Abstract
The discovery of the Ten-Eleven-Translocation (TET) oxygenases that catalyze the hydroxylation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) has triggered an avalanche of studies aiming to resolve the role of 5hmC in gene regulation if any. Hitherto, TET1 is reported to bind to CpG-island (CGI) and bivalent promoters in mouse embryonic stem cells, whereas binding at DNAseI hypersensitive sites (HS) had escaped previous analysis. Significant enrichment/accumulation of 5hmC but not 5mC can indeed be detected at bivalent promoters and at DNaseI-HS. Surprisingly, however, 5hmC is not detected or present at very low levels at CGI promoters notwithstanding the presence of TET1. Our meta-analysis of DNA methylation profiling points to potential issues with regard to the various methodologies that are part of the toolbox used to detect 5mC and 5hmC. Discrepancies between published studies and technical limitations prevent an unambiguous assignment of 5hmC as a ‘true' epigenetic mark, that is, read and interpreted by other factors and/or as a transiently accumulating intermediary product of the conversion of 5mC to unmodified cytosines.
Keywords: DNA demethylation, sixth base, Ten-Eleven-Translocation, 5-hydroxymethylcytosine, 5-methylcytosine
Introduction
In nearly half a century of research and in tens of thousands of publications, the role of the 5-methylcytosine (5mC) DNA modification has been studied in development, differentiation, imprinting, X-inactivation, gene regulation and disease, and still it possesses many riddles. It is often named the fifth base to illustrate its importance and heritability.
The discovery that 5-hydroxymethylcytosine (5hmC) constitutes a small, but significant fraction of cytosines in the DNA derived from Purkinje neurons (Kriaucionis and Heintz, 2009) and the identification of the enzyme converting 5mC to 5hmC—the Ten-Eleven-Translocation (TET) oxidase—(Tahiliani et al, 2009) questions whether methylation of cytosines is the only, or at least the important DNA modification in mammalian genomes and sparked studies to unveil the location and biological function of the mark.
A link between 5hmC and endogenous oxidative stress in DNA of mammalian tissues was first described (Cannon et al, 1988; Boorstein et al, 1989; Cannon-Carlson et al, 1989). Due to technical limitations in analyzing 5hmC, attention faded and the awareness of this modification in mammalian genomes was ‘lost.' In 1993, base J was identified in the nuclear DNA of Trypanosoma brucei (Gommers-Ampt et al, 1993a 1993b). The newly identified TET enzymes are related to the Trypanosoma proteins, JBP1 et JBP2, that belong to the 2 oxoglutarate and Fe(II)-dependent hydroxylases family (Yu et al, 2007; Cliffe et al, 2009). Overexpression of wild-type and mutant TET1 and RNA interference-mediated depletion of endogenous TET convincingly showed that TET catalyzes the conversion of 5mC to 5hmC in cultured cells (Tahiliani et al, 2009; Ito et al, 2010; Koh et al, 2011).
In this Perspective, we will focus on recent genome-wide profiling studies that provide the basis for future functional analysis.
Biological function of TET proteins
The observation that in acute myeloid leukemia (AML), TET1 is an oncofusion partner of the histone H3 Lys4 (H3K4) methyltransferase MLL provided a first link between TET proteins and the epigenome (Ono et al, 2002; Lorsbach et al, 2003). The molecular underpinning of how MLL–TET1 fusion protein contributes to leukemogenesis remained, however, largely unexplored. The identification of TET proteins as oxygenases by the Rao and Heintz laboratories and the discovery that TET2 is frequently mutated in a range of human myeloid malignancies, including myelodysplastic syndromes (Delhommeau et al, 2009; Langemeijer et al, 2009) placed this small family of oxygenases into the limelight. TET2 mutations appear to associate with low 5hmC levels and global hypomethylation (Ko et al, 2010), suggesting that an altered 5hmC status leads to deregulation of important hematopoietic regulators and contributes to malignancy. Mutations of TET2 and the isocitrate dehydrogenase genes IDH1/IHD2 that catalyze the interconversion of isocitrate to α-ketoglutarate appears to be mutually exclusive in AML (Figueroa et al, 2010), consistent with the requirement of the TET enzymes for αKG as substrate.
