Abstract
Intractable epilepsies, that is, seizure disorders that do not respond to currently available therapies, are difficult, often tragic, neurological disorders. Na+ channelopathies have been implicated in some intractable epilepsies, including Dravet syndrome (Dravet 1978), but little progress has been forthcoming in therapeutics. Here we examine a Drosophila model for intractable epilepsy, the Na+ channel gain-of-function mutant parabss1 that resembles Dravet syndrome in some aspects (parker et al. 2011a). In particular, we identify second-site mutations that interact with parabss1, seizure enhancers, and seizure suppressors. We describe one seizure-enhancer mutation named charlatan (chn). The chn gene normally encodes an Neuron-Restrictive Silencer Factor/RE1-Silencing Transcription factor transcriptional repressor of neuronal-specific genes. We identify a second-site seizure-suppressor mutation, gilgamesh (gish), that reduces the severity of several seizure-like phenotypes of parabss1/+ heterozygotes. The gish gene normally encodes the Drosophila ortholog of casein kinase CK1g3, a member of the CK1 family of serine-threonine kinases. We suggest that CK1g3 is an unexpected but promising new target for seizure therapeutics.
Keywords: sodium channel, epilepsy, seizure-suppression, Drosophila
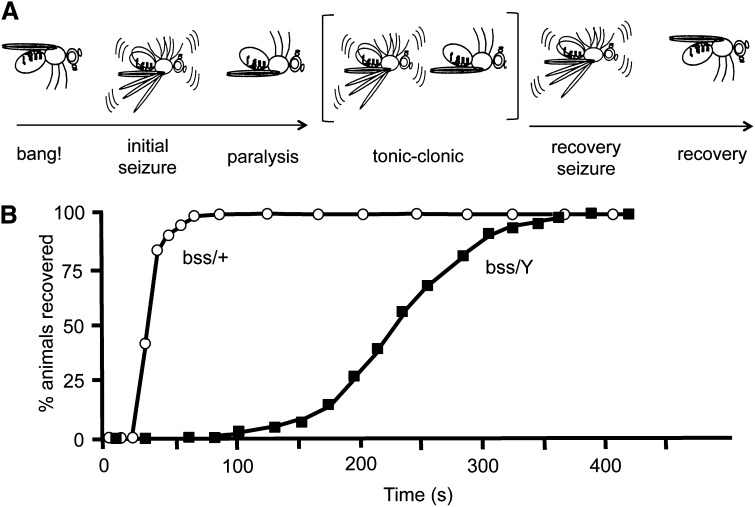
In this study, we examine genetic complexities that underlie seizure-susceptibility by using, as a model, genetic combinations of single-gene mutations in the fruit fly Drosophila: seizure-sensitive, seizure-enhancer, and seizure-suppressor mutations. The study is based on genetic interactions that modify phenotypes in parabss1, a model for intractable epilepsy (parker et al. 2011a). The parabss1 mutant is caused by a gain-of-function mutation in the voltage-gated Na+ channel gene that causes extreme seizure sensitivity. In our Drosophila collection, the parabss1 mutant: (1) displays the lowest threshold to evoked seizure-like activity; (2) exhibits the longest paralytic behavior recovery time with prominent episodes of seizure and paralysis that resemble tonic-clonic-like activity; and (3) is the most difficult mutant to suppress by suppressor mutations or antiepileptic drugs (Pavlidis and Tanouye 1995; Kuebler and Tanouye 2000; Kuebler et al. 2001; Song and Tanouye 2006).
We describe here the results of a search for new enhancers and suppressors of parabss1. Because of the potential biomedical usefulness of some of these observations to intractable epilepsies, we are somewhat more deliberate in our descriptions than is usually customary in Drosophila mutant screens. We further describe identification of charlatan (chn), an enhancer of parabss1, and a parabss1 suppressor named gilgamesh (gish).
Materials and Methods
Fly stocks
Drosophila strains were raised on standard cornmeal-molasses agar medium at room temperature (23−25°). The para gene is located at map position 1−53.5 and encodes a voltage-gated Na+ channel (Loughney et al. 1989; Ramaswami and Tanouye, 1989). The bang-sensitive (BS) allele used in this study, parabss1, previously named bss1, is the most seizure-sensitive of fly mutants, the most difficult to suppress by mutation and by drug, and is a model for human intractable epilepsy (Ganetzky and Wu 1982; Parker et al. 2011a). The parabss1 allele is a gain-of-function mutation caused by a substitution (L1699F) of a highly conserved residue in the third membrane-spanning segment (S3b) of homology domain IV (Parker et al. 2011a). In this study, we use parabss1 and parabss1/+ as genetic backgrounds to screen for enhancers and suppressors of seizure, respectively. The eas gene is located at 14B on the cytological map and encodes an ethanolamine kinase (Pavlidis et al. 1994). The BS allele used in this study is easPC80, which is caused by a 2-bp deletion that introduces a frame shift; the resulting truncated protein lacks a kinase domain and abolishes all enzymatic activity (Pavlidis et al. 1994). Df(2R)Exel7135=51E2-51E11 contains approximately 22 genes. Df(2R)Exel6056=44A4-44C2 contains approximately 39 genes. Df(2R)Exel6078=58B1-58D1 contains approximately 35 genes. UAS-gishRNAi and other UAS-RNAi lines were obtained from the Vienna Drosophila RNAi Center. All other lines, including Gal4 drivers and deletion lines, were obtained from the Bloomington Drosophila Stock Center.
Haplo-deficiency screen for seizure enhancers and suppressors
A screen was designed to detect novel seizure suppressors and enhancers based on haplo-induced changes in parabss1 seizure susceptibility. Using the screen, we examined 200 stocks, each carrying a different Df(2) or Df(3) chromosomal deletion with appropriate CyO, TM3, or TM6 balancer in a parabss1 background. Seizure susceptibility can vary substantially with age, genetic background, and other factors; all comparisons were between age-matched siblings arising from the same cross to minimize variations due to these sources. For Df(2) deletions: female parabss1;+;+ flies were crossed to +/Y;Df(2)/CYO;+ males. Male progeny of the genotype: parabss1/Y;Df(2)/+;+ were tested for enhancement of BS phenotype compared with their sibling controls (parabss1/Y;CYO/+;+). Female progeny arising from the same cross, parabss1/+;Df(2)/+;+, were tested for suppression of the BS phenotype compared with their control siblings (parabss1/+;CYO/+;+). Df(3) deletions were tested similarly. Thus, parabss1/Y;+;Df(3)/+ male flies were examined for enhancement, and parabss1/+;+;Df(3)/+ flies were tested for suppression of BS phenotypes relative to their respective control siblings.
