Summary
Membrane protein structure determination remains a challenging endeavor. Computational methods that predict membrane protein structure from sequence can potentially aid structure determination for such difficult target proteins. The de novo protein structure prediction method, BCL::Fold, rapidly assembles secondary structure elements into 3-dimensional models. Here we describe modifications to the algorithm, named BCL::MP-Fold, in order to simulate membrane protein folding. Models are built into a static membrane object and are evaluated using a knowledge-based energy potential, which has been modified to account for the membrane environment. Additionally, a new symmetry folding mode allows for the prediction of obligate homomultimers, a common property amongst membrane proteins. In a benchmark test of 40 proteins of known structure, the method samples the correct topology in 34 cases. This demonstrates that the algorithm can accurately predict protein topology without the need for large multiple sequence alignments, homologous template structures, or experimental restraints.
Introduction
Membrane proteins (MPs) constitute more than one quarter of the human genome (Fagerberg et al., 2010) and more than one half of all known drug targets (Bakheet and Doig, 2009). However, less than two percent of structures present on the Protein Data Bank (PDB) are MPs, meaning that a large number of MP folds have not yet been characterized (Oberai et al., 2006). Presently, about 100 helical multispan integral MP folds have been determined – about 1200 are expected to exist based on analysis of sequence databases (Hopf et al., 2012). This wide disparity between MP biological relevance and available structural information is in large part due to the specialized biophysical properties that make MPs difficult to express, purify, and study by X-ray crystallography or NMR spectroscopy. Computational techniques can often provide structural insight when experimental data is lacking, however the majority of the effort in this field has been focused on soluble proteins.
There are several successful algorithms for predicting MP transmembrane (TM) spans (Fagerberg et al., 2010), yet predicting MP three dimensional structure remains difficult. One consequence of the low number of MP folds with known structure is that for a larger fraction of MP targets, template-based modeling is not an option. There are a few de novo methods that have been successful in predicting structures for small (< 150 residues) MPs, one example of which is Rosetta-Membrane (Yarov-Yarovoy et al., 2006). However many larger and more complex MPs, such as G-protein coupled receptors (GPCRs), are important both biologically and pharmaceutically, yet remain challenging targets for these methods.
One important caveat is that the accuracy of prediction methods can improve drastically with the incorporation of restraints (Barth et al., 2009). The restraints can be experimental, such as NMR chemical shifts, residual dipolar couplings, and distance restraints (Sanders and Sonnichsen, 2006). Additionally, recent work has demonstrated the power of predicted contact restraints generated from sequence analysis with the accurate prediction of MPs with up to 14 TM segments (Hopf et al., 2012; Nugent and Jones, 2012). Obtaining experimental data or a large number of homologous sequences cannot be guaranteed for any given target; therefore improved algorithms for de novo MP structure prediction would be beneficial for the field.
De novo protein structure prediction methods can be broken down into two components – sampling and scoring. During the sampling phase, the protein model is perturbed in some manner. The protein model is then scored, using a scoring function designed to identify native-like topologies. The bottleneck of de novo protein structure is sampling the large conformational space densely enough so that one model approaches the native conformation to about 2 Å RMSD100 (Carugo and Pongor, 2001). At this accuracy an all-atom energy function can usually distinguish the native-like conformation from alternatives (Bradley et al., 2005). However, without restricting the conformational space through experimental restraints, such sampling densities can only be achieved for very small proteins (<80 residues). Therefore progress in de novo MP structure prediction accuracy is likely to arise from an increase in sampling efficiency. Rosetta uses a fragment assembly approach to replace stretches of 3 or 9 residues with conformations determined from sequence similarity to known structures (Simons et al., 1997). While this method samples local contacts effectively, it is challenged to adequately sample long range contacts, especially for larger proteins (Bonneau et al., 2002; Lange and Baker, 2012). As evidence, modifying the algorithm to allow for random chain breaks improves performance when using long range contacts as restraints (Barth et al., 2009).
BCL::Fold is a novel protein structure prediction method that rapidly assembles secondary structure elements (SSEs) into topologies (Karakas et al., 2012). For soluble proteins, a pool of predicted SSEs is generated using JUFO (Meiler et al., 2001) and PSIPRED (Jones, 1999). The SSEs are then assembled using a Monte Carlo (MC) assembly protocol and evaluated using a consensus knowledge-based scoring function (Woetzel et al., 2012) via the Metropolis criterion. The method achieves prediction accuracies comparable to Rosetta.
We hypothesize that assembly of SSEs is a particular efficient approach to sampling MP topology space: helical MP topology is defined by the relative orientation of the transmembrane helices (TMHs). While there are many variations of the canonical bundle of TMHs (Nugent and Jones, 2011), the fold space of MPs is much reduced when compared to soluble proteins (Oberai et al., 2006). The underlying hypothesis of BCL::Fold is that the interactions between SSEs determine the majority of the protein core and therefore give rise to its thermodynamic stability. For the TMHs in MPs this is particularly true, as the apolar membrane environment promotes formation of secondary structure thereby minimizing the error introduced into BCL::Fold through the absence of loop regions in the initial assembly. In order to test this hypothesis and begin to predict the structure of larger membrane proteins in the absence of experimental data, we have developed BCL::MP-Fold to predict MP topology. The scoring functions were modified to account for the unique properties of the apolar membrane in the amino acid environment potential and an increased radius of gyration along the membrane normal. Three additional scores are introduced to describe the preferred orientation of SSEs with respect to the membrane, penalize connection of two neighboring TMHs that would require passage through the membrane, and assess the agreement of amino acid placement in membrane regions with prediction from sequence. As many MPs are obligate homo-oligomers, a symmetrical folding mode was introduced. The method was benchmarked against 40 MPs of known structure. In 34 cases, BCL::MP-Fold was able to sample the native topology, indicating that assembly of discreet SSEs is a viable strategy in de novo MP protein topology determination.
Results
The BCL::Fold protocol (Karakas et al., 2012) was first updated to accommodate MP structure determination. This involved the addition of additional assembly stages prior to refinement, the incorporation of MP specific scores, and the incorporation of MP specific MC moves. Additionally, symmetric multimer folding was introduced in order to predict homomultimeric MP structure. Finally, the new method was benchmarked against 40 MPs of known structure.