Two studies using conditional knockout of Tet2 provided important insights into the role of TET2 in normal hematopoiesis and malignancies (Moran-Crusio et al, 2011; Quivoron et al, 2011). TET2 loss resulted in expansion of hematopoietic stem and progenitor cell populations directly contributing to myeloproliferation. This is consistent with a role of TET2 disruption (by deletion or sequence mutation) in the pathogenesis of lymphoid as well as myeloid disorders. Moreover, the mutations in both lineages of malignancy are commonly acquired in early hematopoietic progenitors of multi-lineage potential, indicating that the enhanced self-renewal upon TET2 inactivation is an important contributor to transformation. Knockout of Tet1 in embryonic stem cell (ESC) causes a subtle reduction of 5hmC levels in ESC but did not affect pluripotency possibly because of the compensatory action of TET2. Surprisingly, Tet1−/− mice turn out to be viable and fertile (Dawlaty et al, 2011).
In an elegant study, Walter and co-workers revealed the role of 5hmC in genome-wide DNA demethylation in zygotic development. This laboratory had previously shown that the paternal genome in the pronucleus rapidly undergoes active DNA demethylation of 5mC and remains demethylated following several rounds of cell division, while the maternal genome remains methylated even though it is exposed to the same cytoplasmic factors (Oswald et al, 2000). In a recent study, they showed that 5mC is converted into 5hmC in the paternal pronucleus by the TET3 dioxygenase (Wossidlo et al, 2011). Furthermore, they confirmed the role played by PGC7/Stella in blocking/inhibition the TET3-mediated oxidation in the maternal pronucleus (Nakamura et al, 2007; Wossidlo et al, 2011). Inoue and Zhang (2011) further showed that 5hmC of the paternal genome is lost following replication. The stability of the 5hmC mark suggests that it itself may be a functional modification linked to chromatin (re)organization events in early cleavage embryos (Wossidlo et al, 2011). In fact, blocking oxidation of 5mC by TET3 deletion reduced developmental fitness, fetal survival and affected the epigenetic reprogramming of the donor nuclear DNA in somatic cell nuclear transfer (Gu et al, 2011).
Recently, it was found that TET proteins not only have the capacity to oxidize 5mC to 5hmC, but also to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in vitro (He et al, 2011; Ito et al, 2011) and could play a role in DNA demethylation implying that 5hmC, 5fC and 5caC may only be intermediates of the removal of methylated cytosines. Xu and his group showed that T DNA glycosylase (TDG) can specifically recognize and excise 5caC (He et al, 2011); this conversion could trigger TDG initiated by base excision repair (BER) glycosylases that would lead to DNA demethylation. Moreover, studies of nuclear reprogramming showed that activation-induced cytidine deaminase (AID), a member of APOBEC family proteins, mediates deamination of cytosine residues to uracils, which are then repaired by either BER or mismatch repair (Bhutani et al, 2010). Indeed, DNA demethylation was reported in combination with the recruitment of BER enzymes as part of the transcription cycle at target genes such as PS2 (TFF1) in response to ligand activation of the estrogen receptor (Metivier et al, 2008).
