Abstract
Strand-specific RNA sequencing of S. pombe revealed a highly structured programme of ncRNA expression at over 600 loci. Waves of antisense transcription accompanied sexual differentiation. A substantial proportion of ncRNA arose from mechanisms previously considered to be largely artefactual, including improper 3′ termination and bidirectional transcription. Constitutive induction of the entire spk1+, spo4+, dis1+ and spo6+ antisense transcripts from an integrated, ectopic, locus disrupted their respective meiotic functions. This ability of antisense transcripts to disrupt gene function when expressed in trans suggests that cis production at native loci during sexual differentiation may also control gene function. Consistently, insertion of a marker gene adjacent to the dis1+ antisense start site mimicked ectopic antisense expression in reducing the levels of this microtubule regulator and abolishing the microtubule-dependent ‘horsetail’ stage of meiosis. Antisense production had no impact at any of these loci when the RNA interference (RNAi) machinery was removed. Thus, far from being simply ‘genome chatter’, this extensive ncRNA landscape constitutes a fundamental component in the controls that drive the complex programme of sexual differentiation in S. pombe.
Keywords: antisense, meiosis, ncRNA, S. pombe , siRNA
Introduction
Studies in the fission yeast Schizosaccharomyces pombe have done much to inform our view of heterochromatin and its control by the RNA interference (RNAi) machinery (Grewal, 2010). This insight has arisen from the system's reliance upon the creation of heterochromatin at mating type loci, centromeres and telomeres to silence gene expression and generate specialised blocks of chromatin to protect chromosome integrity and facilitate genome transfer. Similar analyses of RNA production, stability and splicing during sexual differentiation suggest that this system will continue to further our understanding of RNA biology (Shimoseki and Shimoda, 2001; Mata et al, 2002; Averbeck et al, 2005; Mata and Bähler, 2006; Xue-Franzen et al, 2006; Moldon et al, 2008; Djupedal et al, 2009; Amorim et al, 2010; Ni et al, 2010; Cremona et al, 2011).
Fission yeast grow in either a haploid or a diploid state (Egel, 2004). Haploid cells express one of the two mating types: plus (P) or minus (M). After each cell division the mating type of one of the two daughter cells switches, generating a mixed population in which each type is equally represented. Starvation of this mixed culture promotes the activation of the HMG-box group transcription factor Ste11 (Sugimoto et al, 1991). A complementary system of pheromone signalling is triggered upon occupancy of cell surface receptors by pheromones produced by cells of the opposite mating type. The subsequent activation of the Byr2/Byr1/Spk1 MAP kinase cascade promotes a cell type-specific transcriptional response (Nielsen, 2004; Mata and Bähler, 2006; Xue-Franzen et al, 2006) that integrates with Ste11 activation to induce sexual differentiation and meiosis (for review, see Harigaya and Yamamoto, 2007).
Cells of opposing mating types grow along pheromone gradients towards one another to conjugate and form a zygote. Zygotes have a choice of two fates. If nutrient provision is restored after conjugation, they embark upon mitotic cell divisions as a diploid cell (Egel, 2004). If starvation persists, sexual differentiation is initiated. Meiotic DNA replication is followed by a phase termed ‘horsetail movement’, in which repeated migration of the nucleus from one end of the cell to the other promotes meiotic recombination. This movement is promoted by differentiation of the microtubule cytoskeleton (Yamamoto et al, 1999; Supplementary Figure S1). This recombination phase is followed by two meiotic divisions, which produce four nuclei that are partitioned into four discrete spores. Spores can remain dormant for extended periods, until germination returns them to a haploid vegetative life cycle. Starvation of a diploid cell expressing both mating types instigates the same programme of sexual differentiation to produce four haploid spores (Egel, 2004).
The RNA binding protein Mei2 controls meiotic commitment (Watanabe and Yamamoto, 1994; Harigaya and Yamamoto, 2007). Mei2 forms a complex with a meiRNA; a ncRNA product of the sme2+ gene at the sme2 locus (Shimada et al, 2003). Mei2 sequesters the Mmi1 protein (Harigaya et al, 2006). Since Mmi1 collaborates with the poly(A) mRNA binding protein Pab2 to block sexual differentiation by targeting meiotic transcripts for destruction during vegetative growth (McPheeters et al, 2009; Yamanaka et al, 2010), sequestration of Mmi1 by Mei2 stabilises these meiotic transcripts and meiosis ensues. Mei2 is inhibited during vegetative growth via phosphorylation by Pat1 kinase (Watanabe et al, 1997). If starved zygotes express both Mat1-Pm and Mat1-Mm products of opposing mating type loci, mei3+ transcription is promoted (Willer et al, 1995; Mata and Bähler, 2006; Xue-Franzen et al, 2006). Since Mei3 is an inactivating pseudosubstrate for Pat1 (McLeod and Beach, 1988; Li and McLeod, 1996), mei3+ expression induces meiosis by relieving Pat1 inhibition of Mei2. An alternative approach to inactivating Pat1 can be achieved via inactivation of the thermosensitive pat1.114 mutant, which induces sexual differentiation (Iino and Yamamoto, 1985; Nurse, 1985). As this induction is largely synchronous within the population, this approach is used widely to generate synchronised meiotic cultures (Mata et al, 2002; Averbeck et al, 2005).
Four waves of transcription accompany sexual differentiation; early Ste11-dependent and Cdc10/Res1/Res2 directed waves of transcription are followed by Mei4 induction of genome segregation genes, before Atf21 and Atf31 instigate the final wave of late transcription (Mata et al, 2002). Further modifications of the meiotic RNA landscape include the stabilisation of RNAs and at least two forms of differential splicing (Averbeck et al, 2005; Moldon et al, 2008; McPheeters et al, 2009; Amorim et al, 2010; Cremona et al, 2011).
Transcriptional and post-transcriptional gene silencing operates through a variety of mechanisms including physical hindrances arising from the collision of polymerases simultaneously transcribing from adjacent convergent loci (Prescott and Proudfoot, 2002; Uhler et al, 2007), and a diversity of more choreographed processes involving RNAi. RNAi describes a collection of processes that regulate gene expression both pre- and post-transcription, and exists in both prokaryotes and eukaryotes. It operates through the destruction or suppression of individual transcripts, identified by small RNAs with complementary sequences to their targets. The biogenesis and function of these small RNA molecules is diverse, and is typified in fungi by the cleavage of dsRNAs by the ribonuclease III, Dicer, with possible amplification through an RNA-dependent polymerase. These associate with a member of the Argonuate family of small RNA binding proteins, and guide them to their targets, where they can exert their effect. This can range from suppression via blocking of the ribosome, poly(A) de-capping or cleavage through endonuclease activity (Ender and Meister, 2010).
