Abstract
Mitochondrial sequence data has contributed to the understanding of historical demography through the application of neutrality tests and coalescent estimators of population growth rates. Characteristics of the mitochondrial genome, such as high mutation rate and lack of recombination, render it particularly suitable for these types of studies. However, selection can also affect patterns of mitochondrial variation. Furthermore, conclusions based on single mitochondrial loci can be sensitive to differences among mitochondrial genes in mutation rate and pattern and levels of homoplasy. We tested the contributions of these factors to patterns of mitochondrial variation in the Caribbean reef fish Halichoeres bivittatus using a multilocus sequence-based approach. Mitochondrial protein-coding loci deviated strongly from a neutral model of evolution and indicated high rates of estimated population growth. In contrast, the mitochondrial control region and a nuclear intron showed little or no deviation from neutrality and low estimated growth rates. The level of variation among loci is inconsistent with a demographic explanation and likely stems from the influence of mutation and selection on the mitochondrial genome. In H. bivittatus, a finding of high rates of population growth is likely an artifact of selection on mitochondrial proteins. This result suggests caution in the interpretation of variation at single mitochondrial loci, and highlights the importance of the use of unlinked nuclear loci to test demographic inferences made from mitochondrial DNA.
Keywords: Population genetics, Caribbean, Halichoeres, Mitochondrial, Selection, Multilocus
1. Introduction
Empirical studies of patterns of variation in DNA sequence data from wild populations have greatly enriched our understanding of ecological and evolutionary processes, including population expansion. Due to favorable properties in most animals such as haploidy, lack of recombination and generally high levels of intraspecific variation, the majority of these sequence-based studies have relied exclusively on markers from the mitochondrial genome (Avise, 2000).
Yet important caveats on the exclusive use of mitochondrial DNA for the interpretation of demographic history have been recognized for some time. First, as mitochondrial markers provide data on only a single pathway of descent among the many possible (Avise, 2000), it is important to test the robustness of inferences from mitochondrial DNA using nuclear markers (Brito and Edwards, 2009). Second, the mitochondrial genome is subject to selection. A recent study on the decoupling of mitochondrial diversity from population size in some taxa was suggested to be due to recurrent selective sweeps of adaptive mutations (Bazin et al., 2006). Negative selection is also expected to have a prominent role in the evolution of animal mitochondrial DNA due to high rates of deleterious mutation (Meiklejohn et al., 2007; Nachman, 1998). Lastly, studies using mitochondrial DNA often rely on data from a single mitochondrial locus. However, mitochondrial genes may differ in mutational pattern and evolutionary rate (Marshall et al., 2009; Rand and Kann, 1998). A distinction of special interest is in the rate of evolution in protein-coding regions of mitochondrial genomes versus non-coding regions. In mammals, high levels of homoplasy were found in the control region, a popular marker for population studies, due to the presence of mutational hotspots (Stoneking, 2000). High levels of variation in site-specific mutation rates in the control region were also found in tropical reef fish, leading to a reduced signal of divergence relative to more slowly evolving genes (McMillan and Palumbi, 1997).
Hence, although patterns of variation at mitochondrial DNA loci are often interpreted in terms of demographic factors, it is also important to test for the contribution of mutation and selection to the creation of patterns of mtDNA variation, which requires multilocus data. In this study we undertake such a test in the fish species Halichoeres bivittatus, an abundant and highly visible inhabitant of Caribbean reefs.
This tropical wrasse species is a particularly interesting study subject in terms of the origins of mitochondrial variation. First, it may have undergone recent population growth. Large-scale climatic disturbances of the Pleistocene have had a marked effect on the genetic structure of higher latitude plant and animal populations (Hewitt, 2000). Such a pattern was not unexpected, given that the current habitats of many of these species overlap or are subsumed by previously glaciated areas. However, the effects of glacial cycles appear to have extended far from the glacial margins, as even species found in tropical environments exhibit the genetic signature of Pleistocene population fluctuations. For example, observed deviations from a neutral model of evolution in coral reef fish mitochondrial sequence data have previously been linked to bottlenecks and population expansions due to the influence of changing sea-level during glacial cycles on Indo-Pacific (Bay et al., 2004; Craig et al., 2007; Fauvelot et al., 2003), and Caribbean (Pruett et al., 2005) coral reef habitats. Second, the large population size of this species should facilitate the action of selection, whether positive or purifying, and selection may explain the lack of correlation between census size and mitochondrial diversity in marine fishes in general (Bazin et al., 2006; Nabholz et al., 2008). Third, adaptation to different thermal regimes may have driven the clade-level mitochondrial divergence between northern and southern populations of H. bivittatus (Rocha et al., 2005), although positive selection has not been explicitly tested for. To discriminate among the multiple factors that may have acted to shape mitochondrial variation in H. bivittatus, we examine patterns of variation at five loci, from both the mitochondrial and nuclear genomes, in both coding and non-coding regions.
2. Materials and methods
2.1. Field collection of samples
To avoid the influence of known population structure on patterns of variation, sampling was dispersed throughout the range of only the southern Caribbean clade of H. bivittatus (Table 1). No evidence for genetic differentiation among sites was found across the sampled region in a previous study (Rocha et al., 2005) or in this study (see Section 3). A single individual of the closely related species Halichoeres garnoti (Barber and Bellwood, 2005) was collected at Grapetree Bay, St. Croix.
Table 1.