Multi-phase MP assembly from SSEs begins with placement of TMHs
BCL::Fold for soluble proteins is broken into two stages: assembly and refinement. Large scale perturbations – such as SSE transformations, SSE addition/deletion, and domain shuffling – are utilized in the assembly stage. The refinement stage focuses on small-amplitude transformations and SSE bending that apply minor modifications to the topology established by the assembly stage.
To enable a subsequent assembly of TMH and non-TMH SSEs, the assembly stage is broken up into five sub-stages. The weight of the clash scores (amino acid and SSE) start at zero in the first stage and are increased linearly to maximum values for the fifth stage. This allows for SSEs to “move through” each other during assembly, allowing for more efficient sampling of distinct topologies. Additionally, two SSE pools are used during the minimization. For the first two stages of refinement, the SSE pool contains only TMHs predicted by SPOCTOPUS (Viklund et al., 2008). This allows for the core of the MP to be assembled without interference from small, amphipathic helices or soluble SSEs which do not determine the overall MP topology. Once the core has been assembled in the second stage, the remaining stages utilize a pool that contains both SPOCTOPUS and JUFO9D (Leman et al., 2012) predictions. This serves two purposes. First, since SPOCTOPUS only predicts TMHs, additional SSEs predicted by JUFO9D can be added at this point. Second, SPOCTOPUS predictions are rather rigid with regards to TMH length; the addition of JUFO9D allows for shrinking/extending of the TMHs based on the predicted probability and the associated knowledge-based (KB) potential. Finally, in the refinement stage small moves optimize SSE interactions and packing. This stage is identical to the one described for soluble proteins, aside from the MP-specific scores and moves described below. The complete assembly and refinement process is repeated 1000 times to generate the final models (Figure S1A).
Three-layer implicit membrane representation with total thickness of 50 Å
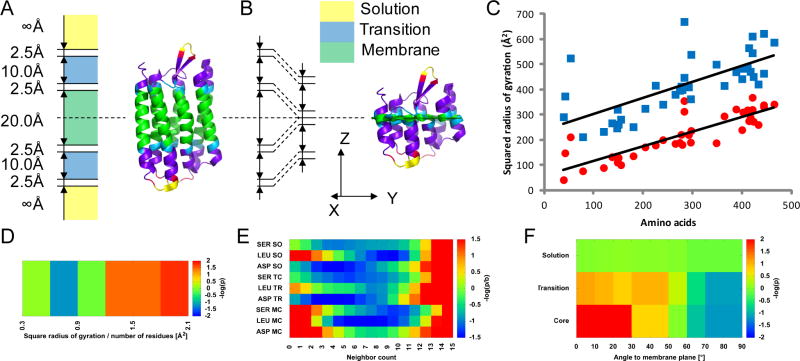
BCL::MP-Fold uses a static, three-layer membrane object (Figure 1A) with scoring functions optimized for each of the three layers. The membrane apolar core is 20 Å thick. A transition region of 10 Å on either side of the membrane core marks the position of lipid head groups. Beyond this transition region, regular potentials for soluble proteins are assumed. To facilitate a smooth transition between the layers, 2.5 Å gaps are introduced between the membrane core and transition region and between the transition region and soluble region. The total membrane thickness is therefore 50 Å. Thickness of membrane core and transition region were held constant for the present study but can be manually adjusted if desired. Bi-cubic splines are used to ensure that all potentials are smooth and continuously differentiable especially along the membrane normal. During the MC assembly process, the membrane is situated along the XY-plane, with the Z-axis as the membrane normal. The minimization starts with the placement of a single, random TMH in the membrane, with its main axis perpendicular to the membrane plane.
Figure 1. MP-specific scoring functions account for the unique transmembrane environment.
A – BCL::Fold uses a static membrane object. The membrane core is 20 Å thick. A 2.5 Å gap separates the core from each 10 Å thick transition region. Another 2.5 Å gap separates the transition region from the solution. B – Protein model after collapsing along the membrane normal. C – Squared radius of gyration values are shown before (blue squares) and after (red circles) collapsing the membrane along the normal. Collapsing the membrane produces more realistic squared radius of gyration values for smaller MPs (< 200 amino acids); the extrapolated line crosses y-axis much closer to 0 after collapsing. D – Radius of gyration using collapsed coordinates along the Z-axis in the membrane core. E – Representative amino acid environment potentials for each environment type; solution (SO), transition (TR) and membrane core (MC). Refer to Figure S2 for the complete plot of potentials. F – SSE alignment relative to the membrane place for each environment type.
BCL::Score – Scoring soluble proteins
BCL::MP-Fold modifies the established soluble scoring function. Here we briefly summarize the theory used to derive the potential. The reader is referred to the BCL::Score manuscript for additional details (Woetzel et al., 2012).
The soluble BCL::Fold method evaluates models using BCL::Score, a knowledge-based potential, derived using Baye’s theorem: . P(struct) is the probability of observing the structure, independent of the sequence. This term describes the relative arrangement of SSEs in space. P(seq|struct) is the probability of observing the sequence for a given structure. This term addresses the likelihood of the placement of specific amino acid types into SSEs. P(seq) is constant for protein structure prediction. Individual knowledge-based scores therefore contribute to one of two terms: 1) P(struct) – structure based or 2) P(seq|struct) – sequence based. All probabilities are converted to potentials using the inverse Boltzmann relation. The final score is a linear combination of each potential and its optimized weight.
MP-specific scores describe rules that govern MP topology
In order to accommodate the unique biophysical properties of the membrane, two existing soluble knowledge-based scores were modified (radius of gyration and amino acid environment) and three new scores were introduced (SSE alignment, MP topology, and environment prediction accuracy). Before describing the scores in detail, the procedure used to evaluate these scores is introduced.
A set of models was created by perturbing a subset of the native structures from the benchmark set described further below (Table 1). These BCL perturbed models were generated in a similar manner as the soluble models (Woetzel et al., 2012). In order for an identical topology to sample various environments, the models were also subjected to two additional global transformations: a random translation along the Z-axis 0 to 5 Å, and a random rotation of 0 to 15° around a random axis in the X–Y plane. The native models were initially oriented in the membrane using PDBTM (Tusnady et al., 2005). Native-like models were defined using three criteria – RMSD100 < 8.0 Å (Carugo and Pongor, 2001), RMSD100_XY < 10.0 Å, and contact recovery (CR) > 25.0 % (Karakas et al., 2012). The RMSD100_XY quality measure was introduced in order to differentiate models with the same topology but sampling different environments. For example, a MP globally translated out of the membrane would have an RMSD100 of 0.0 Å to the native, but when evaluated, would produce a different score due to the different environment. To account for this property, atom coordinates are collapsed onto the membrane plane prior to superimposition. The transformation required to achieve the optimal superimposition is then applied to the original, pre-collapsed structure. In other words, the superimposition of the target onto the native does not change the atom Z-coordinates. The RMSD100 is then calculated normally using the resulting coordinates.
