Abstract
Gynodioecy is a sexual system wherein females coexist with hermaphrodites. It is of interest not only because male-sterile plants are advantageous in plant breeding but also because it can be a crucial step in the evolutionary transition to entirely separate sexes (dioecy) from a hermaphroditic ancestor. The gynodioecious diploid wild strawberry, Fragaria vesca ssp. bracteata (Rosaceae), is a member of a clade with both dioecious and cultivated species, making it an ideal model in which to study the genetics of male sterility. To create a genetic map of F. v. ssp. bracteata, we identified informative polymorphisms from genomic sequencing (3−5x coverage) of two outbred plants from the same population. Using targeted enrichment, we sequenced 200 bp surrounding each of 6575 polymorphisms in 48 F1 offspring, yielding genotypes at 98% of targeted sites with mean coverage >100x, plus more than 600-kb high-coverage nontargeted sequence. With the resulting linkage map of 7802 stringently filtered markers (5417 targeted), we assessed recombination rates and genomic incongruities. Consistent with past work in strawberries, male sterility is dominant, segregates 1:1, and maps to a single location in the female. Further mapping an additional 55 offspring places male sterility in a gene-dense, 338-kb region of chromosome 4. The region is not syntenic with the sex-determining regions in the closely related octoploids, F. chiloensis and F. virginiana, suggesting either independent origins or translocation. The 57 genes in this region do not include protein families known to control male sterility and thus suggest alternate mechanisms for the suppression of male function.
Keywords: pollen, sex chromosome, sex determination, trait mapping, translocations
The evolution of distinct male and female sexes (dioecy) is believed to often begin with the emergence of gynodioecy, the coexistence of females and hermaphrodites, in an ancestrally hermaphroditic population (Charlesworth and Charlesworth 1978; Spigler and Ashman 2012). Thus, the origins of separate sexes and sex chromosomes can be examined with studies of the genetic basis for male sterility and the selective pressures favoring it in naturally gynodioecious species (reviewed in Spigler and Ashman 2012; Charlesworth 2013). The adaptive benefit of male sterility in an otherwise-hermaphroditic individual is puzzling, as first noted by Darwin (1877), and several hypotheses have since been proposed to explain it (reviewed in McCauley and Bailey 2009; Dufay and Billard 2012). Classical hypotheses rest on an adaptive advantage for females, either from greater seed production as a result of their ability to redirect energy from male function or avoid antagonists (Ashman 2002), or from greater seed quality via avoidance of self-fertilization and thus inbreeding depression (Darwin 1877; Lewis 1941; Charnov et al. 1976; Charlesworth and Charlesworth 1978).
In either case, the magnitude of the benefit required for invasion and maintenance of females depends on the underlying genetics of male sterility (reviewed in Dufay and Billard 2012). In addition, gynodioecy can be “stochastic,” where its dynamics are governed largely by meta-population forces of migration and drift (Pannell 1997) as well as the genetic conflict between cytoplasmic male sterility (CMS) mutations that are beneficial to organelles but are not transmitted in the male gametes, and nuclear mutations that restore fertility (Rf) but may come with costs (reviewed in Bailey and Delph 2007; McCauley and Bailey 2009; Touzet 2012; Caruso et al. 2012). These hypotheses are not mutually exclusive, and more than one selective force could contribute to the sex ratio in a population (see Delph and Carroll 2001). Uncovering the genetic basis of male sterility can help to identify the likelihood of transitions to subdioecy (males, females, and hermaphrodites) and dioecy (reviewed in Spigler and Ashman 2012) and reveal new dynamics in the evolution of sex chromosomes (Charlesworth 2013), especially when studies are conducted among closely related species.
The genetic basis for male sterility has been identified in several agricultural and model plant taxa (e.g., Ma 2005; Borg et al. 2009; Wilson and Zhang 2009; Lu et al. 2013), although most of these are rare or induced mutations in otherwise hermaphroditic species. Only a few studies have examined naturally occurring male sterility in wild species (e.g., Dudle et al. 2001; Goldberg et al. 2010; Garraud et al. 2011). Most heavily studied cases are those with cytonuclear sex determination, in which the CMS loci typically encode toxic proteins, often chimeric ATP synthases, and the Rf loci are typically pentatricopeptide repeat (PPR) genes that function to silence the CMS mRNA (Hanson and Bentolila 2004; Wang et al. 2006). Some Rf loci belong to other protein families but still function by blocking the CMS locus (e.g., Matsuhira et al. 2012). Only one copy of an Rf allele is generally necessary for CMS silencing (but see Schnable and Wise 1998; Kohls et al. 2011), so fertility restoration is typically dominant and thus male sterility at Rf loci is typically recessive (Chase 2007). Although there are Rf loci that act recessively in pollen (Wen et al. 2003; Fujii and Toriyama 2009), they act dominantly at the whole-plant level, that is, plants that are heterozygous at these loci are male fertile. Many nuclear male-sterility loci that do not interact with a CMS locus (i.e., are non-Rf loci), are also known in plants, and these have been identified as encompassing diverse gene families such as acyl-CoA synthetase (Wang and Li 2009), plant homeodomain-containing protein (Alves-Ferreira et al. 2007), MYB transcription factor (Durbarry et al. 2005), brassinosteroid signaling genes (Ye et al. 2010), and heat shock proteins (Yang et al. 2009). At most of these loci, male sterility is recessive, although there are known cases of dominant male sterility (Cao et al. 2009) as well as more complex multiallelic systems (Lu et al. 2013). It remains unclear, however, which of these gene families, if any, underlie naturally occurring examples of gynodioecy.
The gynodioecious diploid (2n = 2×=14 chromosomes) wild strawberry Fragaria vesca ssp. bracteata (Rosaceae), native to western North America (Staudt 1989), is an ideal model for studying the genetic basis and evolution of male sterility (Li et al. 2012). It is classified as a subspecies of the otherwise hermaphroditic F. vesca ssp. vesca. However, the chloroplast genomes of Pacific Coast populations of F. v. ssp. bracteata share a closer phylogenetic relationship with several polyploid species than with other F. vesca, suggesting that it is the cytoplasmic donor to this octoploid clade (also known as clade ‘A’; Njuguna et al. 2013). The octoploid clade includes species displaying hermaphroditism (F. iturupensis), subdioecy (F. virginiana), and dioecy (F. chiloensis). These latter two octoploid species are the progenitor species of the cultivated strawberry Fragaria × ananassa. The agriculturally important Rosaceae family also includes other examples of male sterility (e.g., plum, peach and blackberry; Dorsey 1919; Connors 1926; Scott and Weinberger 1938; Crane and Lawrence 1931; Jordano 1993). Previous classical genetic work in octoploid strawberry (F. virginiana and F. chiloensis) and in F. v. ssp. bracteata concluded that female function is determined by a disomically inherited single nuclear locus (Valleau 1923; Ahmadi and Bringhurst 1989) in all three species with male sterility dominant, that is, females are heterozygous and hermaphrodites are homozygous at this locus.
Recent genetic mapping in the octoploid species and their natural hybrid confirms that male sterility is dominant to male fertility but also that it shows linkage to female fertility (Goldberg et al. 2010; Spigler et al. 2011; Govindarajulu et al. 2013). Existing observations support conflicting hypotheses about the evolutionary origin of male sterility. On one hand, the close relationship among several species displaying the rare trait of dominant male sterility suggests a single origin of the trait, with possible translocation (Goldberg et al. 2010). On the other hand, the observation that male sterility maps to different genomic regions in the polyploid Fragaria species (Goldberg et al. 2010; Spigler et al. 2011; Govindarajulu et al. 2013), as well as the existence of hermaphroditic F. iturupensis within an otherwise-sexually dimorphic clade (Njuguna et al. 2013), suggest that male sterility may have evolved several times independently. Moreover, equivocal evidence for a fitness advantage sufficient to maintain females (i.e., high selfing rates in hermaphrodites but no seed quantity advantage and low- and variable-quality advantage in early life stages; Li et al. 2012; Dalton et al. 2013) under the published mode of sex determination (i.e., dominant nuclear male sterility; Ahmadi and Bringhurst 1989) raises the possibility for alternate means of sex determination. Thus, a detailed genetic mapping approach is required to assess the inheritance of sex determination in F. v. ssp. bracteata.