Origins of samples, together with number of haplotypes or alleles sampled from each site for each locus in this study. CR, control region; ATP68, ATPase subunits 6/8; COX1, cytochrome c oxidase subunit 1; S7, S7 ribosomal protein intron 1.
| Locus | ||||
|---|---|---|---|---|
| Sample Site | CR | ATP68 | COX1 | S7 |
| Butler Bay, St. Croix | 10 | 7 | 4 | 8 |
| Northstar Bay, St. Croix | 7 | 6 | 5 | 4 |
| Jack Bay, St. Croix | 17 | 17 | 14 | 24 |
| Grapetree Bay, St. Croix | 10 | 7 | 5 | 2 |
| Ambergris Caye, Belize | 9 | 9 | 5 | 10 |
| South Water Caye, Belize | 11 | 11 | 4 | 12 |
| Cancun, Mexico | 10 | 9 | 7 | 8 |
| San Salvador, Bahamas | 12 | 13 | 10 | 8 |
| Los Roques, Venezuela | 5 | 3 | 3 | 0 |
| Total Hbivittatus | 91 | 82 | 57 | 76 |
| H. garnoti | 1 | 1 | 0 | 0 |
2.2. Laboratory methods
Genomic DNA was extracted from muscle tissue preserved in 70% ethanol using a DNeasy kit (Qiagen, Rockville, MD). Portions of one nuclear and four mitochondrial loci were PCR amplified: control region (CR), cytochrome oxidase subunit 1 (COX1), ATPase sub-units 6 and 8 (ATP6/ATP8), and S7 ribosomal protein intron 1 (S7). Primers for CR were A, K and E (Lee et al., 1995). Primers L8329/ H9076 and L6199/H6855 were initially used for amplification of ATP68 and COX1 (Miya and Nishida, 1999), after which species-specific primers were designed based on acquired sequence for further use (HbivATPF1:5′-GATTGGTGATCCCCAACCACCCC-3′; Hbi-vATPR1: 5′- GGATTAAAGAGGCTAATTGTCTCGAT-3′; HbivCOX1F1: 5′-TCTTCTGGGGTAGAAGCCGGTGC-3′; HbivCOX1R1: 5′-CATTGTAG CGGATGTAAAGTATGC-3′). Reaction conditions for all mitochondrial loci were as follows: 94 °C for 2 min, 35 cycles of 94 °C for 45 s, 50–55 °C for 45 s, 72 °C for 1 min, final extension at 72 °C for 6 min.
A segment of the S7 ribosomal protein intron 1 (S7) was amplified with primers S7RPEX1F and S7RPEX2R (Chow and Hazama, 1998) using the following reaction protocol: 94 °C for 2 min, 40 cycles of 94 °C for 45 s, 58 °C for 45 s, 72 °C for 1 min, final extension at 72 °C for 6 min.
2.3. Sequence assembly and analysis
All PCR products were sequenced with both forward and reverse primers with the exception of COX1, which was sequenced in the forward direction only. Sequences were assembled and processed in Sequencher 4.5 (Gene Codes Corporation, Ann Arbor, MI). For S7 heterozygous sites were called when double peaks occurred in both directions, with secondary peaks at least 60% of primary, using only sequences with high confidence basecalls under Sequencher 4.5 default settings across the analyzed region. We used the Markov chain Monte Carlo method implemented in PHASE 2.1.1 (Stephens et al., 2001) to estimate the phase of haplotypes in the sample from unphased genotypic data. Homozygous individuals, and a small number of cloned alleles, were identified as ‘‘known phase” to improve resolution. For H. garnoti, a homozygous sequence of the S7 first intron (Yaakub et al., 2007) was obtained from Genbank and used in analysis as an outgroup. All sequences were manually aligned in Bioedit 7.0.0 (Hall, 1999). We estimated the minimum number of recombination events using the four-gamete test (Hudson and Kaplan, 1985) in DnaSP 4.0 (Rozas and Rozas, 1999). The population recombination parameter (4Nec) was estimated by γ, and longest non-recombinant intervals were determined, in the program SITES (Hey and Wakeley, 1997). We also tested, via 1000 random permutations of sites, a null model of no recombination under a finite-sites model of sequence evolution (to allow for multiple hits) in LDhat 2.1 (McVean et al., 2002).
We tested for evidence of population structure using pairwise FST values, with significance by comparison to a null distribution determined by 1000 permutations of haplotypes among populations in Arlequin 3.1 (Excoffier et al., 2005). Only locations with five or more sequences for a given locus were used for testing.
Haplotype networks were constructed, and changes at variable sites inferred for each locus under a statistical parsimony criterion (Templeton et al., 1992) with TCS 1.21 (Clement et al., 2000). Measures of sequence diversity were calculated in DNAsp v. 4.0 (Rozas and Rozas, 1999). We tested whether the pattern of observed polymorphism within H. bivittatus is consistent with a neutral Wright-Fisher model using Tajima’s D, Fu and Li’s D* , and Fu’s Fs (Fu, 1997; Fu and Li, 1993; Tajima, 1989).Each of these statistics takes on a negative value with an excess of rare haplotypes or alleles as may occur under scenarios of background selection, selective sweeps, or population expansions.
We also applied Fay and Wu’s H. It is negative when there is an excess of high relative to intermediate frequency derived variants, a pattern argued to be unique to a selective sweep (Fay and Wu, 2000).
The McDonald-Kreitman (MK) test was applied to protein-coding loci (McDonald and Kreitman, 1991), and the neutrality index (NI) was calculated (Rand and Kann, 1996). Hudson-Kreitman-Aguade (HKA) tests (Hudson et al., 1987) were performed in the program HKA to determine whether patterns of polymorphism and divergence varied significantly among loci. We applied a standard Bonferroni correction for multiple tests to control the table-wide error level.
We estimated the population growth parameter g via Markov chain Monte Carlo (MCMC) sampling under an exponential growth model for each locus in a joint analysis in LAMARC 2.1.3 (Kuhner, 2006), accounting for differences in effective population size for markers from either the mitochondrial or nuclear genomes, and allowing for recombination. Best-fit models of sequence evolution for each locus were determined in Modeltest 3.5 (Posada and Crandall, 1998) by the Akaike information criterion (AIC). Nucleotide composition, substitution rate matrices and transition/trans-version ratios (Ti/Tv) were estimated in PAUP 4.04b10 (Swofford, 2002) for each locus as needed to parameterize LAMARC 2.1.3 runs. All LAMARC 2.1.3 outputs are averages from three independent runs of 10 initial chains of 10,000 steps and 2 final chains of 200,000 steps. We assumed equal among-locus mutation rates to allow for direct comparison with single-locus studies and to avoid introducing error caused by using mutation rates estimated from divergence in other reef fish species (Lessios, 2008). Further, differences in mutation rates among loci are small compared to differences among sites within loci, which are accounted for in the substitution models. Lastly, this approach is conservative with respect to differences among mitochondrial and nuclear loci, and significant differences in g among mitochondrial loci are robust if analysis is performed with among-locus rate variation allowed (results not shown).
