Abstract
Physiological barriers, such as the blood-brain barrier and intestinal epithelial barrier, remain significant obstacles towards wider utilization of biopharmaceutical products. Receptor-mediated transcytosis has long been viewed as an attractive means of crossing such barriers, but successful exploitation of this route requires better understanding of the interactions between the receptors and protein-based therapeutics. Detailed characterization of such processes at the molecular level is challenging due to the very large physical size and heterogeneity of these species, which makes use of many state-of-the art analytical techniques, such as high-resolution NMR and X-ray crystallography impractical. Mass spectrometry has emerged in the past decade as a powerful tool to study protein-receptor interactions, although its applications to investigate interaction of biopharmaceuticals with their physiological partners are still limited. We highlight the potential of this technique by considering several recent examples where it had been instrumental for understanding molecular mechanisms critical for receptor-mediated transcytosis of transferrin-based therapeutics.
Keywords: Blood-brain barrier, fusion proteins, growth hormone, intestinal epithelial barrier, lysozyme, protein aggregation, protein-drug conjugate, protein therapeutics, receptor-mediated transcytosis, transferrin, transferrin receptor
1. Targeted delivery of biopharmaceuticals and the problem imposed by physiological barriers
The spectacular success of protein-based therapeutics, such as monoclonal antibodies, in the past decade results from the high degree of selectivity and specificity with which these medicines interact with therapeutic targets in diseased tissues (1–3). However, despite significant investments and extensive R&D efforts there are several areas where the progress has been notably slow, such as development of biopharmaceuticals for oral administration and development of protein drugs targeting pathologies in the central nervous system (CNS). A common problem encountered in these areas is the inability of most proteins to cross physiological barriers, such as the intestinal epithelial barrier and the blood-brain barrier (BBB). An additional challenge for oral biopharmaceuticals is presented by the extreme lability of most proteins in the digestive tract.
1.1. The blood-brain barrier and drug delivery to the central nervous system
Despite being only a small fraction of the total body mass, the brain consumes a disproportionately large part of the cardiac output (ca. 20%), which is delivered via a vast vascular network (4). Brain vasculature is comprised of an estimated 100 billion capillaries, which collectively measure 650 kilometers in length and provide a surface area of nearly 20 m2 (5, 6). Despite such a vast interface, drug delivery to the brain remains a formidable challenge, particularly in the case of protein-based therapeutics. The blood-brain barrier consists of tightly linked cerebrovascular endothelial cells that form a diffusional barrier between circulating blood and the interstitial space of the brain, making the brain practically inaccessible to large or hydrophilic molecules by diffusion. This tight-junction featured barrier dominates the majority of the total capillary surface of the brain, leaving few leaky regions (approximately 1/5,000) to allow entry of even large hydrophilic molecules. In addition to the diffusional barrier that physically limits substance transportation into the brain, the BBB also offers an enzymatic barrier with the presence of intracellular and extracellular enzymes.
While the primary function of the BBB is to protect brain tissue from toxic blood components, as well as from various pathogens, it inevitably presents a grand challenge towards delivery of various drugs to the CNS. It has been estimated that over 98% of all small-molecule drugs and all currently approved biopharmaceuticals do not cross the BBB (7), although endogenous transport systems do offer a possibility of targeted delivery to the CNS for both peptide (8) and protein drugs (9).
1.2. Intestinal epithelial barrier and the problem of oral drug delivery
Another physiological barrier that presents a formidable challenge vis-à-vis greater utilization of biopharmaceutical products in therapeutic practice (particularly in developing biopharmaceuticals for oral administration) is the epithelial intestinal barrier. Above and beyond the inability of the vast majority of macromolecules to cross this barrier, the extreme lability of most proteins in the GI tract effectively eliminates oral administration as a viable option for all protein therapeutics (10). Nevertheless, efforts to overcome these obstacles and establish a viable mechanism of oral delivery (which is often seen as the “holy grail” of protein therapy) still continue (11, 12).
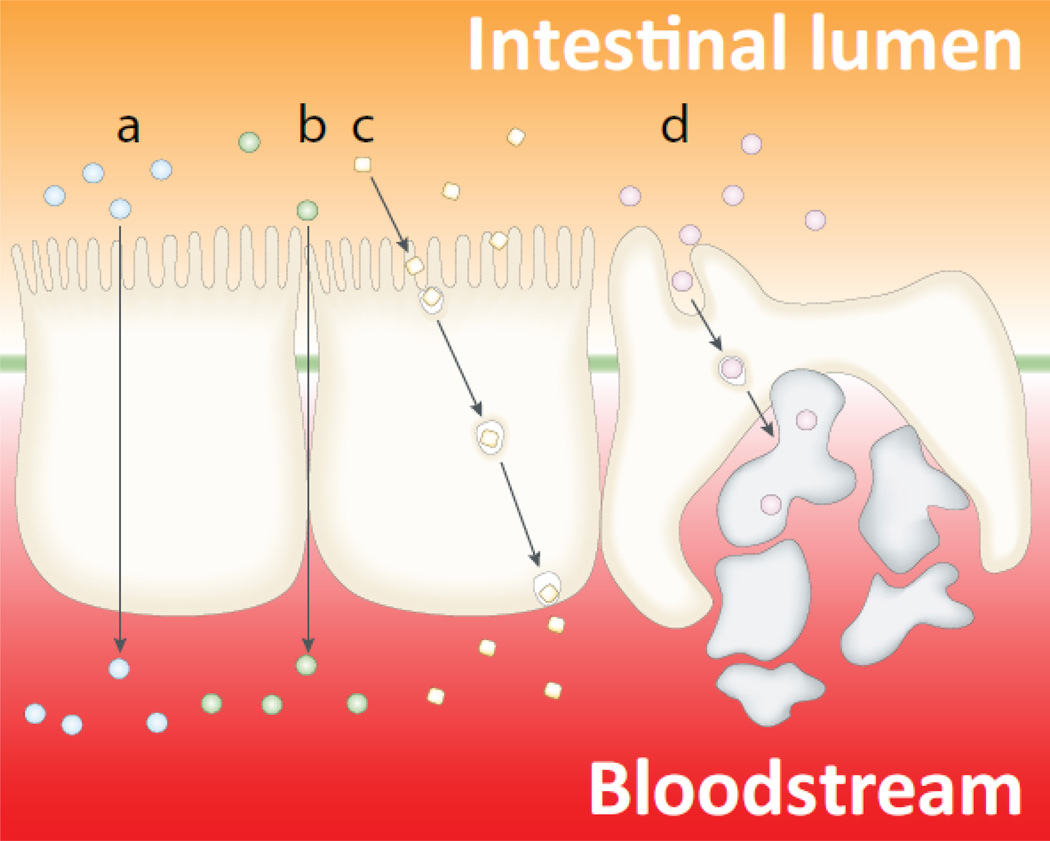
The intestinal epithelial barrier plays an important role in ensuring efficient absorption of nutrients from the GI tract into the circulatory system, while at the same time limiting the ability of pathogens and toxins to enter the bloodstream (13). Similar to the BBB, the intestinal epithelial barrier has anatomical features that tightly control the passage of nutrients and other chemicals, as well as biological molecules (Figure 2). While such tight control has obvious advantages for protecting the organism against toxic chemicals and pathogen invasion, it frequently creates obstacles for drug delivery following oral administration, especially for biopharmaceuticals.
Figure 2.
Schematic depiction of the intestinal epithelium and the pathways available for drug absorption: (a) transcellular pathway through the epithelial cells; (b) paracellular pathway (in between adjacent cells), only small hydrophilic molecules are absorbed through this pathway, and even in these cases the absorption is quite limited because the paracellular pathway comprises a very small percentage of the total epithelial surface area; (c) transcytosis and receptor-mediated endocytosis; and (d) absorption into the lymphatic circulation via M-cells of Peyer’s patches. Adapted with permission from (84).