The RNAi machinery can also promote gene silencing via the establishment of heterochromatin through the induction of histone H3K9 methylation at target loci (Volpe et al, 2002; Grewal, 2010; Allshire, 2011). In S. pombe, dsRNA formed in cis through the expression of transcripts from centromeric repeats (Volpe et al, 2002) and convergent overlapping 3′ untranslated regions (UTRs) (Gullerova et al, 2011; Zhang et al, 2011) is cleaved by Dcr1 and loaded into the single Argonaute family member of fission yeast, Ago1. The RNA-induced initiation of transcriptional gene silencing (TGS) complex, RITS, comprising Ago1, Tas3 and Chp1 is then targeted to homologous regions, where Chp1 binds to H3K9me (Verdel et al, 2004). A second complex, RNA-dependent RNA polymerase (RDRC) is recruited, leading to the synthesis of further dsRNA by one of its constituents, Rdp1, increasing the pool of targeting sRNAs (Motamedi et al, 2004; Colmenares et al, 2007; Simmer et al, 2010). The histone methyltransferase Clr4 is also recruited, as part of the CLRC complex, resulting in further H3K9me, whereby the chromo-domain proteins Swi6, Chp1, Chp2, collaborate to assemble heterochromatin, silencing underlying genes (reviewed extensively in Grewal, 2010; Lejeune and Allshire, 2011); however, Clr4 recruitment can also influence RNAi activity independently of H3K9 methylation (Gerace et al, 2010; Zhang et al, 2011) Furthermore, silencing also occurs in trans via an Rdp1-dependent mechanism that is initiated by dsRNAs in RNA hairpins (Sigova et al, 2004; Simmer et al, 2010).
The number and degree of functional specialisation of Argonaute proteins differs between species; Saccharomyces cerevisiae, for example, has none (Drinnenberg et al, 2009), while C. elegans has 27 (Sigova et al, 2004). RNAi in fission yeast relies on a single Argonaute, a Dicer, and an RNA-dependent polymerase (ago1, dcr1 and rdp1, respectively), making it an excellent model system in which to explore these mechanisms (Grewal, 2010). However, from this simplicity emerges the additional complexity of overlapping pathways and a more generalised role for Ago1. Thus, the Ago1/Dcr1/Rdp1 triad is not only involved in TGS through the RITS complex, but is also implicated in post-transcriptional gene silencing (PTGS). Cross talk also occurs between the heterochromatin pathways and TRAMP surveillance (Bühler et al, 2008; Zhang et al, 2011). Mlo3, a protein required for nuclear export co-precipitates with both members of the TRAMP complex and the histone H3K9 methyltransferase Clr4. In mlo3.Δ cells, elevated levels of antisense transcripts associated with convergent gene pairs are observed, suggesting that Mlo3 may determine whether RNAs are degraded by the exosome or passed through the siRNA pathways. The exosome is also able to promote heterochromatin formation independently of the RNAi machinery (Reyes-Turcu et al, 2011). To further complicate matters, an additional non-canonical RNAi pathway operates independently of Argonaute in N. crassa (Lee et al, 2010).
A significant challenge, therefore, when considering the roles played by TGS and PTGS in coordinating cellular events, is to bring the diversity of different mechanisms by which one RNA molecule can regulate another into a common higher level semantics of gene expression that is independent of the underlying syntax of each individual interaction. This is already demanding given the current level of understanding. The continuing refinement and extension to these pathways (Lejeune and Allshire, 2011) are adding yet more layers of complexity, while suggesting that further study of TGS and PTGS in fission yeast might identify additional mechanisms and pathways.
Around 94% of the S. pombe genome is transcribed (Wilhelm et al, 2008). Using cDNA synthesised from poly(A)-enriched RNA samples, numerous novel ncRNA loci were discovered, and the 5′ and 3′ ends of many other genes were refined (Wilhelm et al, 2008). Although some of these transcripts may encode novel proteins (Bitton et al, 2011), the function of the majority is yet to be determined. We have used strand-specific deep sequencing of RNA, irrespective of poly(A) status, to reveal a highly structured antisense programme that modulates gene expression to dictate cell fate decisions during sexual differentiation.
Results
Generation of a comprehensive RNA database
To gain greater insight into the RNA profile of fission yeast, we sequenced total RNA extracts from both vegetatively growing and sexually differentiating cells following removal of rRNA. The protocol preserved strand specificity. Haploid, wild-type 972 h− cells provided the vegetative growth sample while sexual differentiation was induced in a synchronised manner by temperature inactivation of Pat1 kinase in a pat1.114/pat1.114 diploid strain (IH2912), following a shift from 25 to 32°C. Samples were taken immediately before the temperature shift (0 h) and 3, 5 and 10 h later.
Following alignment, all contiguous stretches in which at least one read was observed in one or more samples were identified and collated to generate a strand-specific map of all transcriptionally active loci in the experiment. We detected 76 315 distinct loci (‘Transcriptional Blocks’; TBlocks) ranging from 50 to 3958 bp in length, covering 65.6% of the genome (ignoring the strand on which transcription occurs). When ‘strandedness’ is considered, 38% of residues are covered. Since our emphasis was on the reliable prediction of novel regulatory loci, we took a conservative approach in which only reads that mapped uniquely to the genome with zero mismatches were used. With less stringent criteria (three mismatches), total coverage increased to 84.4% (genome) and 55.6% (residues), respectively.
Defining the 5′ and 3′ ends of S. pombe genes
Since many ncRNAs function through RNAi, analysis of their role is dependent on a comprehensive, high-resolution map of transcript extent, including their UTRs. We positioned TBlocks relative to known annotation, revealing extensive transcription in intergenic regions (50.5%, 38 522 TBlocks), and 5′ or 3′ extensions to the majority of annotated genes (N=4736; 80.9%; Supplementary Table S1). By considering TBlocks that overlapped with adjacent genes, we were able to systematically demarcate the transcriptional extent of the majority of S. pombe genes (80.5%, 4715), including 26 pseudogenes and 275 ncRNAs. We then amalgamated UTR data and novel extensions of ncRNAs with genome annotations from the Ensembl (Kersey et al, 2009) representation of GeneDB (Hertz-Fowler et al, 2004), to generate the most comprehensive annotation of S. pombe to date (Supplementary Table S2), comprising 10 677 exonic regions (protein-coding and non-coding RNA), 4825 introns, 7287 UTR/extensions and 5834 intergenic regions of varying length. These data are consistent with the recent RNA-Seq derived genome annotations of the Schizosaccharomyces (Rhind et al, 2011). An additional 0.8% of the protein-coding genome, identified through proteomics approaches, was not included in this data set (Bitton et al, 2011). Since our goal was to define the extent of individual genes, reads were pooled from all samples in the experiment to maximise coverage. Since Transcriptional Start Sites (TSSs) and/or Transcriptional Termination Sites (TTSs) of individual transcripts are also of interest, we defined UTRs for each sample independently, using the same methods (Supplementary Table S2). The substantial changes in the length of individual UTRs between samples revealed by this approach, suggest that changes in UTR structure may underpin aspects of the sexual differentiation programme.