3. Results
3.1. Sequence data
A total of 91 control region (CR), 82 ATPase subunit 6/8 (ATP68), and 57 cytochrome c oxidase subunit 1 (COX1) sequences were obtained from H. bivittatus mitochondrial DNA (Table 1). All distinct haplotypes at each mitochondrial locus (Table 2) were submitted to Genbank [Accession Numbers, COX1: HM043290-HM043312; ATP68: HM043394-HM043423; CR: HM043313-HM043393]. A total of 38 S7 ribosomal protein intron 1 (S7) sequences (constituting 76 alleles) were also obtained. All original nuclear sequences complete with IUPAC ambiguity codes were submitted to Genbank [Accession Numbers HM752524-HM752561]. Of 268 phase probabilities calculated across all variable sites in S7, only 10 (3.7%) were below 0.6, a threshold above which haplotypes were correctly inferred computationally, and matched those determined by cloning (Harrigan et al., 2008). No inferred haplotype had more than one phase probability below this threshold, and all instances involved low-frequency variants, with the majority singleton polymorphisms. Excluding low-frequency variants from the nuclear data is problematic, as it could bias against finding a signal of population expansion. We thus present results using the full S7 dataset, accepting minor uncertainty in haplotype reconstruction, as the results should be conservative with respect to the differences among mitochondrial and nuclear loci, a key aspect of this study.
Table 2.
Measures of genetic diversity for mitochondrial and nuclear loci. ATP6 and ATP8 have overlapping reading frames and are considered as a single locus for these measures.Numbers in the labels of the last three rows show the positions of non-recombinant intervals of S7 used for neutrality tests. L, length of sequence; Nhaps, number of distinct haplotypes; h, haplotype diversity; π, nucleotide diversity; SD, standard deviation; k, average number of pairwise differences, α, shape parameter of gamma distribution; nc = not calculated.
| Locus | L | N | Nhaps | h | SD h | π | SD π | k | Model | α |
|---|---|---|---|---|---|---|---|---|---|---|
| CR | 364 | 91 | 81 | 0.995 | 0.003 | 0.01609 | 0.003 | 5.680 | TrN + G | 0.560 |
| ATP68 | 522 | 80 | 30 | 0.756 | 0.052 | 0.00262 | 0.0003 | 1.367 | TVM + I + G | 0.269 |
| COX1 | 511 | 57 | 23 | 0.627 | 0.076 | 0.00203 | 0.006 | 1.039 | HKY | NA |
| S7 | 475 | 76 | 39 | 0.971 | 0.008 | 0.01616 | 0.0007 | 7.676 | TrN + I + G | 0.654 |
| S7 1–142 | 142 | 76 | 6 | 0.493 | 0.064 | 0.00393 | 0.0006 | 0.558 | nc | nc |
| S7 64–181 | 118 | 76 | 8 | 0.701 | 0.046 | 0.00897 | 0.0009 | 1.059 | nc | nc |
| S7 266–328 | 63 | 76 | 9 | 0.697 | 0.034 | 0.02292 | 0.001 | 1.444 | nc | nc |
However, all results for S7 were confirmed by performing neutrality tests and coalescent population growth estimation using a second set in which all 10 individuals with a single site with be-low-threshold phase probability were excluded (N = 56), and a third set in which all individuals with phase probabilities less than 0.8 at any site were excluded (N = 44).
3.2. Population structure
Tests for genetic differentiation among sites in this study corroborated an earlier finding of extensive gene flow among populations of the southern clade of H. bivittatus. Pairwise FST values were uniformly low and insignificant across all loci (Table S1). All subsequent analyses pooled sequences at the clade level.
3.3. Variation at sequenced loci, mutation and recombination
Loci differed in their overall levels of variability. The control region was highly variable, while levels of variation in COX1 and the ATP68 region were substantially lower. The first intron of the S7 ribosomal protein was similar to CR on most measures (Table 2). The presence and magnitude of among-site evolutionary rate variation differed among loci. First, rate variation among sites was included in the best-fit model of sequence evolution for CR, ATP68 and S7, but not for COX1. Second, the level of variation among sites as assessed by the shape parameter of the gamma distribution (α) varied (Table 2). Multiple sites in the control region appeared to be mutational hotspots, with 5–10 changes inferred by parsimony per site. Yet only a single site in ATP68 had more than two inferred changes, while no sites had more than a single inferred change in COX1. As for CR, some sites in S7 had multiple inferred changes. However, the proportion of these hypervariable sites (four or more changes) was substantially lower in S7 (4 in 475 bases) than in CR (15 in 364 bases). The maximum number of inferred changes at a site was also lower for the S7 intron, with only a single site having the maximum number of inferred changes (seven). In the control region six sites had seven or more inferred changes, and two had the maximum 10 inferred changes. Six CR sites were segregating 3 or 4 nucleotides, while one, zero and zero were segregating 3 or 4 nucleotides in ATP68, COX1 and S7 respectively.