Mapping sex-determination loci has been challenging traditionally because of the suppression of recombination on sex chromosomes and the low power of limited-marker, pregenomic studies; therefore, few high-resolution maps have been produced (Charlesworth and Mank 2010; Charlesworth 2013). However, there are several key reasons why high-resolution mapping F. v. ssp. bracteata is now especially feasible. First, suppression of recombination is not expected before the appearance of distinct linked male-function and female-function loci (Charlesworth 2013). Second, the small F. v. ssp. vesca genome has been sequenced and annotated (Shulaev et al. 2011), providing an a priori expectation of marker order as has been used in other reference-guided linkage maps (e.g., Felcher et al. 2012), as well as illuminating translocations and recombination rates that can be important hallmarks of evolving sex determining regions (Charlesworth 2013). Third, new high-throughput approaches such as RAD (restriction-site associated DNA) mapping (Baird et al. 2008), highly parallel single-nucleotide polymorphism (SNP) genotyping (Fan et al. 2003), and targeted capture (Cronn et al. 2012) allow thousands of markers to be readily assessed in multiple individuals. Although sex loci have been mapped with RAD (Anderson et al. 2012) and highly parallel SNP genotyping (Bradley et al. 2011), targeted capture has not been used previously. However, targeted sequence capture presents several advantages for linkage mapping including efficient data generation, the ability to target SNPs known to be informative in the parents, and low individual variation in locus recovery. It also provides a rapid means for comparative analyses of genome structure between related species.
The goals of the study presented here were as follows. First, to genetically map the male-sterility locus in F. v. ssp. bracteata to a narrow genomic region to test for synteny with other Fragaria species, to determine the mode of inheritance, and to identify possible candidate genes. Second, to demonstrate the power of a new technology—targeted capture using RNA baits—for studying genomic variation in plants (Cronn et al. 2012). Third, to compare a high-density F. v. ssp. bracteata linkage map with the F. v. ssp. vesca reference genome (Shulaev et al. 2011) to assess genomic evolutionary phenomena such as recombination rate, which can be unusually high or low near sex-determining regions (Hasselmann and Beye 2006; Charlesworth 2013) but has not been well studied in incipient sex loci, as well as translocations, which can obscure patterns of synteny and contribute to the striking genomic lability in the location of sex-determining loci (Charlesworth and Mank 2010).
Methods
Species description and map cross population
F. vesca ssp. bracteata (Rosaceae) is a herbaceous perennial plant occurring from the Rocky Mountains to the Pacific Northwest and extending from British Columbia to southern Mexico (Staudt 1989). Both parental plants for this study were collected from a single population on Marys Peak, Oregon, (N44°29′18.4″, W123°32′14.7″) and were maintained in the greenhouse at the University of Pittsburgh.
To map male sterility, we created an F1 mapping population by pollinating a female (MRD30) with pollen from a hermaphrodite (MRD60) in 2011. Seeds (N = 127) from this cross were planted in Sunshine germination mix: Fafard #4: sand, and exposed to 14-hr day and 19°/15° day/night temperatures with a relative humidity of 70% in a growth chamber. Seedlings (N = 112) were transplanted into 200-mL pots filled with a 2:1 mixture of Fafard #4 and sand and grown at 21°/15° day/night temperatures and 12-hr daylight. To induce flowering 4 mo after planting, plants were fertilized with bloom booster 10:30:20 N:P:K fertilizer (Scotts Miracle-Gro Company, Marysville, OH) and exposed to a cold/dark treatment with 10-hr day and day/night temperatures of 18°/10° for 3 wk, 18°/4° for 7 wk, and 10°/4° for 2 wk. This treatment was repeated up to two times. All plants were watered and protected from pests as needed.
Scoring male function
Male function was scored on each flowering plant on at least two flowers formed at two separate times. Male function was determined as the presence (fertile) or absence (sterile) of pollen production. Anthers visibly dehiscing pollen were scored as fertile whereas those not dehiscing (i.e., visibly shedding pollen) were confirmed as sterile by observing crushed anther sacs under the microscope. Viability of pollen grains of male-fertile individuals was confirmed via vital stains (e.g., Alexander’s stain; Kearns and Inouye 1993). Six progeny died before flowering and 11 have not yet flowered, so we were able to score male function on a total of 95 progeny.
DNA extraction and library preparation
Total genomic DNA was extracted from ca. 100- to 150-mg sample of fresh leaves from parents (MRD30 and MRD60) and 48 progeny using a modified CTAB procedure (Doyle and Doyle 1987). Extracted DNA samples were quantified using the Qubit dsDNA HS assay (Life Technologies/Invitrogen, Carlsbad, CA). Library construction, solution hybridization, and nucleotide sequencing followed MycroArray MyBait and Illumina protocols, with minor modifications. For library construction, 1−2.5 µg of DNA was sheared using a Diagenode BioRuptor Sonicator for 13 min. After sonication, a small aliquot was assayed by gel electrophoresis, and additional sonication was conducted if the sample did not have a median shear size of approximately 200 bp. Subsequent enzymatic steps used NEBNext (New England Biolabs, Ipswich, MA) reagents. For low-coverage genome sequencing of the paternal parent (MRD60), a custom internal index adapter (Cronn et al. 2008) was used, whereas the target capture libraries (parents and progeny) used external index adapters (Bio Scientific, Austin, TX). The low coverage genome library of the maternal parent (MRD30) was sequenced in one lane of the Illumina GAIIx with 80 bp single end sequencing, whereas MRD60 was sequenced as part of a 13-plex on the Illumina HiSeq 2000 with 101 bp paired end sequencing. For the target capture libraries, agarose gel size-selected fragments (ca. 200-bp inserts) were amplified for nine cycles (minimized to avoid polymerase chain reaction duplicates) using Bio Scientific primers. Following Qubit (Invitrogen/Life Technologies, Eugene, OR) fluorometric quantitation, 30−60 ng of each library was pooled in an 8-plex or 12-plex. Each multiplex pool (400−480 ng) was used in a single solution hybridization (=target enrichment) reaction, and incubated at 65° for 36−40 hr with the RNA baits. After incubation, the target-hybridized biotinylated RNA baits were captured with streptavidin-coated magnetic beads. The captured DNA targets were then eluted in water and amplified by polymerase chain reaction for 9−12 cycles. After purification and quantitation, the target enriched multiplex samples were sequenced as an 8-plex or a 48-plex (Supporting Information, Table S1) with 101-bp single end sequencing on the Illumina HiSeq 2000 at Oregon State University. Sequencing data in FASTQ format was uploaded to SRA (Accession SRP022950).
Genotyping by targeted sequence capture
We filtered Illumina reads by converting bases with Phred quality scores less than 20 to missing (N), and excluding reads with fewer than 75 high-quality (≥20) bases. The low-coverage genomes of the parental plants were mapped using Burrows-Wheeler Aligner, or BWA (Li and Durbin 2009) to the F. v. ssp. vesca reference genome (v. 1.1, based on accession Hawaii 4; Shulaev et al. 2011), masked with RepeatMasker; Smit et al. 1996−2010). This reference genome (hereafter designated FvH4) consists of seven pseudochromosomes assembled from an interspecies linkage map (FvH4_1-FvH4_7) ranging from 21 to 39 Mb in size, as well as a 12-Mb concatenation of unanchored scaffolds (FvH4_0), totaling 207 Mb (Sargent et al. 2011; Shulaev et al. 2011). We converted parental genotypes to vcf format for analysis (Danecek et al. 2011). We identified polymorphic sites in the two parents (SNPs and 1- to 2-bp indels, both coding and noncoding) and designed custom MYbaits (MYcroarray, Ann Arbor, MI) biotinylated RNA baits targeting 6575 of these sites (6376 SNPs and 199 indels; File S1). Each polymorphism is targeted by three 100-bp oligonucleotides, one flanking each side of the polymorphism and one centered on the polymorphism, such that the 100-bp surrounding the polymorphism is targeted twice. We chose polymorphisms that were likely informative (heterozygous in one parent only based on ≥5x coverage in our genomic data), >100 bp away from other polymorphisms, and in regions of intermediate GC content (30–70%) to maximize genotyping reliability. We also minimized marker clustering by excluding sites that were <5 kb from another included polymorphism in one direction and <10 kb from another included polymorphism in the opposite direction.
We randomly chose 48 of the F1 offspring for linkage mapping. Examination of a previous reference genome version (v. 1.0) revealed portions of two scaffolds (scf0513160 positions 1−1,989,403, hereafter scA, 2.0Mb; and scf0513158 positions 2,258,778−4,674,928, hereafter scB, 2.4Mb) that had been left out of FvH4 (v. 1.1). Although we had not designed targeted capture probes for these scaffolds, we mapped our reads to a supplemented reference genome that included these scaffolds, again using BWA (Li and Durbin 2009), and converted genotypes to vcf format for analysis. Because the sum of all baits was less than 1% of the genome, we could sequence targeted regions at very high coverage for very accurate genotyping. Thus, we first filtered for a per-individual depth of at least 20, then we retained genotypes as valid only if the Phred-scaled likelihood was 0 and Phred-scaled likelihood for all other potential genotypes was >40 (likelihood of other genotype < 10−4); otherwise, genotypes were considered missing. To maximize the number of high-quality, true polymorphisms in our linkage map, we only retained putative polymorphisms for linkage mapping if they met several strict criteria, including showing missing genotypes in no more than eight offspring, not showing >85% of offspring with the same genotype, and having informative parental genotypes consistent with segregation in the offspring.