1.3. Receptor-mediated transcytosis as a paradigm of crossing physiological barriers
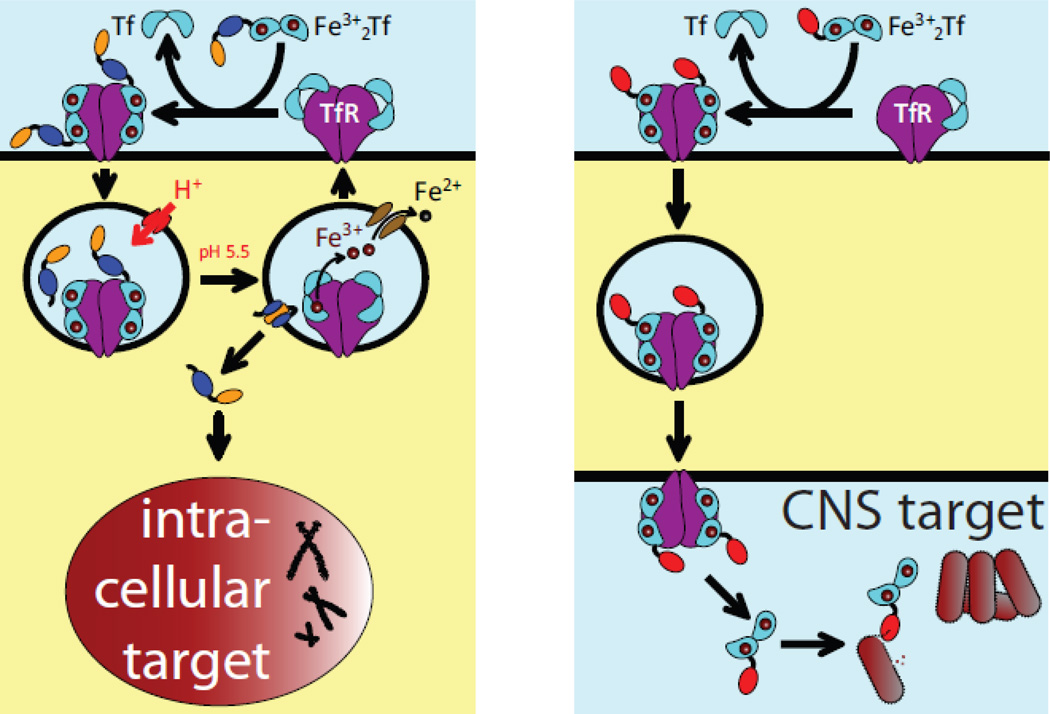
An elegant solution to the physiological barriers’ permeability problem takes advantage of the ability of certain proteins to cross them via receptor-mediated transcytosis. The carrier proteins can be attached to the “payload” proteins either by means of chemical cross-linking (conjugation) or the entire carrier-payload construct can be expressed as a single protein (fusion). One such protein whose ability to enter cells in the process of endocytosis and/or transcytosis can be exploited for targeted drug delivery to specific tissues and/or across the physiological barrier is transferrin (Tf). Tethering a drug payload to Tf has been recognized for some time as an attractive method to enable its precise delivery to the target cells by taking advantage of receptor-mediated endocytosis (14). For example, elevated iron needs of rapidly growing malignant cells make Tf an attractive candidate for targeted transport of anti-cancer drugs to tumors by exploiting Tf receptor (TfR)-mediated endocytosis (Figure 3A). TfR is also one of the abundant proteins in the BBB (15), as well as several other physiological barriers (16), and it is not surprising that Tf has been evaluated as a delivery vehicle for a range of medicines that must cross a certain barrier in order to reach their therapeutic targets. Recent efforts have successfully exploited both barrier-crossing and cell-penetrating capabilities of Tf by designing a dual-targeting nanocarrier for treating brain tumors (17).
Figure 3.
Schematic representation of targeted delivery of Tf-based therapeutic agents to intracellular targets using TfR-mediated endocytosis (a) and across a physiological barrier using TfR-mediated transcytosis (b).
It is clear that the ability to bind TfR is a prerequisite for a physiological barrier crossing by any Tf-based therapeutic agent, and there is a need to develop rapid, sensitive and reliable assays that would allow the receptor association to be screened for a variety of Tf-based drug candidates. This (the ability to bind a receptor being a pre-requisite for successful delivery) is certainly true not only for Tf-based therapeutics, but also for any medicine that utilizes a protein as a means of targeted delivery. The complexity of such biopharmaceutical products made optimization of their therapeutic properties very difficult until recently due to the absence of reliable analytical techniques capable of probing various properties of these products (ranging from structural characterization to monitoring their interactions with physiological partners and therapeutic targets). Recent advances in mass spectrometry (MS) now provide an opportunity to close this gap. MS, and in particular electrospray ionization mass spectrometry (ESI) MS can be used either alone or in combination with classical bioanalytical tools to provide a wealth of information on biopharmaceuticals in general (18, 19), and on complex carrier-payload systems in particular (20). In this review article we discuss recent advances in MS-based approaches to characterize protein/receptor interactions, and demonstrate the analytical power of ESI MS using as examples two specific systems that rely on Tf to traverse both the intestinal epithelial barrier and the BBB.
2. Mass spectrometry-based methods to study protein-receptor interactions
2.1. Monitoring protein-receptor interactions at low resolution: native ESI MS
ESI is a very gentle ionization method, which allows intact biopolymers to be transferred from solution to the gas phase, where their structure can be probed using the vast experimental arsenal accumulated by MS in the past several decades. For example, mass measurement alone (21) frequently allows multiple isoforms of a protein therapeutic to be identified and their relative abundances to be evaluated in a complex mixture without the need for chromatographic separation (22), while the structural characterization of protein therapeutics can be carried out using methods of tandem mass spectrometry either in combination with proteolysis in solution or alone (the so-called “top-down” approach to protein sequencing (23)).
Although the vast majority of MS applications are geared towards analyzing covalent structure of biological molecules (e.g., amino acid sequence and post-translational modifications), it is also possible to preserve non-covalent assemblies formed by proteins and/or other biopolymers in solution (24). Measuring masses of such non-covalent complexes and observing their dissociation pathways in many cases allow their composition and binding stoichiometry to be determined, which is now a primary task of a technique frequently referred to as native ESI MS (25). Although the word “native” may implicitly suggest that the measurements are carried out under physiological (or other therapeutically relevant) conditions, one must realize that ESI MS does place certain restrictions on the nature of the solvent systems that can be used in such experiments. Although it is possible to carry out such measurements in aqueous solutions at physiological pH and ionic strength, only volatile electrolytes (such as ammonium acetate or ammonium bicarbonate) can be used to achieve the desired ionic strength, and common buffers and salts that are typically used to mimic physiological conditions or drug formulations cannot be employed. Nevertheless, numerous studies have demonstrated that volatile electrolytes can be used as a substitute for more common (non-volatile) salts and buffers without having in most cases any negative impact on the stability of protein complexes. Another concern that is frequently expressed in connection with native ESI MS measurements is the possibility of dissociation of non-covalent complexes in the gas phase. Gas phase dissociation of non-covalent assemblies of proteins proceeds via pathways that are very different from those in solution, and the distinction between the two processes can be easily made in most cases (26). Another consideration that must be taken into account when native ESI MS measurements are contemplated relates to the choice of an appropriate mass analyzer. Unlike measurements carried out under denaturing conditions, native ESI MS leads to formation of ions with very low charge density, which appear in the high m/z region of the mass spectrum (27). As a result, some generic mass analyzers (such as quadrupole filters and ion traps) may fail to detect these ions simply because the available m/z range is not sufficient. Typically, time-of-flight mass analyzers provide the most generous coverage in the high m/z range (extending beyond 10,000 m/z units).
A range of antibody/antigen complexes examined by Robinson and co-workers using native ESI MS (28) provided one of the first examples of therapeutically relevant systems where MS provides a direct readout of the composition and binding stoichiometry. This study also demonstrated excellent agreement between the ESI MS and orthogonal methods of analysis (such as analytical ultracentrifugation) when applied to proteins in solution. At the same time, antibody immobilization on the surface of a sensor chip was shown to alter the binding stoichiometry in some cases, an artifact that is obviously avoided by ESI MS, as this technique probes interaction of proteins directly in solution (28). More recently, native ESI MS was used to monitor interactions of an antigenic protein overexpressed in tumor cells with murine and humanized monoclonal antibodies (29), providing not only information about the binding stoichiometry, but also convincing evidence that the intrinsic heterogeneity of the antigen’s disulfide pattern did not affect its recognition by antibodies.
Native ESI MS can also be used to monitor interactions of biopharmaceuticals with their cognate receptors, as was illustrated in a recent study where this technique was employed as a means to monitor interaction between interferon β1a (IFN β1a) and the ectodomains of its cognate receptors IFNAR1 and IFNAR2 (30). In addition to observing formation of both binary (IFNβ1a/IFNAR1 and IFNβ1a/IFNAR2) and ternary (IFNAR1/IFNβ1a/IFNAR2) complexes by ESI MS, the experimental data provided very clear indication that a covalent modification of IFN β1a (alkylation of its only free cysteine residue) resulted in a dramatic decline of the strength of the IFNβ1a/IFNAR1 interaction and, consequently, the stability of the ternary complex (30). This observation suggested that native ESI MS can be used to monitor the effect of chemical modification on the protein’s interaction with its cognate receptor, and provide affinity ranking for different forms of the protein. In some instances native ESI MS can be used to obtain absolute values of dissociation constants for protein/protein interactions (31). In addition to protein/protein association, native ESI MS can also be used to monitor therapeutically relevant interactions of proteins with other biopolymers, such as polysaccharides (32–34) and nucleic acids (35).
Matrix-assisted laser desorption/ionization (MALDI) is another ionization technique capable of producing large macromolecular ions. MALDI MS is generally more sensitive than ESI MS, and is more tolerant to non-volatile electrolytes that are frequently present in biopharmaceutical formulations. However, MALDI usually does not preserve non-covalent complexes due to (i) rapid local heating of the solid sample by the laser beam prior to the ionization event and (ii) presence of large quantities of absorbers of UV radiation (matrix molecules), which typically exert chaotropic effect on proteins. As a result, the protein/protein associations can only be observed by conventional MALDI MS if they are stabilized by chemical cross-linking prior to the analysis (36). An alternative approach relies on using IR radiation to induce desorption and ionization of biomolecules from the solid sample (37). Since water itself has strong absorption bands in the IR region, it can act as a matrix itself, thereby eliminating the chaotropic effect usually associated with the UV matrices. Unfortunately, the measurements are complicated by the fact that the sample has to be kept frozen during the measurements, although a few reports had been published on using IR MALDI to detect non-covalent complexes trapped in ice crystals (38) or frozen glycerol (39).
