, and PAL-1-dependent misexpression of the myogenic promoter HLH-1 in germ cells. We propose that PUF-8 protects GSCs from the influence of somatic differentiation factors such as PAL-1, which are produced in the maternal germline but meant for embryogenesis.
Keywords: Caenorhabditis elegans, PUF-8, Post-transcriptional control, 3′ UTR, Germ cells
Introduction
Members of the PUF family RNA-binding proteins are present in organisms as diverse as yeast, nematodes, insects, human and plants. They regulate a wide range of biological processes. Drosophila Pumilio, the founding member of this family, was originally discovered for its role in embryonic patterning (Lehmann and Nusslein-Volhard, 1987). Two nearly identical Caenorhabditis elegans proteins called FBF-1 and FBF-2 – PUF stands for Pumilio and FBF – were discovered for their role in spermatogenesis-to-oogenesis switch in hermaphrodites (Zhang et al., 1997). Later studies have uncovered PUF function in processes such as the maintenance of germline stem cells (GSCs), meiotic progression of spermatocytes, yeast mating type switch, neuronal excitability, vulva development and adaptation of olfactory sensory neurons (Forbes and Lehmann, 1998; Tadauchi et al., 2001; Crittenden et al., 2002; Schweers et al., 2002; Subramaniam and Seydoux, 2003; Walser et al., 2006; Ariz et al., 2009; Kaye et al., 2009). Of these, promotion of stem cell proliferation appears to be the conserved ancient function of PUF proteins, as their role in this process is conserved from Dictyostelium to mammals (Souza et al., 1999; Xu et al., 2007).
Evidence accumulated so far indicate that PUF proteins function as post-transcriptional regulators. Genetic and biochemical studies have identified a few mRNAs as targets of PUF proteins and established the biological significance of their PUF-mediated control. These studies indicate that PUF proteins influence the expression of a diverse set of mRNAs. For example, mRNAs of proteins involved in transcription activation, cell cycle control, post-transcriptional control and protein phosphorylation have been shown to be regulated by PUF proteins (Murata and Wharton, 1995; Crittenden et al., 2002; Kadyrova et al., 2007; Kaye et al., 2009). They interact with different protein partners to control the expression of different mRNAs. While Pumilio interacts with Nanos and Brain Tumor (Brat) to suppress hunchback mRNA, its suppression of cyclin B mRNA is Brat-independent (Sonoda and Wharton, 1999; Sonoda and Wharton, 2001; Kadyrova et al., 2007). PUF proteins function as positive regulators as well. For example, FBF-1 activates egl-4 expression in the worm olfactory neurons (Kaye et al., 2009). Interestingly, FBF suppresses gld-1 expression in mitotic germ cells – possibly through its interaction with the CCF-1/Pop2 deadenylase – but promotes it in cells entering meiosis, this time probably by interacting with the GLD-2 poly(A) polymerase (Suh et al., 2009). Not surprisingly, PUF proteins employ more than one mechanism to accomplish their function. A conserved mechanism seems to be the recruitment of the deadenylase complex, but interference with translation initiation, mediated via interaction with the initiation factors, has also been observed (Goldstrohm et al., 2006; Goldstrohm et al., 2007; Deng et al., 2008).
Given that PUF proteins influence several different processes, they most likely regulate a large number of mRNAs. However, only a few mRNAs have been identified as real PUF targets. Studies on these few mRNAs have not generated sufficient information for bioinformatic screening of genome sequences for potential PUF targets. Only a short 3′ UTR sequence, UGU, is perfectly conserved in all known PUF targets (Opperman et al., 2005). In addition, no specific RNA secondary structure has emerged as essential for PUF-RNA interaction. A few large-scale biochemical approaches have identified a number of mRNAs, representing most known cellular processes, as potential PUF targets (Gerber et al., 2004; Gerber et al., 2006; Galgano et al., 2008; Morris et al., 2008; Kershner and Kimble, 2010). These need to be tested further to establish that a particular mRNA is indeed regulated by the given PUF protein. More importantly, it is essential to determine the functional significance of such a PUF-mediated control in the actual biological context.
There are 11 PUF family members in C. elegans. Based on sequence similarity, they have been classified into two groups (Wickens et al., 2002). One of them contains two members, PUF-8 and PUF-9, which are more closely related to the insect and vertebrate orthologs than to the other class of worm PUF proteins. PUF-8 is expressed primarily in the mitotic germ cells and mediates several aspects of germ cell development (Ariz et al., 2009). These include prevention of premature proliferation of primordial germ cells, proper localization of germ cells to the somatic gonad, promotion of germline stem cell (GSC) mitosis, sperm-oocyte switch in hermaphrodites and meiotic progression of primary spermatocytes (Subramaniam and Seydoux, 1999; Subramaniam and Seydoux, 2003; Bachorik and Kimble, 2005; Ariz et al., 2009). In addition, PUF-8 functions in the soma as well: it acts as a negative regulator to prevent ectopic vulval differentiation (Walser et al., 2006). So far no targets of PUF-8 have been identified. Its function in GSCs is strikingly similar to its Drosophila ortholog Pumilio, for which only one target, namely Brat, has been reported so far (Harris et al., 2011). Even in this case, whether Pumilio directly interacts with Brat 3′ UTR is not known. Thus, the germline targets of this highly conserved PUF subgroup remains to be discovered.
In an attempt to identify the potential targets of PUF-8, we have isolated C. elegans mRNAs that bind specifically to PUF-8. Of these, 160 are expressed predominantly in the germline. Here we report the detailed characterization of the PUF-8-mediated post-transcriptional control of one such mRNA, namely pal-1, which encodes a somatic transcription factor. Our results demonstrate that PUF-8 directly interacts with the 3′ UTR of pal-1 to suppress its expression in GSCs, and this control is essential to prevent GSCs from expressing the myogenic factor HLH-1, a downstream target of PAL-1. These results indicate that PUF-8 functions as a post-transcriptional repressor to protect GSCs from the influence of somatic factors, such as PAL-1, which are transcribed in the maternal germline but meant for functioning during oocyte maturation and/or embryogenesis.
Materials and methods
C. elegans strains
Worm strains were maintained as described (Brenner, 1974), except that all transgenic lines were kept at 25°C to avoid silencing of transgene expression in the germline (Strome et al., 2001). Introduction of different transgenes into puf-8 mutant background were carried out using standard genetic techniques. The strains used in this study are listed in Table S7.
Protein expression and purification
Complementary DNA (cDNA) corresponding to the RNA-binding region (171-535aa) of PUF-8 was PCR-amplified and inserted at the Sal I and Not I sites of pMAL-c4E, which expresses the inserted ORF as a fusion protein with the maltose-binding protein (MBP) (New England Biolabs). Cloning techniques, including PCR, were carried out following standard protocols (Sambrook et al., 1989). The transformants were grown in LB medium at 37°C until 0.5 OD at 600 nm before induction with 0.05 mM IPTG for 2 h at 16°C. Cells were collected by centrifugation and lysed in lysis buffer [20 mM HEPES (pH 7.4), 0.5 M Na Cl, 5 mM DTT, 0.02% Tween 20, 0.1 mM PMSF] by incubation on ice with 0.5 mg/ml of lysozyme, followed by 3 rounds of freeze-thaw cycles. The lysates were treated with 20 μg/ml of DNase I and cleared by centrifugation. Fusion proteins were purified from clear supernatants by affinity chromatography using HIS-Select Cartridge (Sigma Cat. No. H8286) following manufacturer’s protocols. Purified proteins were concentrated by ultra-filtration, added with glycerol to a final concentration of 50% and stored at −20°C.