Linkage mapping
We used the R package OneMap (Margarido et al. 2007) to generate linkage maps for informative markers in each parent (hereafter referred to as FvbPat for paternal and FvbMat for maternal, or Fvb for the set of linkage groups). Because our markers had known locations on the reference genome (Shulaev et al. 2011), we had an a priori hypothesis for their relative positions, as with other reference-guided linkage maps (e.g., Felcher et al. 2012). Thus, we could justifiably use relatively liberal criteria for linkage map construction, and any features of the linkage map that deviated from the expected pattern, such as the number of linkage groups or marker order within groups, we examined individually for confirmation. Initially we only used markers with no missing genotypes to create the framework linkage maps; markers with missing data were added manually later if possible. We initially used a LOD (logarithm of odds) of 3 to assign markers to linkage groups, and if this resulted in fewer than the expected seven linkage groups we raised the LOD to 4. We used a LOD of 2 to order markers within linkage groups. Markers that could not be placed unambiguously at a LOD of 2 were assigned to the most likely position on the basis of their LOD score. Linkage map results were carefully curated manually to correct for results attributable to genotyping error and to resolve ambiguities. For example, the observation of paired recombination events on either side of a single marker is likely to be a genotyping error rather than a real double recombination event and was treated as such. This approach is conservative because ignoring any true double recombination event on either side of a polymorphism can only decrease our estimates of centimorgan distances slightly and would not qualitatively affect our results. We estimated genotyping error rate using these anomalous genotypes that did not match the expected genotype given the genotype of the same individual at sites upstream and downstream on the linkage map (Figure S1). Using polymorphisms heterozygous in both parents and the locations of markers in FvH4, we aligned the linkage groups between the parents.
We compared the FvbPat and FvbMat linkage maps with FvH4 (visualized with Circos, Krzywinski et al. 2009). We assigned each linkage group a number (1−7) corresponding to the FvH4 pseudochromosome with which it shared the most markers. Interchromosomal translocations were identified as sets of contiguous markers with FvH4 location that did not match the linkage (Fvb) location. Intrachromosomal translocations were identified as sets (≥4) of contiguous markers with the same chromosome number but with an Fvb location that was incompatible with their FvH4 location. Inversions were identified as increases in linkage map distance in the opposite direction from reference genome distance, with at least two observed map locations supporting the directional change, such that a recombination event that reverses that previous recombination event was considered to be a possible double recombinant and not an inversion. To examine variation in recombination rate across the genome, we divided Fvb linkage groups as closely as possible into sections of 1 Mb, a size chosen because our population size would not allow accurate estimates of recombination rate in sections less than 100 kb, and recombination rate might vary substantially across sections of 10Mb (Schnable et al. 1998; Li et al. 2007). We did not attempt to estimate recombination rate across the borders of translocations and inversions, so we assessed contiguous segments defined by these genomic rearrangements, and segments larger than 1 Mb were broken into equal sections less than 1 Mb each. We defined the recombination rate as the ratio of Fvb linkage map distance (cM) to reference genome sequence distance (Mb), following other studies that compare linkage maps and genome sequences between closely related taxa (e.g., Meznar et al. 2010), and thus this represents a rate that assumes congruence in physical distance between Fvb and FvH4. Variation in this rate across the linkage map thus could reflect disparities in genome architecture between the subspecies, i.e., unobserved indels and rearrangements.
Mapping male sterility
Our a priori expectation based on published accounts (Ahmadi and Bringhurst 1989) was that male sterility is determined by a single, perfectly penetrant Mendelian locus, and therefore our sample size would be sufficient (Lander and Botstein 1989) to initially identify genomic regions of major effect consistent with this prediction that could be confirmed and fine-mapped using additional individuals. We coded male sterility as a Mendelian locus (e.g., females AB and males AA), and mapped it along with the targeted capture genotypes using OneMap to determine its position on the linkage map. For confirmation and fine-mapping, we identified nine informative polymorphisms in this region using the low-coverage genome sequences of the parents, and we genotyped these sites using Sanger sequencing in both the phenotyped targeted capture offspring (n = 40) as well as additional offspring from this cross (n = 55) with known sexual phenotypes (primers in Table S2). Genes in the region matching male sterility were identified using the Strawberry Genome Hybrid Gene Model Database, Version 2 (http://strawberrygenome.org).
Results
Genotyping by targeted sequence capture
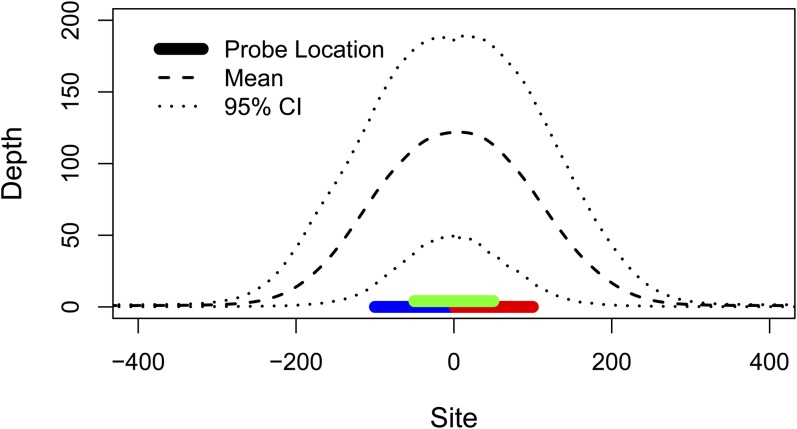
Mean coverage (read depth) of the parental genomes was 3.2x for the paternal parent (MRD60) and 5.4x for the maternal parent (MRD30) across 149 Mb. We observed 184,531 polymorphisms within the parents. In the targeted capture data, mean depth per individual at the 6575 targeted sites was 120x (median depth per individual = 102x; minimum individual depth = 35x; Figure 1, Figure S1). A mean depth of at least 20x was observed at 6414 (98%) of these sites, and 5417 (82%) of these were polymorphic and met our further quality criteria for inclusion in the linkage mapping. An additional 1.92 Mb passed our depth and quality filters, exceeding the number of targeted base pairs by nearly 50% due to coverage in the “splash zones” flanking the targets (Figure 1). Thus, in addition to the targeted polymorphisms, we added 2385 polymorphisms for a total of 7802 high-quality markers for linkage mapping (Figure 2, File S2, File S3). We observed low per-individual rates of both missing data (mean = 2.1%; median = 0.6%) and genotyping errors (mean = 0.2%; median = 0.05%) across the dataset (Figure S1).
Figure 1.
Average targeted capture sequencing depth in all 50 individuals across all 6575 targeted polymorphisms (Table S1, File S1, File S2). Colored bars represent the three probes designed for the targeted site. Dashed and dotted lines depict mean and 95% interval of sequencing depth. Mean sequencing depth of targeted sites was 120x, and high-coverage sequence was obtained even beyond the ends of the probes (mean depth 100 bp from probes = 15x).
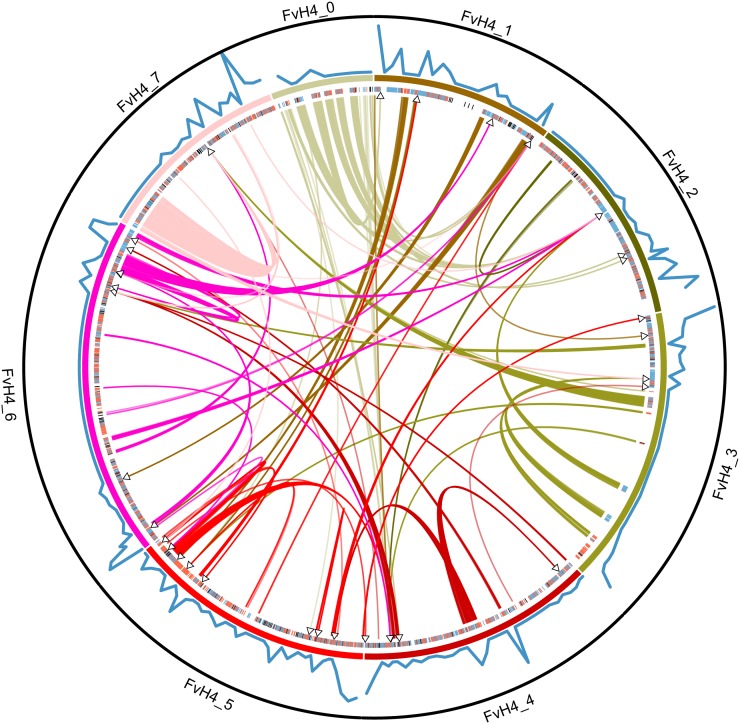
Figure 2.