Changes in transcript levels during sexual differentiation
We considered genome-wide alterations of transcript levels in the different phases of meiosis. We did this by first identifying loci with statistically significant changes in RNA level in at least one sample in the experimental data, and then considering the stages of meiosis for which these changes were most pronounced. Annotation coordinates from our expanded set were used to partition the transcript level data, and RNA abundance for all annotated genes (5857), their corresponding 5857 antisense loci, 4825 introns and 21 199 intergenic TBlocks were computed by counting the number of reads aligning to each locus. Regions without reads were removed, leaving a total of 32 891 expressed regions (Supplementary Table S3).
Transcript levels at 6599 loci changed in at least one sample (G-statistic; False Discovery Rate <5%). In all, 4231 (72.3%) mapped to currently annotated genes, of which 4011 are protein coding (Supplementary Figure S2). Of these, 2089 were found to vary in level by a factor in excess of four-fold (log2>2) between the pat1.114 diploid strain in vegetative growth and the vegetative haploid, 1157 at 3 h, 1611 at 5 h and 2144 at 10 h (all relative to the pat1.114 vegetative diploid; loci may display high fold change in multiple samples, Supplementary Table S4). In all, 162 of the 4231 statistically significant genes have been previously described as MUGs (meiotically upregulated genes) (Mata et al, 2002; Gregan et al, 2005; Martin-Castellanos et al, 2005; Mata and Bähler, 2006; Xue-Franzen et al, 2006), and a significant majority (729/1033) of genes reported by Mata et al as upregulated by at least four-fold in response to meiosis were also found to have significantly increased levels in the RNA-Seq data in the corresponding sample (FDR <0.05; log2 fold change >0), broadly corroborating these previous studies.
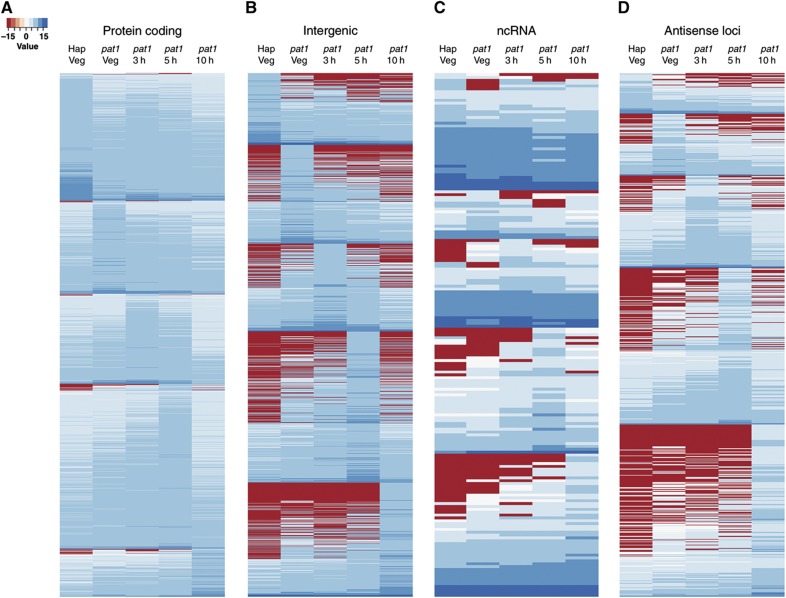
In addition, we found a further 1479 intergenic TBlocks whose levels varied, 80 intronic loci and 809 loci antisense to a known gene. These novel loci exhibited similar temporal ‘expression profiles’ (i.e., transcript levels) to those of previously characterised ‘meiotic’ genes (Figure 1; Mata et al, 2002; Mata and Bähler, 2006; Xue-Franzen et al, 2006), suggesting that they might be under the control of equivalent regulatory mechanisms. The extent of antisense transcription was also striking, and consistent with recent studies (Dutrow et al, 2008; Ni et al, 2010; Quintales et al, 2010; Rhind et al, 2011).
Figure 1.
Non-coding and antisense loci exhibit similar expression profiles to protein-coding genes. Heatmaps showing the expression profiles of (A) 4011 significantly changing protein-coding genes (B) 1479 intergenic regions (C) 183 ncRNA and (D) 809 antisense loci. Expression levels for each locus in each sample were calculated as log2 of the number of sequencing reads starting within that locus, normalised both by the length of the region and by the total number of sequence reads within the sample. Expression data were then grouped according to the sample, in which a given locus displayed the highest expression level.
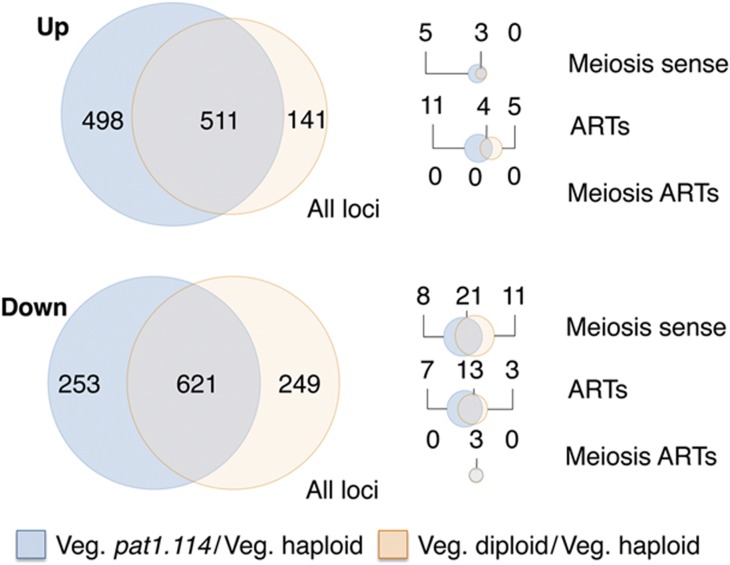
The level of differential gene expression between haploid and pat1.114 diploids in vegetative growth reveals either significant differences between the haploid and diploid state, or a partial defect in pat1.114 at the permissive temperature. We therefore compared the RNA profile of three new data sets of cultures in mid-log phase vegetative growth at 25°C: wild-type 972 h− haploid (IH5974), wild-type ade6.M210/ade6.M216 h−/h+ diploid (IH3365) and pat1.114/pat1.114 ade6.M210/ade6.M216 h−/h+ diploid (IH2912). While the expression profiles for both diploids were similar, more changes were observed in the pat1.114 diploid strain, with 13 additional meiosis-associated transcripts (5 up; 8 down) changing in the pat1.114 diploid, and 11 meiosis-associated transcripts that exhibited reduced levels in the wild-type diploid remaining unchanged in the pat1.114 diploid (Figure 2; Supplementary Table S5), raising the possibility that Pat1 function is partially compromised by the pat1.114 mutation at the permissive temperature, leading to priming of feedback controls that are normally restricted to meiosis.