Affinity purification of mRNA binding to MBP::PUF-8
Total RNA was extracted from wild-type C. elegans using Tri-reagent (Sigma Cat. No. T9424) according to the manufacturer’s protocol and total poly (A)-containing RNA was isolated by using PolyATtract® Systems III (Promega) following the manufacturer’s protocols. For affinity purification with MBP::PUF-8, beads of amylose resin were first washed three times with distilled water, then five times with RNA-binding buffer (RBB) [5 mM HEPES (pH 7.5), 25 mM KCl, 2 mM MgCl2, 1 mM EDTA, 2 mM DTT, 3.5% glycerol, 0.25 mg / ml yeast tRNA]. Washed beads were incubated with MBP::PUF-8 at +4°C for 20 min with gentle agitation. Protein-bound beads were incubated with the total poly (A) RNA in RBB for 20 min at room temperature. After the incubation period, the beads were collected by brief centrifugation and washed five times with RBB. The MBP::PUF-8 protein was eluted from beads with 20 mM maltose and the bound RNA was separated by phenol: chloroform extraction. The RNA was then precipitated and subjected to antisense RNA amplification (see below).
RNA amplification
To obtain sufficient quantities of antisense RNA (aRNA) for microarray hybridization, we performed one round of amplification of the affinity-purified RNA. This amplification was based on the protocol described by Baugh et al (Baugh et al., 2001). Briefly, the affinity-purified RNA was reverse transcribed in the presence of 0.1 μg of (dT)-T7 primer KS2096 (see Table S2 for primer sequence) in 1X reverse transcription buffer (Promega), 5 nmol of dNTPs, 20 U of RNase inhibitor (Fermentas) and 50 U of reverse transcriptase (Promega) in a final volume of 10 μl at 42°C for 60 min. Second-strand synthesis (SSS) was carried out in 50 μl volume with 40 U of DNA polymerase I (Fermentas), 2 U of Escherichia coli RNase H (Fermentas) in 1X DNA polymerase I buffer (Fermentas) simply by adding 40 μl of an ice-cold SSS premix to the heat-inactivated, ice-cold 10 μl reverse transcription reaction and incubating at 15°C for 2 h. The double-stranded (ds) cDNA was blunt-ended usingT4 DNA polymerase, purified using the MinElute Reaction Cleanup Kit following manufacturer’s protocol (Qiagen) and precipitated with sodium acetate / ethanol. The dsRNA pellet was redissolved in sterile water and used as template for in vitro transcription of the antisense strand. In vitro transcription was performed in 50 μl reaction volume containing 200 U of T7 RNA polymerase (Ambion), 10mM NTP mix (Fermentas), and 1× buffer (Ambion) at 37°C for 16 h. Amplified antisense RNA (aRNA) was extracted with acidified phenol, precipitated with ammonium acetate / ethanol and redissolved in sterile water.
Microarray
Florescence labeling of the aRNA and hybridization to C. elegans microarray were performed by the Genome Center at Washington University, St. Louis, MO. Briefly, eight independent hybridizations were performed using RNA from four independent affinity purifications and swapping the fluorescence label between the test and reference samples. Total poly (A) RNA subjected to one round of amplification was used as the reference sample. Statistical analysis of the hybridization results were performed by the same facility at Washington University.
RT-PCR analysis
Both the total and the affinity-purified RNA were reverse transcribed using Mu MLV reverse transcriptase (Fermentas) in a 20-μl reaction volume containing 10 μCi of [α-32P] dCTP as a tracer for quantification purposes. The reaction was carried out at 42°C for 60 min and stopped by heat-inactivation at 70°C for 10 min. Unincorporated nucleotides were removed by spin column chromatography using Sephadex G-50 matrix. The incorporated radioactivity in the purified cDNA was measured using a liquid scintillation counter, and based on the radioactive count, equal amounts of cDNA templates were used for semi-quantitative PCR (see Table S5 for primer sequences). The number of PCR cycles that most accurately reflected the differences in the original template amount was empirically determined for each cDNA, and the amount of PCR products accumulated at the end of that many number of PCR cycles were compared between the test and total RNA samples.
Electrophoretic mobility shift assay
Mobility shift experiments, including preparation of RNA fragments and binding reactions, were performed as described earlier (Jadhav et al., 2008). The sequence of the non-specific RNA used for competition experiments was identical to the one described earlier (Jadhav et al., 2008). Template DNA fragments for in vitro were generated by PCR amplification using appropriate primers from wild-type C. elegans genomic DNA. The T7 promoter sequence was incorporated into DNA templates through the forward PCR primer. Required mutations were also introduced through PCR primers, and confirmed by DNA- sequencing. For testing the activity of purified MBP::PUF-8, primer KS1461 (see Table S2 for primer sequence), which contains the T7 sequence and the 29-nt RNA containing NRE sequence (Opperman et al., 2005), was annealed with primer KS2496 (see Table S2 for primer sequence) and used as template for in vitro transcription of NRE RNA.
Transgenics
We used pKS114 as the vector for testing the regulatory activity of various 3′ UTR sequences. This vector was generated through modifications of pJH4.52, which contains pie-1 promoter, a fusion between GFP and Histone H2B (GFP::H2B), and 3.2 kb downstream of pie-1 STOP codon including the pie-1 3′ UTR (Reese et al., 2000). pJH4.52 was modified to remove the pie-1 3′ UTR (bases 5535 to 5615 in pJH4.52) and include restriction sites (Bsp 120I and Nar I) to facilitate insertion of test 3′ UTR sequences immediately downstream of GFP::H2B and the pie-1 STOP codon. The unc-119-rescuing sequence from pAZ132 was added to the vector using NgoM IV and Sac II sites to finally generate pKS114. The various 3′ UTRs described in this study were PCR-amplified and inserted at the Bsp 120I site of pKS114 and introduced into unc-119(−) strain as described (Jadhav et al., 2008). The 3′ UTR sequence amplified starts from 20 bp upstream of the STOP codon and includes up to 500 bp past the predicted polyadenylation site. The primers used are listed in Table S3.
Mutations in the PUF-8 recognition elements (PRE) of pal-1 3′ UTR were introduced by PCR in the following way. The UTR sequences were PCR-amplified as two separate fragments. The reverse primer of the upstream fragment and the forward primer of the downstream fragment carried the relevant mutated bases (see Table S4 for primer sequences). In addition, these primers contained the Bpi I restriction site, which enables exclusion of the restriction site in the ligated product. In all cases, KS2983, which is at 350 bp upstream of pal-1 STOP codon, was the forward primer for amplification of the upstream fragment, and KS2952, which is downstream of the pal-1 3′ UTR sequence, was the reverse primer for the downstream fragment. The resulting PCR products, which contain the mutated sequences, were digested with Bpi I, ligated and used as a template in a PCR reaction using KS2995 and KS2997 as the forward and reverse primers, respectively. The amplified product was then digested with Bsp 120I and inserted at Bsp 120I site of pKS114. Sequences of all constructs were confirmed by DNA-sequencing.
Microscopy
Worms were examined using Zeiss fluorescence microscope, model Axioskop 2 mot plus and fluorescence images were acquired with Zeiss Axiocam HRm CCD camera. All images were acquired at 400x magnification.
RNAi
Exon regions of the target genes were PCR amplified, inserted into the RNAi feeding vector, pSV2, a modified version of pPR244 vector (Reddien et al., 2005). pSV2 contains the multiple cloning site (MCS) from pBluescript, flanked by T7 promoter sequences, which are again flanked by T7 terminator sequences. The Eco RV site in MCS was digested and added with dT residue at both cut ends using terminal transferase to enable direct cloning of PCR products by the standard TA cloning procedure. These constructs were transformed into E. coli strain HT115 used for RNAi by the feeding procedure (Timmons et al., 2001). To disrupt two genes simultaneously by RNAi, we prepared bacterial lawns containing equal amounts of both bacteria. The efficacy of RNAi was determined by monitoring the known phenotypes. For example, we observed for sperm-only phenotype following fbf-1(RNAi) and the frequency of proximal tumor following puf-8(RNAi) (Crittenden et al., 2002; Subramaniam and Seydoux, 2003).