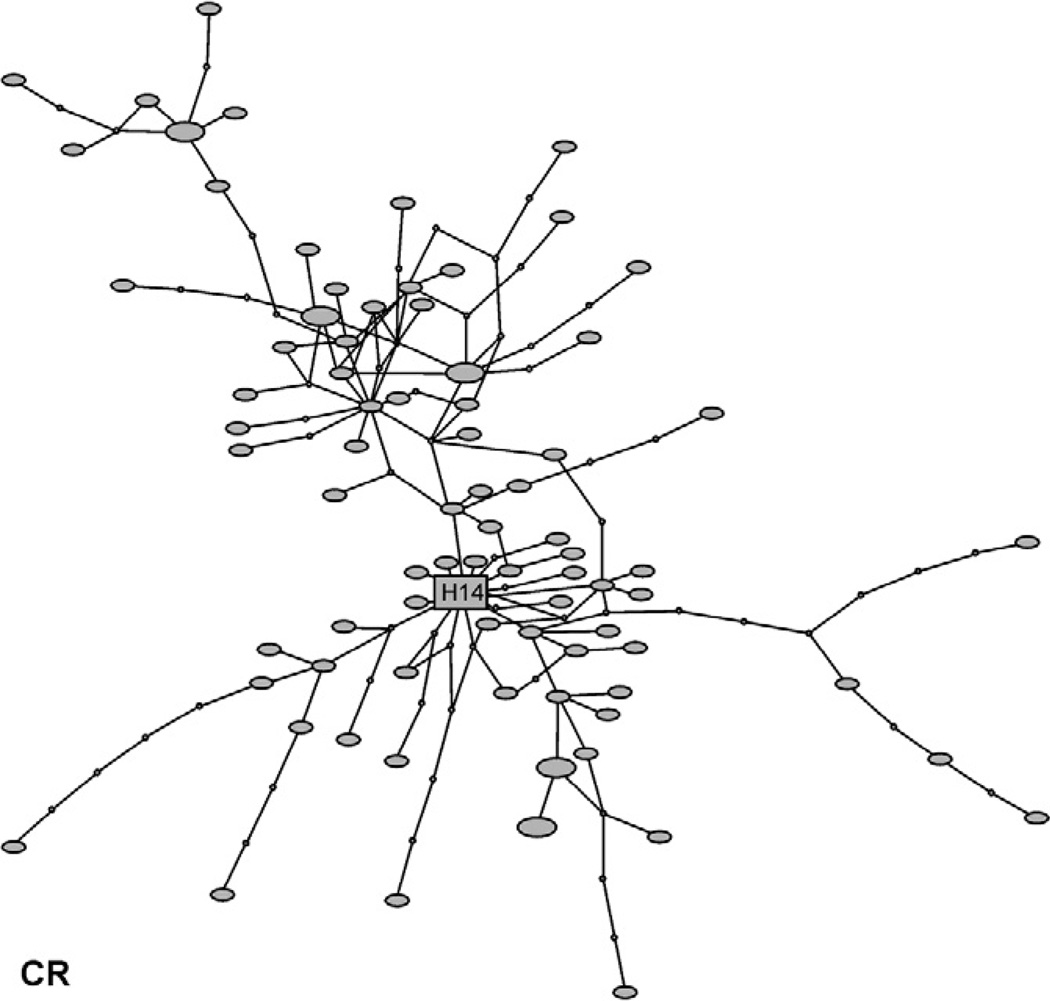
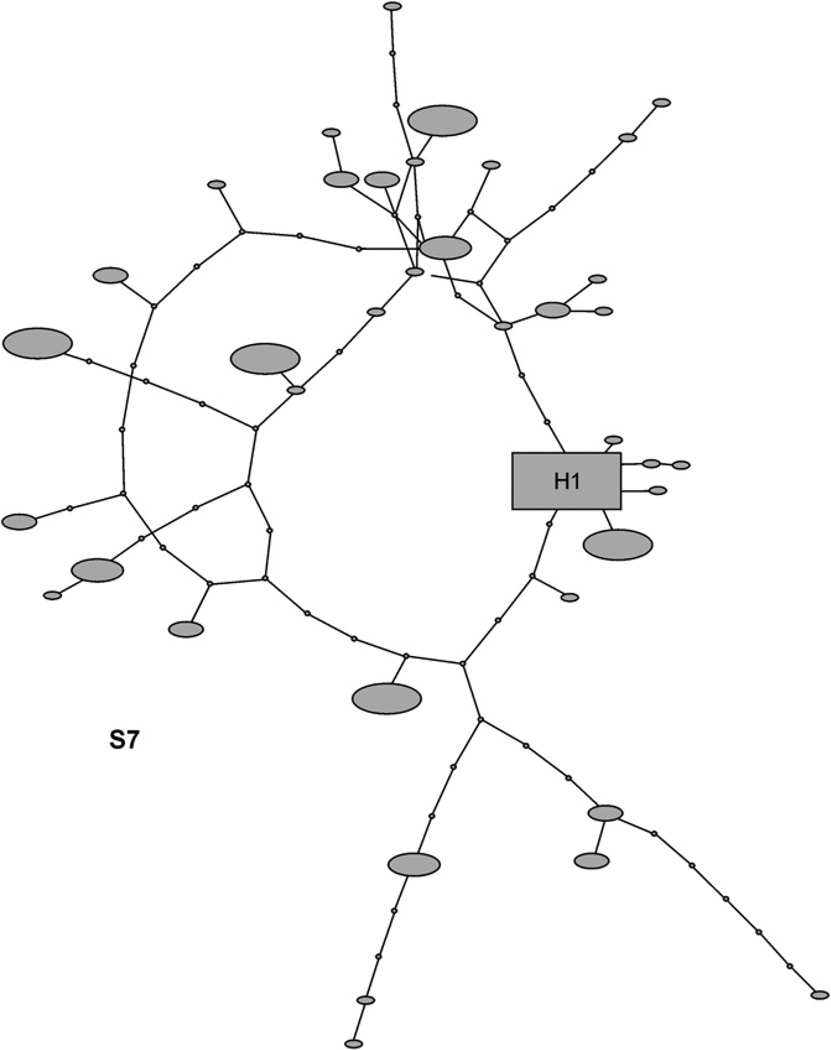
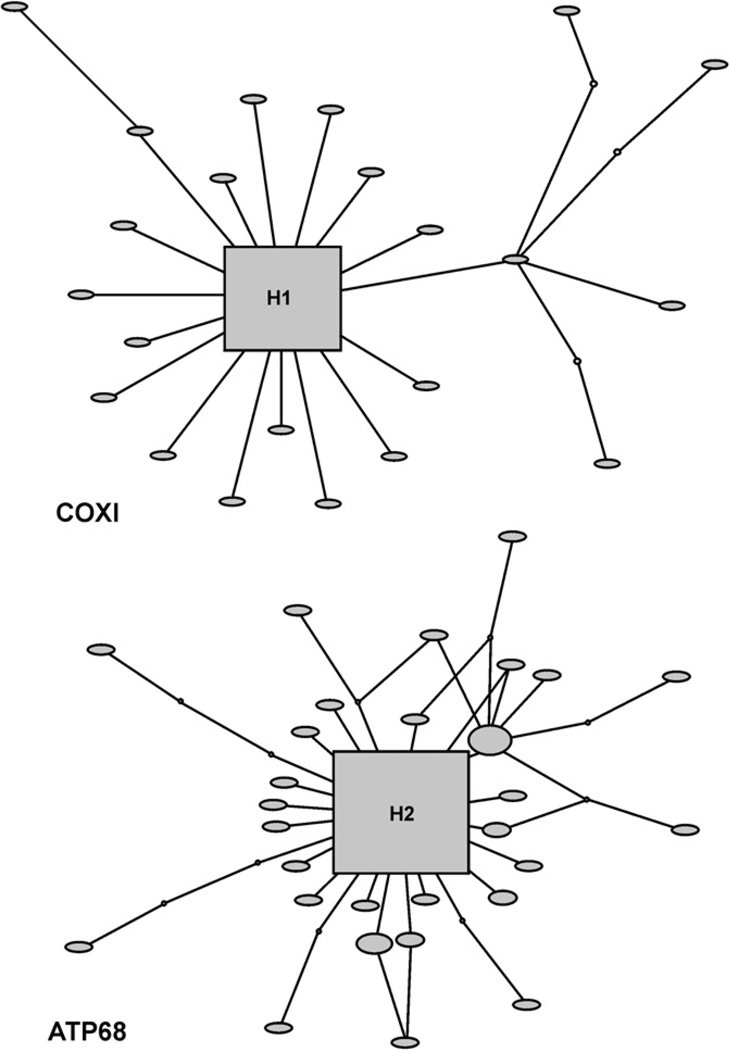
The haplotype networks for CR and S7 were deeply branched, with many haplotypes separated by multiple substitutions (Figs. 1 and 2), and homoplasy was common as a number of closed loops occur. The COX1 and ATP68 networks showed a different pattern, with a single common haplotype and a number of closely related derivatives (Fig. 3). The minimum number of recombination events was estimated at 13 for CR and 9 for S7, while the estimated population recombination parameter (4Nc) for CR was 81.52, and for S7 was 75.18. However, homoplasy evident in the haplotype networks of CR and S7 appeared to be of differential origin, with a mutational basis in CR. Tests for recombination may be misleading when assumptions of the infinite-sites model are violated, as recurrent mutation can produce patterns of variation similar to those under recombination. Hence we tested for recombination by permutation test under a finite-sites model allowing for multiple mutations at a site (McVean et al., 2002). A model with no recombination was rejected (p = 0.002) for S7, but could not be rejected for CR (p = 0.602).
Fig. 1.
Haplotype network for H. bivittatus mitochondrial control region. Square represents the ancestral haplotype, filled circles observed haplotypes, and small open circles inferred intermediate haplotypes. Haplotypes are scaled according to the number of times they occurred in the sample.
Fig. 2.
Network of inferred H. bivittatus nuclear S7 ribosomal protein intron haplotypes. Square represents the ancestral haplotype, filled circles observed haplotypes, and small open circles inferred intermediate haplotypes. Haplotypes are scaled according to the number of times they occurred in the sample.
Fig. 3.
Haplotype network for H. bivittatus COX1 (top) and ATP68 (bottom) gene regions. Square represents the ancestral haplotype, filled circles observed haplotypes, and small circles inferred intermediate haplotypes. Haplotypes are scaled according to the number of times they occurred in the sample.
3.4. Neutrality test results and historical demography