2.2. Characterization of protein-receptor interactions at higher spatial resolution: H/D exchange MS
Although native ESI MS measurements discussed in the preceding section have been very useful as a means of monitoring interactions between protein drugs and their cognate receptors or therapeutic targets, they do not provide detailed structural information beyond identifying the interacting partners and reporting binding stoichiometry. However, interaction of protein drugs with their therapeutic targets and physiological partners can be characterized at much greater detail by another technique, which utilizes hydrogen/deuterium exchange (HDX) in solution with MS detection. HDX MS is a reliable, robust and sensitive technique, which is capable of probing backbone dynamics locally and localizing protein segments involved in binding interfaces. HDX MS probes conformational properties of proteins by measuring the rates of labile hydrogen atoms exchange with the solvent, which are very sensitive to the protein higher order structure. Protons involved in hydrogen bonding or sequestered from solvent (due to being localized either in the protein core or at the protein/protein interface) do not exchange with the solvent as fast as the protons on the protein surface that do not participate in hydrogen bonding network (40). Progress of the exchange reactions can be monitored by monitoring the protein mass change once the exchange is initiated (which is typically done by rapid dilution of the protein solution with D2O-based solvent to replace H2O in the protein environment with D2O). Since each individual proton-to-deuteron exchange event results in a mass increase of 1 Da, MS can be used to detect exchange of even a few protons within the context of a large protein.
Labile hydrogen atoms residing on the protein backbone amides are excellent reporters of higher order structure due to their involvement in H-bonding critical for protein conformation. Contribution of other labile hydrogen atoms (residing on the side chains) to HDX is not very informative and is usually eliminated prior to MS measurements by lowering the solution pH to 2.5–3.0 and temperature to 0–4 °C. These conditions (known as “quench conditions”) minimize the intrinsic (chemical) exchange rate of the backbone amide protons (41). At the same time, labile hydrogen and deuterium atoms on the side chains exchange quickly (42), thereby allowing the global protection of the backbone to be determined without any interference from the side chains. The ability to quench the exchange reactions of backbone amide hydrogens also provides an opportunity to obtain local information on protein backbone protection by carrying out fast proteolysis prior to MS analysis using pepsin or other acidic protease(s) retaining their catalytic activity at pH 2.5–3 (43, 44).
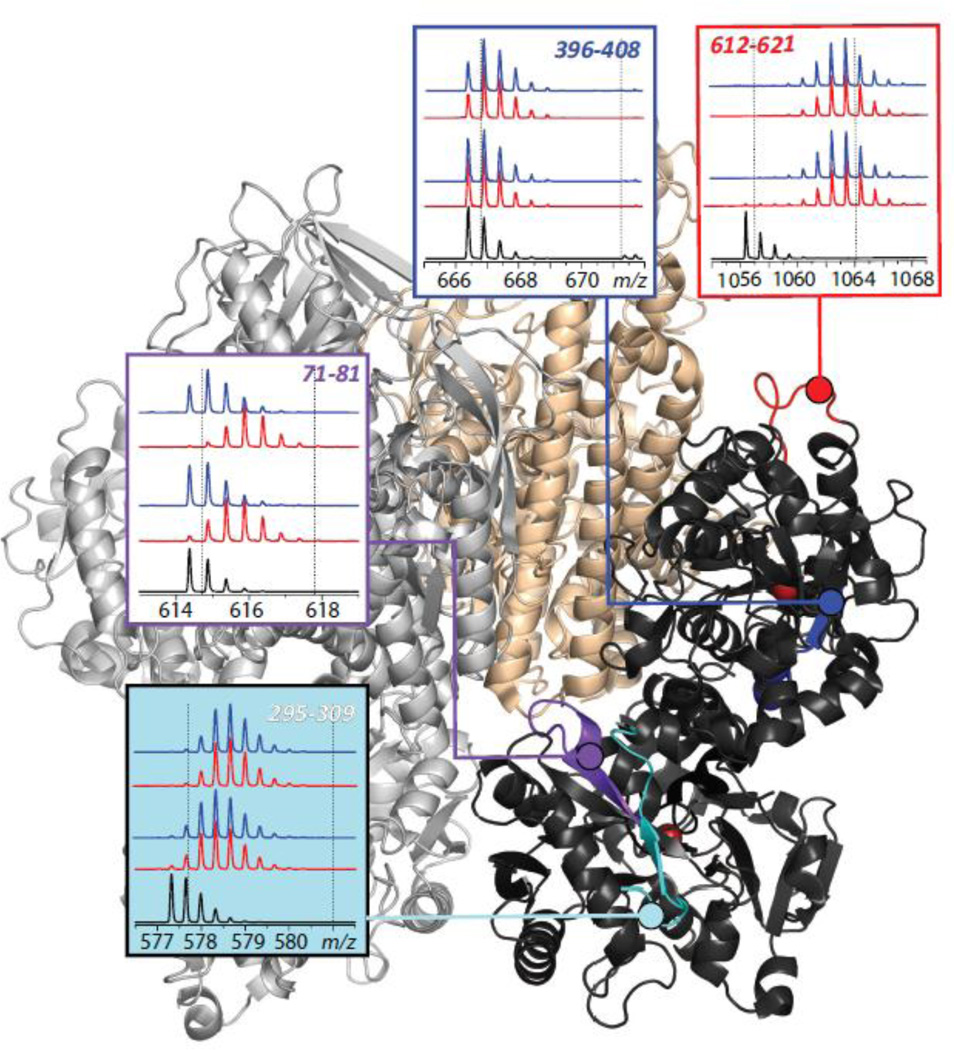
The ability of HDX MS to localize binding interfaces was recognized over a decade ago, leading to a suggestion that it can be used to aid optimization of small molecule drug candidates by providing information on structure and dynamics of the binding site at the therapeutic target (45). HDX MS perhaps is even better suited for mapping protein/protein interfaces, as has been shown by several groups who used this technique to map epitopes in antibody/antigen systems (46–51). An example of using HDX MS to localize a transport protein/cognate receptor binding interface is shown in Figure 4, where the extent of deuterium incorporation in backbone amide groups of Tf is measured following 1 and 10 min of exchange in solution for free and receptor-bound forms of Tf. HDX MS reveals a wide range of backbone protection distributed very unevenly across the polypeptide chain of Tf (e.g., compare evolution of isotopic distributions of peptide ions representing protein segment [396–408] derived from a stable β-strand colored in blue and [612–621] derived from a flexible peripheral loop shown in red). However, exchange patterns of most peptides are not affected by the presence of the receptor, suggesting that local conformational dynamics is not affected by Tf/TfR association. Only few peptides display a difference between the two forms of the protein (e.g., peptide [71–81] in Figure 4), indicating their involvement in the binding interface or manifestation of an allosteric effect.
Figure 4.
Localization of the receptor binding interface within diferric Tf using HDX MS. The panels show isotopic distributions of representative peptic fragments derived from the protein subjected to HDX in the presence (blue) and the absence (red) of the receptor. The black traces at the bottom of each diagram show isotopic distributions of peptic fragments derived from unlabeled protein, and the dotted lines represent the end-points of the exchange reaction. Colored segments within the Tf/TfR complex show location of the peptic fragments. Adapted with permission from (85).
The spatial resolution that can be achieved in HDX MS measurements is typically limited by the size of the peptic fragments. Usually, analysis of the protection patterns of overlapping peptic fragments can improve the resolution, and in some favorable cases amide protection at a single-residue level can be determined (52). An alternative approach to increasing the spatial resolution in HDX MS studies utilizes gas phase fragmentation of peptide ions as a supplement to proteolysis in solution (53, 54). In some instances the proteolytic step can be completely eliminated from the experimental workflow; however, this approached (usually referred to as “top-down” HDX MS or HDX MS/MS) has been successfully applied so far only to proteins of relatively modest size (55, 56).
In addition to structural information on protein drug/target binding, HDX MS methodology can also be used to determine binding affinity (57). Furthermore, HDX MS can also be used to identify protein segments involved in binding to other biopolymers, such as nucleic acids (58), indicating that this technique can become a truly invaluable tool in designing new and enhancing existing biopharmaceutical products including both protein- and oligonucleotide-based drugs and drug candidates.
2.3. Towards mass spectrometry-based quantitation of overcoming physiological barriers: measuring biodistribution of protein therapeutics
Although MS-based methods described in the previous sections provide valuable information on the ability of protein-based drugs to interact with their physiological partners and therapeutic targets, even the most sophisticated in vitro models cannot fully account for the complexity of interaction networks in vivo, and the success of the drug is ultimately decided in clinical studies. Protein-based therapeutics frequently exhibit unexpected behavior in vivo reflecting uniqueness of their interactions with the myriad of other biomolecules including proteases, proteins responsible for immune reaction, etc. (59). Therefore, even though the pharmacokinetic and pharmacodynamic data do not provide direct information on the protein drug interaction with its targets at the molecular level, they are nonetheless critical for understanding how the therapeutic product navigates through complex webs of interactomes at the whole organism level.