Figure 2.
A comparison of gene expression changes between pat1.114 diploid cells and the pat1+ haploid and pat1+ diploid cultures during vegetative growth. Loci that exhibited differential expression identified by Fisher's exact test, FDR <0.05, in comparisons between the following cultures in vegetative growth: haploid (IH5974) versus pat1.114 diploid (IH8814) and haploid (IH5974) versus pat1+diploid (IH3365). Orange set: transcripts with significantly altered levels in the wild-type diploid relative to the haploid (pat1+diploid versus haploid). Blue set: transcripts with significantly altered levels in the pat1.114 diploid relative to the haploid (pat1.114 diploid versus haploid). Data were stratified into upregulated and downregulated sets: All loci: all loci detected in the experiment, Meiosis sense: known meiosis-associated genes. ARTs: antisense regulatory transcripts. Meiotic ARTs: ARTs on the complementary strand to known meiotic genes. All gene lists and corresponding fold changes are provided in Supplementary Table S5.
Enrichment of meiotic loci in the cohort of genes for which levels of antisense exceeded sense during sexual differentiation
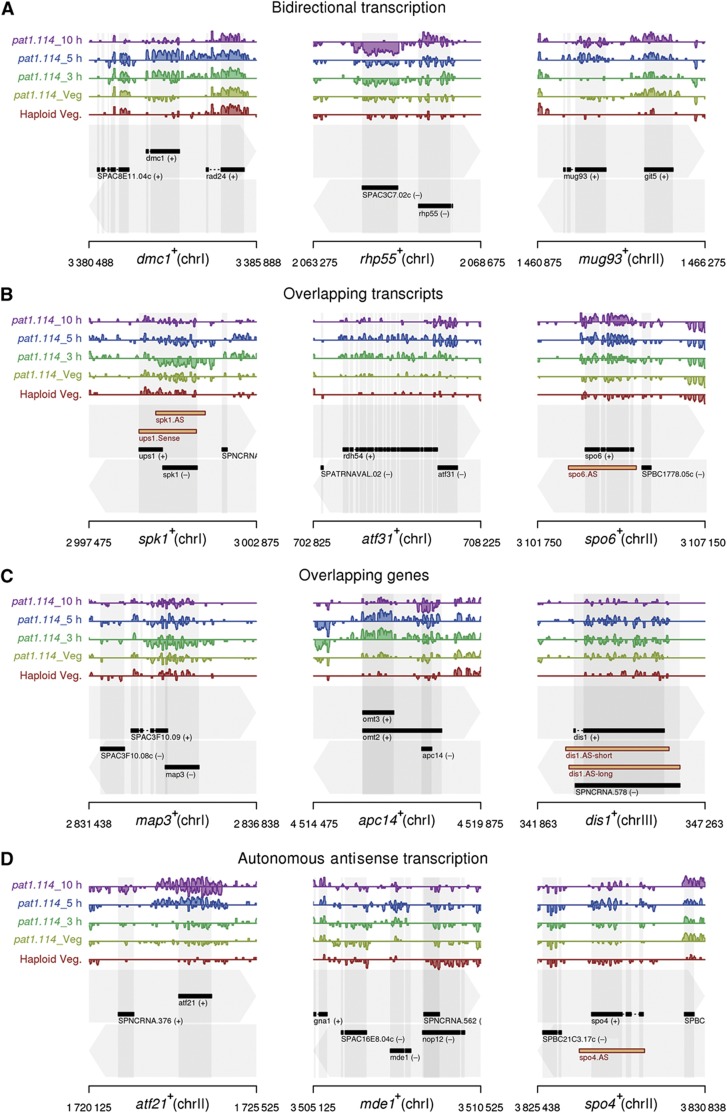
We observed many significant changes in the levels of antisense transcripts opposing a known protein-coding gene. Altered transcript levels are indicative of a variety of mechanisms including bidirectional transcription, run-on transcription from an adjacent, convergent protein-coding locus, and the expression of ncRNAs from independent overlapping loci (Figure 3). Since antisense mechanisms can modulate sense transcript expression through a variety of inhibitory mechanisms (Faghihi and Wahlestedt, 2009), we postulated that waves of antisense expression activated at different stages during meiosis might be regulating protein expression. We computed the relative level of transcripts arising from each strand for all unambiguous antisense/protein-coding transcript pairs, for each of the five samples in the experimental data set (N=4966; of which 4101 were changed significantly in either the sense or the antisense direction). In most cases (N=3747; 91.4%), mRNA levels exceeded their antisense counterparts at all stages. However, for a subset of the data (N=354), greater antisense abundance was observed in at least one sample.
Figure 3.
Examples of antisense transcripts found opposite meiotic and cell-cycle genes. Selected parts of the S. pombe genome. Genome annotation is indicated in the bottom of each panel, with top and bottom strands indicated by the grey arrows in the background. Exons are shown by black rectangles, with direction of transcription indicated by (+) or (−). Normalised, strand-specific, RNA sequencing data on a log2 scale are presented in the tracks above, with forward-strand transcription shown above the centre line of each track, reverse-strand transcription, below. Each track represents a different sample, as indicated. (A) Bidirectional transcription, (B) overlapping transcripts/improper termination, (C) partially/completely overlapping genes and (D) antisense transcription independently from an adjacent locus. The locations of antisense transcripts that were experimentally tested in this study are indicated by the orange boxes in the panels for the spk1+, spo6+, dis1+ and spo4+ loci.
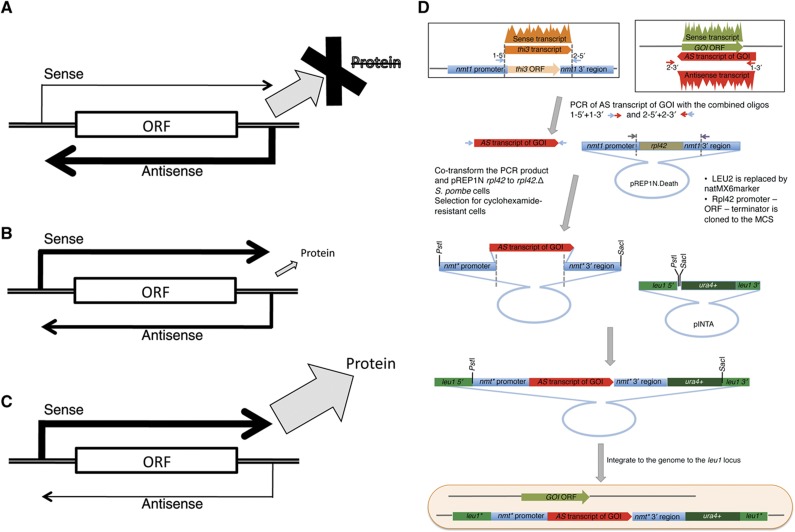
We therefore formed a tentative hypothesis that antisense molecules might act as ‘switches’, suppressing protein production when in excess relative to their target mRNAs, while allowing production when underexpressed. A third class of antisense molecules maintained the sense/antisense ratio close to one (zero in log space). Although speculation, this would be in keeping with a role in maintaining protein homeostasis, in which antisense regulation is used to help keep protein abundance at an appropriate level (Figure 4A–C). A similar model was proposed by Xu et al (2011) in budding yeast while this manuscript was under review.