As was true for mutational pattern, variation existed among loci in the pattern of significance of neutrality tests. All intraspecific tests were significant for mitochondrial protein-coding loci, with ATP68 considered a single region for the purposes of these tests. Only Fu’s Fs was significant for CR (Table 3). For S7, none of the test statistics were significant, allowing for recombination. Although high levels of recombination may increase the power of Tajima’s D to detect population expansions, haplotype based statistics such as Fu’s Fs may have decreased power under recombination (Ramirez-Soriano et al., 2008). Hence we also performed these tests with three non-recombinant intervals from S7, the longest and the most variable based on haplotype and nucleotide diversity. No significant deviations from neutrality were found (Table 3). When the two reduced S7 sets were used, with below-threshold phase probability haplotypes excluded, significance of neutrality tests was unchanged.
Table 3.
Results of neutrality tests and estimates of population growth (g) for mitochondrial and nuclear loci. ATP6 and ATP8 have overlapping reading frames and are considered as a single locus for the purposes of testing. Values of H were calculated using H. garnoti as the outgroup. D, Tajima’s D; D* , Fu and Li’s D* ; Fs, Fu’s Fs; H, Fay and Wu’s H; g, maximum likelihood estimate of population growth parameter. Significant p-values are in bold, nc, not calculated.
| Locus | D | p | D* | p | Fs | p | H | p | g |
|---|---|---|---|---|---|---|---|---|---|
| CR | −1.597 | 0.027 | −2.261 | 0.026 | −123.644 | <0.00001 | −13.653 | 0.011 | 1139.332 |
| ATP68 | −2.556 | <0.00001 | −5.761 | <0.00001 | −38.597 | <0.00001 | −17.784 | <0.00001 | 6747.447 |
| COX1 | −2.593 | <0.00001 | −5.664 | <0.00001 | −30.578 | <0.00001 | −17.127 | <0.00001 | 11327.636 |
| S7 | −0.593 | 0.113 | −0.324 | 0.321 | −17.157 | 0.948 | 2.739 | 0.88 | 154.418 |
| S7 1–142 | −1.011 | 0.186 | 0.034 | 0.478 | −2.452 | 0.08452 | 0.503 | 0.867 | nc |
| S7 64–181 | −0.634 | 0.291 | 0.375 | 0.585 | −2.172 | 0.14661 | 0.704 | 0.840 | nc |
| S7 266–328 | −0.556 | 0.324 | 0.617 | 0.682 | −1.791 | 0.22088 | 0.408 | 0.579 | nc |
Fay and Wu’s H was significant for mitochondrial protein-coding loci using H. garnoti as the outgroup (Barber and Bellwood, 2005). As we did not obtain the 511 bp COX1 fragment for H. garnoti, we arbitrarily used a single COX1 sequence from H. garnoti from a previous study (Yaakub et al., 2007) to perform this test. The region of overlap between the previously sequenced fragment and H. bivittatus sequences from this study was 308 bp. Fay and Wu’s H was not significant for the control region after multiple test correction, nor was it significant for the full sequenced fragment of S7, or the non-recombinant intervals.