TET genome-wide profiles
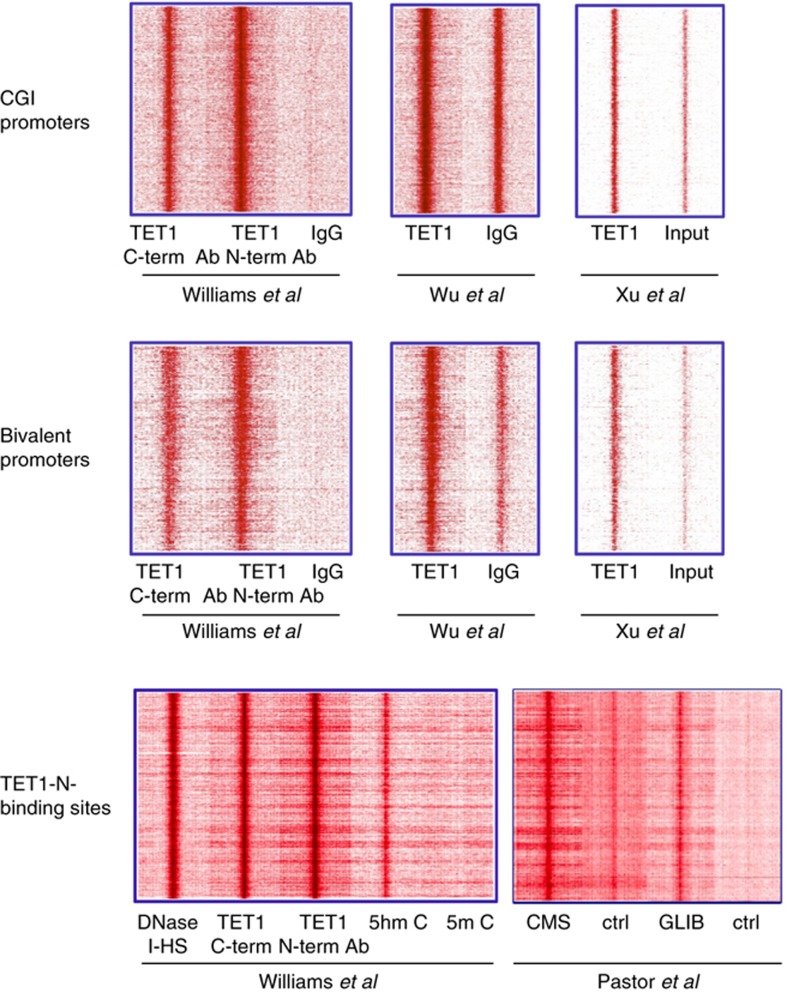
To shed light on the role of TET1 in ES cells, chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) has been used to establish genomic profiles of TET1 (Williams et al, 2011; Xu et al, 2011; Wu et al, 2011b). To rule out possible differences due to mapping algorithms and settings, we remapped the published data from mouse ESC (Williams et al, 2011; Xu et al, 2011; Wu et al, 2011a). TET1 appears to be highly enriched at nearly all CpG-island (CGI) promoters that are hypomethylated (Figure 1 top panel). It was suggested that TET1 may play a role in maintenance of the hypomethylated state (Williams et al, 2011; Xu et al, 2011). However, knockdown of TET1 only led to a small increase in 5mC levels at TET1-binding sites. In the absence of direct and compelling evidence, it remains an open question what role TET1 is playing in this promoter context. Different groups reported that TET1 also binds actively transcribed CpG-poor gene promoters, such as Nanog, Esrrb and Tcl1, whose gene products are pluripotency-related factors (Ito et al, 2010; Ficz et al, 2011; Wu et al, 2011b). However, these findings have not been confirmed following depletion of TET1 and/or TET2 (Koh et al, 2011; Williams et al, 2011). Importantly, in TET1 knockout cells, pluripotency was not affected and expression of the pluripotency markers Oct4, Nanog and Sox2 was not altered (Dawlaty et al, 2011).
Figure 1.
Heatmap representation of TET1 enrichment in CGI promoters (top panel) and bivalent promoters (middle panel). Heatmap representation of TET1 DnaseI-HS (ENCODE-ZhBTc4 cell line), hmC, and mC enrichment in TET1-N-binding sites (bottom panel). All the heatmaps are from +5 to −5 kb flanking TSSs of CGI, bivalent promoters or TET1-N centered binding sites, reads were summed in 100 bp sliding windows. Datasets used in the analysis are indicated.
TET1 is also present at the so-called ‘bivalent' promoters (Figure 1 middle panel) a chromatin state characterized by the presence of H3K4me3, H3K27me3 and PRC2 (Azuara et al, 2006; Bernstein et al, 2006; Mikkelsen et al, 2007; Ku et al, 2008). A direct interaction between TET1 and PRC2 could not be detected (Williams et al, 2011; Wu et al, 2011b). Interestingly, our analysis of the published datasets shows that TET1-binding sites nearly perfectly overlap with DNAseI hypersensitive sites (HS) as determined in the ZhBTc4 ESCs (Levasseur et al, 2008) (Figure 1 bottom panel). These results provide further evidence for a role of TET1 in transcriptional regulation.