Circular map of F. vesca ssp. vesca reference genome (FvH4; pseudochromosomes 0−7 are displayed in a circle) with F. vesca ssp. bracteata genomic data (Fvb) indicated (created with Circos; Krzywinski et al. 2009). Targeted capture markers polymorphic in the mother (blue), father (red), or both (black) are indicated as tick marks just inside the chromosomes. Translocations identified in our Fvb linkage map are indicated by curved links between different regions of FvH4, colored to match their location on FvH4, with arrowheads indicating their estimated position in Fvb. Recombination rate is indicated by a blue line along the outside of the chromosomes. The maximum value observed for recombination rate (20.4 cM/Mb) is indicated by the black circle encompassing the figure. Scaffolds scA and scB not shown.
Linkage mapping
We produced seven paternal linkage groups at LOD 3 for MRD60 (FvbPat_1-FvbPat_7), corresponding to the haploid base number of chromosomes in Fragaria (Figure S2). All linkage groups remained intact at LOD 4, except FvbPat_1 and FvbPat_4 were each split into two groups (FvbPat_1: 36 and 243 markers; FvbPat_4: 437 and 47 markers). At LOD 4, the MRD30 markers yielded seven linkage groups (FvbMat_1−FvbMat_7), matching FvbPat_1−FvbPat_7. Because we expected seven linkage groups in both parents, we retained the linkage groups generated at a LOD of 3 for MRD60 and at a LOD of 4 for MRD30. Despite not being specifically targeted, we observed informative polymorphisms on scA and scB mapping both to FvbPat_4 (4 polymorphisms) and FvbMat_4 (4 polymorphisms) (Table 1, Figure S2, D and K). We could order 2917 polymorphisms (37%) unambiguously within linkage groups at a LOD of 2; other polymorphisms were placed in the most likely position based on LOD score and confirmed by eye.
Table 1. Linkage groups observed.
| Linkage Groupa | Main FvH4 Pseudochromosomeb | Markersc | Size, cMd | Size, Mbe | Map Locations Observedf | Other FvH4 Pseudochromosomesg |
|---|---|---|---|---|---|---|
| FvbPat1 | 1 | 279 | 60.4 | 22.5 | 17 | 0 (53), 6 (32), 7 (1) |
| FvbPat2 | 2 | 649 | 52.1 | 31.7 | 20 | 0 (109), 3 (1), 5 (19), 6 (13), 7 (1) 506 |
| FvbPat3 | 3 | 223 | 50.0 | 28.1 | 19 | 1 (2), 4 (1), 5 (3) |
| FvbPat4 | 4 | 484 | 54.2 | 32.5 | 17 | scA (1), scB (3), 0 (5), 2 (3), 3 (3), 5 (16), 6 (2) |
| FvbPat5 | 5 | 667 | 60.4 | 29.4 | 27 | 0 (2), 1 (35), 3 (9), 4 (19), 6 (6) |
| FvbPat6 | 6 | 1049 | 77.1 | 39.0 | 26 | 1 (60), 3 (12), 4 (33), 7 (2) |
| FvbPat7 | 7 | 590 | 52.1 | 24.3 | 23 | 3 (32), 6 (4) |
| FvbMat1 | 1 | 507 | 47.9 | 22.5 | 22 | 0 (58), 5 (22), 6 (46) |
| FvbMat2 | 2 | 776 | 54.2 | 31.7 | 24 | 0 (111), 3 (29), 5 (1), 6 (26), 7 (1) |
| FvbMat3 | 3 | 358 | 62.5 | 28.1 | 27 | 5 (4), 7 (20) |
| FvbMat4 | 4 | 649 | 58.3 | 32.5 | 18 | scA (1), scB (3), 0 (16), 1 (2), 2 (2), 3 (2), 5 (39), 6 (5) |
| FvbMat5 | 5 | 508 | 62.5 | 29.4 | 25 | 0 (1), 4 (27), 7 (2) |
| FvbMat6 | 6 | 940 | 62.5 | 39.0 | 27 | 1 (56), 3 (13), 4 (29), 5 (1), 7 (4) |
| FvbMat7 | 7 | 600 | 62.5 | 24.3 | 26 | 3 (54), 4 (1), 6 (7) |
Linkage group in F v. ssp. bracteata. Groups beginning with “FvbPat” are paternal, and those with “FvbMat” are maternal.
Pseudochromosome in FvH4 that shares the most markers with each linkage group.
Number of polymorphic markers in the linkage group.
Size of linkage group in centimorgans.
Size of linkage group in megabases.
Number of map locations at which at least one marker was directly observed.
Other FvH4 pseudochromosomes with markers on this linkage group. Number of markers listed in parentheses.
We inferred 392 recombination events between both parents based on 318 mapped locations represented by at least one marker, indicating a genome-wide mean recombination rate of 1.0 cM/Mb (Table 1). Relative to FvH4, we observed 22 inversions, 11 intrachromosome translocations, and 40 interchromosome translocations (shown as curved lines across the middle of the circle in Figure 2, Table S3). We were also able to assign 14 scaffolds from the unmapped FvH4_0 to locations on the Fvb linkage map (Table S3). Of these 87 incongruities between FvH4 and Fvb, 91% were supported by more than one polymorphic marker (median = 18 polymorphisms), suggesting they are unlikely to be mapping errors in Fvb, although some could represent assembly errors in FvH4. Linkage maps developed independently from both parents were very similar, such that inversions and translocations suggested by one map were compatible with the other map, and recombination rates across the genome were correlated between parents (rs = 0.48; P < 10−13; Figure S3). Most (60%) genomic regions showed no recombination in either parent, and those showing recombination often showed it in both parents. Recombination rate varied across the genome, ranging from an observed rate of zero across 23Mb in the central portion of FvbPat_6/FvbMat_6, to more than 20 cM/Mb near the end of FvbPat_7/FvbMat_7. Recombination rate was positively correlated with gene density in the reference genome (rs = 0.52; P < 10−15).
Male sterility mapping
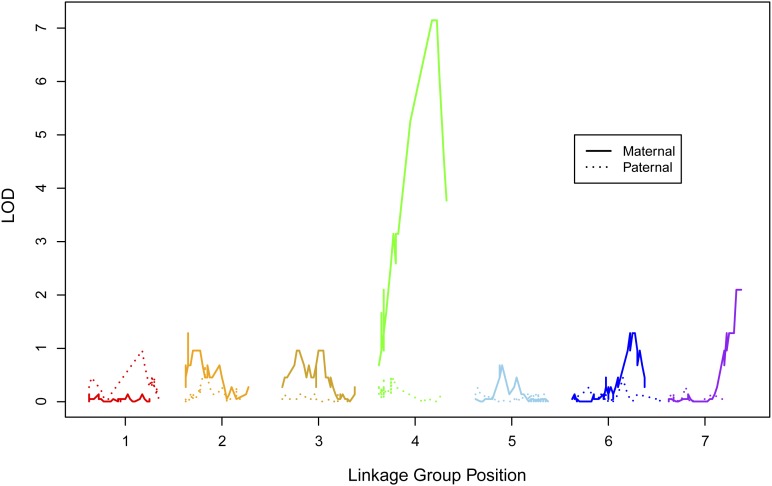
In all 95 progeny that flowered, 49 were hermaphrodite and 46 were female; thus, male function segregated in a 1:1 ratio (χ2 = 0.09; P = 0.75). Of the 48 offspring used in targeted capture linkage mapping, 40 flowered including 22 hermaphrodites and 18 females. Thus, male function also segregated in a 1:1 ratio in this subset (χ2 = 0.4; P = 0.52). Using these 40 targeted capture plants, we unambiguously mapped male sterility to a region near the end of FvbMat_4 (Figure 3, Figure 4). The two closest markers on FvH4 were FvH4_4_26535351 upstream, at which eight individuals mismatched sex (LOD = 3.1), and FvH4_4_26597498 downstream, at which three different individuals mismatched sex (LOD = 7.1). The scaffold scB also mapped in between these same two markers. Marker scB_629161 was the closest match upstream with only three mismatches to sex (LOD = 7.1), and thus we inferred the male-sterility locus to occur between scB_629161 and FvH4_4_26597498. No targeted capture marker showed a perfect match to sex.
Figure 3.
Probability of the sex locus mapping across the genome. A LOD score greater than 3 occurs in one genomic location: near the end of the maternal linkage group 4 (FvbMat_4) (LOD = 7.1 for markers at 22−24 cM, corresponding to markers mapping from 26.6−26.8Mb on FvH4_4). Markers on scB not shown.