Until very recently immunoassays and bioactivity assays were the two main analytical tools supporting the studies of pharmacokinetics of protein drugs, and MS was utilized in only a limited number of niche applications (60). However, the explosive growth of proteomics in recent years, and the central role played by MS in this field (61) have resulted in a dramatic expansion of the scope of MS-based protein quantitation methods in pharmacokinetic studies of protein drugs (62). Discussion of these tools goes beyond the scope of this review, but a detailed account of analytical tools (including MS) used in pharmacokinetic studies of protein drugs is provided in a paper by E. Ezan “Pharmacokinetic studies of protein drugs: past, present and future” in this issue of the journal.
3. Lysozyme-transferrin conjugate as a model protein therapeutic targeted to CNS
The ability of certain endogenous receptors (such as TfR) to assist protein drug delivery across the BBB had been noted nearly two decades ago (63), and exploitation of these properties to design “Trojan horse” protein drugs that enable delivery to the CNS has been a focus of extensive efforts in the past several years. It is not surprising that successful brain targeting exhibited by such biopharmaceuticals is caused in large part by the targeting properties of the transport protein (e.g., by anti-TfR IgG (64)); however, the presence of the payload and its potential to interfere with the receptor binding may also modulate the efficiency of traversing the BBB. ESI MS provides an elegant way to monitor the interactions between the biopharmaceutical product and its cognate receptor and, therefore, can be of great value during the initial stages of the drug design process.
We illustrate this with a Tf-based therapeutic product (LzT, a conjugate of Tf and a bacteriostatic agent lysozyme, Lz); which serves as a model therapeutic targeting pathogens in the CNS (it is being developed as an alternative/complement to antibiotics for treatment/containment of Gram-positive infections in CNS). Lz is an antibacterial enzyme present in a variety of organisms, which exerts its bacteriostatic function by hydrolyzing the β-1,4-glycosidic bond between the N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) residues of peptidoglycans, which results in lysis of bacterial cell walls. Although Lz primarily attacks Gram-positive bacteria, where the peptidoglycan layer is not protected by the outer membrane (as it is in Gram-negative bacteria), certain structural modifications can make it effective against Gram-negative bacteria as well (65). While this protein is widely distributed thoughout the human body, it is not present in cerebrospinal fluid (CSF) of healthy subjects (66) (the detectable levels of lysozyme in CSF is usually associated with various CNS pathologies (67, 68) and is likely to reflect increased permeability of the BBB (68)). Therefore, the ability to deliver Lz “on demand” across the BBB might lead to development of novel effective therapeutic strategies aiming at eradication of Gram-positive infection in the CNS, whose carriers gain access to the brain via a variety of routes (69).
Since Tf is being relied upon as a means of traversing the BBB through receptor-mediated transcytosis, chemical modification of this protein has the potential to interfere with its recognition by TfR and, therefore, hinder transport of the conjugate across the BBB. Conjugation can influence the receptor-binding properties of Tf in two ways. First, chemical modification may have a negative impact on the conformational stability of Tf, triggering unfolding and loss of native structure. Second, even if the native conformation is preserved, the chemical modification of the amino acid residues located at or near the binding interface may obviously have an adverse affect on receptor binding.
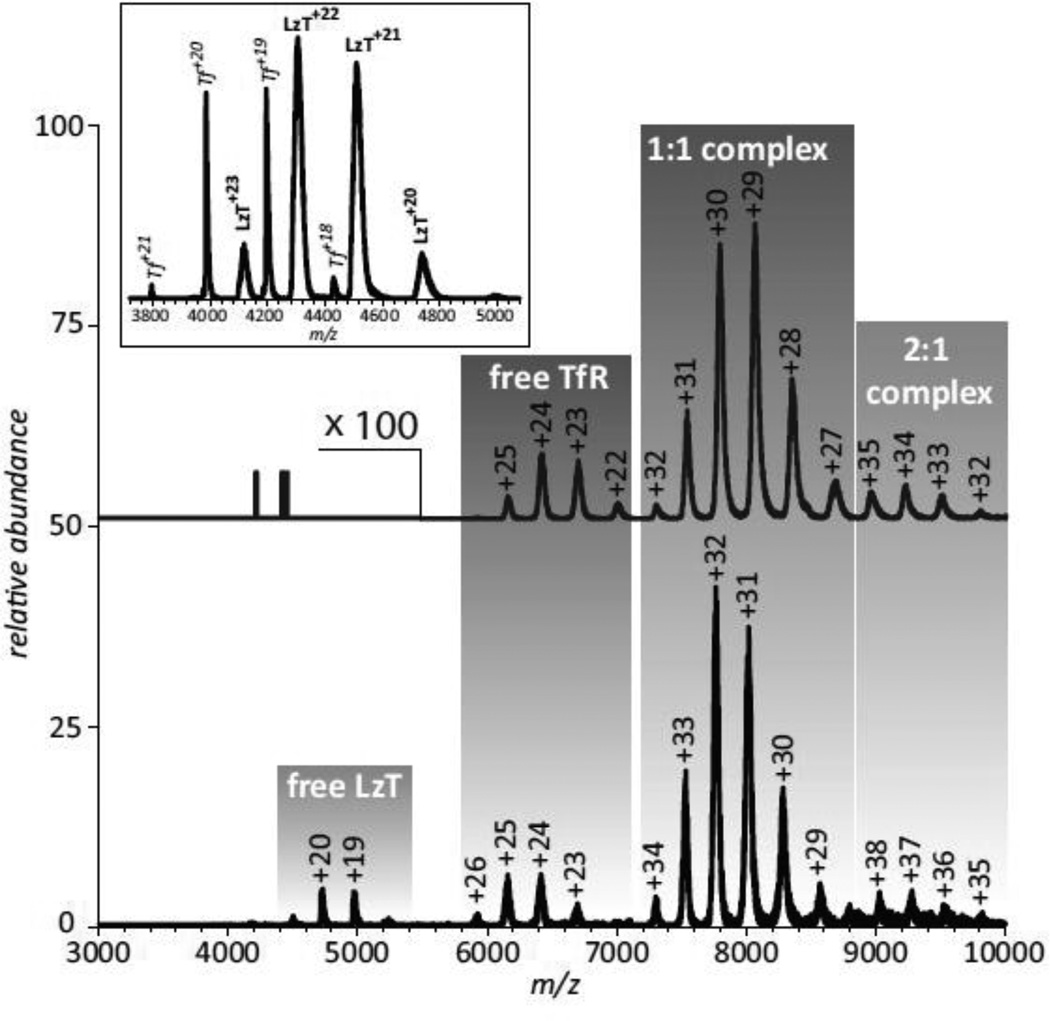
As far as large-scale conformational changes resulting from the conjugation of the payload protein to Tf, no signs of partial or complete unfolding of the protein components are evident in the ESI mass spectrum acquired under near-native conditions (see inset in Figure 5). Indeed, loss of native tertiary structure in solution always manifests itself via the appearance of high charge-density ionic species in the mass spectrum due to the enhanced ability of less compact protein conformations to accommodate a higher number of charges upon transition from solution to the gas phase (27), this is not observed in the mass spectrum of LzT. In fact, the extent of multiple charging of LzT ions is only slightly higher than that of intact Tf (which was added to LzT solution intentionally to enable direct comparison of the charge distributions of ionic species corresponding to these two proteins). However, the LzT ion peaks are noticeably broader compared to those of Tf, indicating significant micro-heterogeneity due to the presence of multiple modifications of the side chains. A more detailed analysis of the mass spectrum reveals very convoluted peak shapes of LzT ions, where the broad ionic mass distribution reflects the presence of either unreacted or dead-ended (hydrolyzed) cross-linking groups on the protein surface (70).
Figure 5.
ESI mass spectra of Tf/TfR (top) and LzT/TfR (bottom) mixtures acquired under near-native conditions (3 μM of each protein in 20 mM ammonium acetate at pH 7.1). Peak labels represent the charge states of all relevant ions. The inset shows a mass spectrum of LzT spiked with intact Tf. Adapted with permission from (70).
The ability of both Tf and Lz to interact with their physiological partners and/or therapeutic targets may be compromised by an unfavorable cross-link location, as well as the presence of multiple nonproductive (unreacted or dead-ended) modifications on the surface of both proteins beyond the crosslink sites. An ESI mass spectrum of the LzT/TfR mixture acquired under near native conditions clearly indicates the receptor recognizes the conjugate, although the binding affinity is diminished compared to intact Tf (Figure 5). Indeed, no ionic signal of unbound Tf is detected in the mass spectrum of the Tf/TfR mixture, consistent with the receptor-binding affinity of Tf being in the sub-μM range (concentration of both proteins in the Tf/TfR mixture was in the low-μM range). At the same time, the presence of a weak, but detectable ionic signal of unbound LzT in the mass spectrum of the LzT/TfR mixture acquired under identical conditions suggests that the receptor binding affinity of the conjugate is in the low-μM range. This affinity range is close to that of intact apo-Tf (71), even though the conjugate was saturated with iron following its isolation from the reaction mixture and its iron status is the same as intact diferric Tf used to acquire the mass spectrum shown at the top of Figure 5.