Figure 4.
Antisense regulatory transcripts (ARTs) modulate protein expression. (A–C) A model in which antisense expression (A) ‘suppresses’, (B) ‘maintains homeostasis’ (C) or ‘permits’ the ability to generate a protein from the gene. (D) A cartoon to illustrate the cloning strategy to express antisense transcripts; for full description, see Materials and methods.
We asked whether ncRNAs tended to oppose genes involved in meiotic pathways. We performed functional term enrichment analysis (Ashburner et al, 2000; Boyle et al, 2004) under extremely stringent settings (Bonferroni adjusted P-value <5 × 10−5; FDR ∼0%), for the 354 genes with greater antisense abundance in one or more samples (Supplementary Table S6). We observed a significant enrichment of meiosis and cell cycle-related Gene Ontology (GO) terms, involving 44/199 proteins annotated to GO terms associated with the meiotic machinery (GO:0007126; GO:0051327; GO:0051321; GO:0000279, Supplementary Table S7). To ensure that this was not a consequence of the underlying structure of a data set, we performed an equivalent analysis using randomly selected sets of genes the same size as the original (354). No term enrichment clusters were obtained using the randomised data (1000 simulations) despite the fact that the simulation was performed using only the subset of protein-coding genes found differentially expressed in our data set (4011). For the majority of these associated meiotic genes (31/44), the highest antisense-sense ratios were recorded in either the vegetative pat1.114 diploid or haploid data sets. This is consistent with the hypothesis that antisense transcripts suppress the impact of these genes in the phases of meiosis at which their gene products are not required.
We refer to the fluctuating antisense transcripts whose levels can, in certain conditions, exceed their sense counterparts, as putative antisense regulatory transcripts (ARTs). We identified 44 ARTs antisense to characterised meiotic loci (Supplementary Tables S6 and S7). Although ARTs account only for a small proportion of the antisense transcription observed in this study (354), many other genes exhibited some antisense transcription (4384 loci with some antisense activity). It is likely that the antisense landscape of the cell will change according to circumstance and developmental context, and that many other ARTs will become operational under different conditions.
Atf21 and Atf31 direct ncRNA production
We asked whether the Atf21 and Atf31 transcription factors that drive fluctuations in sense RNA levels during sexual differentiation (Mata et al, 2002) also impact upon antisense production. Homozygous atf21∷nat and atf31∷kanRpat1.114 diploid strains were induced to promote sexual differentiation by temperature shift alongside a homozygous atf21+atf31+pat1.114 diploid control (Supplementary Figure S3). ‘Bar code’ tags were fused to each of 12 samples before they were pooled and sequenced, as before, and reads mapped uniquely to the genome (Supplementary Table S8). We retrieved data on transcript levels for all regions expressed in at least one of the samples.
A Fisher's exact test was used (Bloom et al, 2009) to derive lists of stage-specific differentially expressed genes likely to have arisen from loss ofAtf21 or Atf31 function. Read counts at each locus for the atf21.Δ pat1.114 and atf31.Δ pat1.114 mutants were systematically compared over the course of sexual differentiation to the equivalent atf21+, atf31+pat1.114 diploid control. Expression levels were altered for a total of 1045 regions in at least one time point in the atf21.Δ data set, while 1356 were found to be significantly changing in atf31.Δ (FDR <0.0125; adjusted according to Storey, 2004). Many of these putative targets were shared (687), of which 550 correspond to protein-coding genes. Separate GO analyses of the two target sets and their intersection revealed significant term enrichment for processes including carbohydrate metabolism, cell wall biogenesis and organisation, and stress responses (Supplementary Table S9). At a more relaxed cutoff (P-value <0.01) terms including sporulation, ascospore formation and sexual reproduction were also enriched (Supplementary Table S9). Since both Atf21 and Atf31 control late meiotic genes (Mata et al, 2002, 2007), these terms are in accordance with the cellular processes (e.g., sporulation), occurring at this time. These data are also consistent with Mata et al (2002) of the 138 genes reported to be dependent on atf21.Δ or atf31.Δ at 15 h following induction of meiosis by starvation, 77 transcripts were found to be changing (log2 fold change >0; FDR <0.05) in the closest equivalent time point in our data sets (10 h following induction of meiosis by Pat1 inactivation). The overlap between the two sets was also found to be statistically significant (P<10−6 by permutation; data not shown).
The overlap in differentially expressed loci in each of the deletion strains supports the documented synergy between atf21+ and atf31+ (Mata et al, 2007). Evidence for synergy is further strengthened by the increased expression of atf21+ in response to atf31+ deletion, and vice versa (Supplementary Figure S3A and B). In total, 1714 regions were flagged as differentially expressed in either deletion background (Supplementary Table S9). These encompass a wide range of RNA species including protein-coding transcripts (1332), unannotated intronic regions (8), pseudogenes (4), annotated ncRNAs (88), TBlocks (197) and antisense loci (85) of which 63 were antisense to protein-coding genes (Supplementary Figure S4). Antisense transcripts opposing the atf21+and atf31+ sense transcripts were also significant, but since these changes were due to the genomic deletions, they were ignored. Similarly, antisense transcription opposing the rdh54 ORF originated from the wild-type atf31+ locus, and was thus not considered further (Supplementary Figure S3B).