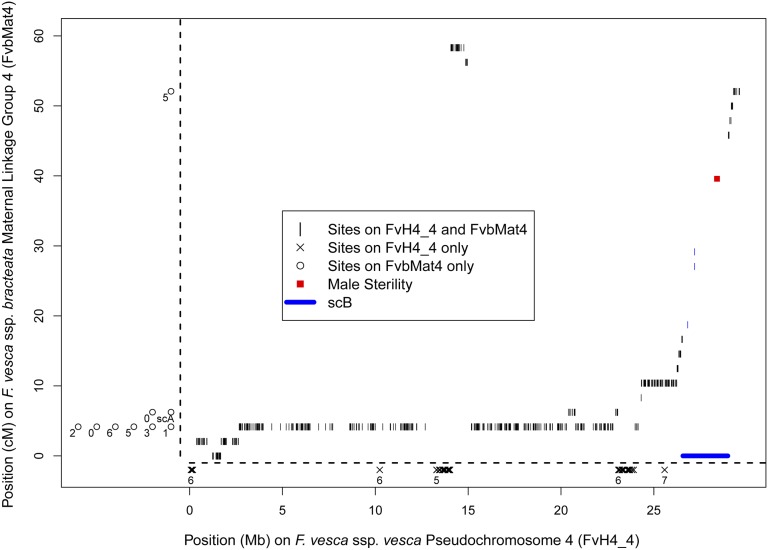
Figure 4.
Maternal linkage map FvbMat_4 aligned to pseudochromosome 4 of the F. vesca ssp. vesca reference genome (FvH4_4). Each short vertical line represents an informative marker in our linkage map. The male-sterility locus resides within the red square that maps to the scaffold scB (thick blue line) which has here been inserted into its inferred location in FvH4_4. There are few informative targeted capture markers on scB (blue vertical lines) because it was not included in probe design. Markers that aligned somewhere in the reference genome other than FvH4_4 are indicated as circles along left side of the figure, numbered according to FvH4 pseudochromosome. Markers that aligned to FvH4_4 but mapped to a different linkage group in Fvb are indicated as x marks along the bottom of the figure, numbered according to FvbMat linkage group. Markers that map to one part of FvbMat_4 but align to a different part of FvH4_4 are within-chromosome translocations, such as the markers at 56−58 cM on FvbMat_4 occurring between 14-15Mb on FvH4_4.
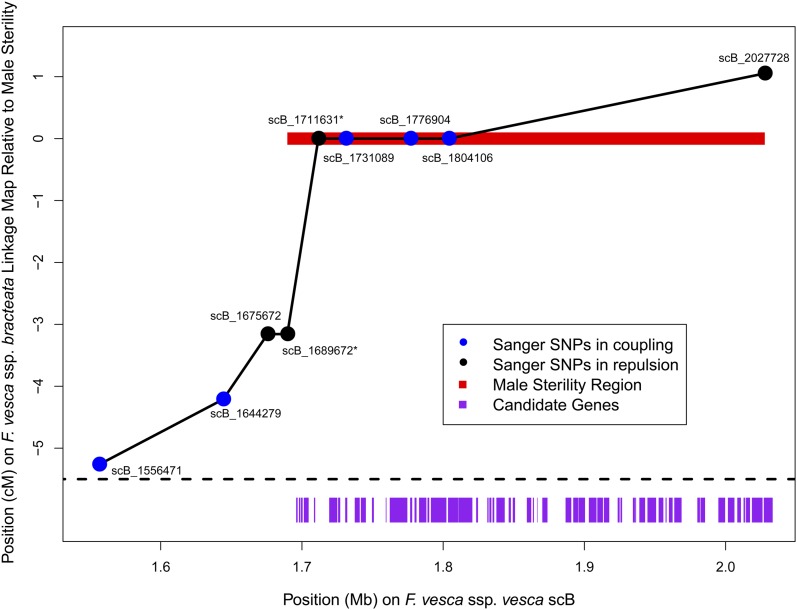
We inferred 14 recombination events on scB in the 48 targeted capture plants, suggesting a recombination rate across scB of 11.8 cM/Mb, and allowing us to conduct additional fine-mapping. To do so we identified nine informative polymorphisms (Figure 5) on scB based on the whole genome sequencing of the parents and genotyped them in the parents and progeny via Sanger sequencing. We genotyped seven of these polymorphisms in all 95 progeny with zero to two missing genotypes per site, and we genotyped two polymorphisms in smaller subsets of progeny that included all potentially recombinant individuals based on the other polymorphisms. Genotypes at four of these polymorphisms showed a perfect match to sex, including three in coupling with male sterility (LOD = 28.6). The polymorphisms on either side of these perfect matches (scB_1689672 with three mismatches, and scB_2027728 with one mismatch) define a maximum candidate region of 338kb containing 57 genes based on FvH4 (Table 2). These genes represent a diverse array of functional roles, but several are known to be expressed in pollen or essential for pollen development, at least in Arabidopsis (Table 2).
Figure 5.
Fine-mapping of the candidate region with Sanger sequencing. Each informative SNP (circle) is plotted according to its position on scB (x-axis) and its mapped position based on observed recombination among 95 offspring (y-axis). SNPs are colored according to whether they are in coupling (i.e., cis; blue) or in repulsion (i.e., trans; black) with the inferred male-sterility locus. Most SNPs were genotyped in the full sample of offspring (n = 93−95), but two SNPs (marked with asterisks) were only genotyped in a subset of individuals including all potential recombinants as inferred from the other SNPs (n = 13). The 338-kb region consistent with a perfect match to sex is shown in red, and genes in this region are indicated with purple rectangles (Table 2).
Table 2. List of 57 genes in the 338kb genomic region on scB (sites 1689672−2027728) that shows a perfect match to sex.
| Gene | Description | Start | End |
|---|---|---|---|
| 04131a | Cytochrome b561 (probable) | 1695908 | 1696955 |
| 04132a | Cytochrome b561 (probable) | 1697882 | 1698854 |
| 04133a | Protein pelota (probable) | 1699354 | 1700487 |
| 04134a | Protein VERNALIZATION INSENSITIVE 3 (probable) | 1701336 | 1704731 |
| 04135 | Zinc finger protein 4 (probable) | 1708545 | 1709366 |
| 04136a | Signal transduction histidine-protein kinase barA (probable) | 1719309 | 1725022 |
| 04137a | Conserved oligomeric Golgi complex subunit 8 (COG complex subunit 8) (probable) | 1725631 | 1727100 |
| 04138 | F-box/kelch-repeat protein At3g06240 (probable) | 1730662 | 1732093 |
| 04139a | Probable receptor protein kinase TMK1, Precursor (putative) | 1737410 | 1740919 |
| 04140a | Probable receptor protein kinase TMK1, Precursor (putative) | 1741738 | 1745245 |
| 04141a | U-box domain-containing protein 21 (probable) | 1749596 | 1750933 |
| 04142 | Zonadhesin, Precursor (probable) | 1759358 | 1759831 |
| 04143a | THO complex subunit 2 (Tho2) (probable) | 1762188 | 1774622 |
| 04144 | Putative FBD-associated F-box protein At5g56690 (probable) | 1776627 | 1778060 |
| 04145a | Probable clathrin assembly protein At4g32285 | 1779727 | 1781103 |
| 04146b | Putative receptor protein kinase ZmPK1, Precursor (probable) | 1782940 | 1788099 |
| 04147 | Dihydroorotase (DHOase) (probable) | 1788929 | 1790128 |
| 04148a | Transmembrane anterior posterior transformation protein 1 homolog (probable) | 1791303 | 1802303 |
| 04149 | Cyclin-SDS (probable) | 1803461 | 1810613 |
| 04150a | Probable leucine-rich repeat receptor-like protein kinase At1g35710, Precursor | 1810803 | 1820658 |
| 04151a | PPPDE peptidase domain-containing protein 2 (probable) | 1823385 | 1824658 |
| 04152a | Uncharacterized protein At4g28440 (similar to) | 1831297 | 1832134 |
| 04153b | Putative F-box/LRR-repeat protein At5g41840 (probable) | 1832760 | 1834112 |
| 04154 | Glycine/sarcosine/betaine reductase complex component C subunit alpha (Protein PC alpha) (probable) | 1834998 | 1836143 |
| 04155a | Ubiquitin carboxyl-terminal hydrolase isozyme L5 (UCH-L5) (probable) | 1837700 | 1843573 |
| 04156 | Protein CCC1 (probable) | 1846416 | 1847932 |
| 04157 | Protein CCC1 (probable) | 1849284 | 1850869 |
| 04158 | Heat stress transcription factor A-8 (AtHsfA8) (similar to) | 1859247 | 1862358 |
| 04159 | Homeobox protein rough sheath 1 (probable) | 1863517 | 1864254 |
| 04160 | Serine/arginine repetitive matrix protein 1 (probable) | 1866464 | 1866799 |
| 04161a | Alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 (AtTPS6) (putative) | 1870217 | 1873846 |
| 06233 | Ribonuclease R (RNase R) (probable) | 1886708 | 1890718 |
| 06234 | Probable S-adenosylmethionine-dependent methyltransferase At5g38100 | 1892129 | 1895522 |
| 06235a | Probable RNA-dependent RNA polymerase 1 (OsRDR1) (similar to) | 1895829 | 1900439 |
| 06236a | tRNA nucleotidyltransferase, mitochondrial, Precursor (probable) | 1903160 | 1908555 |
| 06237a | Probable RNA-dependent RNA polymerase 1 (OsRDR1) (similar to) | 1909159 | 1913119 |
| 06238a | Probable RNA-dependent RNA polymerase 1 (OsRDR1) (similar to) | 1913797 | 1917596 |
| 06239 | Probable RNA-dependent RNA polymerase 2 (OsRDR2) | 1923582 | 1924947 |
| 06240 | Nucleoside diphosphate-linked moiety X motif 17 (Nudix motif 17) (probable) | 1925581 | 1926702 |
| 06241a | Pre-mRNA-splicing factor cwc22 (probable) | 1934341 | 1935780 |
| 06242 | Shikimate kinase (SK) (probable) | 1936002 | 1936594 |
| 06243a | Probable RNA-dependent RNA polymerase 1 (OsRDR1) (similar to) | 1938940 | 1942992 |
| 06244a | Aspartate-semialdehyde dehydrogenase (ASA dehydrogenase) (probable) | 1944681 | 1950781 |
| 06245 | Two-component response regulator ARR14 (similar to) | 1952583 | 1955894 |
| 06246 | tRNA modification GTPase mnmE (probable) | 1957499 | 1958195 |
| 06247a | Adagio protein 3 (FBX2a) (putative) | 1959870 | 1962462 |
| 06248a,b | Dynamin-related protein 1C (putative) | 1963652 | 1968778 |
| 06249 | F-box protein At3g59000 (probable) | 1979978 | 1981804 |
| 06250 | hypothetical protein | 1982266 | 1985447 |
| 06251 | Myosin-7 (MyHC-beta) (probable) | 1994893 | 1999664 |
| 06252 | Integrator complex subunit 11 (Int11) (probable) | 2001649 | 2006256 |
| 06253a | E3 ubiquitin-protein ligase At4g11680 (probable) | 2008307 | 2010848 |
| 06254 | cGMP-gated cation channel alpha-1 (CNG-1) (probable) | 2012734 | 2013688 |
| 06255 | Farnesyltranstransferase, Precursor (similar to) | 2014248 | 2014574 |
| 06256a | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, Precursor (putative) | 2014656 | 2017275 |