Table 1.
Benchmark statistics and results. The reported RMSD100 values are reported after sorting the models by either RMSD100 (a) or score (b). The RMSD100 is calculated over all Cα atoms in native SSEs.
| BCL::MP-Fold | Rosetta-Membrane | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| PDB | TMH | Domain | Residues | Subunits | RMSD100 a | Scoreb | RMSD100 a | Scoreb | ||||
|
| ||||||||||||
| Best | Top 5% | Best | Top 5% | Best | Top 5% | Best | Top 5% | |||||
|
Traditional folding
| ||||||||||||
| 2BG9 | 3 | A: 211-301 | 91 | 1 | 2.8 | 3.4 | 9.9 | 6.7 | 3.9 | 5.5 | 10.7 | 10.2 |
| 1NKZ | 2 | A: 2-53; B: 1-41 | 93 | 1 | 4.3 | 4.6 | 11.2 | 8.3 | ||||
| 2L35 | 3 | A: 1-63; B: 1-32 | 95 | 1 | 3.1 | 3.7 | 17.2 | 9.7 | ||||
| 2KSF | 4 | 396-502 | 107 | 1 | 3.9 | 4.5 | 5.1 | 5.6 | 5.6 | 6.9 | 10.5 | 10.9 |
| 1J4N | 3 | 4-119 | 116 | 1 | 4.9 | 5.9 | 9.6 | 9.0 | 4.5 | 6.5 | 9.7 | 10.2 |
| 3SYO | 2 | 76-197 | 122 | 1 | 5.2 | 6.3 | 9.7 | 10.0 | 6.4 | 7.9 | 11.0 | 11.2 |
| 1PY6 | 4 | 77-199 | 123 | 1 | 3.9 | 4.7 | 5.4 | 6.4 | 2.2 | 3.2 | 2.8 | 5.7 |
| 2PNO | 4 | A: 2-131 | 130 | 1 | 5.0 | 6.7 | 5.4 | 8.6 | 4.0 | 5.1 | 9.4 | 8.2 |
| 2BL2 | 4 | 12-156 | 145 | 1 | 2.9 | 3.8 | 6.7 | 7.3 | 2.2 | 3.1 | 3.5 | 5.6 |
| 2K73 | 4 | 1-164 | 164 | 1 | 4.7 | 5.9 | 10.1 | 9.1 | 3.4 | 5.2 | 8.7 | 8.4 |
| 1RHZ | 5 | A: 23-188 | 166 | 1 | 6.7 | 8.0 | 9.9 | 10.4 | 6.0 | 7.9 | 9.3 | 11.7 |
| 1IWG | 5 | 330-497 | 168 | 1 | 4.3 | 5.6 | 8.5 | 8.3 | 5.1 | 6.9 | 8.5 | 9.3 |
| 3P5N | 6 | 10-188 | 179 | 1 | 5.8 | 7.4 | 8.3 | 9.8 | 5.2 | 6.9 | 12.0 | 9.6 |
| 2IC8 | 6 | 91-272 | 182 | 1 | 6.0 | 7.2 | 9.5 | 9.3 | 5.2 | 7.2 | 10.1 | 10.5 |
| 2YVX | 5 | A: 284-471 | 188 | 1 | 5.1 | 6.9 | 9.2 | 9.4 | 5.7 | 7.6 | 8.9 | 10.5 |
| 1PV6 | 6 | 1-190 | 189 | 1 | 5.7 | 6.8 | 10.6 | 9.4 | 6.2 | 7.6 | 10.0 | 10.4 |
| 1OCC | 5 | C: 71-261 | 191 | 1 | 4.6 | 5.9 | 8.5 | 8.0 | 7.3 | 9.2 | 10.8 | 11.3 |
| 2NR9 | 6 | 4-195 | 192 | 1 | 5.7 | 7.2 | 8.7 | 9.5 | 5.6 | 7.6 | 10.5 | 10.5 |
| 4A2N | 5 | 1-192 | 192 | 1 | 4.3 | 6.2 | 8.1 | 8.8 | 5.0 | 7.1 | 9.6 | 10.3 |
| 1KPL | 7 | 31-233 | 203 | 1 | 8.7 | 10.5 | 14.4 | 12.5 | 11.1 | 13.2 | 15.4 | 16.1 |
| 2ZW3 | 4 | A: 2-217 | 216 | 1 | 4.0 | 5.1 | 9.2 | 8.1 | 6.7 | 8.4 | 13.5 | 11.1 |
| 2BS2 | 5 | C: 21-237 | 217 | 1 | 5.4 | 6.9 | 11.0 | 9.2 | 6.0 | 8.5 | 13.0 | 10.9 |
| 1L0V | 6 | C: 20-130; D: 10-119 | 221 | 1 | 5.2 | 7.2 | 9 | 9.4 | ||||
| 2KSY | 7 | 1-223 | 223 | 1 | 5.1 | 6.3 | 9.3 | 8.6 | 4.6 | 5.7 | 6.1 | 9.1 |
| 1PY6 | 7 | 5-231 | 227 | 1 | 4.8 | 5.9 | 6.1 | 8.4 | 3.3 | 5.8 | 8.4 | 8.7 |
| 3KCU | 7 | 29-280 | 252 | 1 | 7.3 | 8.5 | 11.2 | 10.5 | 7.2 | 9.3 | 11.0 | 11.4 |
| 1FX8 | 7 | 6-259 | 254 | 1 | 6.4 | 7.6 | 9.3 | 9.8 | 8.9 | 10.4 | 11.8 | 12.4 |
| 1U19 | 7 | 33-310 | 278 | 1 | 5.3 | 6.6 | 8.9 | 8.8 | 9.7 | 12.7 | 10.7 | 15.2 |
| 1OKC | 6 | 2-293 | 292 | 1 | 7.1 | 8.2 | 9.9 | 10.3 | 10.3 | 11.7 | 12.5 | 13.5 |
| 3KJ6 | 7 | A: 35-346 | 311 | 1 | 5.9 | 7.4 | 10.5 | 10.0 | 8.0 | 10.5 | 13.6 | 13.2 |
| 3B60 | 6 | A: 10-328 | 319 | 1 | 9.5 | 10.8 | 12.4 | 13.2 | 6.8 | 8.0 | 10.4 | 10.0 |
| 3HD6 | 12 | 6-448 | 403 | 1 | 7.2 | 8.2 | 11.0 | 10.3 | 8.0 | 9.5 | 11.9 | 11.7 |
| 3GIA | 12 | 3-435 | 433 | 1 | 9.6 | 10.7 | 13.4 | 12.6 | 11.6 | 13.9 | 14.2 | 15.8 |
| 3O0R | 12 | B: 10-458 | 449 | 1 | 6.9 | 8.2 | 10.2 | 10.3 | 6.8 | 8.6 | 12.3 | 11.1 |
| 2XUT | 12 | A: 13-500 | 488 | 1 | 7.7 | 9.0 | 10.2 | 11.4 | 9.4 | 11.1 | 13.6 | 13.3 |
| 3HFX | 12 | 12-504 | 493 | 1 | 8.9 | 9.7 | 13.1 | 11.4 | 11.7 | 13.5 | 15.3 | 15.4 |
| 1YEW | 14 | A: 151-225; B: 7-244; C: 45-259 | 528 | 1 | 9.7 | 11.5 | 14.1 | 13.3 | ||||
| 2XQ2 | 15 | A: 9-573 | 565 | 1 | 8.2 | 10.1 | 12.2 | 12.1 | 11.6 | 13.2 | 12.6 | 15.4 |
| mean | 7 | 242 | 1 | 5.8 | 7.1 | 9.9 | 9.6 | 6.6 | 8.4 | 10.6 | 11.1 | |
| std. dev. | 3 | 130 | 0 | 1.9 | 2.0 | 2.5 | 1.8 | 2.7 | 2.8 | 2.8 | 2.6 | |
|
| ||||||||||||
|
Multimer folding
| ||||||||||||
| 2HAC | 1 | -3-30 | 33 | 2 | 1.0 | 1.3 | 3.5 | 3.7 | ||||