Results
Identification of the potential mRNA targets of PUF-8
We chose an affinity chromatography-based approach for the isolation of potential targets of PUF-8. For this, we expressed the PUF domain (171-535aa) of PUF-8 in bacteria as a fusion protein with the maltose-binding protein (MBP), and used amylose resin bound with this fusion protein as the affinity matrix. In electrophoretic mobility shift assays (EMSA), MBP::PUF-8 fusion protein, retarded the mobility of an RNA bearing the Nanos response element (NRE) of hunchback 3′ UTR, which is a well-established target of Drosophila Pumilio (Fig. 1A). This retardation was sequence-specific, indicating that the MBP::PUF-8 produced in E. coli can be used to affinity-purify mRNAs that contain NRE-like sequences. Using this approach, we purified mRNAs that bound to MBP::PUF-8 from the total mRNA pool isolated from wild-type adult hermaphrodites, and identified them through microarray hybridization. Little or no mRNA bound to MBP alone, and therefore, total mRNA was used as the reference (see Materials and methods). To generate a list of mRNAs that were significantly enriched following affinity purification, intensities of hybridization signals of the test and reference probes were compared by paired, two-class Significance-Analysis-of-Microarrays (SAM) method and false discovery rates (FDRs) were determined for each mRNA (Tusher et al., 2001). Data were converted to log2 ratios (test / reference) to determine the number of folds of enrichment. We short-listed 347 mRNAs as potential targets of PUF-8 by applying a cut-off filter of 1% FDR and at least 3-fold enrichment in the affinity-purified pools compared to the reference. For 60 randomly-selected mRNAs, we validated the microarray results by two repeats of semi-quantitative RT-PCR. Thirty transcripts showed higher amplification with the affinity-purified fraction in both repeats, and all showed similar results in at least one of the two repeats (Fig. 1C and data not shown).
Fig. 1.
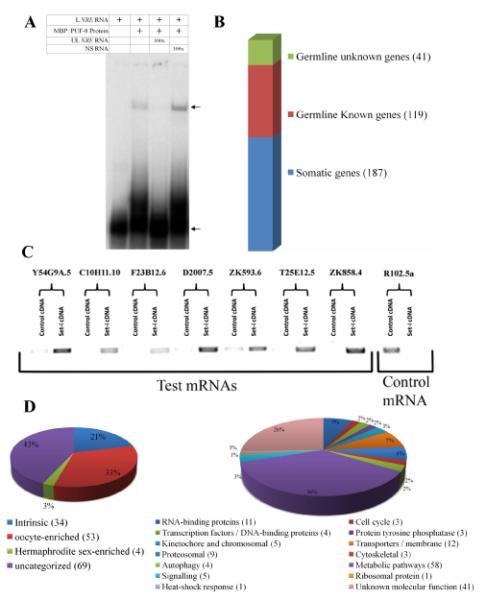
Identification of PUF-8-associated mRNAs. (A) Electrophoretic mobility patterns of radiolabeled NRE RNA in the presence of components indicated at the top. L NRE RNA – radiolabeled, 29-nt RNA bearing the Nanos response element, UL NRE RNA – same RNA but without radiolabeling, NS RNA – unlabeled non-specific RNA, 100x – molar concentration of the unlabeled RNA is about 100-fold higher than the radiolabeled RNA. The top arrow points to the MBP::PUF-8-NRE RNA complex and the bottom one indicates the free NRE RNA. (B) Soma-germline classification of the mRNAs that are at least 3-fold enriched in the fraction that affinity-purified with MBP::PUF-8 (see Materials and methods and the text for details). (C) Reverse transcription-PCR amplification of a few affinity-purified mRNAs from total mRNA (control cDNA) or from the affinity-purified fraction. Names of these mRNA are shown on top. Control mRNA – an mRNA that did not show enrichment with MBP::PUF-8 in the microarray hybridizations. (D) Classification of the germline mRNAs that affinity-purified with MBP::PUF-8 based on spatial distribution pattern (Left), or biochemical functions as annotated at Wormbase (www.wormbase.org).
Since our focus was the germline targets of PUF-8, we further short-listed these 347 mRNAs based on whether they have been previously known to be expressed in germ cells. We used annotations available in the microarray-based expression database generated by Kim et al (Kim et al., 2001) and the in situ hybridization-based expression database generated by the Kohara laboratory (Nematode Expression Pattern DataBase (NEXTDB), http://nematode.lab.nig.ac.jp). Of the 347 mRNAs, 187 are expressed only in somatic tissues, whereas 160 are expressed in the germline and/or soma (Fig. 1B and Table S1). Results of an earlier study indicate that 34 of the 160 are germline-intrinsic, 53 are oocyte-enriched and 4 are hermaphrodite sex-enriched (Reinke et al., 2004). The remaining 69 have not been assigned to any specific group (Fig. 1D and Table S1). Of the 160 germline-expressed genes, no functional information is available for 41 genes and the remaining 119 come from several functional groups. The largest group comprises of 58 genes that code for enzymes and other proteins involved in metabolic pathways of all types (Fig. 1D and Table S1). Transporters and proteins (12 genes) functioning in membrane trafficking constitute the second major group. Eleven mRNAs of this short list encode potential RNA-binding proteins, including proteins such as NOS-1, POS-1 and SPN-4 that have been well established to play important roles in germ cell development (Subramaniam and Seydoux, 1999; Tabara et al., 1999; Ogura et al., 2003). Other members of this group include CAR-1, which is involved in physiological apoptosis in germ cells and cytokinesis in embryos (Audhya et al., 2005; Boag et al., 2005), and ALY-2, which is essential for normal hermaphroditism by controlling tra-2 mRNA export from nucleus (Kuersten et al., 2004). In addition, the short list of potential PUF-8 targets includes cell-cycle components (3), constituents of the proteosome (9), members of the kinetochore and chromosomal components (5), proteins involved in autophagy (4), protein tyrosine phosphatases (3), Ras-like small GTPases (3), and transcription factors (4). In summary, the affinity purification-microarray hybridization strategy has identified mRNAs with a broad range of biological functions as potential targets of PUF-8.
PUF-8 mediates post-transcriptional control via the 3′ UTRs of several mRNAs in mitotic germ cells
Results presented above suggest that PUF-8 may bind to the identified mRNAs. However, they do not reveal whether PUF-8 indeed interacts with, and regulates the expression of, these mRNAs in worm germ cells. To address this, we employed a transgene-based assay that tests the post-transcriptional control activity of 3′ UTRs in vivo. In this assay, the GFP::H2B reporter fusion is placed upstream of the selected 3′ UTR such that the entire fusion – GFP::H2B-3′ UTR – is produced as a single transcript. Expression in the germline is achieved by using pie-1 promoter, which has been shown earlier to drive transgene expression in the germline (Reese et al., 2000). This reporter system has been successfully used earlier to assay the regulatory activity of various 3′ UTRs in the C. elegans germline (D’Agostino et al., 2006; Merritt et al., 2008; Merritt and Seydoux, 2010). Using this assay, we tested the 3′ UTR activity of about 10% (17) of the 160 germline-enriched mRNAs identified above as potential targets of PUF-8. For four of them, pal-1, pos-1, puf-5 and spn-4, GFP::H2B-3′ UTR reporter fusions were already available from the germline 3′ UTR reporter library developed by Seydoux and colleagues (Merritt et al., 2008). We prepared similar constructs for 13 additional genes and generated transgenic lines by microparticle bombardment. These include six genes – ubc-6, ubc-16, ubc-18, uev-1, T01C3.3 and C06A5.8 – of the proteosomal group and seven genes, namely car-1, hip-1, kca-1, mex-1, C56C10.10, F30F8.3 and Y54E5A.7, which were selected randomly.