5hmC state maps
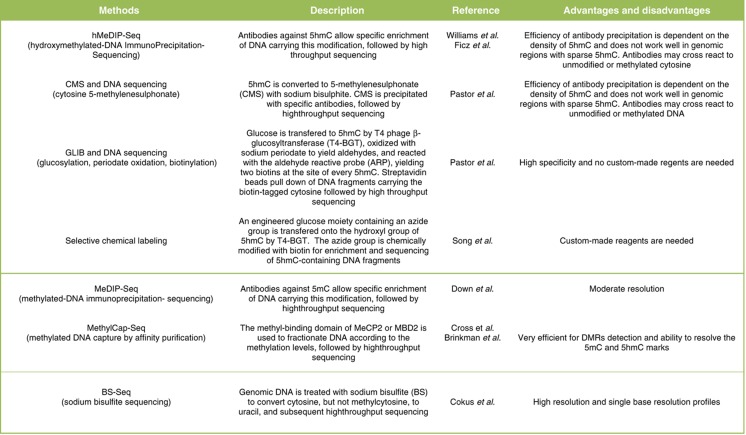
The discovery of 5hmC spurred efforts to detect and profile 5hmC. Methods based on affinity enrichment through methylcytosine-specific protein domains (Cross et al, 1994) or through antibody-mediated immunoprecipitation as in MeDIP (methylated-DNA immunoprecipitation-sequencing; Weber et al, 2005) that efficiently enrich for 5mC-containing DNA, do not pull down 5hmC (Huang et al, 2010; Jin et al, 2010; Nestor et al, 2010). Furthermore, sodium bisulfite treatment of DNA (Clark et al, 1994) does not distinguish between 5mC and 5hmC (Huang et al, 2010; Jin et al, 2010; Nestor et al, 2010). In a relatively short time, an entire toolbox has been developed to distinguish 5mC from 5hmC. Many of these methods only detect and quantitate 5hmC levels, while others have coupled their method to massive parallel sequencing to study 5hmC localization on a genomic scale (Table I).
Table 1. The 5hmC and 5mC toolbox.
 |
The first genome-wide 5hmC profile was generated for mouse cerebellum DNA using selective chemical labeling and affinity enrichment involving an azide-modified glucose transfer to 5hmC by β-glucosyltransferase (βGT) and click chemistry to couple a biotin derivative used for subsequent enrichment of modified 5hmC (Song et al, 2010). 5hmC appeared to be enriched in gene bodies, regions proximal to the transcription start sites (TSSs) as well as transcription end sites of highly expressed genes suggestive of a role in activation and/or maintenance of gene expression. Several genome-wide studies used hMeDIP-seq (hydroxymethylated-DNA immunoprecipitation-sequencing), a method adapted from MeDIP-seq (Weber et al, 2005; Down et al, 2008; Butcher and Beck, 2010) using antibodies raised against 5hmC. Notwithstanding the use of the same antibody source, the conclusions of these studies are not entirely concordant (see below).
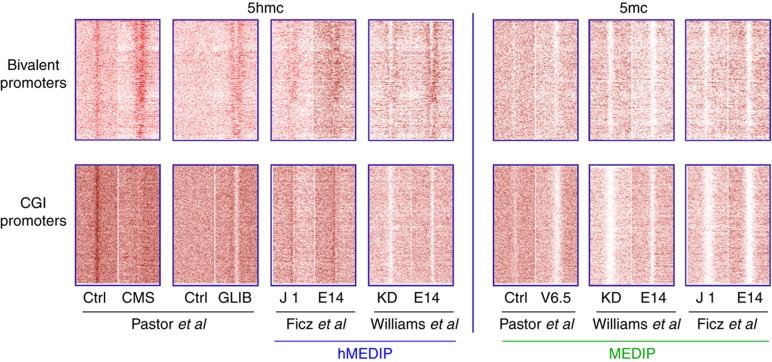
In a first comparison, we computed intensity plots of genomic locations that are occupied by TET1 (Figure 2). At bivalent promoters, a very clear enrichment of 5hmC is detected in GLIB (glucosylation, periodate oxidation, biotinylation), an approach involving βGT conversion (Pastor et al, 2011). A good enrichment is also attained by CMS (cytosine 5-methylenesulfonate) that elegantly makes use of sodium bisulfite conversion of 5hmC to CMS (Hayatsu and Shiragami, 1979) and immunoprecipitation with an antibody raised against CMS (Ko et al, 2010; Pastor et al, 2011). The signal over background is much less pronounced in the various hMeDIP-seq assays. In all studies, MeDIP-seq displays a clear depletion of 5mC at bivalent loci (Figure 2).