Behavior and electrophysiology
Behavioral testing for BS paralysis was performed on flies 2−3 d after eclosion, as described previously (Kuebler and Tanouye 2000). Flies were anesthetized with CO2 before collection and tested the following day. For testing, 15−20 flies were placed in a food vial and stimulated mechanically with a VWR vortex mixer at maximum speed for 10 sec. For analysis, recovery time was measured for each fly from the end of the vortex stimulation until it resumed an upright standing position. Mean recovery time (MRT) was the average time taken for a fly exhibiting BS behavior to recover in a population. Pools of flies are combined (in total, n ≈ 100 for each genotype). For the purposes of comparisons, these are expressed here as normalized mean recovery time (nMRT), which is the MRT of the experimental flies divided by MRT of their control siblings. For genotypes that display only partial penetrance of BS paralysis, only those flies that displayed paralysis were used for recovery time analysis. A simpler measure of recovery time is RT50 (50% recovery time), the time at which half of BS flies have recovered from paralysis. RT50 was used in some analyses and especially to facilitate initial identification of enhancers and suppressors.
In vivo recording of seizure-like neuronal activity and seizure threshold determination in adult flies was performed as described previously (Kuebler and Tanouye 2000; Lee and Wu 2002). Flies 2−3 d posteclosion were mounted in wax on a glass slide, leaving the dorsal head, thorax, and abdomen exposed. Stimulating, recording, and ground metal electrodes were made of uninsulated tungsten. Seizure-like activity was evoked by high-frequency electrical brain stimulation (0.5-ms pulses at 300 Hz for 400 ms) and monitored by dorsal longitudinal muscle recording. During the course of each experiment, the giant fiber circuit was monitored continuously as a proxy for holobrain function. For each genotype tested, n ≥ 10, and unless otherwise noted, all flies were female. Comparisons of paralytic recovery time and seizure threshold were Student t-test. For all figures, error bars represent SEM, and statistical significance is indicated by *P < 0.01 and **P < 0.0001.
Results
Screening for parabss1 enhancers with deficiencies
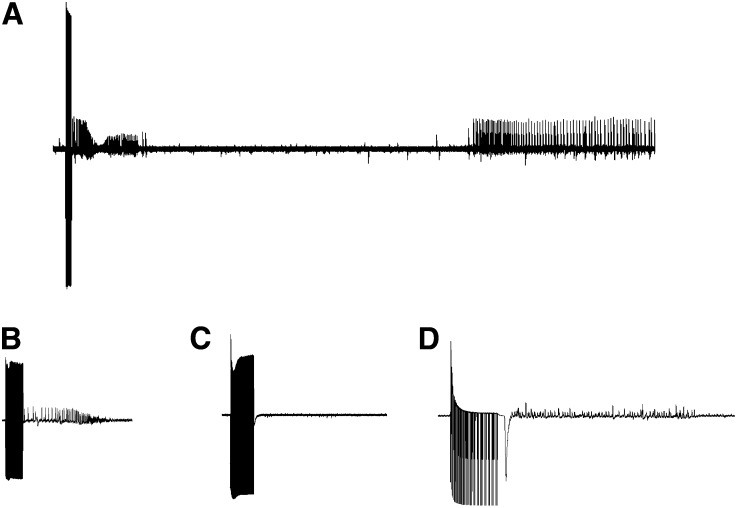
The parabss1 mutant displays phenotypes that are similar to other mutants of the BS paralytic class such as easPC80, sdaiso7.8, and tko25t (Ganetzky and Wu 1982; Royden et al. 1987; Pavlidis et al. 1994; Zhang et al. 2002), albeit more severe. BS seizure-like behaviors and paralysis are observed in response to mechanical shock (“a bang”) (Figure 1). The time of BS paralysis for parabss1 is much longer than for other mutants and exhibits unusual tonic-clonic-like behaviors. For example, total paralytic time for parabss1 is about 240 sec, longer than for sdaiso7.8, which is about 25 sec (Zhang et al. 2002; Parker et al. 2011a). The parabss1 mutant also has a low threshold for seizure-like activity evoked by high-frequency electrical stimulation (HFS) of the brain. For example, seizure threshold for parabss1 is 3.2 ± 0.6 V HFS, lower than the threshold for sdaiso7.8, which is 6.2 ± 0.8 V HFS; wild-type Canton-Special flies have a seizure threshold of 30.1 ± 3.8 V HFS, for comparison (Figure 2) (Kuebler et al. 2001).
Figure 1.
Behavior phenotypes for parabss1 mutants. (A) Illustration depicting stereotype behavioral phenotype of parabss1 flies subjected to a mechanical shock (10-sec vortex: “bang!”): initial seizure-like behavior, followed by complete paralysis and then a tonic/clonic period that is unique to parabss1 and not evident in other BS mutant genotypes. One clonus-like event is depicted, but the number can vary, as can the duration of the period. The tonic/clonic-like period is followed by a recovery seizure, and the fly then recovers. Not depicted is a quiescent period of variable duration often observed between the recovery seizure and recovery, as well as the refractory period during which flies are resistant to further seizures that occurs immediately following recovery. (B) Recovery times from behavioral paralysis for parabss1/Y hemizygous males (labeled “bss/Y”) is substantially longer than for parabss1/+ heterozygous females (labeled “bss/+”). For the enhancer screen described in the text, heterozygous deletions were selected that prolonged the parabss1/Y recovery time compared to sibling controls. For the suppressor screen described in the text, heterozygous deletions were selected that reduced the percentage of parabss1/+ females paralyzed by the mechanical shock compared to sibling controls. (Figure adapted from Parker et al. 2011a).
Figure 2.
Electrophysiology phenotype of parabss1 mutants. Seizure-like electrical activity in parabss1 and wild-type flies. The mutant fly is more susceptible to seizures and has a lower threshold. (A) Seizure-like activity displayed at a slow sweep speed showing initial seizure, period of synaptic failure, and recovery seizure. (B) Seizure-like activity is evoked by 4-V HFS stimulus and displayed at high sweep speed. The mutant is susceptible to low-voltage evoked seizures indicating extreme seizure-sensitivity. (C) A low-voltage 4 V HFS stimulus delivered to a wild-type fly is ineffective at eliciting seizure-like activity because it is below the seizure threshold. (D) A greater voltage 30-V HFS stimulus delivered to a wild-type fly elicits seizure-like activity because it is above threshold for seizure initiation.