Expression pattern of these 17 transgenes could be broadly classified into four groups: Transgenes with kca-1, pal-1, pos-1, puf-5, C56C10.10, F30F8.3 and T01C3.3 3′ UTRs expressed the GFP::H2B reporter strongly in oocytes, but not in mitotic or early pachytene regions. With mex-1, ubc-6, ubc-16, C06A5.8, and Y54E5A.7 3′ UTRs, reporter expression was observed in oocytes and the meiotic region, but was either absent or very low in the mitotic germ cells. Interestingly, the expression patterns of these two groups are complementary to that of a PUF-8::GFP transgene, which is expressed in a gradient fashion with its level stronger in the mitotic germ cells of the distal gonad than the developing gametes at the proximal end (Ariz et al., 2009). The third group, comprising car-1, hip-1, ubc-18 and uev-1 3′ UTRs, expressed GFP::H2B ubiquitously in the germline (Fig. 2 and data not shown). The transgene containing spn-4 3′ UTR forms the last group, which expressed in the distal region and oocytes, but not in the middle meiotic zone [(Merritt et al., 2008) and data not shown].
Fig. 2.
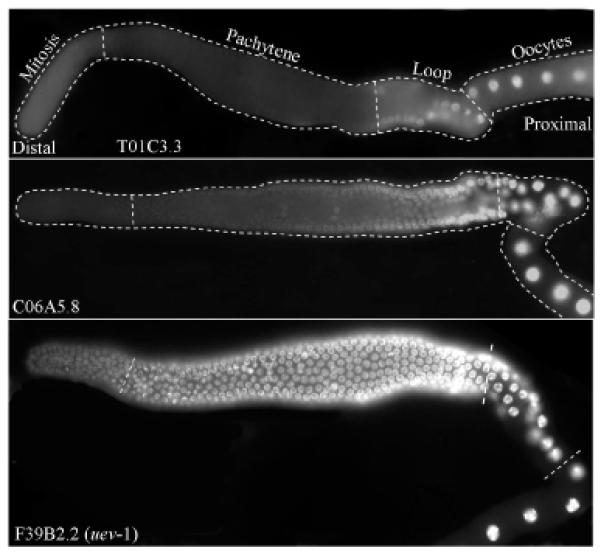
Expression patterns of 3′ UTR fusions. Fluorescence photomicrographs of adult hermaphrodite gonads showing GFP::H2B expressed under control of the indicated 3′ UTRs. Each of these three is representative example of the three classes of expression patterns observed: class I – No expression in the mitotic and pachytene zones. Expression begins only from the loop region; class II – No expression in the mitotic zone. Expression begins from the second half of the pachytene zone; and class II – Expression in all the zones.
To test whether any of these expression patterns were mediated by PUF-8, we examined them in PUF-8-depleted gonads. In contrast to the wild-type expression patterns described above, mex-1, pal-1, pos-1, ubc-16, C06A5.8 and F30F8.3 3′ UTR fusions expressed GFP::H2B in the distal mitotic germ cells in PUF-8-depleted worms (Fig. 3A). In the case of pal-1, similar distal misexpression was observed using a transgene that contained the coding sequence and the 3′ UTR of pal-1 in the gonads of puf-8(RNAi) worms as well as worms homozygous for puf-8(ok302), which is a null allele (Fig. 3C) (Subramaniam and Seydoux, 2003). In contrast, the expression of hip-1 3′ UTR transgene, which was observed throughout the wild-type gonad, was significantly reduced in the distal mitotic cells in puf-8(RNAi) gonads (Fig. 3B). Expression patterns of the other ten 3′ UTR fusions were unaffected by the depletion of PUF-8 (data not shown). Thus, the reporter expression mediated by 7 out of 17 3′ UTRs (about 40%) tested are influenced by PUF-8. These results strongly suggest that PUF-8 may post-transcriptionally control the expression of several mRNAs in the mitotic germ cells, by acting through their 3′ UTRs. While it seems to primarily function as a negative regulator, at least in the case of hip-1, PUF-8 functions as a positive regulator as well.
Fig. 3.
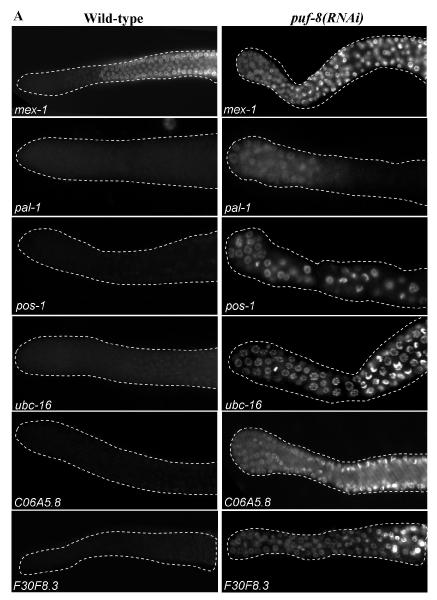
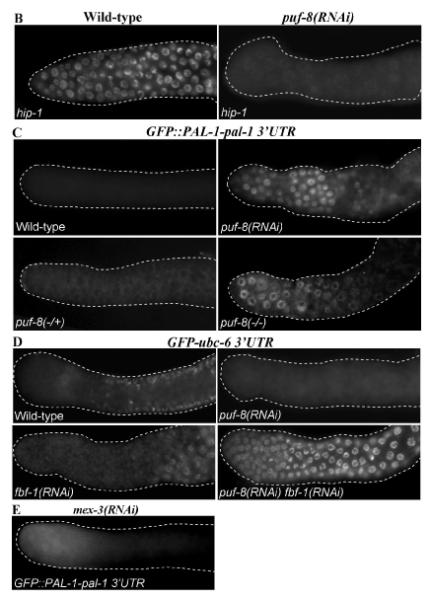
PUF-8 regulates transgene expression mediated by 3′ UTRs of several germline mRNAs. Fluorescence photomicrographs of distal part of C. elegans adult hermaphrodite gonad. In (A) and (B), expression patterns of GFP::H2B under the control of the indicated 3′ UTRs in the wild-type and puf-8(RNAi) gonads are shown. (A) Negative regulation by PUF-8; (B) positive regulation by PUF-8. (C) Expression pattern of the GFP::PAL-1-pal-1 3′ UTR transgene in the indicated RNAi or genetic background are shown: puf-8(−/+) – heterozygous for the null allele, ok302; puf-8(−/−) – homozygous for ok302. (D) Expression pattern of ubc-6 3′ UTR transgene in the indicated RNAi background, which reveals the redundant control by FBF and PUF-8. (E) Expression pattern of GFP::PAL-pal-1 3′ UTR in mex-3(RNAi) gonad.
PUF-8 and FBF redundantly control the expression mediated by ubc-6 3′ UTR in mitotic germ cells
Yeast three-hybrid studies have shown that PUF-8 preferentially binds to an 8-nt consensus sequence of UGUMHRDW, where M is A or C, H is A, U, or C, R is A or G, D is A, U, or G, and W is A or T (Opperman et al., 2005). Except ubc-18 3′UTR, all 3′ UTRs tested above contain at least one copy of this consensus sequence. In addition, six of them, namely kca-1, puf-5, ubc-6, C06A5.8, C56C10.10 and T01C3.3, contain the FBF recognition element (FBE), UGUDHHAU, where D is A, U, or G and H is A, U, or C, as well (Bernstein et al., 2005; Kershner and Kimble, 2010). Significantly, both FBFs and PUF-8 are expressed in the distal gonad, and FBF-1 and PUF-8 function redundantly to control the sperm-oocyte switch in hermaphrodites (Zhang et al., 1997; Lamont et al., 2004; Bachorik and Kimble, 2005; Ariz et al., 2009). Therefore, to test whether FBF and PUF-8 redundantly control the expression of any of these mRNAs, we depleted both proteins by RNAi in transgenic worms carrying the corresponding 3′ UTR fusions. As described above, the C06A5.8 3′ UTR fusion misexpressed the reporter in PUF-8-depleted worms, but was not affected by FBF depletion (data not shown). In contrast to the wild-type and either single RNAi, ubc-6 3′ UTR fusion strongly misexpressed the reporter in distal germ cells of the fbf-1(RNAi) puf-8(RNAi) worms (Fig. 3D), indicating that ubc-6 mRNA, which encodes an E2 ubiquitin-conjugating enzyme (Jones et al., 2002), may be post-transcriptionally suppressed by both FBF and PUF-8 in a redundant manner in distal germ cells. The other four 3′ UTR fusions were unaffected by the double RNAi (data not shown).