| 2KIX | 1 | 1-33 | 33 | 4 | 2.2 | 2.4 | 5.7 | 4.2 | ||||
| 1NKZ | 2 | A: 2-53;B: 1-41 | 93 | 9 | 4.1 | 4.7 | 8.1 | 8.4 | ||||
| 3SYO | 2 | 76-197 | 122 | 4 | 9.3 | 9.6 | 10.7 | 10.5 | ||||
| 2PNO | 4 | A: 2-131 | 130 | 3 | 3.3 | 5.2 | 10.5 | 8.1 | ||||
| 2BL2 | 4 | 12-156 | 145 | 10 | 2.8 | 3.6 | 5.7 | 5.4 | ||||
| 2YVX | 5 | A: 284-471 | 188 | 2 | 4.3 | 5.9 | 9.5 | 8.4 | ||||
| 2ZW3 | 4 | A: 2-217 | 216 | 6 | 4.8 | 5.6 | 7.9 | 8.0 | ||||
| 1FX8 | 7 | 6-259 | 254 | 4 | 10.8 | 11.3 | 11.7 | 12.0 | ||||
| 3B60 | 6 | A: 10-328 | 319 | 2 | 7.9 | 9.2 | 11.3 | 11.1 | ||||
| 3HD6 | 12 | 6-448 | 403 | 3 | 5.8 | 7.4 | 9.0 | 9.5 | ||||
| 3HFX | 12 | 12-504 | 493 | 3 | 8.7 | 9.6 | 10.9 | 11.3 | ||||
| mean | 5 | 202 | 4 | 5.4 | 6.3 | 8.7 | 8.4 | |||||
| std. dev. | 4 | 143 | 3 | 3.1 | 3.1 | 2.6 | 2.8 | |||||
The ability for each score to select for native-like models is determined by its enrichment (Woetzel et al., 2012), which is its ability to select the 10% native-like models out of the given pool. Maximal enrichment is 10.0, no enrichment is 1.0, and enrichment values less than 1.0 indicate the score disfavors native-like models.
Modified radius of gyration measure deflates membrane core region
For soluble proteins, the normalized radius of gyration of a model, NRgyr = (Rgyr)2/length, is evaluated using a knowledge-based potential such that P(struct) ≅ P(NRgyr) (Woetzel et al., 2012). In order to allow for TMHs to span the membrane without inflating the Rgyr score, the coordinates are collapsed along the membrane normal by an amount equal to one half of the membrane core thickness, 10 Å, prior to the NRgyr calculation (Figure 1B). The NRgyr is then evaluated using the KB potential (Figure 1D), as for soluble proteins. The rationale for this procedure is that minimizing radius of gyration is an effect seen in soluble proteins caused by the (1) overall drive to bury hydrophobic amino acids and (2) maximize constructive interactions of side chains. This first component is absent once an amino acid is embedded in the membrane core, i.e. there is no force to compact the MP along the membrane normal. As the second component is still present, compacting forces in the membrane plane are still active. Collapsing the membrane is particularly important for smaller proteins (< 200 residues), which would otherwise have an unnaturally high Rgyr2 value (Figure 1C). The average enrichment values for the radius of gyration score was 1.4 for each criteria. The percentage of models with a Z-score > 1.0 were 56%, 61%, and 67% for the RMSD100, RMSD100_XY, and CR criteria, respectively.
Amino acid environment potential reflects apolar membrane core region
The amino acid environment potential describes the preference for a particular amino acid type to be exposed or buried: , where aai is the residue type, and ei is the modified neighbor count. The MP amino acid environment score function is determined identically to its soluble counterpart, except that the procedure is executed separately for the three membrane regions: membrane core, transition, and solution. For residues in the gap regions, an average of the score in the neighboring regions is computed. Exposure is represented by neighbor count, such that a low neighbor count corresponds to a high solvent accessible surface area (Durham et al., 2009). Figure 1E displays representative potentials, and the complete potentials can be found in Figure S2. As expected, the polar serine and the negatively charged aspartate both favor buried environments (higher neighbor counts) in the membrane core relative to solution. Additionally, the hydrophobic leucine, has the opposite property, favoring a more exposed environment in the membrane core. The average enrichment values for the amino acid environment score were 2.7, 2.7, and 3.0 for the RMSD100, RMSD100_XY, and CR criteria, respectively. The percentage of models with a Z-score > 1.0 were 88%, 100%, and 100%.