Even this lower receptor affinity should be sufficient for transient binding to TfR at the cell surface, and may in fact prove beneficial for dissociation from TfR upon crossing the BBB. In fact, a recent report noted that decreased TfR affinity enhances the brain uptake of anti-TfR antibodies, as the high receptor affinity proteins tend to remain associated with the BBB without being released on its abluminal side (72). Therefore, optimization of the therapeutic properties of complex Tf-based drugs may require modulation of their receptor-binding properties, and native ESI MS will undoubtedly be a very effective analytical tool supporting these efforts. Furthermore, native ESI MS may also provide another contribution critical for the success of the new therapies, i.e. monitoring the ability of the transporter-payload conjugate to interact with the intended therapeutic targets or their mimics, generating biochemical information important for the selection of promising candidates and optimizing the conjugation protocols in preclinical studies (70).
4. Transferrin-based fusion protein as a model of a biopharmaceutical designed for oral delivery: mass spectrometry reveals the critical role of protein aggregates
Despite the significant efforts invested in developing oral biopharmaceutical products, no orally available protein drug is currently on the market, and the list of promising candidates remains disappointingly short (10, 11). One obvious reason for this rather modest record of success is the fact that a successful biopharmaceutical designed for oral administration not only has to traverse the intestinal epithelial barrier, but also survive exposure to the GI environment, and in particular to the extreme conditions inside the stomach. A rare success story in this field is a recently designed growth hormone/transferrin fusion protein (GHT), which has been shown to be able to overcome the obstacles faced by oral biopharmaceuticals, and demonstrated biological activity following oral administration (73). Although the presence of TfR throughout the GI tract has been firmly established (74), it remained unclear until recently whether the growth hormone segment of GHT interferes with its ability to bind TfR. This question can now be easily addressed using a straightforward ESI MS approach described in the previous section for LzT. However, these straightforward measurements fail to explain another remarkable feature of GHT, namely its ability to evade hydrolysis in the stomach prior to interacting with TfR and subsequent crossing of the intestinal epithelial barrier. Furthermore, the measurable activity of GHT following oral administration clearly suggested that the payload protein is also protected from degradation in the stomach, leading to speculative suggestions that the transporter also acts as a payload “protector” in the harsh GI environment (75). This conjecture, however, runs contrary to the known fact that Tf is effectively proteolyzed by pepsin even in mildly acidic solutions (76), thereby failing to protect even itself, let alone its companion.
The apparent success of GHT in crossing the intestinal epithelial barrier may provide important clues towards successful design of the next generation of oral biopharmaceuticals, a consideration that warranted detailed investigation of various aspects of GHT structure and behavior that are critical for its ability to not only avoid the common fate of the vast majority of proteins in the extreme environment of stomach, but also to transport itself and its therapeutic cargo across the intestinal epithelial barrier (20). Analysis of GHT by a combination of size exclusion chromatography (SEC) and ESI MS revealed that a significant population existed in an oligomeric state in addition to the anticipated monomer. The soluble oligomeric fraction was separated from GHT monomer by SEC, and its physical size was estimated to be in the 3,000–10,000 Å range using dynamic light scattering.
Stability of both GHT monomer and the oligomeric fraction in the stomach-like environment was investigated by subjecting these two species (as well as a Tf control) to hydrolysis under strongly acidic conditions in the presence of pepsin at 37 °C. The control Tf sample was degraded very quickly, consistent with the previous reports of its susceptibility to efficient digestion with pepsin, and GHT monomers displayed only slightly higher stability. The oligomeric species, however, was remarkably stable and displayed only a negligible decrease in abundance over the first 5 hours of digestion. The remarkable stability of this species is certainly related to its high degree of oligomerization, as the tight packing of proteins shields those of them localized at the core of the aggregate from the hydrolytic attack, and even the damage to the protein molecules located on the periphery of the aggregate is likely to be greatly reduced due to the steric hindrance.
Survival of GHT oligomers in the stomach is necessary, but not sufficient for eliciting the desired therapeutic action following oral administration. Passage across the intestinal epithelial barrier would only become possible for such species if they can be recognized by TfR present on the inner lining of the GI tract. While the receptor recognition by the monomeric form of GHT can be easily monitored using a native ESI MS (similar to that discussed in the preceding section for the LzT/TfR interaction), a different approach is needed to establish the ability of large GHT oligomers to associate with TfR. Indeed, the very large size and heterogeneity of the oligomeric species make direct application of ESI MS impractical even in the absence of the receptor, let alone its putative complex with TfR. SEC alone is not very informative either, as the very large physical size of oligomeric GHT species causes them to elute with the column void, and a shift to an earlier retention is simply not possible even if oligomers do associate with TfR.
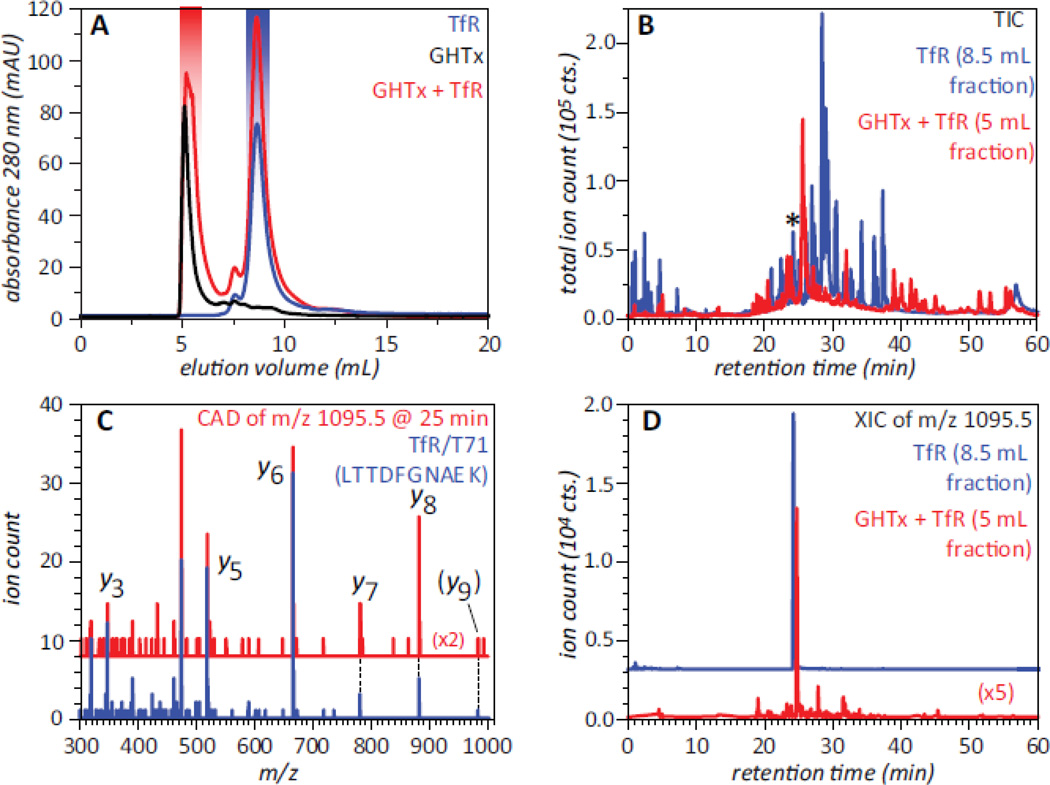
Even though neither ESI MS nor SEC alone can address the question of whether the large soluble aggregates of GHT can bind to TfR, a combined use of these two techniques provides a definitive answer to this question. Addition of the GHT oligomeric species to TfR solution followed by isolation of the early eluting peak and bottom up analysis of its contents (e.g., proteolytic digestion of the material with subsequent MS analysis of the fragment peptides and a database search to identify the precursor proteins) reveals the presence of TfR in this fraction. This process is illustrated in Figure 6, where LC/MS/MS analysis of the tryptic digest of the void volume fraction positively identified several proteolytic fragments derived from TfR in addition to the fragment peptides matching various GHT segments. A closer look at all proteolytic fragments of TfR identified in the digest of the void volume fraction by the database search confirms the assignments, as their MS/MS patterns and LC elution times closely match those of the tryptic fragments derived from the reference TfR sample (e.g., see blue and red traces for a tryptic fragment T71 in Figure 6). Since free TfR elutes in SEC at a much later time compared to the void volume fraction, its association with GHT oligomers is the only possible explanation that can account for its presence in the fraction that represents the protein species too large to be retained on the size-exclusion gel.
Figure 6.
SEC of GHT oligomers, TfR and their mixture (A), and total ion chromatograms (B) of tryptic digests of the SEC fractions of TfR (blue trace) and the TfR/GHT oligomer mixture (red trace) highlighted in panel A. Panels C and D illustrate detection of TfR in the early-eluting SEC fraction of GHTx/TfR mixture (highlighted with red in panel A): single-scan MS/MS spectrum (C) and extracted ion chromatogram (D) of a tryptic peptide eluting at 25 min (marked with asterisk in panel B). The blue trace in panel C shows the reference MS/MS spectrum of a tryptic fragment T71 (LTTDFNAEK) of TfR, and the blue trace in panel D corresponds to this peptide derived from TfR in the absence of GHT oligomers (SEC fraction highlighted with blue in panel A). Adapted with permission from (20).