Figure 2.
Heatmap representation of 5hmC and 5mC enrichment at CGI promoters and bivalent promoters depicted as in Figure 1.
The intensity plots of CGI promoters reveal discordance between the published datasets. Whereas a very slight enrichment is seen in the Ficz et al data depletion is observed in the other hMeDIP-seq profiles. Also in GLIB and CMS, 5hmC is depleted over the TSS (Figure 2). The origin of the ‘background' signal in the CMS control—essentially a bisulfite sequencing profile—is not clear at present.
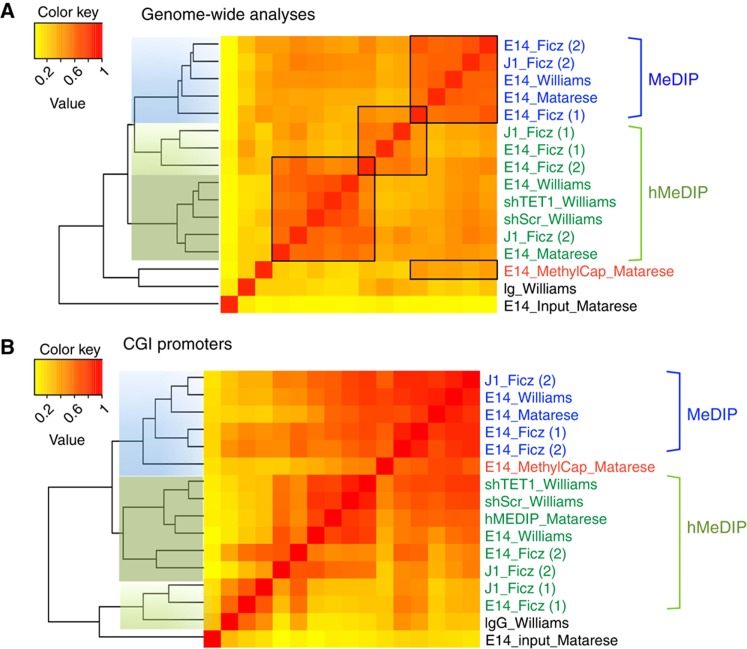
To gain insight into the differences, we performed a genome-wide sliding window approach and pairwise R2 correlations. We included three of our own datasets: MeDIP-, hMeDIP- and MethylCap-seq of E14 ESC (for more details on methods, data processing and analysis see Supplementary information). It should be noted that the same antibody source was used in these MeDIP and hMeDIP studies, respectively. The GLIB and CMS profiles are excluded from the pairwise correlations because of the intrinsic difference in the data structure. The heatmap reveals that the MeDIP-seq data group together in a distinct cluster whereas the hMeDIP-seq splits up into two clusters (Figure 3a), suggesting that the hMeDIP approach is not yet technically mature and/or sufficiently standardized. This becomes even more apparent in the clustering of CGI promoters (−500 to +500 bp around the TSS). The heatmap is rather disorganized in hMeDIP profiles and they do not cluster together (Figure 3b); some of the hMeDIP profiles correlate with ChIP's performed with IgG. The clustering is likely driven by background as foreground signals are largely absent (see also Figure 1). In the absence of clear proof that 5hmC is present/elevated at CGI promoters, the model that TET1 clears CGI promoters from 5mC by converting it into 5hmC (Ficz et al, 2011) needs to be taken with great caution.
Figure 3.
Hierarchical clustering of pairwise R2 correlation on 5hmC and 5mC genome-wide profiling studies in mouse ESCs. (A) Hierarchical clustering and heatmap representation in the full genome (1 kb sliding windows) (B) CGI promoters (±500 bp sliding window from TSS).
Asymmetric 5(h)mC?