TMH alignment displays strong preference to be parallel with respect to membrane normal
During the BCL::MP-Fold minimization, SSEs can be rotated in any orientation. In order to select for models with TMHs properly spanning the membrane, an SSE alignment score was introduced. This score evaluates the measured angle, θ, of the SSE to the membrane plane given the environment type (Figure 1F), such that . TMHs tend to span the membrane at a 70° – 90° angle to the membrane plane. The average enrichment values for the SSE alignment score were 2.2, 2.3, and 2.4 for the RMSD100, RMSD100_XY, and CR criteria, respectively. The percentage of models with a Z-score > 1.0 were 89%, 94%, and 94%.
MP topology score penalizes loop connections between TMHs that require passage through the membrane
TM loops (coils) are rare, short, and typically associated with reentrant regions and functional sites in kinked TMHs (Kauko et al., 2008). Since BCL models do not contain loops during minimization, a penalty score is needed to disfavor topologies that would require long membrane-spanning loop regions to connect two TMHs. While the soluble protein loop scores are still utilized for MP folding, they only ensure that the loop can be closed, not that the loop will be in a favorable environment. The MP topology score introduces a penalty when the C-terminus of a TMH is not on the same side of the membrane as the N-terminus of the subsequent TMH, for example. In practice, this is achieved by grouping alternating TMHs, and determining whether the termini lie on the same side of the membrane. We therefore evaluate and where zc,i is the Z-coordinate of C-terminus of TMHi and zn,i+1 is the Z-coordinate of N-terminus of TMHi+1. SSEs that do not span the membrane, such as amphipathic or soluble helices predicted by JUFO9D are excluded from this analysis. The average enrichment values for the MP topology score were 4.8, 4.2, and 2.5 for the RMSD100, RMSD100_XY, and CR criteria, respectively. The percentage of models with a Z-score > 1.0 were 100% for each of the three criteria.
Agreement of amino acid placement in membrane regions predicted from sequence
Both SPOCTOPUS and JUFO9D predict the environment type (membrane core versus soluble region) as well as the secondary structure – SPOCTOPUS only predicts TMHs while JUFO9D gives a nine state prediction, with three secondary structure types (helix, coil, strand) and three environment types (membrane core, transition region, soluble region). Models are therefore scored both on secondary structure and environment prediction. The secondary structure agreement score is calculated in an identical manner as for soluble proteins with soluble prediction methods (Woetzel et al., 2012). The membrane placement prediction agreement is scored analogously, where the probability of each residue to be observed in the current environment type given the SPOCTOPUS and JUFO9D predictions is calculated: .
The average enrichment values for the environment prediction score were 3.0, 2.9, and 2.3 for the RMSD100, RMSD100_XY, and CR criteria, respectively for JUFO9D, and 2.8, 2.8, and 2.7 for OCTOPUS. The percentage of models with a Z-score > 1.0 were 100%, 100%, and 88% for JUFO9D, and 100% for all three criteria for OCTOPUS.
Efficient sampling of MP topologies through a tailored set of MC moves
The soluble BC::Fold method utilizes a total of 107 MC moves (Karakas et al., 2012). They are classified into six categories: 1) adding SSEs, 2) removing SSEs, 3) swapping SSEs, 4) single SSE moves, 5) SSE-pair moves, and 6) moving domains. An additional move category, global transformations, containing two moves, was added for membrane folding in order to effectively sample the different membrane environments (Figure S1B). First, a global translation of 2 to 10 Å along the membrane normal allows for the protein to move in and out of the membrane. Second, a rotation around a random vector with the membrane plane samples different orientations within the membrane. These perturbations preserve protein structure and therefore have no impact on previous components of the BCL::Fold scoring function. They alter the five new/modified scores for MPs.
Multimerization achieved by replicating single subunit prior to scoring
In order to facilitate prediction of homomultimeric MPs, folding with cyclic symmetry was introduced. All perturbations are performed on a single subunit, and the subunit is then replicated around an axis of symmetry prior to scoring. The total number of subunits is defined by the user prior to the minimization. Additional perturbations are required to sample varying subunit interfaces (Figure S1C). First, a random global rotation of the primary subunit allows for a new interface to be sampled. Second, a global translation towards or away from the symmetry axis changes the compactness of the multimer. Third, a domain-swap perturbation is also added that exchanges one SSE with its counterpart in an adjacent subunit.
Benchmark set of 40 MPs of known structure
A benchmark set of 40 MPs of known structure was created to assess the protein structure prediction method. The set contains all proteins from the Rosetta-Membrane benchmark (Yarov-Yarovoy et al., 2006) as well as additional proteins that are both structurally and functionally diverse (Table 1). The proteins range from 91 to 565 residues and from 2 to 15 TMHs. Twelve of the proteins form obligate multimers containing 2 to 10 subunits, resulting in 66 to 1479 total residues. 1000 models were generated for each protein using the BCL::MP-Fold algorithm. Two runs were performed for multimeric proteins – structures were predicted of just the protomer using the standard method, and the structure of the complete multimer was also predicted taking advantage of the cyclic symmetry in the multimeric folding mode. Proteins 2HAC and 2KIX were only folded as multimers since they each contain only one TMH per subunit. The quality of SSE placement is assessed using the RMSD100 criteria. A subset of the results is shown in Figure 2, and the complete results can be found in Figure S3.
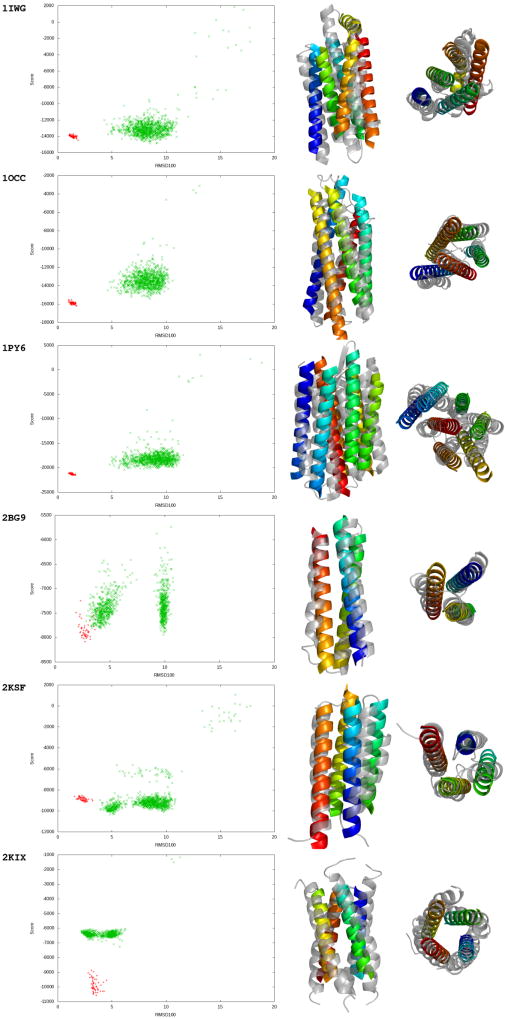
Figure 2. Gallery of select benchmark results.