Both PUF-8 and MEX-3 contribute to pal-1 3′UTR-mediated suppression in mitotic germ cells
Two other RNA-binding proteins, namely GLD-1 and MEX-3, have been shown to suppress pal-1 mRNA in the meiotic zone and oocytes, respectively (Hunter and Kenyon, 1996; Mootz et al., 2004). Of these two, MEX-3 is expressed in the distal mitotic cells as well (Ciosk et al., 2004). To test whether MEX-3 contributes to pal-1 suppression in these cells, we disrupted its activity by RNAi. As shown in Fig. 3E, we could readily detect GFP in the distal part of the gonad, although the signal was not as strong as puf-8(−) worms. In the distal gonad, MEX-3 expression is limited only to the mitotic germ cells (Ciosk et al., 2004). Consistent with this, GFP misexpression was strictly confined to this region only in mex-3(RNAi) worms (compare Fig. 3A and E). Thus, in the case of pal-1 mRNA, both PUF-8 and MEX-3 are essential to suppress its expression in mitotic germ cells. As the mex-3(−) puf-8(−) double mutant gonads contain very few germ cells, we were not able to confidently determine whether MEX-3 and PUF-8 function additively to suppress pal-1 3′ UTR transgene.
PUF-8 control of pal-1 prevents ectopic expression of HLH-1
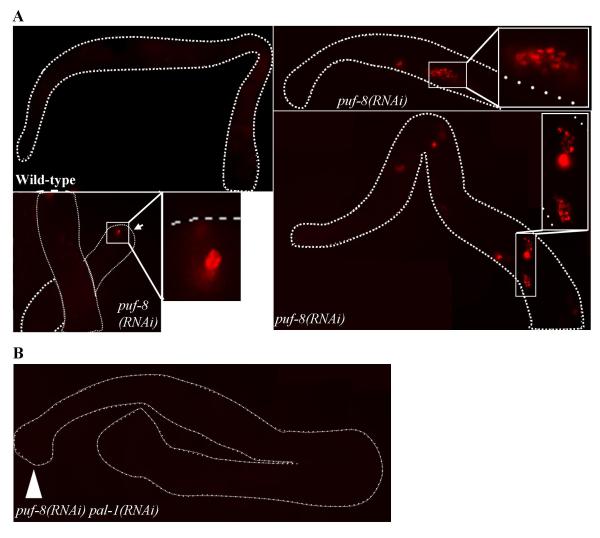
Next, we wanted to determine the biological significance of the PUF-8-mediated post-transcriptional suppression. One way to investigate this is to check whether the removal of the misexpressed target mRNA rescues, at least partially, some of the phenotypic defects observed in the puf-8 mutant. These phenotypes include the size of the distal mitotic region, sperm / oocyte switch, embryonic lethality at 20 °C, frequency of male progeny at 20 °C and sterility at 25 °C (Subramaniam and Seydoux, 2003; Bachorik and Kimble, 2005; Ariz et al., 2009). Unfortunately, we were unable to detect any such rescue following RNAi of any of the six mRNAs suppressed by PUF-8. This is presumably because the phenotypic defects observed in the puf-8 mutant results from the misregulation of multiple genes. Alternatively, it is possible that additional, protein-level regulation may operate to ensure suppression of these genes even when the controls at the RNA level fail. However, since removal of PUF-8 leads to misexpression of the transgene containing the PAL-1 coding sequence (Fig. 3), it is unlikely that a protein-level control operates in the case of pal-1. Consequently, we chose an alternative approach to investigate the consequences of pal-1 misexpression. PAL-1 functions as a transcription factor and activates the transcription of its downstream targets such as hlh-1 (Hunter and Kenyon, 1996; Lei et al., 2009). hlh-1 encodes the worm ortholog of the myogenic regulatory factor (MRF) HLH-1/MyoD, and is normally expressed in the embryonic muscle lineage (Krause et al., 1990). However, production of PAL-1 protein in meiotic germ cells, where its mRNA is normally not expressed, has been known to activate hlh-1expression ectopically in germ cells (Ciosk et al., 2006). Therefore, we decided to check if hlh-1 is similarly misexpressed in puf-8 mutant germ cells. We monitored hlh-1 expression using a transgenic line that expresses the HIS-24::mCherry reporter fusion under the control of hlh-1 promoter . This transgene has been shown earlier to reflect the endogenous expression pattern of hlh-1 (Murray et al., 2008). Consistent with this, we did not observe any expression of this transgene in the germline of wild-type worms (n = 257). In contrast, HIS-24::mCherry could be detected in the germ cells of about 42% of puf-8(RNAi) gonads (n = 64) (Fig. 4A). Expression was mostly observed in the cells of central and proximal regions of the puf-8 mutant tumorous gonads (Fig. 4A). However, in 7% of these gonads, expression was observed in a few cells near the distal region as well (Fig. 4A). To determine if the expression of hlh-1 reporter in cells lacking PUF-8 was dependent on PAL-1, we depleted PAL-1 as well in these cells through RNAi. As shown in Fig. 4B, HIS-24::mCherry could not be detected in any of the double RNAi worms that we examined (n = 56), which clearly shows that the activation of hlh-1 in PUF-8-depleted germ cells is indeed due to the misexpression of PAL-1.
Fig. 4.
PUF-8-mediated suppression of pal-1 prevents ectopic expression of HLH-1. (A) Expression pattern of hlh-1, one of the transcription targets of PAL-1, in wild-type and puf-8(RNAi) gonads. Activation of hlh-1 transcription has been visualized using HIS-24::mCherry reporter fusion expressed under the control of hlh-1 promoter. The three puf-8(RNAi) gonads presented here show hlh-1 promoter activity in the different regions of the puf-8(RNAi) germline. mCherry-positive cells are shown at a higher magnification in the insets. Arrow in the lower panel points to the distal end of the gonad. (B) Expression pattern of hlh-1, monitored as described in (A), in puf-8(RNAi) pal-1(RNAi) hermaphrodites. Arrow head indicates the distal end of the gonad.