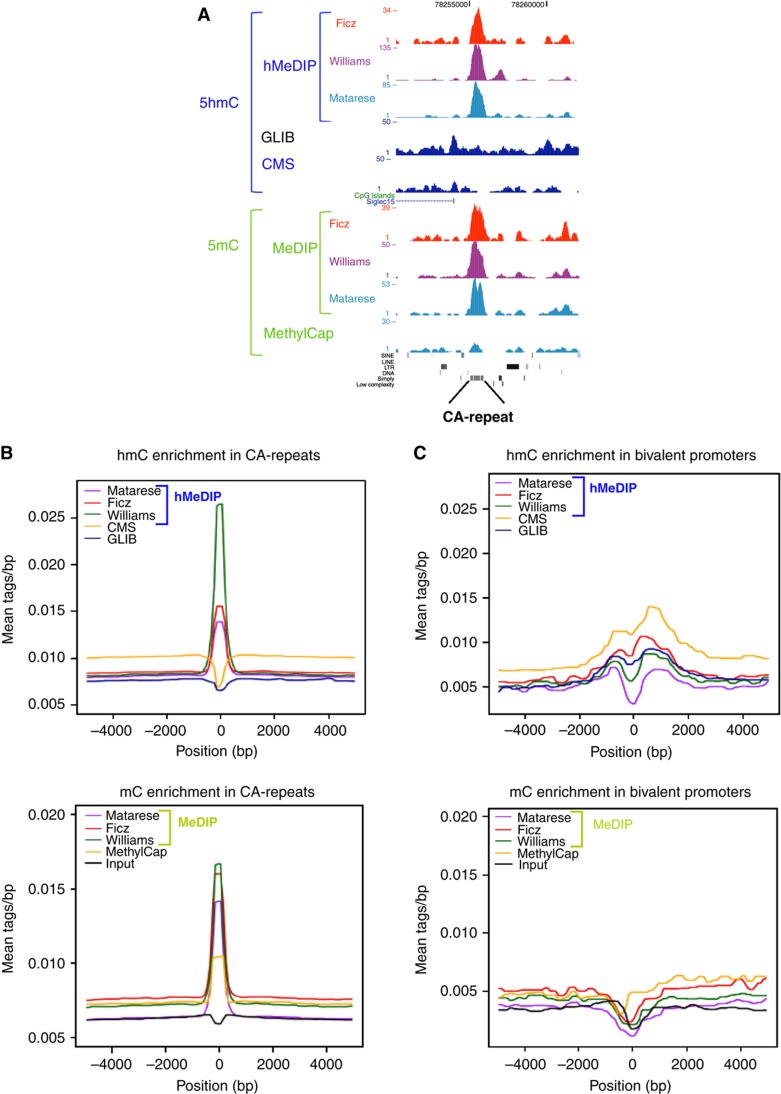
In the genome-wide comparison, the profile generated by MethylCap-seq (Brinkman et al, 2010) though having a different data structure shows a correlation with the MeDIP (boxed in Figure 3). It should be noted that binding of the MBD in MethylCap requires symmetrically methylated CpG (Nan et al, 1993). In immunoprecipitation with 5mC or 5hmC antibodies, asymmetrically methylated DNA can also be pulled down as the immunoprecipitation is performed on denatured, single-stranded DNA. Indeed, visual inspection of the profiling data in a genome browser showed high enrichment of CA-repeats (Figure 4a) and CT-repeats (data not shown) in MeDIP and hMeDIP but not in MethylCap. Genome-wide analysis revealed a very prominent enrichment for CA- and CT-repeats (Figure 4b). The DNA strands containing the CA- or CT-sequence, but not their complementary strands, are highly preferentially enriched and sequenced. The question arises whether this prominent enrichment is a proof of 5(h)mC at CA- and CT-repeats or whether these regions are precipitated because of cross-reactivity of the antibodies with unmethylated cytosines. Deep sequencing of bisulfite converted DNA uncovered the presence of asymmetric non-CpG methylation at least in human ES cells (Ramsahoye et al, 2000; Lister et al, 2009). Because the bisulfite approach does not discriminate between 5mC and 5hmC, the intriguing question is whether asymmetric modification of the 5mC as well as 5hmC type indeed occur in the CA- and CT-repeats in the genome and what is the role of such an asymmetric distribution.
Figure 4.
5hmC and 5mC distributions at CA simple repeats and bivalent promoters. (A) Genome browser view of a CA simple repeat element with read distributions for 5hmC and 5mC using datasets as indicated. (B) 5hmC and 5mC distributions at CA simple repeats and in (C) bivalent promoters. All the distribution plots are from +5 to −5 kb flanking CA simple repeats centered (B) or TSSs bivalent promoters (C), reads were summed in 100 bp sliding windows.