Left column – BCL score versus RMSD100 plots of 1000 generated models. The 1000 models generated by BCL::MP-Fold for the prediction are shown in green. In red, 50 minimizations of the native model using only the refinement stage are shown. Middle column – Side view of best model sampled. Right column – Overhead view of best model sampled. The bottom result, 2KIX, was generated using multimer folding. For the gallery of the complete benchmark results, please see Figure S3.
Among the single subunit predictions, the average RMSD100 to the native of the best model sampled is 5.8 ± 1.9 Å (Table 1). In 32 of the 38 cases (84%), the correct topology is sampled using the criteria of RMSD100 < 8.0 Å. When large proteins containing twelve or more TMHs are excluded, the correct topology is sampled in 29 out 31 cases (94%). In 27 of the 38 cases (71%), the top 5% of models by BCL score contain the correct topology. This changes to 25 out of 31 cases (81%) when the large proteins are excluded.
The two proteins with 7 TMHs or fewer that BCL::MP-Fold was unable to sample the correct topology were a subdomain of the H+/Cl− transporter (Dutzler et al., 2002) (1KPL) and the ABC transporter MsbA (Ward et al., 2007) (3B60). The H+/Cl− transporter was part of the Rosetta-Membrane benchmark (Yarov-Yarovoy et al., 2006), and the method performed similarly poorly. As the authors mention, this protein has a particularly complex topology, compounded by the SPOCTOPUS method only predicting 5 of the 7 transmembrane spans; the fourth and sixth TMHs are missed. JUFO9D successfully predicts these missing TMHs, but since these predictions are not added to the SSE pool until the third assembly stage, the BCL::MP-Fold method has difficulty placing them correctly amongst the five previously placed TMHs. When folding is initiated with the JUFO9D-predicted TMHs, the correct topology is sampled; the RMSD100 of the best model is then 7.1 Å. MsbA is a particularly difficult target. First, the TMHs are extremely long (40+ residues), whereas the SPOCTOPUS predicted TMHs only span 21 residues each. Second, the protein is flexible, with a large ligand-induced conformational shift. Third, MsbA is a homodimer, which makes it a better target for the multimeric predictions discussed below.
For the multimeric predictions, the average RMSD100 (over the entire multimer) to the native of the best model sampled is 5.4 ± 3.1 Å (Table 1). This corresponds to 9 out of 12 cases (75%) where the correct topology is sampled. In the same 9 of 12 cases (75%), the top 5% of models by BCL score contain the correct topology. MsbA was one of the successfully sampled multimer targets, with the best model sampled having an RMSD100 to the native of 7.7 Å.
Despite having only two TMHs per subunit, the tetrameric topology of GIRK2 K+ channel protein (Whorton and MacKinnon, 2011) (3SYO) was not correctly predicted by BCL::MP-Fold. In particular, the current version of BCL::MP-Fold cannot adequately form the ion conducting channel since it largely contains loop residues. Additionally, the TMHs pass through the membrane at an angle of approximately 50°, rather than the more commonly observed and thus more energetically favorable 70° – 90° (Fig. 2C). The tetrameric glycerol conducting channel (Fu et al., 2000) (1FX8) was also a challenging multimeric target. In the native tetramer, the core is dominated by interactions between TMH 2 and TMH 6, whereas in the best BCL model, the core only contains TMH 2.
Discussion
BCL::Fold has been updated via MP-specific energy potentials and MC moves to accurately predict MP topology from sequence. Correct topologies were sampled for 32 of 38 protomer benchmark targets and 9 of 12 multimer benchmark targets. These results demonstrate the viability of discontinuous TMH placement to predict MP topology.
BCL::MP-Fold produces accurate models for targets of varying size, complexity, and SSE content
The BCL::MP-Fold MP algorithm was able to sample the native topology of 2XUT, a 12 TM, 488 residue protein. The algorithm failed to predict the correct topology for the two larger proteins, 3HFX and 2XQ2, which places the upper limit for single subunit predictions at around 500 residues. This is a considerable achievement for de novo MP structure prediction, as these larger proteins are typically excluded from benchmark sets. For example, the largest Rosetta-Membrane target, 1U19, contains 278 residues. As expected, leveraging oligomeric symmetry greatly extends the upper limit; the decameric 2BL2 contains 1450 residues, and the best BCL model has an RMSD100 of 2.8 Å to the native.
One of the design goals when developing BCL::Fold was to facilitate sampling of non-local contacts, resulting in more accurate sampling of proteins of high contact order. For targets, containing less than 400 residues, this is the case (Figure 3A); BCL::MP-Fold accurately samples native topologies with NCO values ranging from 0.15 to 0.50. For targets larger than 400 residues, this is no longer the case, with the highest NCO value sampled being 0.23. Thus the current algorithm is capable of sampling proteins with a large number of residues (> 400) or a high NCO (> 0.25), but not both. In order to keep sampling relatively rapid, each stage terminates after a maximal number of MC steps – 2000 for each assembly stage and 4000 for the refinement stage. Sampling for these large, complex proteins would likely benefit from increased MC steps, as stages terminated at the maximum 54 ± 2% of the time for these three proteins (2XQ2, 3HFX, and 3GIA), versus 44 ± 7% of the time over the entire benchmark.
Figure 3. Accurate models are produced for targets of varying size, complexity, and SSE content.