| 06257a | Nuclear pore complex protein Nup155 (probable) | 2018622 | 2026133 |
| 06258 | Protein VAC14 homolog (probable) | 2027060 | 2033300 |
Genes with Plant Ontology categorization under “pollen” based on expression data in Arabidopsis (Schmid et al. 2005).
Genes with putative functional roles in pollen development or recognition, highlighted in the Discussion section.
Discussion
In the process of mapping male sterility in F. v. ssp. bracteata, we have generated a high-density linkage map which we can compare to the F. vesca reference genome. One striking result from this comparison is the large number of well-supported translocations and inversions (Figure 2, Table S3). We cannot determine from our data whether these are real evolutionary differences, errors in the genome assembly, or rearrangements that resulted from the hybrid nature of the cross on which the FvH4 pseudochromosome assembly is based (F. vesca × F. bucharica). Some rearrangements correspond to a complete single scaffold, consistent with assembly error, while others do not (Table S3), so it is likely that our observations include both assembly errors and real structural rearrangements. Observed recombination rate varied across the genome (Figure 2, Figure S2) and was correlated with gene density as is common in plants (Schnable et al. 1998; Li et al. 2007) and other taxa (Nam and Ellegren 2012). Because of potential chromosomal rearrangements between FvH4 and Fvb, the observed recombination rate is likely influenced by both the true rate of crossing over and differences in genome architecture between the subspecies. Most chromosomes contained a wide region (~10 Mb or more) showing no recombination (Figure S2), possibly the result of centromeres or similar recombination-suppressing chromatin structure, mirroring patterns seen in a previous F. vesca linkage map (Sargent et al. 2012). Although we endeavored to find markers spaced evenly across the genome, we were not able to generate many probes for the middle of FvH4_3, and those that we did use turned out to map to other Fvb linkage groups (Figure 2). Apparently the two parents happen to be homozygous for large haplotype blocks throughout this portion of the chromosome.
Our approach uses high-throughput genotyping of thousands of polymorphisms to map a Mendelian trait with a linkage cross, and thus leverages the combined strengths of both genomics and classical genetics. As with other recent applications of targeted sequence capture (Bi et al. 2012; Del Viso et al. 2012; Nadeau et al. 2012; Salmon et al. 2012; Tennessen et al. 2012), we have shown that MYcroarray MYbaits are highly effective in genotyping a large number of samples at thousands of sites. We have multiplexed 12 Illumina libraries in a single target enrichment (hybridization) reaction, and sequenced 48 target-enriched libraries in a single HiSeq lane, yielding high coverage sequence of nearly 1% of the genome for all individuals (Table S1). Few studies have used targeted capture for trait mapping (but see Del Viso et al. 2012), in contrast to other, more heavily used high-throughput technologies like RAD mapping and highly parallel SNP genotyping (e.g., Bradley et al. 2011; Anderson et al. 2012). Our results indicate that a targeted capture approach should be successful in future studies of linkage and comparative analyses of other genome-wide patterns.
We have mapped male sterility in F. v. ssp. bracteata to a single genomic region which in F. v. ssp. vesca is approximately 338kb and harbors 57 genes (Figure 5, Table 2). In Arabidopsis, homologs of many of these genes are important in pollen development. For example, a dynamin-related GTPase similar to gene06248 is required for plasma membrane maintenance in maturing pollen (Kang et al. 2003; Backues et al. 2010), and an F-box/LRR-repeat protein similar to gene04153 is required for pollen mitosis II (Gusti et al. 2009). Many genes in this region are known to be expressed in Arabidopsis pollen, including PPPDE protein, RNA-dependent RNA polymerase 1, and Aspartate-semialdehyde dehydrogenase (Table 2; Schmid et al. 2005; Wang et al. 2008). Only a single gene in this region appears in a recent curated database of genes involved in plant male reproduction (Cui et al. 2013): gene04146, encoding a putative receptor protein kinase structurally similar to S-loci, and thus potentially involved in pollen recognition. Notably absent are gene families known to cause male sterility in other species, such as PPR proteins (Hanson and Bentolila 2004; Wang et al. 2006), peptidase M48 proteins (Matsuhira et al. 2012), or acyl-CoA synthetases (Wang and Li 2009).
Because male sterility is dominant, we do not expect the causal polymorphism to be a simple loss of function in a gene essential for pollen development. Rather, a single allele in heterozygous state must somehow actively suppress pollen maturation. Although there are several promising candidates, the causal gene cannot be conclusively determined with our current data. It is also possible that the causal gene or its genomic location is unique to F. v. ssp. bracteata, such that it is not identifiable from the genes in this region of the reference genome. Additional experimental evidence to pinpoint the causal gene could include further fine-mapping with larger sample sizes, genetic knockdown/knockout experiments, genetic expression data for anthers or pollen, and/or complete sequencing of the candidate region in F. v. ssp. bracteata.
Male sterility is dominant, as seen in other Fragaria species, but its chromosomal location is not syntenic with the sex-determining regions of octoploid Fragaria (Goldberg et al. 2010; Spigler et al. 2011; Govindarajulu et al. 2013). There are three possible explanations for these observations. The first explanation is that the gene has been translocated at least twice among F. v. ssp. bracteata, F. virginiana, and F. chiloensis. Although some observed “translocations” (Figure 2, Table S3) may be genome assembly artifacts, our results suggest a dynamic ongoing restructuring of Fragaria genomes and support the possibility that the male-sterility locus could have translocated. A double translocation of the same gene would be notable but consistent with a high rate of translocation for sex determining regions (Charlesworth and Mank 2010) and polyploid genomes (Lim et al. 2008; Gaeta and Pires 2010). The second explanation is that the relatively unusual trait of dominant male sterility has arisen multiple times independently in Fragaria. This interpretation would suggest that there are many possible biochemical mechanisms to disrupt pollen development (see Diggle et al. 2011, and references in introduction) in a dominant fashion. Perhaps some of these mechanisms are more likely to persist for long time periods, or under certain ecological contexts, and thus conducive to the eventual evolution of subdioecy and dioecy (Spigler and Ashman 2012), while others are more short-lived and are lost with a reversion to total hermaphroditism (Dufay and Billard 2012). Repeated independent evolution of dominant male sterility in Fragaria but not other taxa would suggest that some unique aspect of Fragaria genetics, evolutionary history or ecology favors these mutations. The third possibility is that male sterility evolved only once in Fragaria, such that the biochemical mechanism is the same in all three species, but the genetic control of this trait has shifted over time to other, unlinked genes that regulate the original gene encoding male sterility. This rapid turnover of the sex determination region may occur in taxa that do not have conserved sex chromosomes (van Doorn and Kirkpatrick 2007).