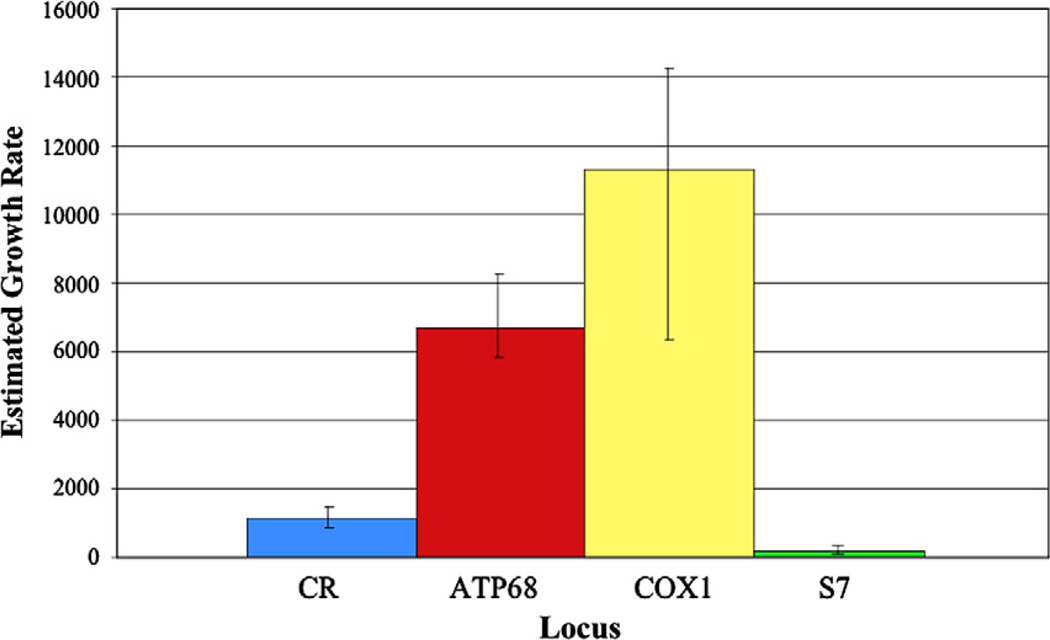
Furthermore, results indicated orders of magnitude differences in the estimated population growth rate (g) between mitochondrial loci and S7, with 95% confidence intervals on g in mitochondrial genes not overlapping those from S7 (Fig. 4). While this is true for all mitochondrial loci, the effect is most pronounced in comparisons between S7 and mitochondrial coding regions. For example, the maximum likelihood estimate of g for COX1 was 73 times that of S7 (Table 3). Substantial variation also occurred between mitochondrial genes. The mean estimated g for COX1 was nearly twice that of ATP68, although the lower 95% confidence limit for COX1 overlapped the mean estimated g for ATP68. Mean estimated growth rates for ATP68 and COX1 were approximately 6–10 times greater than the mean estimated g for CR. There was no overlap in 95% confidence intervals for estimated growth rates between CR and the protein-coding loci. When the two above-threshold phase probability sets of S7 were analyzed, estimates of g varied only minimally from the full set, with strongly overlapping confidence intervals for the different datasets (results not shown).
Fig. 4.
Maximum-likelihood estimated growth rates under an exponential growth model from LAMARC 2.1.3. Error bars indicate 95% confidence intervals on estimates.
3.5. Polymorphism and divergence
Purifying selection on amino-acid altering mutations was evident in the mitochondrial coding sequence data. No nonsynonymous polymorphisms occurred in COX1 within H. bivittatus and no nonsynonymous fixed differences occurred to the outgroup H. garnoti over the 308 bp fragment, or to Halichoeres poeyi, or the distantly related Indo-Pacific Halichoeres melanurus (Barber and Bellwood, 2005) using the 511 bp fragment. As the ATP68 region sequenced contains reading frames from both ATP6 and ATP8, we considered them separately for the following analysis. The MK test for the 396 bp ATP6 open reading frame was not significant (G = 1.663, p = 0.197). The NI for ATP6 was 3.077. The level of variation at synonymous sites (π = 0.011) was an order of magnitude higher than at nonsynonymous sites (π = 0.0003). The MK test for ATP8 (131 bp reading frame) was not significant (G = 0.100, p = 0.752), although NI for ATP8 was 0.667 in comparison to H. garnoti. As in ATP6, the level of variation at synonymous sites in ATP8 (π = 0.003) was an order of magnitude higher than that of nonsynonymous sites (π = 0.0003). All nonsynonymous polymorphisms in ATP6/ATP8 were singletons.
In contrast to expectations under a neutral model, patterns of polymorphism and divergence varied between coding and non-coding loci. For sites in all three mitochondrial protein-coding genes, there were more fixed differences to H. garnoti than polymorphic sites. The opposite was true for the non-coding CR and S7, which had more polymorphic sites than fixed differences (Table S2). Yet an HKA test for all 5 loci was not significant (Sum of deviations = 6.059, df = 4, p = 0.144). However, the presence of low-frequency deleterious variants may obscure the signature of adaptive evolution in procedures that examine patterns of polymorphism and between-species divergence (Fay et al., 2001), and there was pronounced variability in the frequency of polymorphisms at different loci. The majority of polymorphisms in COX1 (23 of 25 in the 511 bp fragment, 16 of 16 in the 308 bp fragment), ATP6 (18 of 25) and ATP8 (4 of 4) were singletons. This was not true for S7 where 11 of 46 were singletons or for CR (19 of 52 singletons). If we conservatively remove only singletons at all loci, under the assumption that they represent deleterious mutations, the HKA test for all loci was significant (Sum of deviations = 23.797, df = 4, p < 0.00001). Deviations from expected values under a constant-rate neutral model occurred primarily at mitochondrial loci, with an excess of polymorphism and deficit in divergence in the control region and an excess of divergence and deficit in polymorphism at the protein-coding loci all contributing substantially. Deviations from neutrality at the nuclear S7 locus were minor.