A – NCO versus amino acids. Each point represents the best model produced for a particular target. Single subunit predictions are circles and multimer predictions are squares. Both use the following coloring scheme based on RMSD100: RMSD100 < 6.0 Å (green), 6.0 Å ≤ RMSD100 < 8.0 Å (light green), 8.0 Å ≤ RMSD100 < 10.0 Å (yellow), 10.0 Å ≤ RMSD100 (red). Proteins with multiple chains in the protomer are excluded since the contact order is undefined. For multimers, the contact order reported is for the protomer while the amino acid count is the total for the complete multimer. B – Q3 versus SSE content. The plotted Q3 value is the average of the JUFO9D and SPOCTOPUS predictions. SSE content is the percentage of residues that are part of a helix or strand. C – RMSD100 of top 5% of best models versus amino acids. BCL::MP-Fold shown as black triangles and Rosetta-Membrane as circles. D – RMSD100 was computed for BCL::MP-Fold and Rosetta-Membrane to the SSEs of the native models. The value for the top 1% of models ± S.D. sampled is plotted for each method. The dashed line at 8 Å indicates the arbitrary cutoff for “native-like” quality.
The folding algorithm is also accurate for targets covering a wide range of SSE content (from 70% to 92%) and SSE prediction accuracy (average Q3 values from 0.60 to 0.90) (Figure 3B). Not surprisingly, the method fails for protein 1YEW, which has the lowest SSE content at 66% and average Q3 value at 0.50 of any protein in the benchmark set. Nonetheless, Figure 3 demonstrates that the BCL::MP-Fold algorithm is robust and is typically able to compensate for inaccuracies in secondary structure prediction.
BCL::MP-Fold samples more native-like topologies than Rosetta-Membrane
Rosetta is a well established de novo protein structure prediction method that has performed well during several iterations of the Critical Assessment of protein Structure Prediction (CASP) (Raman et al., 2009). In 2006, the method was updated to predict membrane protein structure (Yarov-Yarovoy et al., 2006). In order for a direct comparison between BCL::MP-Fold and Rosetta-Membrane, Rosetta 3.4 was used to predict structures for each of the single chain proteins in the benchmark set (Table 1, Figures 3C and 3D). In order to directly compare BCL and Rosetta models, the RMSD100 values are calculated only over Cα atoms in native SSEs. The average RMSD100 to native of the best model sampled by Rosetta-Membrane is 6.6 ± 2.7 Å, compared to 5.9 ± 1.8 Å for BCL::MP-Fold for those same proteins. The correct topology (RMSD100 < 8.0 Å) was sampled in 25 out of the 34 cases (74%) by Rosetta-Membrane, and in 29 cases (85%) by BCL::MP-Fold.
Limitations include amphipathic and bent helices
BCL::MP-Fold models do not typically accurately represent amphipathic and bent helices. Amphipathic helices are difficult to model for two reasons. First, placing the helix parallel to a TMH maximizes packing and results in a favorable SSE packing score. In the correct orientation, parallel to the membrane, SSE packing is reduced and no other component of the consensus energy function compensates. Second, the TMH alignment potential in the transition region is very similar to that observed in the membrane core (Figure 1F). TMHs tend to span both the membrane core and the transition, thus the bulk of the counts in the transition region come from TMHs and not amphipathic helices. Thus placement of a short, amphipathic helix parallel to a TMH in the transition region minimizes both the SSE packing and TMH alignment scores. An additional KB potential specific for amphipathic helices would likely be needed to accurately predict these helices in future iterations of BCL::MP-Fold.
The BCL::MP-Fold algorithm starts with an idealized, perfectly straight, pool of SSEs. There are bending moves during the simulation that bend the SSEs. Even so, the current algorithm does not adequately capture the kinks and bends that are commonly see in native TMHs. This limitation can be overcome with increased probabilities for the bending MC moves or more sophisticated bend moves that perturb several phi/psi angles simultaneously by fitting to observed TMH fragments.
Contact recovery can be improved
For soluble protein structure prediction using BCL::Fold, accurately predicted models tend to have low RMSD100 values and high contact recovery values (Karakas et al., 2012). This is not observed for MP structure prediction, however. When CR > 20% is considered to be “native-like”, the native conformation is only sampled in 61% of the single subunit predictions, compared to 84% using the RMSD100 < 8.0 Å criterion. This suggests that although the TMHs span the membrane in the correct places, they are not necessarily rotated correctly, with normally contact-forming, buried residues instead being solvent or membrane exposed. This property is due to the MP amino acid environment potentials, which consistent with the generated statistics and the underlying biophysics, are less discriminating than the soluble potentials. For example, when scoring a soluble protein, helix rotations that place hydrophobic residues in the core of the protein will be scored favorably. When scoring a MP, on the other hand, rotations that bury hydrophobic residues would be scored similarly to those rotations that exposed the hydrophobic residues. Addition of a knowledge-based exposure score would likely improve TMH rotations. This would involve implementing a method to predict individual residue exposure (such as solvent accessible surface area) from sequence in a manner similar to secondary structure and environment prediction by JUFO9D.
Enhanced accuracy with restraints
Experimental or predicted restraints can dramatically improve MP structure prediction accuracy by limiting the potential conformational space (Barth et al., 2009; Hopf et al., 2012; Nugent and Jones, 2012). This should hold for BCL::MP-Fold as well since the consensus scoring function is highly adaptable and can be modified to incorporate restraints. In fact, BCL::Fold has already been successfully applied to proteins with medium resolution cryo-electron microscopy data (Lindert et al., 2009). Efforts are currently underway to incorporate additional restraint types such as NMR, EPR, SAXS and contacts.
Experimental Procedures
MP databank
A databank of diverse MP structures was created in order to create statistics for the MP-specific knowledge-based potentials. A subset of known structures from the PDB were selected using the culling server, PISCES (Wang and Dunbrack, 2005). The following criteria were used to select proteins: sequence identity less than 25%; resolution of less than 3.0 Å, R-value of less than 0.3, sequence length of at least 40 residues. Only those structures determined by X-ray crystallography were considered. The final list contained 111 MPs and 175 chains. PDBTM (Tusnady et al., 2005) was used to properly orient each structure into the membrane.
Radius of gyration
As for soluble proteins (Woetzel et al., 2012), the radius of gyration for MPs is calculated using the formula, , and is normalized, . For MPs, however, the Rgyr term is calculated using a collapsing normal. All Cβ atoms within the membrane core have their Z-coordinate set to zero. Atoms outside the membrane core are translated toward the membrane plane by a distance of 10 Å, which corresponds to one half the membrane core thickness. Statistics for NRgyr were collected over the MP databank in order to generate the knowledge-based potential.