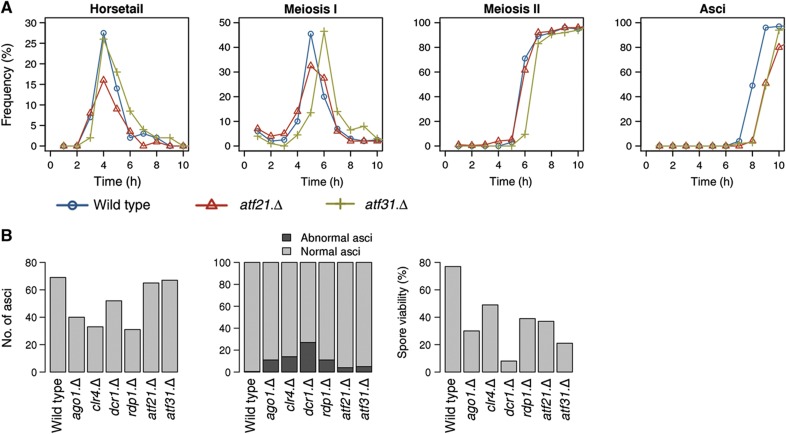
Overall, transcripts from 185 non-protein coding loci had significantly altered levels in the knockout studies, demonstrating a role for these transcription factors in their regulation. Loss of either transcription factor had only modest impact upon meiotic progression (Figure 5A). Atf21 deficiency led to a minor delay in ascus formation while removal of atf31+ delayed the onset of meiosis I, and subsequent events by 30 min, without affecting the profile of these subsequent phases (Figure 5A). Thus, the impact of removing these transcription factors upon ncRNA production is unlikely to arise as a secondary consequence of altered meiotic progression. Rather, it shows that a considerable proportion of ncRNA production during sexual differentiation is dependent on Atf21 and Atf31. The markedly reduced viability of spores arising from meiosis conducted in the absence of either transcription factor suggests that Atf21- and Atf31-mediated events have important roles in ensuring successful sexual differentiation (Figure 5B; Morita et al, 2011). It is currently unclear whether Atf21 and Atf31 modulate these events via their transcriptional activity, or because they can emulate the ability of the related transcription factor Atf1 to seed heterochromatin (Jia et al, 2004).
Figure 5.
The impact of the RNAi machinery and Atf21 and Atf31 transcription factors on the fidelity of meiotic progression. (A) pat1.114 atf21+atf31+ (IH2912), pat1.114 atf21.Δ (IH8832) and pat1.114 atf31.Δ (IH8814) homozygous diploid strains were grown to mid-log phase and shifted from 25 to 32°C stained with DAPI at the indicated intervals to score the frequency of each meiotic phase. (B) Sexual differentiation of the indicated haploid h90 strains was induced by nitrogen starvation of mid-log phase liquid cultures by plating onto MSL plates. Samples were processed for DAPI staining 24 h later to score the indicated phenotypes. In all, 10 000 spores were plated to assess spore viability.
Using expression data from all 12 samples, plus wild-type haploid and diploid strains, we were able to re-identify 73% of ARTs (257/354) from the initial sequencing data set, and to identify an additional 282 statistically significant ARTs (G-test FDR <0.05; Supplementary Table S10). GO analysis of all 636 ARTs revealed enrichment of the same meiotic terms as before (identical parameters) with the involvement of 14 more members of the meiotic machinery 58/199 (Supplementary Table S7). The differences between the first and the second set of ARTs are likely to have arisen not only as a consequence of experimental effects (although concordance between replicates was high; Supplementary Figure S5), but also because the gene deletions disrupt the transcriptional pattern of the cells, resulting in the transition of antisense transcripts into ARTs and vice versa.
Antisense regulation and cell fate
We next tested the model in Figure 4A–C. Our experimental design was based upon the following assumption: if changes to antisense/sense transcript ratio during a particular phase of sexual differentiation regulate protein expression, then the continued presence of the antisense throughout this specific phase should abolish protein function during this phase of the differentiation programme. The nmt1+/thi3+ gene has no antisense transcript, and the levels of sense transcription do not fluctuate during sexual differentiation (Supplementary Figure S3C). We therefore used this widely exploited repressible promoter (Maundrell, 1990) to study the functional impact of the production of antisense transcripts for loci that are known to modulate cell fate. Two types of loci were selected: isolated genes for which the ART is generated by a gene-specific event (dis1+ and spo4+) and genes with an ART generated by run through transcription of a sense transcript on the opposite strand (spk1+ and spo6+). In each case, our cloning strategy swapped the entire nmt1+/thi3+ transcript (5′ UTR+ORF+3′ UTR) for the entire ncRNA in a cassette encompassing the entire nmt1+/thi3+ locus. This cassette was integrated into the genome at the leu1 locus (Figure 4D). Thus, for each particular target, the normal, controlled production of ncRNA from the endogenous locus was supplemented by continuous production, in trans, of the same antisense ncRNA transcript from a single ectopic locus upon removal of thiamine.
Antisense ncRNA control of pheromone signalling via modulation of Spk1
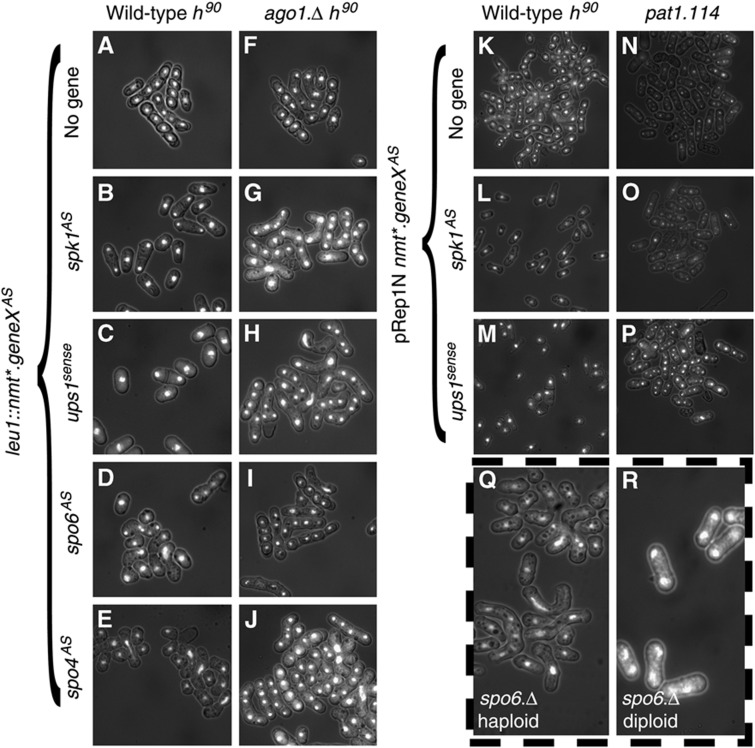
The MAP kinase Spk1 acts within the pheromone-signalling cascade that drives transcription of the Mat1-Pc and Mat1-Mc mating type genes. Removal of Spk1 abolishes mating and the ensuing sexual differentiation. The antisense transcription that opposes the sense spk1+ transcript originated from improper termination of the sense ups1+ transcript on the opposite strand (Figure 3B, left locus). Thus, when addressing the consequences of spk1AS production, it was important to establish whether any phenotypes arising from elevating spk1AS arose from increased Ups1 or downregulated Spk1 function. We therefore expressed each of the two antisense transcripts from the expression cassette at the leu1 locus; the natural, full-length, ups1+ transcript (ups1sense) and a truncated version that was restricted to the portion of the ups1+ transcript that overlapped with the spk1+ sense transcript (spk1AS; Figure 3B left locus, orange transcripts). Constitutive production of either transcript from the ectopic locus abolished mating of a wild-type h90 strain (Figure 6B and C; Supplementary Figure S6A, B and G). Importantly, induction of either spk1AS or ups1sense had no impact upon the ability of pat1.114 cells to execute a haploid meiosis (Figure 6K–P) indicating that spk1AS production can specifically perturb the Spk1 pheromone response pathway, rather than blocking sexual differentiation per se.