PUF-8 physically interacts with pal-1 3′ UTR in vitro
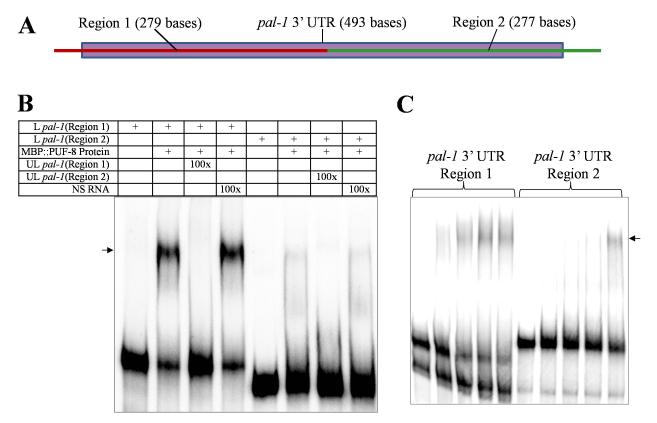
There are at least two explanations possible for the PUF-8-mediated repression of pal-1 mRNA: 1) as suggested by microarray and RT-PCR, PUF-8 directly interacts with pal-1 mRNA to suppress its expression, and 2) PUF-8 associates with, or regulates the expression of, another protein that controls pal-1 mRNA through direct physical interaction. To distinguish between these two mechanisms, we tested whether the bacterially-expressed MBP::PUF-8 fusion could bind to pal-1 3′ UTR in gel shift assays described above (see Materials and methods also). The 3′ UTR of pal-1 is comparatively large, so for the ease of performing this assay, we divided it into two roughly equal parts and named them as Region 1 and Region 2 (Figs. 5A, S1). As shown in Fig. 5B, the electrophoretic mobility of both parts of pal-1 3′ UTR was retarded by MBP::PUF-8 in a sequence-specific manner. Gel shift experiments with varying concentrations of MBP::PUF-8 indicate that the Region 1 of pal-1 3′ UTR has higher affinity than Region 2 for binding to MBP::PUF-8 (Fig. 5C). In these experiments, MBP alone did not affect the mobility of either part of pal-1 3′ UTR (data not shown). These results demonstrate that PUF-8 is capable of direct interaction with pal-1 3′ UTR. Unfortunately, due to lack of an anti-PUF-8 antibody suitable for co-immunoprecipitation, we have not been able to test whether PUF-8 binds in vivo to pal-1 3′ UTR. Presumably due to low levels of transgene expression, we were also unable to immunoprecipitate a PUF-8::GFP fusion protein, which rescues loss of puf-8 (Ariz et al., 2009), using anti-GFP antibody.
Fig. 5.
PUF-8 binds to pal-1 3′ UTR in vitro. (A) Schematic illustration of pal-1 3′ UTR showing the two regions used in gel mobility shift assays. The dotted bar represents the full-length 3′ UTR; Red line on the left part corresponds to Region 1 and the green line on the right is Region 2. See Fig. S1 for the complete sequence of pal-1 3′ UTR. (B) Electrophoretic mobility patterns of radiolabeled Region 1 and Region 2 of pal-1 3′ UTR RNA in the presence of MBP:PUF-8. L pal-1 – radiolabeled pal-1 3′ UTR; UL pal-1 – unlabeled pal-1 3′ UTR; NS RNA – unlabeled non-specific RNA; and 100x – number of times molar excess over L pal-1. (C) Electrophoretic mobility shift of the radiolabeled Region 1 and Region 2 of pal-1 3′ UTR RNA incubated with increasing concentrations of MBP::PUF-8. In both cases, the protein concentrations were: 1ane 1 – no protein; lane 2 – 0.02 nM, lane 3 – 0.04 nM, lane 4 – 0.06 nM and lane 5 – 0.08 nM. In both (B) and (C), the shifted band is indicated by the arrow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A specific sequence element within pal-1 3′ UTR is critical for suppression in mitotic germ cells as well as for in vitro interaction with PUF-8
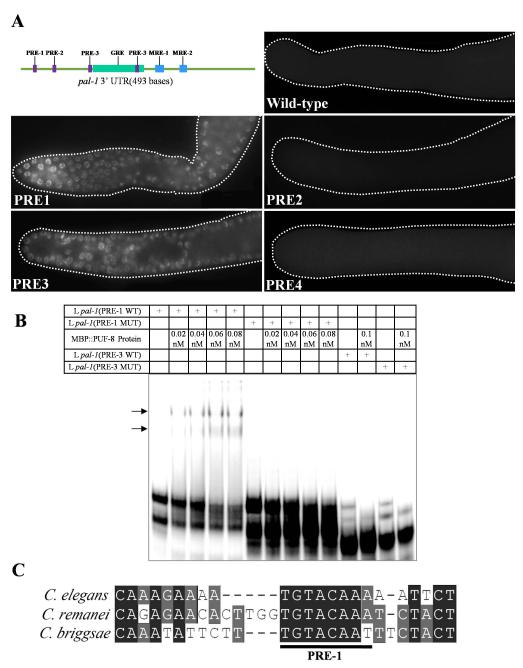
To identify specific sequences within pal-1 3′ UTR that are critical for PUF-8-mediated regulation, we examined the Region 1 sequence for recognizable features. Earlier studies have shown that a three-base sequence, UGU, is absolutely essential for high-affinity binding of PUF proteins (Opperman et al., 2005). We found four such UGU sequences within Region 1 of pal-1 3′ UTR, and named them as PRE-1, PRE-2, PRE-3 and PRE-4 (PRE stands for PUF-8 recognition element) in the order of their occurrence starting from the stop codon (Fig. 6A). To determine whether any of these four PREs has a regulatory role, we mutated each PRE in the GFP::H2B-pal-1 3′ UTR transgene by substituting 16 nucleotides – 3 nucleotides upstream of UGU, UGU itself and 10 downstream nucleotides – with 8 repeats of the dinucleotide AC, and generated transgenic lines expressing the mutant versions of the transgene. As shown in Fig. 6A, mutations in PRE-1 and PRE-3 resulted in GFP::H2B expression throughout the distal mitotic zone, whereas mutations in PRE-2 and PRE-4 didn’t affect the normal pattern of expression. Based on these results, we conclude PRE-1and PRE-3 of pal-1 3′ UTR are critical for the suppression of pal-1 in the distal germline.
Fig. 6.
Same sequence element within pal-1 3′ UTR is essential for post-transcriptional suppression in vivo and PUF-8 binding in vitro. (A) Top left: Schematic illustration of pal-1 3′ UTR showing the positions of the four potential PUF recognition elements (PREs) relative to other known cis-elements. GRE is GLD-1 recognition element (Mootz et al., 2004), and MRE-1 and -2 are the two MEX-3 recognition elements (Pagano et al., 2009). See Fig. S1 for the complete sequences of pal-1 3′ UTR and the three cis-elements. The other panels are fluorescence micrographs of the distal gonad of worms expressing GFP::H2B under the control of pal-1 3′ UTR bearing mutations in the indicated regions. In each case, four flanking nucleotides and the 8-nt PRE itself were replaced by eight repeats of the dinucleotide AC. (B) Electrophoretic mobility shifts by MBP::PUF-8 of the wild-type and mutant versions of radiolabeled 29-nt RNA encompassing PRE-1 or PRE-3. In both cases, the first 4 nucleotides of the PRE, UGUA, were changed to ACAU. Shifted bands are indicated by arrows. (C) Sequence alignment of pal-1 3′ UTR around the PRE-1 region from C. elegans, C. remanei, and C. briggsae.
Since PUF-8 binds in vitro to Region 1 of pal-1 3′ UTR, which contains the above described PREs, and the removal of PUF-8 also results in a similar pattern of misexpression, we tested whether any of these PREs is critical for interaction with PUF-8 using gel shift assays (see Materials and methods). While a 29-nt part of Region 1 containing PRE-1 showed a shift with MBP::PUF-8, a similar-length RNA from the region with PRE-3 did not (Fig. 6B) (see Table S4 for RNA sequence). Mutation of just the UGUA sequence alone to ACAU abolished the mobility shift completely, indicating that it is the PRE in the first RNA fragment that is critical for interaction with PUF-8. Significantly, PRE-1 element has been well conserved in the pal-1 3′ UTR of three Caenorhabditis species, namely C. elegans, C. briggsae, and C. remanei (Fig. 6D), for which this sequence information is available. Therefore, PUF-8 may similarly regulate pal-1 expression in the other two species as well, and the PUF-8-mediated repression of pal-1 in mitotic germ cells may be an evolutionarily conserved function.