Despite the existing severity of parabss1 phenotypes, we explored the possibility that these might be exacerbated further by enhancer mutations. We have previously found that recovery time from BS paralysis for parabss1 varies with genetic background, age, and other factors (parker et al. 2011a). The length of time required for recovery appears to be primarily dependent on the number of bouts of tonic-clonic-like activity. We exploited this in an initial screen, investigating the possibility that potential enhancers may reside in chromosomal segments made haploid by deletions, and these would become manifest by a change in the time required to recover from BS paralysis. We then examined enhancers for effects on other parabss1 phenotypes. We measured BS paralytic recovery times in parabss1/Y; Df/+ flies compared with their control siblings of genotype parabss1/Y; Balancer/+ (Table 1, File S1). Several deficiency chromosomes consistently showed increased recovery times for parabss1 males (Table 1). For example, Df(2R)Exel7135 had a MRT of 363 s for experimental males, compared with 234 sec for their sibling controls yielding an nMRT of 1.55. Other notable deficiencies included: Df(2R)Exel6078 and Df(2R)Exel6056 with nMRTs of 2.27 and 2.53, respectively. Here we focus on Df(2R)Exel7135 as representative of our findings on parabss1 enhancers.
Table 1. Chromosomal deletions that enhance the behavioral bang-sensitive (BS) paralytic phenotype of parabss1/+ flies.
| Deficiency | Experimental (Df) MRT (s) | Control (Balancer) MRT (s) | nMRT |
|---|---|---|---|
| Df(2R)Exel7135 | 363 | 234 | 1.55 |
| Df(2R)Exel6078 | 306 | 135 | 2.27 |
| Df(2R)Exel7094 | 232 | 102 | 2.27 |
| Df(2R)Exel6071 | 217 | 118 | 1.84 |
| Df(2R)Exel6056 | 215 | 85 | 2.53 |
Values of the length of time that hemizygous parabss1/Y males remained paralyzed are depicted as MRT. To minimize the effects of genetic background, experimental males of the general genotype: parabss1/Y;Df/+ were compared directly with sibling control brothers arising from the same cross (genotype: parabss1/Y;Balancer/+). The ratio of MRT for experimental males with that of their control siblings is listed as nMRT. MRT, mean recovery time; nMRT, normalized mean recovery time.
Reduced expression of charlatan (chn) contained in the Df(2R)Exel7135 chromosomal segment enhances parabss1 BS paralysis but not seizure threshold
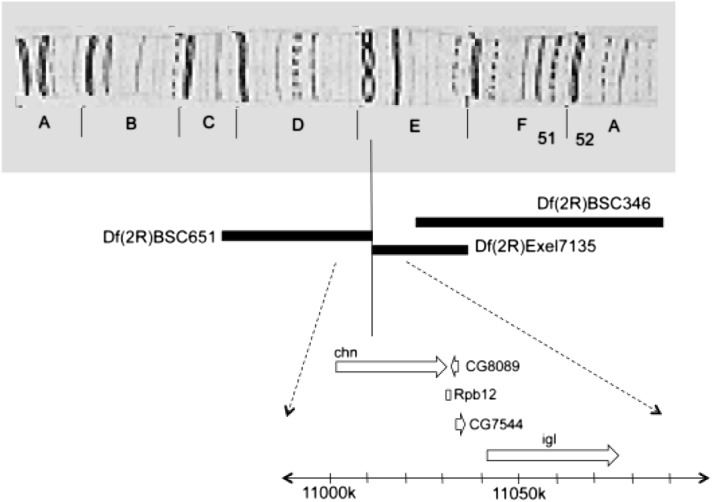
The Df(2R)Exel7135 deficiency is a deletion spanning from 51E2 to 51E11 on chromosome 2R and contains approximately 22 genes. Deletion analysis further limited this segment to 51E2 to 51E7 on the basis of observations that the parabss1/Y recovery time is not enhanced by the heterozygous Df(2R)BSC346/+ (51E7-52C2) but is enhanced by Df(2R)BSC651/+ (51C5-51E2) (Figure 3, File S4). We found that BS enhancement in the segment is accounted for by reduced expression of the charlatan (chn) gene. The gene is broken by the 51E2 breakpoints of Df(2R)Exel7135 and Df(2R)BSC651 and is the only apparent gene affected by both rearrangements. Further identification of chn as an enhancer of parabss1/Y is by UAS-chnRNAi. Flies of the genotype ELAV-Gal4C155 parabss1/Y;UAS-chnRNAi/+ show increased BS recovery times with an MRT of 261.9 ± 17.1 sec compared with 105.6 ± 9.4 sec for their ELAV-Gal4 parabss1/Y;+/+ sibling controls for an nMRT of 2.48 (P < 0.001) (File S4).
Figure 3.
Chromosomal segment deleted in Df(2R)Exel7135. The upper panel of the figure depicts region 51 of the polytene chromosome. The chn gene is disrupted by the distal breakpoint of Df(2R)BSC651 and the proximal breakpoint of Df(2R)Exel7135; both rearrangements enhance BS paralytic recovery time in parabss1 hemizygotes. The BS paralytic recovery time phenotype is not enhanced by the Df(2R)BSC346.
The chn gene encodes an NRSF/REST transcriptional repressor of neuronal-specific genes (Escudero et al. 2005; Tsuda et al. 2006; Yamasaki et al. 2011). It has not been previously identified in seizure susceptibility or electrical excitability. Surprisingly, the enhancement of parabss1 by chn was limited to BS paralysis recovery time phenotype, that is, an increase in the severity of this phenotype; there was no apparent enhancement of the other major phenotype: threshold for evoked seizure. For example, flies of the genotype ELAV-Gal4C155 parabss1/Y;UAS-chnRNAi/+ have a seizure threshold of 3.32 ± 0.47 V HFS, similar to the threshold of 3.87 ± 0.53 V HFS (P = 0.46) for their sibling controls (File S3). Flies of the genotypes ELAV-GAL4C155/Y;UAS-chnRNAi/+ and Df(2R)Exel7135/Cyo exhibited no bang sensitivity, indicating that chn enhances seizure severity without being a bang-sensitive mutant itself (File S4). These findings are consistent with the results of Df(2R)Exel7135 and all of the other enhancers identified in this screen: the enhancers increased BS paralysis time to recovery but did not reduce seizure threshold in electrophysiology tests.