4. Discussion
In H. bivittatus, as in other tropical reef fish species, we find significant departures from a neutral model of evolution in the spectrum of mitochondrial variation. Such a departure may occur under scenarios of background selection (Charlesworth et al., 1993), selective sweeps (Maynard Smith and Haigh, 1974) or demographic expansion (Fu, 1997). While this last explanation has been favored to explain patterns of mitochondrial variation in tropical reef fishes (Bay et al., 2004; Chen et al., 2008; Craig et al., 2007; Fauvelot et al., 2003), patterns of mitochondrial variation in H. bivittatus do not appear to be easily explained by a bottleneck or an expansion of population size, which should similarly affect all genes in both mitochondrial and nuclear genomes. The strong signature of nonneutrality and high estimated population growth in H. bivittatus, in contrast, appears restricted to mitochondrial protein-coding loci. The control region showed inconsistent results across neutrality tests, and no neutrality tests were significant for a nuclear marker (S7). Estimated population growth for S7 in this study is far lower than for mitochondrial genes, and this difference is of too great a magnitude to be explained by the expected fourfold difference in effective population size between genomes. There is also a large discrepancy in estimated growth rate among mitochondrial genes, which cannot be explained by differences in effective population size. Hence, we must turn to other forces, specifically mutation and selection, to explain the strong variation in results among marker genes, and genomes, observed in this study.
The influence of purifying selection is pronounced in the mitochondrial genome of H. bivittatus, as is clear from the dearth of nonsynonymous polymorphism or divergence. This finding is supported by reanalysis of cytochrome b sequences from an earlier study (Rocha et al., 2005), which showed zero nonsynonymous polymorphisms in H. bivittatus and zero fixed differences to H. garnoti. Purifying selection can lead to a reduction in variability at linked neutral sites (Charlesworth et al., 1993), which may contribute to the reduced variation in H. bivittatus mitochondrial protein-coding regions. It can also have a pronounced effect on the distribution of mutations along a gene genealogy when mutation rates are high, and multiple linked deleterious mutations occur, as is likely for mitochondrial DNA. This shift in mutation distribution can lead to significant deviations from neutral expectations, and to significant biases in coalescent-based estimations of population parameters (Seger et al., 2010; Williamson and Orive, 2002). Hence purifying selection against deleterious amino-acid altering mutations would seem a likely candidate explanation for the strong nonneutral and population growth signature in mitochondrial protein-coding genes.
However, the power to reject neutrality requires deleterious mutations to be segregating in the sample, and segregating neutral mutations can obscure the signal of purifying selection (Williamson and Orive, 2002). If we assume that only nonsynonymous mutations are selected against, we should have low power to detect deviations from neutrality at the coding loci, as few nonsyn-onymous polymorphisms segregate. An alternative is that the frequency spectrum of synonymous mutations varies from neutral expectations because some synonymous mutations are themselves deleterious. The vast majority of synonymous mutations in the H. bivittatus COX1 and ATP68 genes are singletons, a pattern that contrasts strongly with non-coding polymorphism in both the mitochondrial and nuclear genomes, where singleton polymorphisms are in the minority. There is some bias towards particular codons within codon families in COX1, ATP6 and ATP8 (data not shown), suggesting that some synonymous mutations may be selected against, although it seems unlikely that this can account for the large observed deviations from neutrality. However, the number of codons sampled to date is small and additional sequencing of mitochondrial loci from H. bivittatus could help to resolve the importance of this phenomenon.
The significant values of Fay and Wu’s H for both mitochondrial protein-coding genes and the excess of fixations at these loci are difficult to explain by purifying selection alone, and suggest that positive selection could have a role in determining the pattern of variation in H. bivittatus mitochondrial DNA. However, an excess of amino acid fixations is clearly not driving this pattern, at least based on the protein-coding loci sampled in this study or a previous study (Rocha et al., 2005). Yet, in the absence of recombination, the entire mitochondrial genome may act as a linkage group, and the possibility remains that amino acid changes have recently been driven towards or to fixation at other loci, leading to a signature in linked variation (Maynard Smith and Haigh, 1974). As levels of mitochondrial polymorphism are substantial, any selective sweep that might have occurred was not recent, or was incomplete.
Although its exact nature remains to be clarified, it is clear that selection has affected H. bivittatus mitochondrial DNA, and apparently high rates of population growth estimated from H. bivittatus protein-coding loci may be due largely to selection. This finding is consistent with a number of studies on the role of selection, both negative and positive, in shaping mtDNA variation in a wide range of animal species (Bazin et al., 2006; Meiklejohn et al., 2007; Rand and Kann, 1996). As the action of selection on mtDNA may compromise estimation of effective population size and inferences of historical changes in this parameter (Hurst and Jiggins, 2005), it should be considered when making inferences from mitochondrial DNA sequence data, particularly from individual protein-coding loci.
Mutational effects also appear to influence the pattern of variation at mitochondrial loci. While part of the same linkage group as the protein-coding loci, the control region lacks a signature of positive selection, and in fact appears to have an excess of polymorphism that contributes substantially to the significance of the HKA test. Although ‘‘recombination” was apparent in this region, this result was not significant under a model allowing for recurrent mutation. The hypermutability of this region leads to substantial homoplasy that may be driving the decoupling of its pattern of polymorphism and divergence from that at coding sites and obscuring the signature of mitochondrial selection as assessed by most neutrality tests. In contrast, hypermutational effects on patterns of sequence variation, which resemble recombination, might also be contributing to the inflated negative value of Fs for the control region, mimicking the effects of demographic expansion for this statistic, which is haplotype-based and sensitive to recombination (Ramirez-Soriano et al., 2008). Control region hypermutability has been documented in tropical reef fish (McMillan and Palumbi, 1997), and has been implicated as responsible for apparent recombination in mammalian mitochondrial DNA (Stoneking, 2000). In spite of the likely strong influence of mutation on patterns of control region variation in many species, it is still commonly used as a marker for population dynamics. These results suggest that caution is warranted in demographic inference from control region data.
In H. bivittatus, while selection and hypermutation confound estimation of historical demography from mitochondrial DNA, the nuclear S7 intron sequenced for this study showed no evidence for selection, and has few hypervariable sites, yet does indicate a weakly positive population growth rate. Using a mutation rate for this locus derived from Caribbean reef fish (Lessios, 2008), we estimate the time to 1% of current population size as approximately 10 million years before present under an exponential growth model as a proxy for the onset of population expansion using this locus (Wares and Cunningham, 2001). Although crude, this estimate greatly predates Pleistocene glaciation, invoked as a common factor in modulating population sizes of other reef fish through sea level change (Fauvelot et al., 2003), and would suggest an alternative history for H. bivittatus. However, it is clear from this study that we have learned more about among-gene variation in the mutation-selection balance than we have about the historical demography of Halichoeres, and further sampling of unlinked nuclear loci is necessary to confirm this result. It is clear that Pleistocene impacts on tropical reef fish populations need more careful study before cause and effect are clearly established, including further studies of multilocus variation.