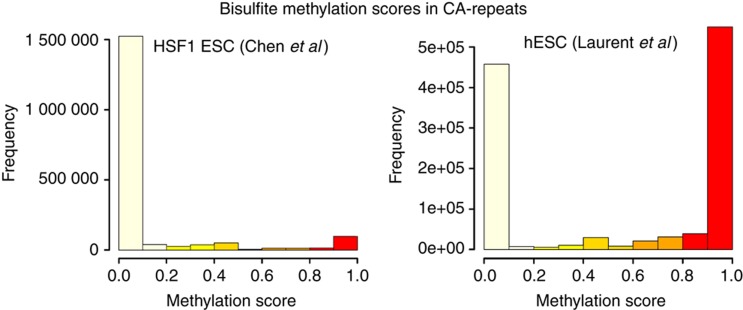
The extent to which the CA- and CT-repeats are enriched in MeDIP and hMeDIP is however puzzling; the average tag density of peaks over CA-repeats is higher than over bivalent loci (Figure 4c), suggesting that the repeats may be extensively (hydroxyl)methylated in mouse ESC. In striking contrast with the high enrichment in (h)MeDIP, genome-wide deep sequence analysis of bisulfite converted DNA from human ESCs show that symmetric CpG methylation is much more abundant than non-symmetric CA or CT modification (Lister et al, 2009; Laurent et al, 2010; Chen et al, 2011). Our re-analysis of bisulfite deep sequencing data (see also Supplementary information) shows that CA- and CT-repeats are predominantly unmodified in HFS1 ESCs (Chen et al, 2011), whereas in WA09 hESCs (Laurent et al, 2010), modified and unmodified cytosine are present at roughly equal level in the repeats (Figure 5). While the basis of the discrepancy between the two lines is unclear, these BS-seq data reveal that cytosine modification in repeats are present at least in some human ESCs. Given these large differences, extrapolation of the data obtained with human to mouse ESC is premature and awaits the availability of BS-seq profiles in mouse ESC lines.
Figure 5.
Methylation score distribution in CA-repeats from bisulfite-seq studies in human ESCs. Datasets used in the analysis are indicated. Only cytosines with a coverage higher than 1 are included.
The reasons underlying the surprisingly prominent enrichment in (h)MeDIP remain unclear. It could very well be due to cross-reactivity of the antibodies with unmethylated cytosine. Dot blot analysis revealed selectivity for the respective modified cytosine; however, these experiments were not performed with synthetic oligonucleotides with high cytosine content or with CA- and CT-repeats. If cross-reactivity indeed plays a role, one would predict that genomic regions with high local cytosine density such as in CA- and CT-repeats might be efficiently immunoprecipitated. The extend of 5mC and in particular 5hmC is very low as compared with unmethylated cytosine; therefore, immunoprecipitation of the much more abundant unmodified cytosines in regions of high cytosine density could be substantial. The lack of signal in GLIB and CMS at CA- and CT-repeats indicates but does not proof that the signals in the CA- and CT-repeats is at least in part due to cross-reactivity.
To resolve this ambiguity, parallel BS-seq and (h)MeDIP analysis need to be performed on the same cell lines/strains, but it is beyond the scope of this Perspective to include such extensive analysis. Definitive proof awaits systematic comparison and benchmarking of the different methods.
Concluding remarks
Exciting and unexpected new observations appear at a dazzling speed and change our perspectives on the function of cytosine modifications. It is still early times though to propose unifying models and mechanisms. One likely scenario is that the TET enzymes are involved in removal of 5mC and maintenance of the unmethylated state. The conversion of 5mC to 5hmC by TET enzymes would be the first step maybe followed by further conversion into 5fC and 5caC. Several reports suggest that TET enzymes indeed act along with BER pathways to promote active DNA demethylation. The knockout of TDG revealed its active role in the demethylation process (Cortellino et al, 2011) and a very recent study reports that 5caC form is specifically recognized and excised by TDG (He et al, 2011). The presence of 5hmC at bivalent loci and DNaseI HS might imply that simultaneous removal as well as renewed deposition of 5mC by DNMTs is taking place and that the kinetics of the removal of 5hmC is slow, resulting in accumulation of 5hmC. The presence of TET1 at CGI promoters and the contrasting absence or the very, very low signal of 5hmC (Ficz et al, 2011) might simply be explained by a faster kinetics of 5hmC conversion/removal by BER or TDG than the deposition of 5mC and it conversion to 5hmC.