Screening for parabss1 suppressors with deficiencies
The parabss1 mutant is severely seizure sensitive: phenotypes are difficult to suppress by antiepileptic drug feeding and Drosophila seizure-suppressor mutations thus far identified have been ineffective at alleviating parabss1 phenotypes. The parabss1 mutation is semidominant with seizure-like behaviors, and BS paralysis reduced in heterozygous parabss1/+ flies, but still present at high penetrance (>95%) (Figure 1) (Ganetzky and Wu 1982; Parker et al. 2011a). We exploited this feature to screen for suppressor mutations inferring that heterozygotes would provide a genetic background that is sensitized for detecting putative suppressors. As an initial screen, we investigated the possibility that potential suppressors may reside in chromosomal segments made haploid by deletions and that these would become manifest by a change in BS paralysis. That is, we compared parabss1/+; Df/+ females with their control sisters of genotype parabss1/+; Balancer/+ for differences in the percentage of flies undergoing BS paralysis. Several deletion chromosomes consistently reduced the BS phenotype in parabss1/+ females (Table 2, File S1). For example, only 13% of parabss1/+; Df(3R)ED10639/+ females showed BS paralysis compared with their sibling controls, an apparent phenotypic suppression of approximately 87%. Other notable deletions included Df(2R)Exel6285 and Df(3L)ED4502 that caused 97% and 93% suppression, respectively. Here, we focus on Df(3R)ED10639 as representative of our findings on parabss1 suppressors.
Table 2. Chromosomal deletions that revert the behavioral bang-sensitive (BS) paralytic phenotype of parabss1/+ flies.
| Deficiency | BS |
|---|---|
| Wild type | 0.00 |
| Df(2R)Exel6285 | 0.03 |
| Df(3L)ED4502 | 0.07 |
| Df(3R)ED10639 | 0.13 |
| Df(3L)ED224 | 0.19 |
| Df(3L)ED201 | 0.29 |
| Df(3L)ED4502 | 0.42 |
| Df(2R)BSC427 | 0.49 |
| Df(3R)ED5518 | 0.50 |
| Df(3L)ED4486 | 0.50 |
| parabss1/+ | 0.95 |
Ordinarily, approximately 95% of parabss1/+ flies show a BS paralytic phenotype: paralysis aftermechanical stimulation. Wild-type flies never show BS paralysis. The number of flies showing BS paralysis is greatly reduced by the deficiency chromosomes listed in the table. Flies tested carried the heterozygous deficiency and were of the general genotype: parabss1/+; Df/+. In all cases, to control for genetic background, experimental flies were compared directly with sibling control flies arising from the same cross (genotype: parabss1/+; Balancer/+).
Reduced expression of gilgamesh (gish) contained in the Df(3R)ED10639 chromosomal segment suppresses parabss1/+ BS paralysis
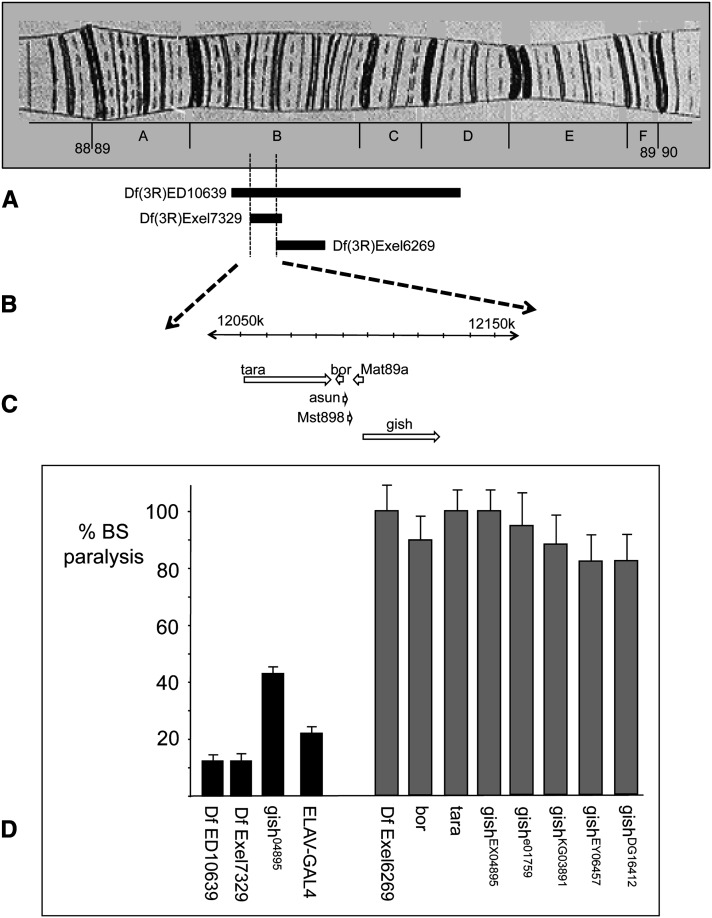
The Df(3R)ED10639 deficiency is a deletion spanning from 89B7 to 89D5 and contains approximately 57 genes. In this section, we describe analyses showing that parabss1/+ suppression in the segment is accounted for by reduced expression of the gilgamesh (gish) gene (Figure 4). parabss1/+ BS suppression phenotype was mapped to a small region on chromosome 3R between 89B9 and 89B12 using overlapping deficiencies. In particular, localization of the suppression phenotype is based on its inclusion in the Df(3R)Exel7329 deletion, which affects the number of animals paralyzed (Figure 4) (89B9-89B13), and its exclusion from the Df(3R)Exel6269 deletion which has no effect on paralysis (Figure 4) (89B12-B18). This localization is consistent with the combined findings from other overlapping deletions in the region (Figure 4).
Figure 4.
Suppression of parabss1/+ BS paralytic phenotype by a heterozygous chromosomal segment deleted in 89B. (A) Depicted is polytene chromosome map of region 89 on 3R. (B) The segment deleted in Df(3R)ED10639 causes suppression of parabss1/+ BS paralysis, as described in the text. Also, Df(3R)Exel7329 causes suppression but Df(3R)Exel6269 does not. The breakpoints of these rearrangements delimit a small region (89B9 to 89B12) responsible for seizure suppression. (C) Six genes are contained in the 89B9 to 89B12 chromosomal segment including tara, bor, and gish. (D) BS paralytic phenotypes (% BS paralysis) of several genotypes in a parabss1/+ background, as described in the text. Genotypes showing BS suppression are depicted as black bars; gray bars are used in genotypes showing no suppression. In each case, the experimental genotype shown is normalized relative to sibling controls. Df ED10639 is the genotype parabss1/+; Df(3R) ED10639/+ showing 13% BS paralysis (87% suppression of BS phenotype). This indicates the apparent presence of a gene that acts as a haplo-seizure suppressor. Df Exel7329 is parabss1/+;Df(3R)Exel7329/+ showing 13% BS paralysis and providing one boundary for suppressor location at 89B9 based on inclusion within the deleted segment. Df Exel6269 is parabss1/+;Df(3R)Exel6269/+ showing 100% BS paralysis and providing a second boundary for suppressor location at 89B12 based its exclusion from the deletion. Flies that are parabss1/+;borc05496/+ and parabss1/+;tara1/+ (labeled bor and tara) show no suppression with 91% and 100% BS paralysis, respectively. Flies that are parabss1/+;gish04895/+ (labeled gish04895) show 43% BS paralysis, indicating suppression of the BS paralytic phenotype. Flies that are parabss1/+;gishEX04895/+ (labeled gishEX04895) are a line with a remobilized, precise excision of the gishEX04895 P-element; they show no suppression with 98% BS paralysis. Flies that are ELAV-Gal4C155 parabss1/+; UAS-gishRNAi/+ (labeled ELAV-GAL4) show 25% BS paralysis indicating suppression of the BS paralytic phenotype. Several gish alleles as heterozygotes show no suppression of parabss1/+ BS paralytic phenotypes. Thus, gishe01759/+, gishDG16412/+, gishKG03891/+, gishEY06457/+ heterozygous combinations in a parabss1/+ background show 95%, 88%, 84%, and 83% BS paralysis, respectively.