The recombination rate across scB (11.8 cM/Mb) is substantially greater than the genome-wide average (1.0 cM/Mb). The proportion of coding sites in scB (25.9%) is also greater than the genome-wide average (19.5%), although the recombination rate at scB is high even among gene-dense regions (mean = 6.0 cM/Mb for regions with >25% coding sites). We observed three recombination events in the relatively short (22 kb) genomic distance between scB_1689672 and scB_1711631, suggestive of a local recombination hotspot (144 cM/Mb; Figure 5). It is possible that the male-sterility locus was more likely to evolve in a high-recombination region because that is where relatively more genes are located. Alternatively, these results may indicate that there are large cryptic insertion(s) in scB in F. v. spp. bracteata, possibly containing the candidate gene, which would be consistent with dynamic chromosomal restructuring near sex-determining regions (Charlesworth and Mank 2010).
Male sterility in our F. v. spp. bracteata cross segregates in a Mendelian fashion and maps to the nuclear genome and thus does not conform to described features of CMS. Furthermore, several lines of evidence point away from the sterility locus being an Rf locus. First, male sterility in F. v. ssp. bracteata is dominant, which is not typical for Rf loci. The gene presumably performs some function which is lost in the recessive allele. If the function degrades CMS mRNAs, and a single copy of an allele is sufficient to perform the function, then heterozygotes should be fertile, as seen in other species (Hanson and Bentolila 2004; Wang et al. 2006). Therefore, it is more plausible that the biochemical role of the male-sterility locus is to directly suppress, not restore, pollen development. Second, although F. v. ssp. bracteata may differ somewhat from the reference genome, there is no evidence of PPR genes or other members of known Rf gene families in the genomic interval that shows a perfect match to sex. The evidence is more consistent with non-Rf nuclear determination of gynodioecy in this population of F. v. ssp. bracteata as described by Ahmadi and Bringhurst (1989) and suggests that ecological studies of fitness advantage need to consider whole life time components of quantity or quality or biotic interactions (Li et al. 2012; Dalton et al. 2013). Given the potential spatiotemporal heterogeneity of sex determination (Pannell 1997; Olson et al. 2006), however, our results do not preclude the possible role of CMS (or cyto-nuclear interactions) in other populations of F. v. ssp. bracteata or even in the recent evolutionary history of this population.
To complete our understanding of the evolution of sex determination in Fragaria, several additional steps will be necessary. Further fine-mapping and functional analyses are needed to identify the causal genes in F. v. ssp. bracteata and its congeners. In addition, studies with additional individuals and with F. v. ssp. bracteata from other populations will help evaluate the species-wide importance of the locus mapped in this study and determine if other independent loci are also involved in the control of male function. Our success with target capture in this study suggests that it will be feasible to generate high-density maps in other Fragaria, including polyploids, for comparative studies. Inferring the molecular evolutionary history of the causal genes for male sterility may reveal how and when natural selection has acted to shape sexual variation in this genus.
Supplementary Material
Acknowledgments
We thank J.-M. Rouillard for assistance with target enrichment bait design; R. Cronn for target enrichment advise and use of laboratory space; M. Parks and K. Weitemier for laboratory assistance; S. Straub and M. Parks for help with sequencing of MRD parents; M. Dasenko for Illumina sequencing; C. Sullivan, S. Jogdeo, and M. Peterson for bioinformatics support; and K. Schuller, M. Koksi, and R. Dalton for help scoring and maintaining the map cross. This work was supported by the National Science Foundation to T.L.A. (DEB 1020523) and A.L. (DEB 1020271).
Footnotes
Communicating editor: J. D. Faris
Literature Cited
- Ahmadi H., Bringhurst R. S., 1989. Genetics of sex expression in Fragaria species. Am. J. Bot. 78: 504–514 [Google Scholar]
- Alves-Ferreira M., Wellmer F., Banhara A., Kumar V., Riechmann J. L., et al. , 2007. Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. L., Rodríguez Marí A., Braasch I., Amores A., Hohenlohe P., et al. , 2012. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS ONE 7: e40701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashman T.-L., 2002. The role of herbivores in the evolution of separate sexes from hermaphroditism. Ecology 83: 1175–1184 [Google Scholar]
- Backues S. K., Korasick D. A., Heese A., Bednarek S. Y., 2010. The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22: 3218–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. F., Delph L. F., 2007. A field guide to models of sex-ratio evolution in gynodioecious species. Oikos 116: 1609–1617 [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K., Vanderpoo D., Singhal S., Linderoth T., Moritz C., et al. , 2012. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics 13: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M., Brownfield L., Twell D., 2009. Male gametophyte development: a molecular perspective. J. Exp. Bot. 60: 1465–1478 [DOI] [PubMed] [Google Scholar]
- Bradley K. M., Breyer J. P., Melville D. B., Broman K. W., Knapik E. W., et al. , 2011. An SNP-based linkage map for zebrafish reveals sex determination loci. G3 (Bethesda) 1: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Somers D. J., Fedak G., 2009. A molecular marker closely linked to the region of Rht-D1c and Ms2 genes in common wheat (Triticum aestivum). Genome 52: 95–99 [DOI] [PubMed] [Google Scholar]
- Caruso C. M., Case A. L., Bailey M. F., 2012. The evolutionary ecology of cytonuclear interactions in angiosperms. Trends Plant Sci. 17: 638–643 [DOI] [PubMed] [Google Scholar]
- Charnov E. L., Maynard Smith J., Bull J. J., 1976. Why be a hermaphrodite? Nature 263: 125–126 [Google Scholar]
- Charlesworth D., 2013. Plant sex chromosome evolution. J. Exp. Bot. 64: 405–420 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997 [Google Scholar]
- Charlesworth D., Mank J. E., 2010. The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics 186: 9–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase C. D., 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 23: 81–90 [DOI] [PubMed] [Google Scholar]
- Connors C. H., 1926. Sterility in peaches. Mem. Hort. Soc. N. Y. 313: 215–221 [Google Scholar]
- Crane M. B., Lawrence W. J. C., 1931. Inheritance of sex, colour and hairiness in the raspberry, Rubus idaeus L. J. Genet. 24: 243–255 [Google Scholar]
- Cronn R., Liston A., Parks M., Gernandt D. S., Shen R., et al. , 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R., Knaus B. J., Liston A., Maughan P. J., Parks M., et al. , 2012. Targeted enrichment strategies for next-generation plant biology. Am. J. Bot. 99: 291–311 [DOI] [PubMed] [Google Scholar]
- Cui X., Wang Q., Yin W., Xu H., Wilson Z. A., et al. , 2013. PMRD: a curated database for genes and mutants involved in plant male reproduction. BMC Plant Biol. 12: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton R., Koski M., Ashman T.-L., 2013. Maternal sex and inbreeding effects under varied environmental conditions in gynodioecious Fragaria vesca subsp. bracteata. Ann. Bot. DOI:10.1093/aob/mct120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C., 1877. The Different Forms of Flowers on Plants of the Same Species. The University of Chicago Press, Chicago [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph L. F., Carroll S. B., 2001. Factors affecting relative seed fitness and female frequency in a gynodioecious species, Silene acaulis. Evol. Ecol. Res. 3: 487–505 [Google Scholar]
- Del Viso F., Bhattacharya D., Kong Y., Gilchrist M. J., Khokha M. K., 2012. Exon capture and bulk segregant analysis: rapid discovery of causative mutations using high-throughput sequencing. BMC Genomics 13: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P. K., Di Stilio V. S., Gschwend A. R., Golenberg E. M., Moore R. C., et al. , 2011. Multiple developmental processes underlie sex differentiation in angiosperms. Trends Genet. 27: 368–376 [DOI] [PubMed] [Google Scholar]
- Dorsey M. J., 1919. A study of sterility in the plum. Genetics 4: 417–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15 [Google Scholar]
- Dudle D. A., Mutikainen P., Delph L. F., 2001. Genetics of sex determination in the gynodioecious species Lobelia siphilitica: evidence from two populations. Heredity 86: 265–276 [DOI] [PubMed] [Google Scholar]
- Dufay M., Billard E., 2012. How much better are females? The occurrence of female advantage, its proximal causes and its variation within and among gynodioecious species. Ann. Bot. (Lond.) 109: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbarry A., Vizir I., Twell D., 2005. Male germ line development in Arabidopsis: duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 137: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. B., Oliphant A., Shen R., Kermani B. G., Garcia F., et al. , 2003. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 68: 69–78 [DOI] [PubMed] [Google Scholar]