Importantly, the monomeric form of the fusion protein is also recognized by the receptor, but it shows only marginal anti-hydrolytic stability in the stomach-like environment, and it seems unlikely to make a significant contribution to the observed therapeutic activity of the fusion protein followed its oral administration, pointing at the critical role of GHT aggregation for achieving therapeutic objectives. Although the current paradigm attaches an unquestionably negative connotation to a wide range of protein aggregation phenomena, particularly in the biopharmaceutical arena, examples begin to emerge where aggregation of protein drugs can be used to enhance their therapeutic properties (77). The conclusions derived from the analysis of stability and receptor-binding properties of GHT aggregates carried out with a combination of SEC and ESI MS further question the negative stigma traditionally attached to the aggregation processes by clearly demonstrating that they may be exploited in the design of efficient orally administered protein therapeutics (20).
5. Current challenges in the field and future directions
The utility of ESI MS for characterizing interactions of biopharmaceutical products with cognate receptors highlighted in this paper underscores the fact that this technique is poised to become a very valuable addition to the analytical toolbox facilitating the development of new safe and efficient therapies. However, there still are serious challenges that must be addressed before this technique truly becomes a routine analytical tool for evaluating interactions of protein drugs with their physiological partners and therapeutic targets. One such challenge is posed by the extreme heterogeneity frequently displayed by many biopharmaceutical products caused by post-translational modifications (PTMs). While non-enzymatic PTMs accumulating as a result of protein degradation are obviously random, even biologically relevant enzymatic PTMs do not have the same level of precise control that makes the process of translation from genetic material so perfectly reproducible. Glycosylation is one particular form of enzymatic PTMs that is frequently observed in therapeutic proteins, and the structural heterogeneity exhibited by glycoproteins depends on the extent of glycosylation. Typically, dealing with even large proteins (over 100 kDa) having < 5% total carbohydrate content by mass is not problematic, while glycosylation levels exceeding 10% of the total protein mass pose a formidable challenge to ESI MS-based analysis. Another common source of structural heterogeneity that is present in many biopharmaceutical products is “designer” PTMs, such as protein PEGylation.
Recently, extensive efforts have been directed towards developing robust analytical methods capable of dealing with highly heterogeneous systems. For example, a combination of complexity reduction (by mass selecting a narrow population of glycoprotein or PEGylated protein ions) and gas phase chemistry (charge reduction via electron transfer or electron capture) has been shown to be extremely useful in dealing with extensively glycosylated therapeutic proteins (78). This approach has also been used successfully to study binding of highly glycosylated proteins to their physiological partners, e.g. hemoglobin capturing by haptoglobin (78). Another MS-based technique that might become very useful in dealing with highly heterogeneous systems is ion mobility (IM) MS. While the majority of its current applications are geared towards providing information on geometry of macromolecular ions in the gas phase, the potential utility of IM-MS as a means of reducing complexity of heterogeneous systems by providing an additional separation stage prior to MS detection is frequently overlooked. The ability of this technique to separate various isoforms of both carbohydrates (79) and synthetic polymers (80) is well-documented, and in fact has already been used to facilitate the analysis of covalent structure of large glycoproteins (81) and protein-polymer conjugates (82).
In recent years we have witnessed an explosive growth in the use of ESI MS in various fields of biophysics and structural biology, and the biotechnology sector was not an exception to this trend. Without a doubt, this technique is and will continue to be making important contributions in various stages of design of protein drugs, and catalyzing further progress in the field of biotechnology.
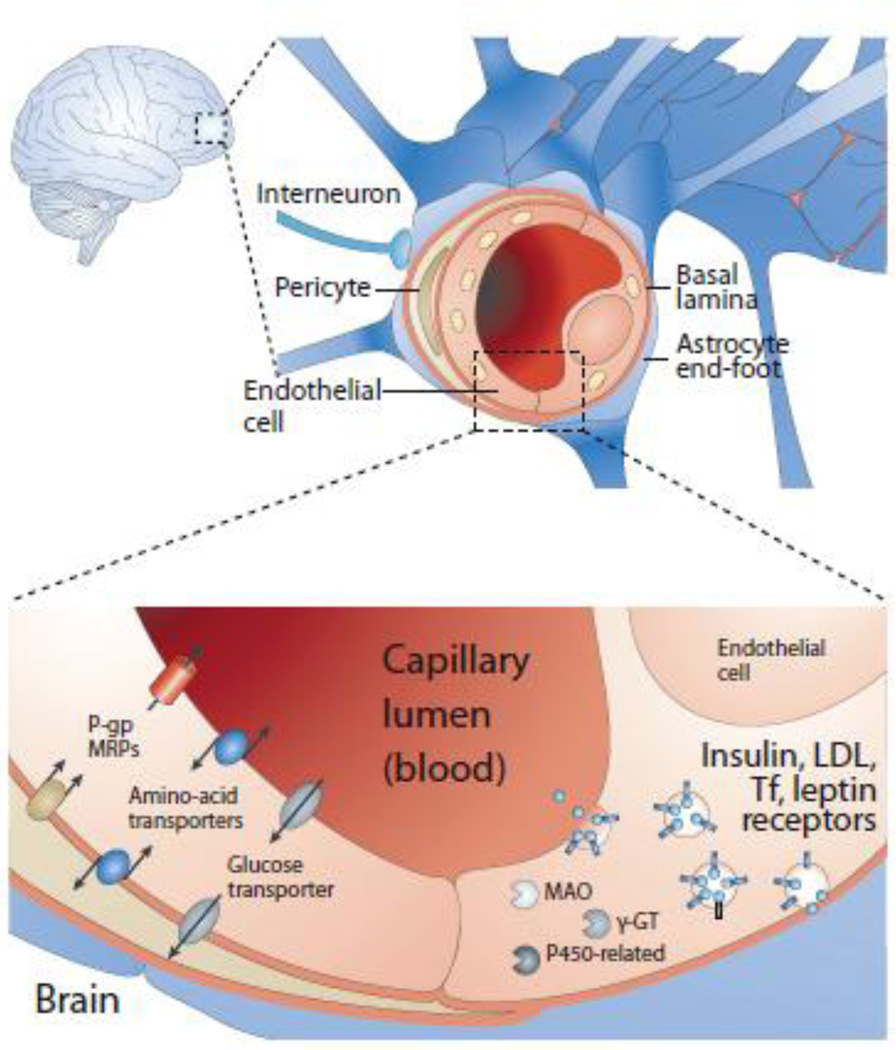
Figure 1.
A schematic diagram representing the blood-brain barrier (top), and the transport of proteins from luminal side of BBB (capillary) to abluminal side (brain) via receptor mediated endocytosis. Adopted from (83).
Acknowledgements
This work was supported by a grant R01 GM061666 from the National Institutes of Health. The authors also express gratitude to our collaborators Prof. Anne B. Mason (University of Vermont College of Medicine), Prof. Wei-Chiang Shen (Univ. of Southern California School of Pharmacy), Drs. Philip Savickas and John Thomas (Shire Human Genetic Therapies, Lexington, MA) and Drs. Damian Houde and Steven Berkowitz (Biogen IDEC, Cambridge, MA) for many helpful discussions.