The 89B9-89B12 segment contains six genes (Figure 4). We found that an allele of belphegor (bor), parabss1/+;borc05496/+, which showed similar BS paralysis compared with control siblings (9% reduction in BS paralysis), did not appear to cause suppression based on flies of the genotype: Also, an allele of taranis (tara) did not appear to cause suppression based on flies of the genotype parabss1/+;tara1/+, with BS paralysis similar to their sibling controls (0% reduction in BS paralysis). In contrast, an allele of gilgamesh (gish) caused substantial suppression based on flies of the genotype parabss1/+;gish04895/+, which showed a 57% reduction in BS paralysis compared with their parabss1/+;TM3/+ control siblings (File S4).
The gish gene
The gish gene of Drosophila is homologous to mammalian casein kinase CK1g3, both members of the CK1 family of serine-threonine kinases (Zhai et al. 1995). The Drosophila gene is approximately 30 kb and alternatively spliced to express 12 different isoforms in four main classes (Hummel et al. 2002; Tan et al. 2010). These arise from two initiation sites: two classes of long transcript (~3 kb) arise from an upstream initiation site; two classes of short transcript (~2.5 kb) from a downstream initiation site (Hummel et al. 2002; Tan et al. 2010). The gish04895 mutation is a P-element insertion in exon 2, present in long, but not short gish transcripts. Reverse-transcription polymerase chain reaction analysis (Tan et al. 2010) has shown that long gish transcripts are apparently undetectable in gish04895 mutants. Interestingly, in contrast, short transcripts appear to be more abundant in gish04895 mutant than in wild-type flies (Tan et al. 2010). In the present experiments, gish04895 acts as a recessive lethal, in contrast to previous reports, suggesting that it is a viable (Tan et al. 2010). We are unclear on the reasons for this apparent difference in viability. We find that precise excision of the gish04895 P-element completely reverted the BS suppressor phenotype (Figure 4, File S2, File S4), restored viability, but did not appear to revert the male sterility phenotype seen among gish mutant alleles (Castrillon et al. 1993).
Identification of gish as a parabss1/+ BS suppressor by mutant analysis was supported further by RNAi analysis. Flies of the genotype ELAV-Gal4C155 parabss1/+;UAS-gishRNAi/+ showed a 75% reduction in BS paralysis compared with their ELAV-Gal4C155 parabss1/+;+/+ control siblings, showing that BS suppression occurred when gish expression was reduced in all neurons with the ELAV-Gal4 pan-neuronal driver (File S4). We propose that gish is a suppressor of parabss1/+ based on reversion of phenotypes by gish04895/+, by ELAV-Gal4C155-driven UAS-gishRNAi, by Df(3R)ED10639/+, and by Df(3R)Exel7329/+. Several mutant alleles of gish that failed to suppress parabss1/+ BS paralytic phenotypes were also found in these analyses. Thus, suppression was not observed for 3 P-element mutations with inserts in the second intron of gish which is spliced out of the long transcripts (genotypes: parabss1/+;gishKG03891, parabss1/+;gishDG16412, and parabss1/+;gishEY06457) (Figure 4). No suppression was seen in parabss1/+;gishe01759/+ flies, which has an insert upstream of the first transcript initiation site (Figure 4, File S4).
The gish04895 mutation raises the threshold for evoked seizures in parabss1/+ flies
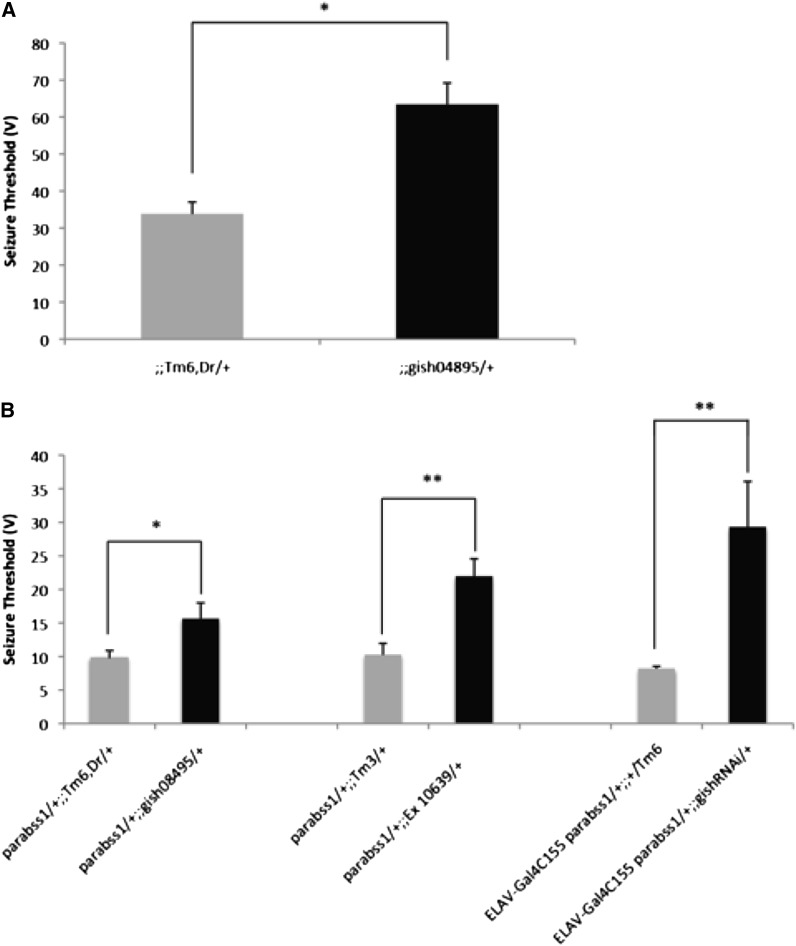
The mutation gish04895 is a recessive lethal. As a heterozygote, in a wild-type background, it displays a seizure-resistant phenotype. Thus, the seizure threshold of gish04895/+ flies is about twice that of wild-type Canton-Special flies, 63.4 ± 5.8 V HFS and 33.8 ± 3.2 V HFS, respectively (Figure 5). The gish04895/+ flies have no other apparent phenotypes: their electrophysiology, behavior, and morphology are all wild type.