Figure 6.
Antisense RNA production perturbs Spk1 and Spo4/Spo6 function in an Ago1-dependent manner. (A–J) The indicated antisense RNA transcripts were induced by derepression of the nmt* promoter in the indicated h90 genetic backgrounds by removal of thiamine. Seventy-two hours later, the nitrogen source was removed to induce sexual differentiation. Twelve hours later, cells were subjected to DAPI staining to give the DAPI/DIC images in the respective panels. (K–P) Expression of the nmt* promoter on the indicated plasmids in either wild-type h90 pat1.114 strains was derepressed for 48 h before starvation by removal of nitrogen source (h90 strains K–M) or temperature shift from 25 to 32°C (pat1.114 strains N–P). The panels show DAPI/DIC images of fields of cells. (Q) Sexual differentiation was induced in haploid h90spo6.Δ cells by nitrogen starvation, 24 h later, staining of the cells/zygotes/asci with DAPI indicated that ablation of Spo6 function in h90 cells compromised meiotic progression in an identical manner to spo6AS. (R) pat1.114 spo6.Δ homozygous diploid (IH8833) cells 24 h after meiotic induction by shift from 25 to 32°C. Loss of Spo6 function in the context of a pat1.114-induced meiosis contrasted with the natural mating involving pheromone signalling as it did not perturb the rate of execution of early events, but blocked transit of meiosis II.
Further, the ability of constitutive expression of the natural ups1+ transcript to block meiotic commitment (Figure 6C; Supplementary Figure S6G), clearly demonstrates that that 3′ gene extensions can result in functional suppression of the gene on the complementary strand.
Antisense ncRNA control of meiotic progression via modulation of Spo4/Spo6 kinase
spo4+ and spo6+ encode the catalytic and regulatory subunits of a kinase that is reported to be required for progression through meiosis II (Nakamura et al, 2000, 2002). Expression of the spo6+ antisense transcript (spo6AS) in trans from the cassette in the leu1 locus (3′ extension of SPBC1778.05c; Figure 3B, right) impaired meiotic progression. While the majority of wild-type h90 control cells had formed asci 24 h after plating onto MSA medium, many spo6AS expressing zygotes were at much earlier meiotic stages, such as horsetail movement. Abnormal meiotic chromosome segregation was also visible (Figure 6D; Supplementary Figure S7B). Similar perturbations were seen upon expression of the spo4+ antisense transcript (spo4AS) from the ectopic expression cassette at the leu1+ locus (Figure 6E; Supplementary Figure S8B).
These consequences of spo6AS and spo4AS expression differed from the reported arrest during meiosis II arising from disruption of either the spo4+ or spo6+ locus (Nakamura et al, 2000, 2002). We therefore constructed a strain in which the spo6+ coding sequences were completely replaced with the marker kanR, and subjected a spo6∷kanR h90 strain to the same mating test used for the h90 strains above. The meiotic profile of the spo6∷kanR h90 strain (Figure 6Q; Supplementary Figure S7C) essentially emulated that of h90 cells expressing spo4AS or spo6AS (Figure 6Q; Supplementary Figures S7B and S8B). The original assessment of spo4.Δ and spo6.Δ disruption phenotypes was conducted in a pat1.114 diploid meiosis (Nakamura et al, 2000, 2002), rather than the natural mating employed here. Subjection of a spo6.Δ pat1.114 diploid strain (IH8833) derived from our spo6∷kanR deletion, to a temperature shift revealed the same block to meiotic progression at meiosis II as reported previously (Figure 6R; Supplementary Figure S7D). Critically, spo6AS induction in the context of a pat1.114 diploid meiosis completely phenocopied the change in spo6∷kanR phenotype expression brought about by changing the method of meiotic induction to pat1.114 induction; antisense induction now blocked meiotic progression at meiosis II (Supplementary Figure S7E). Thus, Spo4/Spo6 kinase appears to execute more extensive functions than previously appreciated. Specifically, the requirements for Spo4/Spo6 kinase differs between natural and pat1.114 induced meioses, a view that is reinforced by recent studies of Spo4- and Spo6-deficient strains (Arai et al, 2010; Asakawa et al, 2010).
We conclude that the generation of spo4/spo6 antisense transcripts had the same impact upon sexual differentiation as ablation of the kinase by gene deletion, indicating that they effectively abolished the function normally derived from these loci. The shared context specificity of the spo6.Δ and spo6AS phenotypes strongly suggests that antisense production induces a locus-dependent rather than generic, non-specific, modulation of gene function.
dis1AS RNA controls the differentiation of the microtubule cytoskeleton that drives horsetail movement
dis1+ encodes a member of the chTOG family of microtubule polymerases that modulates spindle function during vegetative growth (Nabeshima et al, 1995; Garcia et al, 2001; Nakaseko et al, 2001; Al-Bassam and Chang, 2011). Two factors prompted us to study the impact of the dis1+ antisense transcript (dis1AS) function: the independence of dis1AS production from transcription at adjacent loci (Figure 3C, right locus) and the availability of Dis1 antibodies to monitor the gene product (Nabeshima et al, 1995).