Amino acid environment
The amino acid environment potential is derived in the same manner as for soluble proteins (Woetzel et al., 2012). The potential is split into three components based on environment type – membrane core, transition, and solution. Statistics for the neighbor count for each residue type are then collected for each of the three environment types in order to generate the separate potentials. Histogram counts are normalized across membrane regions for each residue type. If the residue is in a gap region, the score is calculated as a cosine weighted average of the two adjacent environments based on the Z-coordinate of the Cβ atom.
SSE Alignment
Separate potentials are generated for the membrane core, transition, and soluble environment types. Helical SSEs in the BCL can be represented as overlapping ideal fragments of five residues (Woetzel et al., 2012). Each fragment is evaluated based on the angle between its main axis and the membrane plane given the environment type. In a similar manner to the amino acid environment potential, fragments in gap regions are scored as a cosine weighted average of the two adjacent environments based on the Z-coordinate of the center of the fragment. The score for a given SSE is simply the sum of its fragment scores.
Membrane topology
A set of predicted SSEs making up the transmembrane domain of the protein is provided using SPOCTOPUS. The helical transmembrane segments of the protein are assumed to have an anti-parallel topology. During folding, the current SSEs in the model are matched to the associated transmembrane SSE expected from prediction. The N- terminal Cα atom coordinates for each transmembrane helix are put into one of two groups according to which termini should be on the same side of the membrane. The group is determined by the SPOCTOPUS predicted topology. The membrane center has a z-coordinate of zero, so the side of the membrane a terminus lies on is given by the sign of the z-coordinate. Next, within a group, the coordinates are pairwise compared, and a penalty is given for every pair of coordinates that reside on opposite sides of the membrane. This selects for correctly placing N-termini that should be on the same side of the membrane. Lastly, coordinates are pairwise compared between groups, and a penalty is given for every pair of coordinates that reside on the same side of the membrane. This selects for correctly placing N-termini that should be on opposite sides of the membrane. The above procedure is repeated for C-terminal Cα coordinates to give the final total penalty score.
Environment prediction agreement
The environment prediction agreement is scored analogously to the secondary structure prediction agreement (Woetzel et al., 2012). Briefly, , where pEnv,i is the probability of the observed environment in the model for residue i, μEnv is the mean probability for an accurately predicted environment type, and σEnv is the standard deviation for an accurately predicted environment type.
Benchmark
1000 models were created for each protein in the benchmark set. SSE pools were generated using the BCL application BCL::SSEPool (Karakas et al., 2012). The initial pool was generated using SPOCTOPUS (Viklund et al., 2008), and the final pool included both SPOCTOPUS and JUFO9D predictions. Example score weight sets, stage files, and command lines for both BCL::SSEPool and BCL::MP-Fold can be found in the supplementary information.
Availability
BCL::MP-Fold is implemented as part of the BioChemical Library, a suite of software currently under development in the Meiler laboratory (www.meilerlab.org). BCL software, including BCL::MP-Fold, is freely available for academic use.
Supplementary Material
Highlights.
BCL::Fold is a novel de novo membrane protein structure prediction algorithm.
Monte Carlo minimization is combined with a knowledge-based energy function.
34 out of 40 native membrane protein topologies were sampled in a benchmark test.
Acknowledgments
The authors thank Stephanie DeLuca for assistance with Rosetta-Membrane. We thank the Vanderbilt University Center for Structural Biology computational support team for hardware and software maintenance. We also thank the Vanderbilt University Advanced Computing Center for Research and Education for computer cluster access and support. Work in the Meiler laboratory is supported through NIH (R01 GM080403, R01 MH090192, R01 GM099842) and NSF (Career 0742762).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakheet TM, Doig AJ. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci U S A. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Ruczinski I, Tsai J, Baker D. Contact order and ab initio protein structure prediction. Protein Sci. 2002;11:1937–1944. doi: 10.1110/ps.3790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- Carugo O, Pongor S. A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci. 2001;10:1470–1473. doi: 10.1110/ps.690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham E, Dorr B, Woetzel N, Staritzbichler R, Meiler J. Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J Mol Model. 2009;15:1093–1108. doi: 10.1007/s00894-009-0454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Jonasson K, von Heijne G, Uhlen M, Berglund L. Prediction of the human membrane proteome. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Karakas M, Woetzel N, Staritzbichler R, Alexander N, Weiner BE, Meiler J. BCL::Fold - De Novo Prediction of Complex and Large Protein Topologies by Assembly of Secondary Structure Elements. PLoS One. 2012;7:e49240. doi: 10.1371/journal.pone.0049240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauko A, Illergard K, Elofsson A. Coils in the membrane core are conserved and functionally important. J Mol Biol. 2008;380:170–180. doi: 10.1016/j.jmb.2008.04.052. [DOI] [PubMed] [Google Scholar]
- Lange OF, Baker D. Resolution-adapted recombination of structural features significantly improves sampling in restraint-guided structure calculation. Proteins. 2012;80:884–895. doi: 10.1002/prot.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman JK, Mueller R, Karakas M, Woetzel N, Meiler J. Simultaneous prediction of protein secondary structure and trans-membrane spans. Proteins: Structure, Function, and Bioinformatics. 2012 doi: 10.1002/prot.24258. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindert S, Staritzbichler R, Wotzel N, Karakas M, Stewart PL, Meiler J. EM-fold: De novo folding of alpha-helical proteins guided by intermediate-resolution electron microscopy density maps. Structure. 2009;17:990–1003. doi: 10.1016/j.str.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J, Muller M, Zeidler A, Schmaschke F. Generation and evaluation of dimension-reduced amino acid parameter representations by artificial neural networks. J Mol Model. 2001;7:360–369. [Google Scholar]
- Nugent T, Jones DT. Membrane protein structural bioinformatics. J Struct Biol. 2011 doi: 10.1016/j.jsb.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Nugent T, Jones DT. Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis. Proc Natl Acad Sci U S A. 2012;109:E1540–1547. doi: 10.1073/pnas.1120036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberai A, Ihm Y, Kim S, Bowie JU. A limited universe of membrane protein families and folds. Protein Sci. 2006;15:1723–1734. doi: 10.1110/ps.062109706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44(Spec No):S24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H, Bernsel A, Skwark M, Elofsson A. SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- Wang G, Dunbrack RL., Jr PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33:W94–98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woetzel N, Karakas M, Staritzbichler R, Muller R, Weiner BE, Meiler J. BCL::Score-Knowledge Based Energy Potentials for Ranking Protein Models Represented by Idealized Secondary Structure Elements. PLoS One. 2012;7:e49242. doi: 10.1371/journal.pone.0049242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.