Figure 5.
Suppression of seizure threshold by gish04895 and Df Ed10639. Seizure-like activity was recorded in flies of different genotypes. Depicted are the relative HFS voltages required to evoke seizure-like activity at threshold. Loss-of-function mutations of gish suppress seizure-sensitivity in parabss1 heterozygotes, indicated by an increase in seizure threshold voltage compared to controls. In each case, experimental flies are compared with controls that are siblings arising from the same cross in order to minimize genetic background differences. (A) Seizure threshold of gish04895/+ compared with the wild type. The heterozygous mutant gish04895/+ has a slightly greater voltage at threshold suggesting that it is a seizure-resistant mutation. (B) Seizure thresholds of parabss1 heterozygotes in different seizure-suppressor backgrounds. Experimental gish04895/+ flies were of the genotype parabss1/+; gish04895/+ and had a greater seizure threshold than their control siblings (genotype: parabss1/+; TM6,Dr/+), indicating seizure-suppression. Experimental Df Ed10639/+ flies were of the genotype parabss1/+; Df(3R)Ed10639/+ and had a greater seizure threshold than their control siblings (genotype: parabss1/+; TM3/+) indicating seizure suppression. Experimental ELAV-Gal4-driven gishRNAi flies were of the genotype ELAV-Gal4C155 parabss1/+; UAS-gishRNAi/+ and had a higher seizure threshold than their control siblings (genotype: ELAV-Gal4C155 parabss1/+; TM6/+) indicating seizure-suppression.
Seizure-suppression for gish is seen with flies of the genotype: parabss1/+; gish04895/+, which show a seizure threshold of 15.6 ±2.42 V HFS, which is greater than the threshold of their parabss1/+;TM6/+ control siblings (9.8 ± 1.09 V HFS seizure threshold; Figure 5). This seizure-suppression is caused by a loss of gish function as seen most clearly in deletion flies: parabss1/+;Df(3R)ED10639/+ show a seizure threshold nearly in the wild-type range (22.0 ± 2.62 V HFS; Figure 5). Their parabss1/+;TM3/+ siblings show a low seizure threshold (10.3 ± 1.73 V HFS). The loss of gish function finding was confirmed further by RNAi analysis. Flies of the genotype ELAV-Gal4C155 parabss1/+; UAS-gishRNAi/+ showed an increased seizure threshold of 29.28 ± 6.78 V HFS compared with their ELAV-Gal4C155 parabss1/+; +/Tm6 control siblings (8.19 ± 0.355 V HFS; Figure 5, File S3).
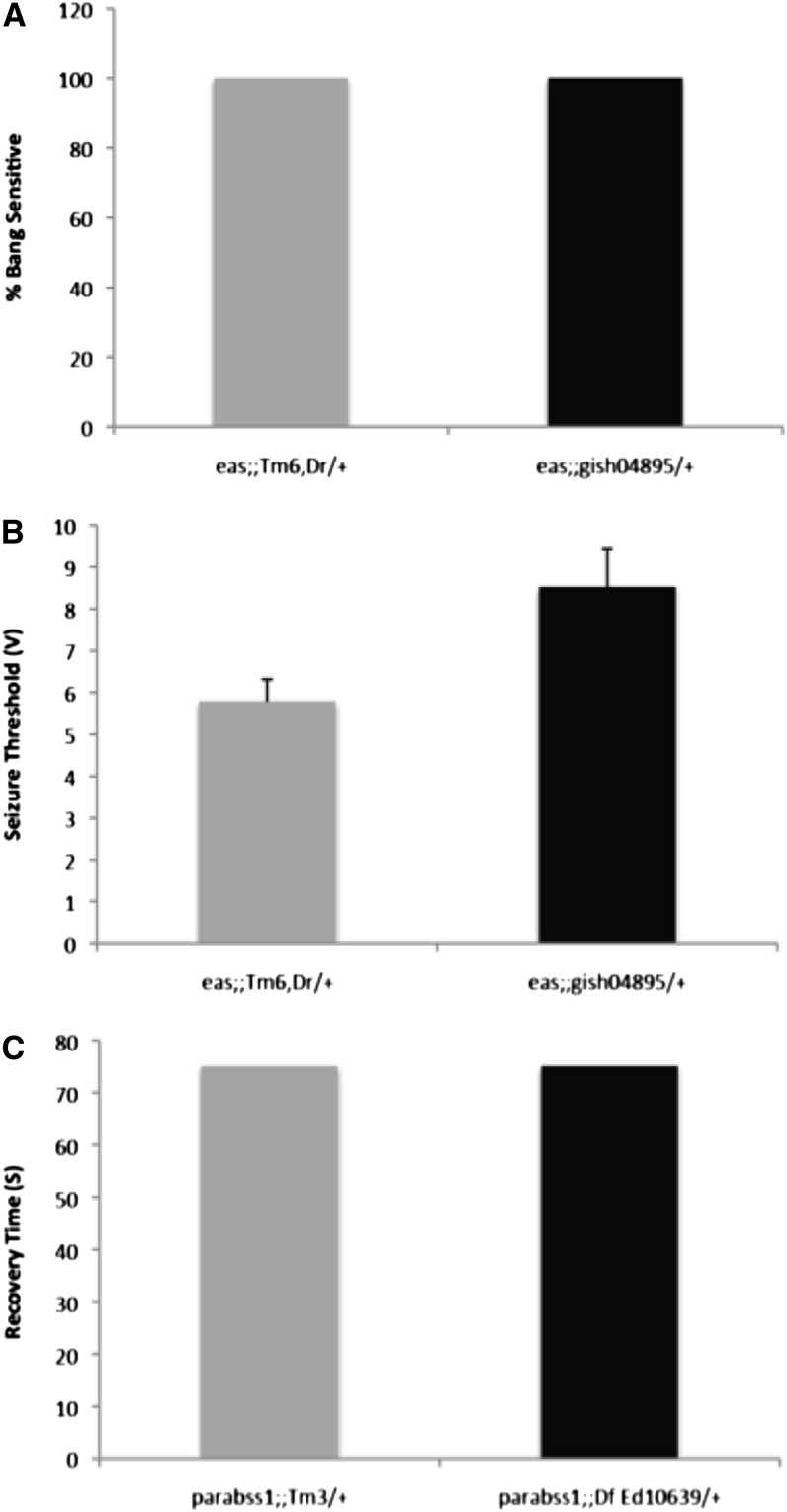
Seizure suppression by gish is specific to parabss1/+ heterozygotes
Seizure suppressor mutations that have been identified previously have been general suppressors, each suppressing several Drosophila BS mutants. In contrast, gish04895/+ suppression is found here to be specific: it appears to only suppress parabss1/+ heterozygotes. We tested for gish04895/+ suppression against BS mutant, eas: gish was ineffective as a suppressor. Thus, eas mutants showed 100% BS paralysis in a gish04895/+ background; electrophysiology also showed minimal increases in seizure threshold (Figure 6, File S3, File S4). We also find that gish/+ does not suppress phenotypes of parabss1 homozygous females and parabss1/Y hemizygous males. Thus, parabss1 homozygotes and hemizygotes showed 100% BS paralysis in a gish background: BS paralysis could not be suppressed by gish04895/+, by Df(3R)ED10639/+, or by UAS-gishRNAi. In addition, a Df(3R)ED10639/+ background caused no reductions of BS paralytic recovery time in parabss1 homozygotes and hemizygotes, a phenotype of parabss1 that is ordinarily easier to suppress than BS paralysis (Figure 6, File S4).