5. Conclusions
The use of multilocus sequence data provides evidence that the effects of selection and mutation on variation at mitochondrial loci in the Caribbean reef fish H. bivittatus are pronounced. These effects lead to patterns similar to that found under strong population growth, and might have led to spurious demographic inference if single mitochondrial loci had been studied. This finding highlights the importance of the use of unlinked nuclear loci to test inferences made from mitochondrial DNA. The results presented here raise the possibility of low rates of population growth for H. bivittatus, and suggest that the influence of Pleistocene glaciation on tropical reef fish populations may be complex and variable.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Craig Layman for assistance in sample collection, Mark Bertness, IZE, Genaissance Pharmaceuticals, NSF (DEB 0108500 and 0343464 to DMR) and NOAA-NERRS and Rhode Island EPSCoR fellowships to RAH for funding.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ympev.2010.07.014.
Contributor Information
Robert A. Haney, Email: robert.a.haney@gmail.com.
Brian R. Silliman, Email: brs@ufl.edu.
David M. Rand, Email: David_Rand@brown.edu.
References
- AvisePhylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press; [Google Scholar]
- Barber PH, Bellwood DR. Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Mol. Phylogenet. Evol. 2005;35:235–253. doi: 10.1016/j.ympev.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Bay LK, Choat JH, van Herwerden L, Robertson DR. High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): evidence of an unstable evolutionary past? Mar. Biol. 2004;144:757–767. [Google Scholar]
- Bazin E, Glemin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- Brito P, Edwards SV. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica. 2009;135:439–455. doi: 10.1007/s10709-008-9293-3. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SP, Liu T, Li ZF, Gao TX. Genetic population structuring and demographic history of red spotted grouper (Epinephelus akaara) in South and East China Sea. Afr. J. Biotechnol. 2008;7:3554–3562. [Google Scholar]
- Chow S, Hazama K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 1998;7:1255–1256. [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Craig MT, Eble JA, Bowen BW, Robertson DR. High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae) Mar. Ecol. Prog. Ser. 2007;334:245–254. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. Online. [PMC free article] [PubMed] [Google Scholar]
- Fauvelot C, Bernardi G, Planes S. Reductions in the mitochondrial DNA diversity of coral reef fish provide evidence of population bottlenecks resulting from Holocene sea-level change. Evolution. 2003;57:1571–1583. doi: 10.1111/j.0014-3820.2003.tb00365.x. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. Bioedit sequence alignment editor. Department of Microbiology, North Carolina State University. 1999 http://www.mbio.ncsu.edu/BioEdit/BioEdit.html.
- Harrigan RJ, Mazza ME, Sorenson MD. Computation vs. Cloning: evaluation of two methods for haplotype determination. Mol. Ecol. Resour. 2008;8:1239–1248. doi: 10.1111/j.1755-0998.2008.02241.x. [DOI] [PubMed] [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. P. Roy. Soc. B-Biol. Sci. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner MK. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Conroy J, Howell WH, Kocher TD. Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 1995;41:54–66. doi: 10.1007/BF00174041. [DOI] [PubMed] [Google Scholar]
- Lessios HA. The great American schism: divergence of marine organisms after the rise of the Central American isthmus. Annu. Rev. Ecol. Evol. S. 2008;39:63–91. [Google Scholar]
- Marshall HD, Coulson MW, Carr SM. Near neutrality, rate heterogeneity, and linkage govern mitochondrial genome evolution in Atlantic Cod (Gadus morhua) and other gadine fish. Mol. Biol. Evol. 2009;26:579–589. doi: 10.1093/molbev/msn279. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh Locus in Drosophila . Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McMillan WO, Palumbi SR. Rapid rate of control-region evolution in Pacific butterflyfishes (Chaetodontidae) J. Mol. Evol. 1997;45:473–484. doi: 10.1007/pl00006252. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM. Positive and negative selection on the mitochondrial genome. Trends. Genet. 2007;23:259–263. doi: 10.1016/j.tig.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Miya M, Nishida M. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar. Biotechnol. 1999;1:416–426. doi: 10.1007/pl00011798. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Mauffrey JF, Bazin E, Galtier N, Glemin S. Determination of mitochondrial genetic diversity in mammals. Genetics. 2008;178:351–361. doi: 10.1534/genetics.107.073346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW. Deleterious mutations in animal mitochondrial DNA. Genetica. 1998;103:61–69. [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pruett CL, Saillant E, Gold JR. Historical population demography of red snapper (Lutjanus campechanus) from the northern Gulf of Mexico based on analysis of sequences of mitochondrial DNA. Mar. Biol. 2005;147:593–602. [Google Scholar]
- Ramirez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A. Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics. 2008;179:555–567. doi: 10.1534/genetics.107.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA. Genetica. 1998;103:393–407. [PubMed] [Google Scholar]
- Rocha LA, Robertson DR, Roman J, Bowen BW. Ecological speciation in tropical reef fishes. P. Roy. Soc. B-Biol. Sci. 2005;272:573–579. doi: 10.1098/2004.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolutionanalysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Seger J, Smith WA, Perry JJ, Hunn J, Kaliszewska ZA, Sala LL, Pozzi L, Rowntree VJ, Adler FR. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics. 2010;184:529–545. doi: 10.1534/genetics.109.103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am. J. Hum. Genet. 2000;67:1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP Phylogenetic analysis using parsimony. (* and other methods) Sunderland, MA: Sinauer Associates, Inc; 2002. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonu-clease mapping and DNA sequence data.III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wares JP, Cunningham CW. Phylogeography and historical ecology of the North Atlantic intertidal. Evolution. 2001;55:2455–2469. doi: 10.1111/j.0014-3820.2001.tb00760.x. [DOI] [PubMed] [Google Scholar]
- Williamson S, Orive ME. The genealogy of a sequence subject to purifying selection at multiple sites. Mol. Biol. Evol. 2002;19:1376–1384. doi: 10.1093/oxfordjournals.molbev.a004199. [DOI] [PubMed] [Google Scholar]
- Yaakub SM, Bellwood DR, van Herwerden L. A rare hybridization event in two common Caribbean wrasses (genus Halichoeres; family Labridae) Coral Reefs. 2007;26:597–602. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.