However, TET1 could also play a regulating function in addition to its enzymatic role. Given the association of TET1 with the Sin3-complex, TET1 may contribute to repression of transcription. Alternatively, the co-recruitment of (co)factors to CGI promoters may or may not lead to conversion into and/or accumulation of formyl- (fC) and/or caC instead of 5hmC. Proof of the accumulation of fC and/or caC at CGI promoters awaits development of methods for their detection.
An important question is whether readers of 5hmC exist that could shield the 5hmC mark from enzymatic removal and that could translate/interpret its presence into a biological action as can be expected from a ‘true' epigenetic mark. Affinity capture experiments as performed for the histone methyl mark readers (Vermeulen et al, 2010) will likely uncover whether such 5hmC readers exist. Until then, the role of 5hmC as a new epigenetic mark—the ‘sixth base'—that is read and interpreted by other factors or merely a removal intermediate remains an open question.
Acknowledgments
We thank Hendrik Marks for the MethylCap data and our colleagues for discussion and critical reading of the manuscript. This work was funded by ZonMW, partners of the ERASysBio+ initiative supported under the EU ERA-NET Plus scheme in FP7. The hMedip-, Medip- and MethylCap-seq data reported in this study have been deposited in the NCBI GEO SuperSeries GSE31343.
Footnotes
The authors declare that they have no conflict of interest.
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM (2010) Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein RJ, Chiu LN, Teebor GW (1989) Phylogenetic evidence of a role for 5-hydroxymethyluracil-DNA glycosylase in the maintenance of 5-methylcytosine in DNA. Nucleic Acids Res 17: 7653–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG (2010) Whole-genome DNA methylation profiling using MethylCap-seq. Methods 52: 232–236 [DOI] [PubMed] [Google Scholar]
- Butcher LM, Beck S (2010) AutoMeDIP-seq: a high-throughput, whole genome, DNA methylation assay. Methods 52: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon-Carlson SV, Gokhale H, Teebor GW (1989) Purification and characterization of 5-hydroxymethyluracil-DNA glycosylase from calf thymus. Its possible role in the maintenance of methylated cytosine residues. J Biol Chem 264: 13306–13312 [PubMed] [Google Scholar]
- Cannon SV, Cummings A, Teebor GW (1988) 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun 151: 1173–1179 [DOI] [PubMed] [Google Scholar]
- Chen PY, Feng S, Joo JW, Jacobsen SE, Pellegrini M (2011) A comparative analysis of DNA methylation across human embryonic stem cell lines. Genome Biol 12: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22: 2990–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R (2009) JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res 37: 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D et al. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP (1994) Purification of CpG islands using a methylated DNA binding column. Nat Genet 6: 236–244 [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F et al. (2009) Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301 [DOI] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R et al. (2008) A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD et al. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers-Ampt JH, Teixeira AJ, van de Werken G, van Dijk WJ, Borst P (1993a) The identification of hydroxymethyluracil in DNA of Trypanosoma brucei. Nucleic Acids Res 21: 2039–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers-Ampt JH, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P (1993b) beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T brucei. Cell 75: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Hayatsu H, Shiragami M (1979) Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry 18: 632–637 [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A (2010) The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5: e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y (2011) Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet Proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP (2010) Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A (2010) Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH (2009) Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842 [DOI] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL (2010) Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH (2008) Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev 22: 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR (2003) TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 17: 637–641 [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD et al. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71 [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A (1993) Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res 21: 4886–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor C, Ruzov A, Meehan R, Dunican D (2010) Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques 48: 317–319 [DOI] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y (2002) LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 62: 4075–4080 [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J (2000) Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W et al. (2011) TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38 [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 97: 5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C (2010) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 29: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–962 [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J (2011) 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2: 241. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y (2011a) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y (2011b) Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, van Luenen HG, Rigden DJ, Sabatini R, Borst P (2007) The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res 35: 2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]