Figure 6.
Suppression of seizure sensitivity by gish is specific to parabss1 heterozygotes. (A) The percentage of eas flies showing a bang sensitive paralytic behavioral phenotype is not reduced by gish04895/+. Paralysis is 100% of flies in both experimental (genotype: eas; gish04895/+) and control siblings (genotype: eas; TM6, Dr/+) genotypes. (B) Electrophysiological recording shows that the seizure threshold of eas is a little greater in a gish04895/+ background (genotype: eas; gish04895/+), but there is no significant suppression compared with control siblings (genotype: eas; TM6, Dr/+). (C) Recovery time of parabss1 homozygotes and hemizygotes is not altered by gish loss-of-function. Depicted are recovery times compared between parabss1; Df(3R)Ed10639/+ experimental flies and their control siblings (genotype: parabss1; TM3/+).
Seizure suppression by gish does not appear to be dependent on Wg/Wnt signaling
The prickle gene functions in noncanonical Wg/Wnt signaling, and mutations have been found to cause myoclonic seizures in humans and BS paralytic behavior in Drosophila (Tao et al. 2011). CK1g casein kinases subserve a large number of cellular processes with diverse substrates (Knippschild et al. 2005), and one prominent role for gish is to phosphorylate arrow, a co-receptor for Wg (Zhang et al. 2006). To test whether seizure suppression by gish might be via Wg signaling, we examined other components of the pathway by RNAi. To test arrow loss-of-function, flies of the genotype ELAV-Gal4C155 parabss1/+;; UAS-arrRNAi/+ showed a slightly lower, but not significant percentage of BS paralysis compared with control ELAV-Gal4C155 parabss1/+; +/Tm6 flies (data not shown, File S4). To test Wg and pangolin loss-of-function, flies of the genotypes ELAV-Gal4C155 parabss1/+; UAS-WgRNAi/+ and ELAV-Gal4C155 parabss1/+; UAS-panRNAi/+ were comparatively equal in percentage of BS paralysis as their ELAV-Gal4C155 parabss1/+; tft/+ controls (data not shown, File S4). Thus, we conclude that seizure suppression by gish is not directly linked to Wg/Wnt signaling.
Discussion
In the present article, we examine severe seizure phenotypes and explore the possibility that severity may be modulated by genetics. We use as substrate the Drosophila parabss1 mutation a channelopathy affecting the voltage-gated Na+ channel. Severe seizure sensitivity is observed in parabss1 mutants, severity that is unresponsive to available drug treatment. In addition, parabss1 has not responded to seizure suppressor mutations identified in screens based on the Drosophila mutants eas and sda. The present study is based on an unbiased, forward genetics screen for mutations that interact with parabss1 by either exacerbating seizure phenotypes (seizure enhancer mutations) or reducing the severity of phenotypes (seizure suppressor mutations).
The search for parabss1 enhancers and -suppressors identified several candidates. Analysis of chn was representative of an enhancer. We found that the time of paralysis of parabss1 individuals was increased (the phenotype screened for), but there was otherwise no obvious enhancement of seizure-sensitivity or severity. Behavioral phenotypes of parabss1 generally resemble those of other BS mutants: all BS mutants are behaviorally similar in initial seizure, initial paralysis, and recovery seizure (Parker et al. 2011a). Unlike other BS mutants, initial paralysis in parabss1 homozygotes is followed by an extended period of tonic/clonic-like activity, resembling activity observed in several human epilepsies (Parker et al. 2011a). During this period in parabss1, the fly is mainly quiescent, resembling a tonic phase. The quiescence is broken up by multiple bouts of clonus-like activity. Because of its period of tonic/clonic-like activity, bss1 recovery time is much longer than for other BS mutants such as sda or eas (Parker et al. 2011a). It is this recovery time, the tonic/clonic period, that is extended by the chn enhancer mutation. A surprise to us was that there was no chn enhancement of the other major parabss1 phenotype: a low electrophysiology seizure threshold. Also, the chn mutation is the only seizure enhancer that we have identified thus far, that does not cause any BS phenotypes (Glasscock and Tanouye 2005; Hekmat-Scafe et al. 2006).
Analysis of gish was representative of a parabss1 suppressor. We found that seizure sensitivity of heterozygous parabss1/+ individuals was greatly reduced by gish loss-of-function mutation and by RNAi. Also, electrophysiological threshold is increased, a further indication that seizure-susceptibility has been reduced in parabss1/+ individual flies. The parabss1 mutant has been exceptionally difficult to suppress. Previously, we have identified 13 seizure-suppressor mutations that suppress the BS behavioral phenotypes of sda and eas mutants, and raise the electrophysiology seizure threshold, often to nearly wild-type levels (reviewed in Parker et al. 2011b). However, seizure suppressors identified heretofore have been ineffective at suppressing parabss1 phenotypes. Seizure suppression by gish loss-of-function mutations reported here is unusual in several respects. It is the only seizure suppression that is effective in reverting parabss1 phenotypes, although it is effective only with heterozygotes, and not homozygotes or hemizygotes. Surprisingly, the seizure suppression is ineffective with sda and eas mutants. Previously, we had attributed this simply to different seizure-sensitive mutants being more or less refractory to suppression. The present results suggest, however, that there may be a fundamental difference between sda and eas mutants, on the one hand, and parabss1 on the other. The nature of the difference remains unclear, at present, but parabss1 seems somehow to be special. We suspect that this could be because of something special about the voltage-gated Na+ channel, the gain-of-function nature of the parabss1 mutation, or both. Also, somewhat perplexing is the reason why parabss1 phenotypes might be suppressed by gish mutations, how a loss of casein kinase function can interfere with voltage-gated Na+ gain-of-function. Also, we do not yet know whether the presence of normal Na+ function (in the heterozygote) is a strict requirement for the suppression of the gain-of-function mutation. The gish mutations do not appear to otherwise be seizure-suppressor mutations, as judged by their lack of effectiveness with sda and eas, but their suppression of parabss1 is pretty remarkable.