ABBREVIATIONS
- BBB
Blood-brain barrier
- CNS
central nervous system
- CSF
cerebrospinal fluid
- ESI
electrospray ionization
- GHT
growth hormone-transferrin fusion protein
- HDX
hydrogen/deuterium exchange
- Lz
lysozyme
- LzT
lysozyme-transferrin conjugate
- MS
mass spectrometry
- Tf
transferrin
- TfR
transferrin receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knäblein J, editor. Modern Biopharmaceuticals: Design, Development and Optimization. Vol. 1-4. Weinheim: WILEY-VCH Verlag GmbH & Co; 2005. [Google Scholar]
- 2.Walsh G. Pharmaceutical biotechnology : concepts and applications. Chichester, England ; Hoboken, NJ: John Wiley & Sons; 2007. [Google Scholar]
- 3.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 4.Carvey PM, Hendey B, Monahan AJ. The blood-brain barrier in neurodegenerative disease: a rhetorical perspective. Journal of Neurochemistry. 2009;111:291–314. doi: 10.1111/j.1471-4159.2009.06319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeken JF, Loscher W. The blood-brain barrier and cancer: transporters, treatment, and Trojan horses. Clin. Cancer Res. 2007;13:1663–1674. doi: 10.1158/1078-0432.CCR-06-2854. [DOI] [PubMed] [Google Scholar]
- 6.Pardridge WM. Blood-brain barrier drug targeting: the future of brain drug development. Mol. Interv. 2003;3:90–105. doi: 10.1124/mi.3.2.90. [DOI] [PubMed] [Google Scholar]
- 7.Pardridge WM. Blood-brain barrier delivery. Drug Discov. Today. 2007;12:54–61. doi: 10.1016/j.drudis.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 8.McGonigle P. Peptide therapeutics for CNS indications. Biochemical Pharmacology. 2012;83:559–566. doi: 10.1016/j.bcp.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Pardridge WM, Boado RJ, Wittrup KD, Gregory LV. Methods Enzymol. Academic Press; 2012. Reengineering biopharmaceuticals for targeted delivery across the blood/brain barrier; pp. 269–292. [DOI] [PubMed] [Google Scholar]
- 10.Morishita M, Peppas NA. Is the oral route possible for peptide and protein drug delivery? Drug Discov. Today. 2006;11:905–910. doi: 10.1016/j.drudis.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Shen W-C. Oral peptide and protein delivery: unfulfilled promises? Drug Discov. Today. 2003;8:607–608. doi: 10.1016/s1359-6446(03)02692-8. [DOI] [PubMed] [Google Scholar]
- 12.Moeller EH, Jorgensen L. Alternative routes of administration for systemic delivery of protein pharmaceuticals. Drug Discov. Today. 2008;5:e89–e94. doi: 10.1016/j.ddtec.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Scaldaferri F, Pizzoferrato M, Gerardi V, Lopetuso L, Gasbarrini A. The Gut Barrier: New Acquisitions and Therapeutic Approaches. J. Clin. Gastroenterol. 2012;46:S12–S17. doi: 10.1097/MCG.0b013e31826ae849. [DOI] [PubMed] [Google Scholar]
- 14.Daniels TR, Delgado T, Helguera G, Penichet ML. The transferrin receptor part II: targeted delivery of therapeutic agents into cancer cells. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Enerson BE, Drewes LR. The rat blood-brain barrier transcriptome. J Cereb Blood Flow Metab. 2005;26:959–973. doi: 10.1038/sj.jcbfm.9600249. [DOI] [PubMed] [Google Scholar]
- 16.Russell-Jones G. Intestinal receptor targeting for peptide delivery: an expert's personal perspective on reasons for failure and new opportunities. Ther. Deliv. 2011;2:1575–1593. doi: 10.4155/tde.11.129. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, He H, Jia X, Lu W-L, Lou J, Wei Y. A dual-targeting nanocarrier based on poly(amidoamine) dendrimers conjugated with transferrin and tamoxifen for treating brain gliomas. Biomaterials. 2012;33:3899–3908. doi: 10.1016/j.biomaterials.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: Mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol. Adv. 2012;30:210–222. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat. Rev. Drug Discov. 2012;11:527–540. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bobst CE, Wang S, Shen W-C, Kaltashov IA. Mass spectrometry study of a transferrin-based protein drug reveals the key role of protein aggregation for successful oral delivery. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13544–13548. doi: 10.1073/pnas.1206924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carini M, Regazzoni L, Aldini G. Mass Spectrometric Strategies and Their Applications for Molecular Mass Determination of Recombinant Therapeutic Proteins. Curr. Pharm. Biotechnol. 2011;12:1548–1557. doi: 10.2174/138920111798357348. [DOI] [PubMed] [Google Scholar]
- 22.Rosati S, Thompson NJ, Barendregt A, Hendriks LJA, Bakker ABH, de Kruif J, Throsby M, van Duijn E, Heck AJR. Qualitative and semiquantitative analysis of composite mixtures of antibodies by native mass spectrometry. Analytical chemistry. 2012;84:7227–7232. doi: 10.1021/ac301611d. [DOI] [PubMed] [Google Scholar]
- 23.Bondarenko PV, Second TP, Zabrouskov V, Makarov AA, Zhang ZQ. Mass measurement and top-down HPLC/MS analysis of intact monoclonal antibodies on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 2009;20:1415–1424. doi: 10.1016/j.jasms.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Heuvel RHvd, Heck AJ. Native protein mass spectrometry: from intact oligomers to functional machineries. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Abzalimov RR, Frimpong AK, Kaltashov IA. Gas-phase processes and measurements of macromolecular properties in solution: On the possibility of false positive and false negative signals of protein unfolding. Int. J. Mass Spectrom. 2006;253:207–216. [Google Scholar]
- 27.Kaltashov IA, Abzalimov RR. Do ionic charges in ESI MS provide useful information on macromolecular structure? J. Am. Soc. Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Oda M, Uchiyama S, Robinson CV, Fukui K, Kobayashi Y, Azuma T. Regional and segmental flexibility of antibodies in interaction with antigens of different size. FEBS J. 2006;273:1476–1487. doi: 10.1111/j.1742-4658.2006.05168.x. [DOI] [PubMed] [Google Scholar]
- 29.Atmanene Cd, Wagner-Rousset E, Malissard M, Chol B, Robert A, Corvaía N, Dorsselaer AV, Beck A, Sanglier-Cianfeírani S. Extending Mass Spectrometry Contribution to Therapeutic Monoclonal Antibody Lead Optimization: Characterization of Immune Complexes Using Noncovalent ESI-MS. Analytical Chemistry. 2009;81:6364–6373. doi: 10.1021/ac9007557. [DOI] [PubMed] [Google Scholar]
- 30.Kaltashov IA, Bobst CE, Abzalimov RR, Berkowitz SA, Houde D. Conformation and dynamics of biopharmaceuticals: transition of mass spectrometry-based tools from academe to industry. J. Am. Soc. Mass Spectrom. 2010;21:323–337. doi: 10.1016/j.jasms.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeri Erba E, Barylyuk K, Yang Y, Zenobi R. Quantifying protein-protein interactions within noncovalent complexes using electrospray ionization mass spectrometry. Anal. Chem. 2011;83:9251–9259. doi: 10.1021/ac201576e. [DOI] [PubMed] [Google Scholar]
- 32.Crown SE, Yu Y, Sweeney MD, Leary JA, Handel TM. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J. Biol. Chem. 2006;281:25438–25446. doi: 10.1074/jbc.M601518200. [DOI] [PubMed] [Google Scholar]
- 33.Harmer NJ, Robinson CJ, Adam LE, Ilag LL, Robinson CV, Gallagher JT, Blundell TL. Multimers of the fibroblast growth factor (FGF)-FGF receptor-saccharide complex are formed on long oligomers of heparin. Biochem. J. 2006;393:741–748. doi: 10.1042/BJ20050985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abzalimov RR, Dubin PL, Kaltashov IA. Glycosaminoglycans as naturally occurring combinatorial libraries: Developing a mass spectrometry-based strategy for characterization of anti-thrombin interaction with low molecular weight heparin and heparin oligomers. Anal. Chem. 2007;79:6055–6063. doi: 10.1021/ac0710432. [DOI] [PubMed] [Google Scholar]
- 35.Gordiyenko Y, Robinson CV. The emerging role of MS in structure elucidation of protein-nucleic acid complexes. Biochem.l Soc. Trans. 2008;036:723–731. doi: 10.1042/BST0360723. [DOI] [PubMed] [Google Scholar]
- 36.Pimenova T, Pereira CP, Schaer DJ, Zenobi R. Characterization of high molecular weight multimeric states of human haptoglobin and hemoglobin-based oxygen carriers by high-mass MALDI MS. J. Sep. Sci. 2009;32:1224–1230. doi: 10.1002/jssc.200800625. [DOI] [PubMed] [Google Scholar]
- 37.Berkenkamp S, Karas M, Hillenkamp F. Ice as a matrix for IR-matrix-assisted laser desorption/ionization: Mass spectra from a protein single crystal. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7003–7007. doi: 10.1073/pnas.93.14.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen LRH, Strupat K, Hillenkamp F. Analysis of Quaternary Protein Ensembles by Matrix Assisted Laser Desorption/Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1997;8:1046–1052. [Google Scholar]
- 39.Kirpekar F, Berkenkamp S, Hillenkamp F. Detection of Double-Stranded DNA by IRand UV-MALDI Mass Spectrometry. Anal. Chem. 1999;71:2334–2339. doi: 10.1021/ac990018w. [DOI] [PubMed] [Google Scholar]
- 40.Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 41.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dempsey CE. Hydrogen exchange in peptides and proteins using NMR-spectroscopy. Progr. Nucl. Magn. Res. Spectrosc. 2001;39:135–170. [Google Scholar]