- Felcher K. J., Coombs J. J., Massa A. N., Hansey C. N., Hamilton J. P., et al. , 2012. Integration of two diploid potato linkage maps with the potato genome sequence. PLoS ONE 7: e36347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Toriyama K., 2009. Suppressed expression of Retrograde-Regulated Male Sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc. Natl. Acad. Sci. USA 106: 9513–9518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., 2010. Homoeologous recombination in allopolyploids: the polyploid ratchet. New Phytol. 186: 18–28 [DOI] [PubMed] [Google Scholar]
- Garraud C., Brachi B., Dufay M., Touzet P., Shykoff J. A., 2011. Genetic determination of male sterility in gynodioecious Silene nutans. Heredity 106: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. T., Spigler R. B., Ashman T.-L., 2010. Comparative genetic mapping points to different sex chromosomes in sibling species of wild strawberry (Fragaria). Genetics 186: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu R., Liston A., Ashman T.-L., 2013. Sex-determining chromosomes and sexual dimorphism: insights from genetic mapping of sex expression in a natural hybrid Fragaria × ananassa subsp. cuneifolia. Heredity 110: 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusti A., Baumberger N., Nowack M., Pusch S., Eisler H., et al. , 2009. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 4: e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. R., Bentolila S., 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16: S154–S169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann M., Beye M., 2006. Pronounced differences of recombination activity at the sex determination locus of the honeybee, a locus under strong balancing selection. Genetics 174: 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordano P., 1993. Pollination biology of Prunus mahaleb L. – deferred consequences of gender variation for fecundity and seed size. Biol. J. Linn. Soc. Lond. 50: 65–84 [Google Scholar]
- Kang B. H., Rancour D. M., Bednarek S. Y., 2003. The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J. 35: 1–15 [DOI] [PubMed] [Google Scholar]
- Kearns C. A., Inouye D. W., 1993. Techniques for Pollination Biologists. University Press of Colorado, Niwot, CO [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohls S., Stamp P., Knaak C., Messmer R., 2011. QTL involved in the partial restoration of male fertility of C-type cytoplasmic male sterility in maize. Theor. Appl. Genet. 123: 327–338 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D., 1941. Male-sterility in natural populations of hermaphrodite plants. New Phytol. 40: 56–63 [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hsia A. P., Schnable P. S., 2007. Recent advances in plant recombination. Curr. Opin. Plant Biol. 10: 131–135 [DOI] [PubMed] [Google Scholar]
- Li J., Koski M. H., Ashman T.-L., 2012. Functional characterization of gynodioecy in Fragaria vesca ssp. bracteata (Rosaceae). Ann. Bot. (Lond.) 109: 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. Y., Soltis D. E., Soltis P. S., Tate J., Matyasek R., et al. , 2008. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS ONE 3: e3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Liu J., Xin Q., Wan L., Hong D., et al. , 2013. A triallelic genetic male sterility locus in Brassica napus: an integrative strategy for its physical mapping and possible local chromosome evolution around it. Ann. Bot. (Lond.) 111: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., 2005. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Margarido G. R. A., Souza A. P., Garcia A. A. F., 2007. OneMap: software for genetic mapping in outcrossing species. Hereditas 144: 78–79 [DOI] [PubMed] [Google Scholar]
- Matsuhira H., Kagami H., Kurata M., Kitazaki K., Matsunaga M., et al. , 2012. Unusual and typical features of a novel restorer-of-fertility gene of sugar beet (Beta vulgaris L.). Genetics 192: 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley D. E., Bailey M. F., 2009. Recent advances in the study of gynodioecy: the interface of theory and empiricism. Ann. Bot. (Lond.) 104: 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meznar E. R., Gadau J., Koeniger N., Rueppell O., 2010. Comparative linkage mapping suggests a high recombination rate in all honeybees. J. Hered. 101(Suppl 1): S118–S126 [DOI] [PubMed] [Google Scholar]
- Nadeau N. J., Whibley A., Jones R. T., Davey J. W., Dasmahapatra K. K., et al. , 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K., Ellegren H., 2012. Recombination drives vertebrate genome contraction. PLoS Genet. 8: e1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njuguna W., Liston A., Cronn R., Ashman T.-L., Bassil N., 2013. Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol. Phylogenet. Evol. 66: 17–29 [DOI] [PubMed] [Google Scholar]
- Olson M. S., Graf A. V., Niles K. R., 2006. Fine scale spatial structuring of sex and mitochondria in Silene vulgaris. J. Evol. Biol. 19: 1190–1201 [DOI] [PubMed] [Google Scholar]
- Pannell J., 1997. The maintenance of gynodioecy and androdioecy in a metapopulation. Evolution 51: 10–20 [DOI] [PubMed] [Google Scholar]
- Salmon A., Udall J. A., Jeddeloh J. A., Wendel J. F., 2012. Targeted capture of homoeologous coding and noncoding sequence in polyploid cotton. G3 (Bethesda) 2: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent D. J., Kuchta P., Lopez-Girona E., Zhang H., Davis T. M., et al. , 2011. Simple sequence repeat marker development and mapping targeted to previously unmapped regions of the strawberry genome sequence. Plant Genome 4: 165–177 [Google Scholar]
- Sargent D. J., Passey T., Surbanovski N., Lopez Girona E., Kuchta P., et al. , 2012. A microsatellite linkage map for the cultivated strawberry (Fragaria × ananassa) suggests extensive regions of homozygosity in the genome that may have resulted from breeding and selection. Theor. Appl. Genet. 124: 1229–1240 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., et al. , 2005. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Wise R. P., 1998. The molecular basis of cytoplasmic male sterility. Trends Plant Sci. 3: 175–180 [Google Scholar]
- Schnable P. S., Hsia A. P., Nikolau B. J., 1998. Genetic recombination in plants. Curr. Opin. Plant Biol. 1: 123–129 [DOI] [PubMed] [Google Scholar]
- Scott D. H., Weinberger J. H., 1938. Inheritance of pollen sterility in some peach varieties. Proc. Am. Soc. Hort. Sci. 45: 229–232 [Google Scholar]
- Shulaev V., Sargent D. J., Crowhurst R. N., Mockler T. C., Folkerts O., et al. , 2011. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 1996–2010 RepeatMasker Open-3.0. Available at: http://www.repeatmasker.org Accessed: June 19, 2013.
- Spigler R. B., Lewers K. S., Ashman T.-L., 2011. Genetic architecture of sexual dimorphism in a subdioecious plant with a proto-sex chromosome. Evolution 65: 1114–1126 [DOI] [PubMed] [Google Scholar]
- Spigler R. B., Ashman T.-L., 2012. Gynodioecy to dioecy: are we there yet? Ann. Bot. (Lond.) 109: 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt G., 1989. The species of Fragaria, their taxonomy and geographical distribution. Acta Hortic. 265: 23–33 [Google Scholar]
- Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., et al. , 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzet P., 2012. Mitochondrial genome evolution and gynodioecy, pp. 71–98 in Advances in Botanical Research, Vol. 63, edited by Marechal-Drouard L. Academic Press Ltd-Elsevier Science, London [Google Scholar]
- Valleau W. D., 1923. The inheritance of flower types and fertility in the strawberry. Am. J. Bot. 10: 259–274 [Google Scholar]
- van Doorn G. S., Kirkpatrick M., 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912 [DOI] [PubMed] [Google Scholar]
- Wang X. L., Li X. B., 2009. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. Plant J. 57: 473–486 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zou Y., Li X., Zhang Q., Chen L., et al. , 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W. Z., Song L. F., Zou J. J., Su Z., et al. , 2008. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Ruesch K. L., Ortega V. M., Kamps T. L., Gabay-Laughnan S., et al. , 2003. A nuclear restorer-of-fertility mutation disrupts accumulation of mitochondrial ATP synthase subunit alpha in developing pollen of S male-sterile maize. Genetics 165: 771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Z. A., Zhang D. B., 2009. From Arabidopsis to rice: pathways in pollen development. J. Exp. Bot. 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Yang K. Z., Xia C., Liu X. L., Dou X. Y., Wang W., et al. , 2009. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 57: 870–882 [DOI] [PubMed] [Google Scholar]
- Ye Q., Zhu W., Li L., Zhang S., Yin Y., et al. , 2010. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 107: 6100–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.