In summary, removal of the trans-acting factor PUF-8, or the mutation of the cis-element PRE-1, both result in similar effects – misexpression in mitotic germ cells – on the expression of pal-1 3′ UTR-mediated expression. Although, due to technical reasons, we have not been able to demonstrate in vivo interaction of PUF-8 with pal-1 3′ UTR (see above), it is unlikely that the mutation of a short RNA sequence would purely coincidentally affect in vitro interaction with PUF-8 and in vivo expression in a manner similar to PUF-8 removal. If pal-1 misexpression in puf-8(−) worms were due to potential transdifferentiation or tumorigenesis of germ cells, and not due to direct post-transcriptional control, then cis-element mutations, which have been observed in otherwise normal germ cells, would not have affected pal-1 3′ UTR-mediated transgene expression. Further, puf-8(−) germ cells do possess the germ cell marker P granules (Ariz et al., 2009), and therefore, we do not think these cells have completely lost their germ cell identity. We conclude PUF-8 suppresses pal-1 expression in distal mitotic germ cells by acting via pal-1 3′ UTR and suggest it does so by directly binding to the PRE-1 sequence.
Discussion
In this study, we have identified 160 germline-expressed mRNAs as potential targets of PUF-8. We have tested the regulatory activity of seventeen of their 3′ UTRs, and found that PUF-8 could mediate post-transcriptional regulation through seven of them in the mitotic germ cells. While six are negatively regulated by PUF-8, expression of one, hip-1, is promoted by PUF-8. Thus, PUF-8 functions as both a positive and negative regulator. We have investigated the role of PUF-8 in the post-transcriptional control of one mRNA, namely pal-1, in detail using a combination of in vivo transgene-based assay and in vitro biochemical binding reactions, and obtained compelling evidence that PUF-8 suppresses pal-1 expression by directly binding to specific 3′ UTR sequences. We have also uncovered the functional significance of this control: in the absence of PUF-8, PAL-1 is misexpressed in distal germ cells, which activates the expression of HLH-1, a muscle-specific transcription factor. We propose pal-1 suppression by PUF-8 is essential to prevent these cells from entering into somatic programs such as muscle differentiation.
Identification of potential targets of PUF-8
So far, five studies have reported genome-wide screens for mRNA targets of PUF proteins (Gerber et al., 2004; Gerber et al., 2006; Galgano et al., 2008; Morris et al., 2008; Kershner and Kimble, 2010). In all these cases, the PUF protein has been purified from whole organism- or cell-extracts and the co-purified mRNAs have been identified. The number of mRNAs thus isolated ranges from 40 to 224 in the case of five yeast PUF proteins, and from 500 (for human PUM2) to 1350 (for C. elegans FBF) in metazoans. Thus, the 347 mRNAs identified here is, by comparison, appears less. We think this is due to differences in the approaches, rather than due to a real difference in the number of targets of other metazoan PUFs and PUF-8. First, in the absence of any known PUF-8 target, we applied 3-fold enrichment and an FDR of 1% as the cut-off, which is more stringent than the 2.25% FDR applied for FBF (Kershner and Kimble, 2010). At around 1.3% FDR, the number of potential targets identified for FBF is 332 (Kershner and Kimble, 2010), which is comparable to the 347 that we have obtained for PUF-8. Second, our approach, which is based on the in vitro interaction of purified RNAs and PUF-8, would have most likely isolated only the direct targets. In contrast, the earlier approach might have isolated indirect targets as well. Those RNAs that weakly associate with PUF in the presence of other proteins, or bind to other proteins that interact with PUF are possible indirect targets. Consistent with this, about 80% of mRNAs in our short list contain at least one copy of the consensus sequence recognized by PUF-8. For FBF, a similar value has been reported only for the top 314 of its potential targets (Kershner and Kimble, 2010). In addition, since we used bacterially-expressed protein, our approach would have missed mRNAs whose binding may require some post-translational modification on PUF-8. On the other hand, since our approach does not depend on whether a given mRNA is expressed in the same cellular compartment as PUF-8 or not, it might have isolated some non-target mRNAs merely because they are capable of interacting with PUF-8 in vitro. In the case of germline, we circumvented this problem by using information available in two expression databases to identify the germline-expressed mRNAs in the short list. Our GFP-3′ UTR fusion experiments show that the 3′ UTRs of 7 out of 17 (40%) such potential targets can confer PUF-8-mediated control in the germline. This suggests that the short list of mRNAs identified in the current study is significantly enriched for real targets of PUF-8. Due to potential redundant control (see ubc-6 example discussed below), this estimate is likely an underestimate.
Targets of FBF and PUF-8
Comparison of the potential targets of PUF-8 and FBF reveals these two proteins may have several common as well as distinct sets of target mRNAs. Sixty of the 160 germline targets of PUF-8 (37%) are present in the FBF’s list as well (Table S1). Significantly, 18 of them are present among the top 314 targets of FBF, and 16 contain both PRE and FBE sequences (Table S6). Our 3′ UTR fusion results indicate that the ubc-6 mRNA is one such common target redundantly controlled by PUF-8 and FBF. On the other hand, 26% of PUF-8 targets (40 out of 160) do not find a place in the FBF list even when we go down the list up to the lowest rank, which is 4722. These observations may account for the ability of FBF and PUF-8 to participate in some processes in a redundant fashion, while functioning largely in a non-redundant manner. For instance, both function redundantly to promote sperm-oocyte switch in hermaphrodites (Bachorik and Kimble, 2005), although it is not clear whether they accomplish this through common targets. However, in the case of GSC maintenance, FBF functions by preventing meiotic entry (Crittenden et al., 2002), whereas PUF-8 functions by directly promoting GSC mitosis (Ariz et al., 2009). Similarly, while PUF-8 is essential for progression of primary spermatocytes through meiosis (Subramaniam and Seydoux, 2003), spermatogenesis is not affected in fbf mutants (Crittenden et al., 2002). Similar redundant and distinct functions have been observed even among the nearly identical FBF-1 and FBF-2: both function redundantly to suppress fem-3 expression, but function distinctly to determine the size of the mitotic region (Lamont et al., 2004). Thus, PUF proteins in worms seem to have duplicated and evolved to expand the number of mRNAs –by extension, the number of biological processes – controlled by them. From this stand point, it is intriguing how the number of PUF proteins dwindled later in evolution – only one in Drosophila and two in mammals.
Post-transcriptional regulation of pal-1
PAL-1 is a homeodomain transcription factor similar to Caudal. It is essential for posterior patterning of the C. elegans embryo, where it specifies the fate of posterior blastomeres C and D (Hunter and Kenyon, 1996). PAL-1 activity is restricted to these blastomeres through post-transcriptional repression by MEX-3, which is expressed in the anterior cells, and through an unknown mechanism mediated by SKN-1, a transcription factor present in the posterior cells (Hunter and Kenyon, 1996). pal-1 mRNA is present throughout the germline, including developing oocytes, and all blastomeres of the early embryo. Therefore, repression of pal-1 mRNA begins well before embryogenesis, presumably to ensure strict asymmetric distribution of PAL-1 protein in the embryo. First, GLD-1, a STAR domain RNA-binding protein, silences pal-1 mRNA in the distal arm of the gonad. Second, MEX-3 takes over this suppression from GLD-1 in the proximal gonad, where GLD-1 is not present and MEX-3 expression begins (Mootz et al., 2004). However, GLD-1 is not present in GSCs, which are present at the very distal end of the gonad (Jones et al., 1996). Although MEX-3 is expressed in GSCs (Ciosk et al., 2004), it has not been clear so far which protein controls pal-1 mRNA in these cells. Our results clearly show that it is PUF-8 that is primarily responsible for pal-1 suppression in GSCs. In puf-8(−) gonads, the misexpression of pal-1 3′ UTR transgene is limited only to the distal mitotic zone (Fig. 3A second panel), which indicates that the GLD-1 control of pal-1 is still intact in these worms. Thus, the repression of pal-1 mRNA is maintained all the way from GSCs until the 4-cell-stage embryo through the sequential actions of three different RNA-binding proteins, namely, PUF-8 in the mitotic zone, GLD-1 in the meiotic zone and, finally MEX-3 in oocytes, zygote and anterior blastomeres of the early embryo.