It is clear from this study that gish is capable of suppressing parabss1/+ phenotypes and from other deletions identified in our screen that additional suppressor mutations may be found. The parabss1 mutant has been presented as a model for human intractable epilepsy, especially Dravet syndrome (Dravet 1978), a Na+ channelopathy (Parker et al. 2011a). The findings presented here on gish suppression of parabss1 suggest a compelling novel approach for developing options for intractable epilepsy therapeutics depending on exactly how well parabss1 models Dravet syndrome or other intractable epilepsies and how well these findings transfer to mammalian models. At present, available data show that the parabss1 model is a good one. Further experiments of this type as well as the isolation of new suppressors may bring us closer to unraveling the complexity of seizure disorders, especially intractable disorders.
Supplementary Material
Acknowledgements
This study was supported by awards from the McKnight Foundation and the National Institutes of Health (NS31231) to M.A.T. We thank the members of the Tanouye lab for helpful discussions throughout the project.
Footnotes
Communicating editor: H. D. Lipshitz
Literature Cited
- Castrillon D. H., Gönczy P., Alexander S., Rawson R., Eberhart C. G., et al. , 1993. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravet C., 1978. Les épilepsies graves de l’enfant. Vie Med 8: 543–548 [Google Scholar]
- Escudero L. M., Caminero E., Schulze K. L., Bellen H., Modolell J., 2005. Charlatan, a Zn-finger transcription factor, establishes a novel of regulation of the proneural achaete/scute genes of Drosophila. Development 132: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Ganetzky B., Wu C. F., 1982. Indirect suppression involving behavioral mutants with altered nerve excitability in Drosophila melanogaster. Genetics 100: 597–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E., Tanouye M. A., 2005. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics 169: 2137–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe D. S., Lundy M. Y., Ranga R., Tanouye M. A., 2006. Mutations in the K+/Cl− co-transporter gene kazachoc (kcc) increase seizure susceptibility in Drosophila. J Neurosci 26: 8943–8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T. S., Attix S., Gunning D., Zipursky S. L., 2002. Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron 33: 193–203 [DOI] [PubMed] [Google Scholar]
- Knippschild U., Gocht A., Wolff S., Huber N., Lohler J., Stoter M., 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal 17: 675–689 [DOI] [PubMed] [Google Scholar]
- Kosoff E., 2011. Intractable childhood epilepsy: choosing between the treatments. Semin. Pediatr. Neurol. 18: 145–149 [DOI] [PubMed] [Google Scholar]
- Kuebler D., Tanouye M. A., 2000. Modifications of seizure susceptibility in Drosophila. J. Neurophysiol. 83: 998–1009 [DOI] [PubMed] [Google Scholar]
- Kuebler D, Zhang H. G., Ren X., Tanouye M. A., 2001. Genetic suppression of seizure susceptibility in Drosophila. J. Neurophysiol. 86: 1211–1225 [DOI] [PubMed] [Google Scholar]
- Lee J., Wu C. F., 2002. Electroconvulsive seizure behavior in Drosophila: analysis of the physiological repertoire underlying a stereotyped action pattern in bang-sensitive mutants. J. Neurosci. 22: 11065–11079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Kreber R., Ganetzky B., 1989. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 58: 1143–1154 [DOI] [PubMed] [Google Scholar]
- Parker L, Padilla M., Du Y., Dong K., Mark A. Tanouye, 2011a Drosophila as a model for epilepsy: bss is a gain-of-function mutation in the para sodium channel gene that leads to seizures. Genetics 187: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L., Howlett I. C., Rusan Z. M., Tanouye M. A. 2011b Seizure and epilepsy: studies of seizure-disorders in Drosophila. Int.Rev. Neurobiol. 99: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Tanouye M. A., 1995. Seizures and failures in the giant fiber pathway of Drosophila bang-sensitive paralytic mutants. J. Neurosci. 15: 5810–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P., Ramaswami M., Tanouye M. A., 1994. The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79: 23–33 [DOI] [PubMed] [Google Scholar]
- Ramaswami M., Tanouye M. A., 1989. Two sodium channel genes in Drosophila: implications for channel diversity. Proc. Natl. Acad. Sci. USA 86: 2079–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royden C. S., Pirrotta V., Jan L. Y., 1987. The tko locus, site of a behavioral mutation in D. melanogaster, codes for a protein homologous to prokaryotic ribosomal protein S12. Cell 51: 165–73 [DOI] [PubMed] [Google Scholar]
- Song J., Tanouye M. A., 2006. Seizure suppression by shakB2, a gap junction mutation in Drosophila. J. Neurophysiol. 95: 627–635 [DOI] [PubMed] [Google Scholar]
- Tan Y., Yu D., Pletting J., Davis R. L., 2010. Gilgamesh is required for rutabaga-independent olfactory learning in Drosophila. Neuron 67: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Manak J. R., Sowers L., Mei X., Kiyonari H., et al. , 2011. Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am. J. Hum. Genet. 88: 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda L., Kaido M., Lim Y.-M., Kato K., Aigaki T., Hayashi S., 2006. An NRSF / REST-like repressor downstream of Ebi / SMRTER/ Su(H) regulates eye development in Drosophila. EMBO J. 25: 3191–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki Y., Lim Y. M., Niwa N., Hayashi S., Tsuda L., 2011. Robust specification of sensory neurons by dual functions of charlatan, a Drosophila NRSF/REST-like repressor of extramacrochaetae and hairy. Genes Cells 16: 896–909 [DOI] [PubMed] [Google Scholar]
- Zhai L., Graves P. R., Robinson L. C., Italiano M., Culbertson M. R., et al. , 1995. Casein kinase I gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J. Biol. Chem. 270: 12717–12724 [DOI] [PubMed] [Google Scholar]
- Zhang H., Tan J., Reynolds E., Kuebler D., Faulhaber S., et al. , 2002. The Drosophila slamdance gene: a mutation in an aminopeptidase can cause seizure, paralysis and neuronal failure. Genetics 162: 1283–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jia J., Wang B., Amanai K., Wharton K. A., et al. , 2006. Regulation of wingless signaling by the CKI family in Drosophila limb development. Dev. Biol. 299: 221–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.