- 43.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 45.Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell. Biochem. 2001;37S:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 46.Baerga-Ortiz A, Hughes CA, Mandell JG, Komives EA. Epitope mapping of a monoclonal antibody against human thrombin by R/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Protein Sci. 2002;11:1300–1308. doi: 10.1110/ps.4670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu JR, Witcher DR, White MA, Wang XL, Huang LH, Rathnachalam R, Beals JM, Kuhstoss S. IL-1 beta epitope mapping using site-directed mutagenesis and hydrogen - Deuterium exchange mass spectrometry analysis. Biochemistry. 2005;44:11106–11114. doi: 10.1021/bi0505464. [DOI] [PubMed] [Google Scholar]
- 48.Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q, Willison LN, Tripathi P, Sathe SK, Roux KH, Emmett MR, Blakney GT, Zhang HM, Marshall AG. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2011;83:7129–7136. doi: 10.1021/ac201501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandit D, Tuske SJ, Coales SJ, Yen S, Liu A, Lee JE, Morrow JA, Nemeth JF, Hamuro Y. Mapping of discontinuous conformational epitopes by amide hydrogen/deuterium exchange mass spectrometry and computational docking. J. Mol. Recogn. 2012;25:114–124. doi: 10.1002/jmr.1169. [DOI] [PubMed] [Google Scholar]
- 51.Obungu VH, Gelfanova V, Rathnachalam R, Bailey A, Sloan-Lancaster J, Huang L. Determination of the mechanism of action of anti-FasL antibody by epitope mapping and homology modeling. Biochemistry. 2009;48:7251–7260. doi: 10.1021/bi900296g. [DOI] [PubMed] [Google Scholar]
- 52.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Structure and properties of α-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rand KD, Zehl M, Jensen ON, Jorgensen TJD. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 54.Huang RYC, Garai K, Frieden C, Gross ML. Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemistry. 2011;50:9273–9282. doi: 10.1021/bi2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan J, Han J, Borchers CH, Konermann L. Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. J. Am. Chem. Soc. 2009;131:12801–12808. doi: 10.1021/ja904379w. [DOI] [PubMed] [Google Scholar]
- 56.Abzalimov RR, Kaplan DA, Easterling ML, Kaltashov IA. Protein conformations can be probed in top-down HDX MS experiments utilizing electron transfer dissociation of protein ions without hydrogen scrambling. J. Am. Soc. Mass Spectrom. 2009;20:1514–1517. doi: 10.1016/j.jasms.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tu TT, Dragusanu M, Petre BA, Rempel DL, Przybylski M, Gross ML. Protein-peptide affinity determination using an H/D exchange dilution strategy: Application to antigen-antibody interactions. J. Am. Soc. Mass Spectrom. 2010;21:1660–1667. doi: 10.1016/j.jasms.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sperry JB, Shi X, Rempel DL, Nishimura Y, Akashi S, Gross ML. A mass spectrometric approach to the study of DNA-binding proteins: Interaction of human TRF2 with telomeric DNA. Biochemistry. 2008;47:1797–1807. doi: 10.1021/bi702037p. [DOI] [PubMed] [Google Scholar]
- 59.Lin JH. Pharmacokinetics of biotech drugs: Peptides, proteins and monoclonal antibodies. Curr. Drug Metab. 2009;10:661–691. doi: 10.2174/138920009789895499. [DOI] [PubMed] [Google Scholar]
- 60.Baumann A. Early development of therapeutic biologics - pharmacokinetics. Curr. Drug Metab. 2006;7:15–21. doi: 10.2174/138920006774832604. [DOI] [PubMed] [Google Scholar]
- 61.Fenselau C. A review of quantitative methods for proteomic studies. J. Chromatogr. B. 2007;855:14–20. doi: 10.1016/j.jchromb.2006.10.071. [DOI] [PubMed] [Google Scholar]
- 62.Ezan E, Dubois M, Becher F. Bioanalysis of recombinant proteins and antibodies by mass spectrometry. Analyst. 2009;134:825–834. doi: 10.1039/b819706g. [DOI] [PubMed] [Google Scholar]
- 63.Shin SU, Friden P, Moran M, Olson T, Kang YS, Pardridge WM, Morrison SL. Transferrin-antibody fusion proteins are effective in brain targeting. Proc. Natl. Acad. Sci. U. S. A. 1995;92:2820–2824. doi: 10.1073/pnas.92.7.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou QH, Boado RJ, Pardridge WM. Selective plasma pharmacokinetics and brain uptake in the mouse of enzyme fusion proteins derived from species-specific receptortargeted antibodies. J. Drug Target. 2012;20:715–719. doi: 10.3109/1061186X.2012.712132. [DOI] [PubMed] [Google Scholar]
- 65.Ibrahim HR, Aoki T, Pellegrini A. Strategies for new antimicrobial proteins and peptides: Lysozyme and aprotinin as model molecules. Curr. Pharm. Des. 2002;8:671. doi: 10.2174/1381612023395349. [DOI] [PubMed] [Google Scholar]
- 66.Stoop MP, Coulier L, Rosenling T, Shi S, Smolinska AM, Buydens L, Ampt K, Stingl C, Dane A, Muilwijk B, Luitwieler RL, Smitt P, Hintzen RQ, Bischoff R, Wijmenga SS, Hankemeier T, van Gool AJ, Luider TM. Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol. Cell. Proteomics. 2010;9:2063–2075. doi: 10.1074/mcp.M110.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mishra OP, Batra P, Ali Z, Anupurba S, Das BK. Cerebrospinal fluid lysozyme level for the diagnosis of tuberculous meningitis in children. J. Trop. Pediatr. 2003;49:13–16. doi: 10.1093/tropej/49.1.13. [DOI] [PubMed] [Google Scholar]
- 68.Rosenling T, Stoop MP, Attali A, Aken Hv, Suidgeest E, Christin C, Stingl C, Suits F, Horvatovich P, Hintzen RQ, Tuinstra T, Bischoff R, Luider TM. Profiling and identification of cerebrospinal fluid proteins in a rat EAE model of multiple sclerosis. J. Proteome Res. 2012;11:2048–2060. doi: 10.1021/pr201244t. [DOI] [PubMed] [Google Scholar]
- 69.Peppard WJ, Johnston CJ, Urmanski AM. Pharmacologic options for CNS infections caused by resistant Gram-positive organisms. Expert. Rev. Anti Infect. Ther. 2008;6:83–99. doi: 10.1586/14787210.6.1.83. [DOI] [PubMed] [Google Scholar]
- 70.Nguyen SN, Bobst CE, Kaltashov IA. Mass spectrometry-guided optimization and characterization of a biologically active transferrin-lysozyme model drug conjugate. Mol. Pharm. 2013 doi: 10.1021/mp400026y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leverence R, Mason AB, Kaltashov IA. Noncanonical interactions between serum transferrin and transferrin receptor evaluated with electrospray ionization mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8123–8128. doi: 10.1073/pnas.0914898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu YJ, Zhang Y, Kenrick M, Hoyte K, Luk W, Lu Y, Atwal J, Elliott JM, Prabhu S, Watts RJ, Dennis MS. Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002230. 84ra44. [DOI] [PubMed] [Google Scholar]
- 73.Amet N, Wang W, Shen WC. Human growth hormone-transferrin fusion protein for oral delivery in hypophysectomized rats. J Control Release. 2010;141:177–182. doi: 10.1016/j.jconrel.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banerjee D, Flanagan PR, Cluett J, Valberg LS. Transferrin receptors in the human gastrointestinal tract. Relationship to body iron stores. Gastroenterology. 1986;91:861–869. doi: 10.1016/0016-5085(86)90687-6. [DOI] [PubMed] [Google Scholar]
- 75.Melanie B. Transferrin' the load. Nat. Rev. Drug Discov. 2005;4:537–537. [Google Scholar]
- 76.Frimpong AK, Abzalimov RR, Eyles SJ, Kaltashov IA. Gas-phase interferencefree analysis of protein ion charge-state distributions: detection of small-scale conformational transitions accompanying pepsin inactivation. Anal. Chem. 2007;79:4154–4161. doi: 10.1021/ac0704098. [DOI] [PubMed] [Google Scholar]
- 77.Johnston KP, Maynard JA, Truskett TM, Borwankar AU, Miller MA, Wilson BK, Dinin AK, Khan TA, Kaczorowski KJ. Concentrated dispersions of equilibrium protein nanoclusters that reversibly dissociate into active monomers. ACS Nano. 2012 doi: 10.1021/nn204166z. in press. [DOI] [PubMed] [Google Scholar]
- 78.Abzalimov RR, Kaltashov IA. Electrospray ionization mass spectrometry of highly heterogeneous protein systems: Protein ion charge state assignment via incomplete charge reduction. Anal. Chem. 2010;82:7523–7526. doi: 10.1021/ac101848z. [DOI] [PubMed] [Google Scholar]
- 79.Fenn LS, McLean JA. Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys. Chem. Chem. Phys. 2011;13:2196–2205. doi: 10.1039/c0cp01414a. [DOI] [PubMed] [Google Scholar]
- 80.Trimpin S, Plasencia M, Isailovic D, Clemmer DE. Resolving oligomers from fully grown polymers with IMS-MS. Anal. Chem. 2007;79:7965–7974. doi: 10.1021/ac071575i. [DOI] [PubMed] [Google Scholar]
- 81.Damen C, Chen W, Chakraborty A, van Oosterhout M, Mazzeo J, Gebler J, Schellens J, Rosing H, Beijnen J. Electrospray ionization quadrupole ion-mobility time-of-flight mass spectrometry as a tool to distinguish the lot-to-lot heterogeneity in N-glycosylation profile of the therapeutic monoclonal antibody trastuzumab. J. Am. Soc. Mass. Spectrom. 2009;20:2021–2033. doi: 10.1016/j.jasms.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 82.Bagal D, Zhang H, Schnier PD. Gas-phase proton-transfer chemistry coupled with TOF mass spectrometry and ion mobility-MS for the facile analysis of poly(ethylene glycols) and PEGylated polypeptide conjugates. Anal. Chem. 2008;80:2408–2418. doi: 10.1021/ac7020163. [DOI] [PubMed] [Google Scholar]
- 83.Cecchelli R, Berezowski V, Lundquist S, Culot M, Renftel M, Dehouck MP, Fenart L. Modelling of the blood-brain barrier in drug discovery and development. Nat. Rev. Drug Discov. 2007;6:650–661. doi: 10.1038/nrd2368. [DOI] [PubMed] [Google Scholar]
- 84.Goldberg M, Gomez-Orellana I. Challenges for the oral delivery of macromolecules. Nat. Rev. Drug Discov. 2003;2:289–295. doi: 10.1038/nrd1067. [DOI] [PubMed] [Google Scholar]
- 85.Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: Is there a need for a top-down approach? Anal. Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]