Interestingly, both PUF-8 and MEX-3 are essential for complete suppression of pal-1 mRNA in GSCs. Removal of MEX-3 alone also leads to some misexpression of the reporter in a few cells at the distal end, although this misexpression is not as dramatic as in puf-8(−) gonads (Compare the Fig. 3A second panel, 3C and 3E). While MEX-3 is strictly confined to the first half of the distal mitotic zone (Ciosk et al., 2004), PUF-8 expression extends into the early meiotic cells in a distal-to-proximal gradient fashion (Ariz et al., 2009). These expression patterns are reflected in the extent of derepression of pal-1 3′ UTR-controlled transgene expression: in mex-3(−) worms, it is observed only in a few cells at the distal end, whereas it extends up to the distal part of the meiotic zone as well in puf-8(−) worms. It is not clear at the moment how PUF-8 and MEX-3 both contribute to pal-1 control in the same cells. One possibility is that they both function independently by binding to their respective recognition sequences in the pal-1 3′ UTR (Fig. 6A top left panel), and contribute to pal-1 suppression in an additive manner. Alternatively, they may function as partners. For example, the 3′ UTR binding, interaction with the translational machinery, or any other intermediate factor of one is dependent on the other. We favor the first model for the following reasons: one, both proteins bind to distinct parts of pal-1 3′ UTR independently in in vitro binding experiments [Fig. 6 and (Pagano et al., 2009)]. Two, as described above, the extent of derepression of pal-1 3′ UTR-mediated transgene expression mirrors the distribution patterns of PUF-8 and MEX-3, which is unlikely if the suppression were to be dependent on both proteins. Third, MEX-3 suppresses pal-1 mRNA in oocytes, where PUF-8 is either not present or present in very low levels (Ariz et al., 2009).
FBF does not seem to control pal-1 translation. pal-1 mRNA ranks low (#3584) in the list of FBF’s potential targets, and does not contain the FBE consensus sequence (Kershner and Kimble, 2010). The closest to this sequence in pal-1 3′ UTR is PRE-1 – differing only in the last base, A instead of U, which is critical for PUF-8 binding as well as for translational suppression in the distal gonad. Although we have not tested whether FBF binds to pal-1 3′ UTR, pal-1 3′ UTR transgene expression was not affected by the fbf-1(ok91) fbf-2(q704) double mutant (K. Pushpa and K. Subramaniam, unpublished observation).
PAL-1 activates hlh-1, the worm homolog of MyoD, by directly binding to an hlh-1 enhancer (Lei et al., 2009). Consistently, we find that the puf-8(−) germ cells misexpress HLH-1 in a PAL-1-dependent manner. Similar PAL-1-dependent misexpression of HLH-1 has been observed earlier in the loop region of gonads lacking GLD-1 and MEX-3 (Ciosk et al., 2006). Surprisingly, only a few puf-8(−) germ cells misexpressed HLH-1, although we saw GFP::H2B-pal-1 3′ UTR misexpression in all distal germ cells in puf-8(−) worms. An obvious explanation is that a protein-degradation mechanism ensures absence of PAL-1 even when the RNA-level controls fail. However, since we saw misexpression of the GFP::PAL-1 protein fusion as well in puf-8(−) worms (Fig. 3B), this possibility is unlikely. Alternatively, PAL-1 misexpression alone may not be sufficient for hlh-1 activation, and the other required factors may not be present in mitotic germ cells, or misexpressed in all puf-8(−) germ cells. Even in the case of gld-1(−) mex-3(−) mutant, muscle transdifferentiation is dependent on meiotic entry (Ciosk et al., 2006). Another surprising observation is the presence of HLH-1-postive cells in more proximal parts of the puf-8(−) gonad, where PUF-8 level is significantly low. A potential explanation would be that these are cells in which the misexpressed PAL-1 persisted until meiotic entry, at which stage they might have become competent for PAL-1-mediated activation of HLH-1.
Given that the posterior blastomeres do not transcribe new mRNAs (Seydoux and Fire, 1994; Seydoux et al., 1996) and that the asymmetric distribution of PAL-1 in the embryo is essential for posterior patterning (Hunter and Kenyon, 1996), the importance of maternal production of pal-1 mRNA and its post-transcriptional control in oocytes and early embryo can be readily appreciated. However, while the oocytes actively transcribe new mRNAs during their early phase of meiosis, it is puzzling why pal-1 mRNA is present throughout the germline, starting all the way from the GSCs and is translationally kept quiescent until the 4-cell stage embryo by the sequential actions of three spatially-restricted RNA-binding proteins. This is probably the case with many other maternally-inherited mRNAs as well – about 33% of the PUF-8’s potential targets belong to the oogenesis-enriched class (Fig. 1 and Table S1). As many of these mRNAs are detected in the germline of very early larva that have not yet begun meiosis, we do not think their presence at the distal is due to the diffusion of proximally-produced mRNAs in the syncytial gonad to the distal region. Probably the transcriptional output of the early oocytes is not sufficient to meet the demands of the maturing oocytes, and the syncytial nature of the gonad is perhaps to draw the transcriptional output from many nuclei to meet this demand. This underscores the importance of RNA-binding proteins and post-transcriptional regulation in the functioning of germline, and probably explains why the well conserved PUF family members may control a large number of mRNAs.
Supplementary Material
Acknowledgements
We thank Peter Reddien and Sanchez Alvarado for the pPR244 plasmid vector; C. elegans Genetics Center (CGC) for worm strains; The Wellcome Trust, London, UK for a SRF to K.S.; and the Council of Scientific and Industrial Research, Government of India for a SRF to R.M. We appreciate the comments of two anonymous examiners of R.M.’s PhD thesis, which were very useful in writing this paper.
References
- Ariz M, Mainpal R, Subramaniam K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 2009;326:295–304. doi: 10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachorik JL, Kimble J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10893–10897. doi: 10.1073/pnas.0504593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag PR, Nakamura A, Blackwell TK. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- D’Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell. 2011;20:72–83. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–1812. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Jones D, Crowe E, Stevens TA, Candido EP. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L’Etoile ND. A 3′UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Krause M, Fire A, Harrison SW, Priess J, Weintraub H. CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell. 1990;63:907–919. doi: 10.1016/0092-8674(90)90494-y. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Segal SP, Verheyden J, LaMartina SM, Goodwin EB. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol. Cell. 2004;14:599–610. doi: 10.1016/j.molcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature. 1987;329:679–693. [Google Scholar]
- Lei H, Liu J, Fukushige T, Fire A, Krause M. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development. 2009;136:1241–1249. doi: 10.1242/dev.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Seydoux G. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development. 2010;137:1787–1798. doi: 10.1242/dev.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol. Cell. Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. 2003;130:2495–2503. doi: 10.1242/dev.00469. [DOI] [PubMed] [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat. Struct. Mol. Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, Essien KI, Ryder SP. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20252–20257. doi: 10.1073/pnas.0907916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza GM, da Silva AM, Kuspa A. Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development. 1999;126:3263–3274. doi: 10.1242/dev.126.14.3263. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 2003;13:134–139. doi: 10.1016/s0960-9822(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tadauchi T, Matsumoto K, Herskowitz I, Irie K. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Battu G, Hoier EF, Hajnal A. Distinct roles of the Pumilio and FBF translational repressors during C. elegans vulval development. Development. 2006;133:3461–3471. doi: 10.1242/dev.02496. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Xu EY, Chang R, Salmon NA, Reijo Pera RA. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol. Reprod. Dev. 2007;74:912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.