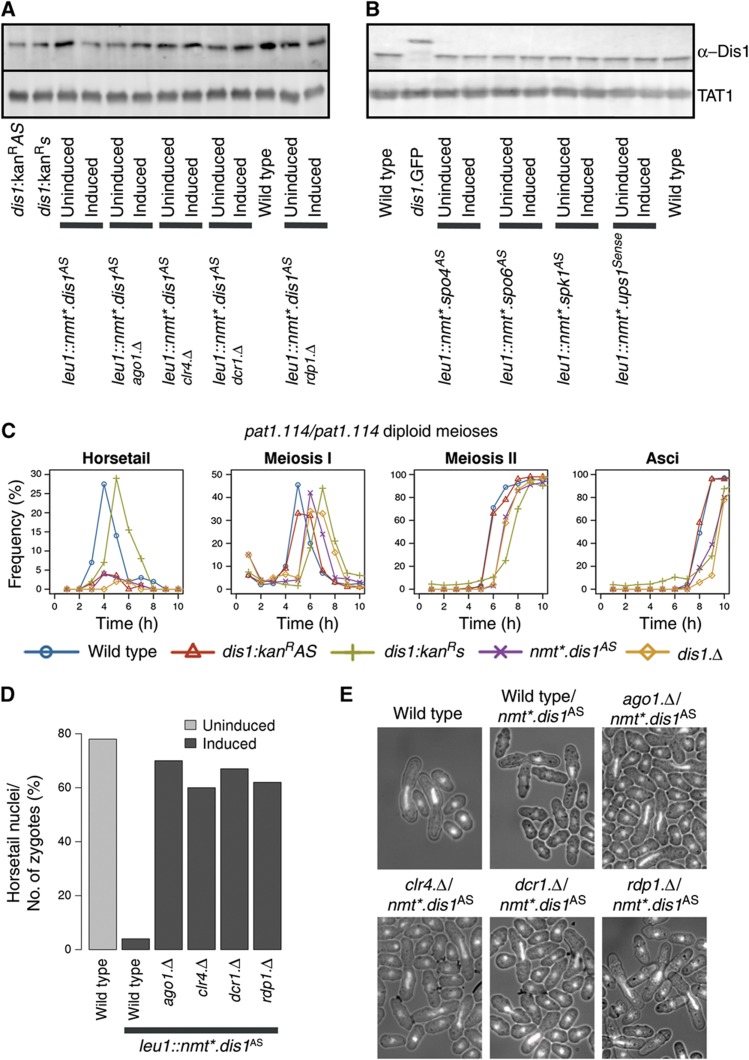
dis1AS is an annotated ncRNA molecule (SPNCRNA.578). Since our data suggested that the transcript is slightly shorter than the documented assignment (Figure 3C, right locus), we tested both the short (our prediction) and the long versions (dis1AS-short and dis1AS-long, respectively) for their ability to alter Dis1 protein levels when expressed from pRep1 multicopy vector (Supplementary Figure S9A). As expression of either transcript reduced protein level and impaired meiotic progression to the same degree (Supplementary Figure S9A), we selected the long version (dis1AS-long) for integration into the leu1 locus under the control of the nmt1+ promoter. Induction of this antisense transcript led to a significant decrease in Dis1 protein levels (Figure 7A). ncRNA production in trans from this ectopic locus during sexual differentiation induced by either temperature shift of a pat1.114 diploid strain or starvation of a homothallic h90 strain reduced protein levels and virtually abolished the stage of meiotic progression that relies upon differentiation of the microtubule cytoskeleton: horsetail movement (Figure 7C; Supplementary Figures S1, S10 and S12B). As Dis1 function has not been specifically linked to this stage of sexual differentiation before, we asked whether Dis1 protein was indeed required for horsetail movement, by monitoring the meiotic progression of a pat1.114/pat1.114 dis1.Δdis1.Δ diploid strain shifted to 32°C. Strikingly, it was (Figure 7C, orange diamonds). We conclude that correct antisense ncRNA control of Dis1 protein levels is critical for the differentiation of the microtubule cytoskeleton that drives horsetail movement.
Figure 7.
dis1AS control of horsetail movement is reliant upon RNAi and Clr4. dis1AS production was induced for 48 h in the indicated strains and samples processed for blotting to monitor protein levels either before or after induction (A, B) or DAPI staining 10 h after the shift to score horsetail nuclei (D, E). (C) pat1.114 h90 diploid strains either homozygous for the kanR marker insertions (red triangles and green crosses), or the leu1∷nmt*dis1ASura4+ expression cassette (induced for 48 h) were subjected to temperature shift from 25 to 32°C to induce sexual differentiation and the samples subjected to DAPI staining to score the frequency of the indicated phenotypes at the indicated intervals.
Insertion of a marker gene immediately upstream of the dis1AS start site phenocopies ectopic antisense production
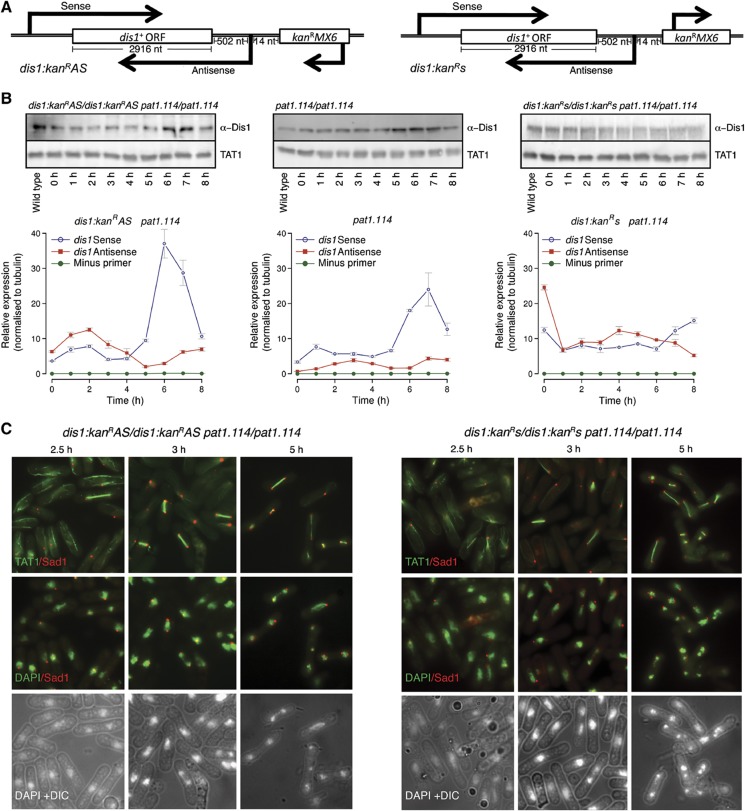
We reasoned that dis1 antisense transcript expression may be influenced by sequences located the 5′ end, upstream of the antisense transcription start site (i.e., at the 3′ region of the sense gene). For example, if antisense production is driven by TATA box-mediated polII or polIII transcription, then the insertion of a marker between the 5′ regulatory regions and the start site may alter this control. We therefore integrated the kanRMX6 marker 14 nucleotides before the start site of the dis1+ antisense transcript, such that the marker was either transcribed in the same orientation as the dis1+ antisense transcript (dis1:kanRAS; back into the gene) or that of the sense (dis1:kanRs; away from the gene) (Figure 8A). Dis1 levels were reduced by marker insertion in the dis1:kanRAS but not in the dis1:kanRs orientation (Figure 8B). Cultures of diploids homozygous for both pat1.114 and dis1:kanRAS or pat1.114 and dis1:kanRs (IH9675 and IH9758, respectively) were shifted from 25 to 32°C to induce meiosis. There was a considerable reduction in protein levels when the marker was in the antisense orientation. Critically, the reduction in Dis1 levels arising from the introduction of the marker reading back into the gene (dis1:kanRAS) mimicked in trans antisense production and the removal of dis1+ coding sequences (Figure 7C) in ablating horsetail movement in these pat1.114 driven meioses (Figures 7C and 8C; Supplementary Figure S1). This abolition of horsetail movement did not arise from a general perturbation of meiotic progression as transit through subsequent meiotic stages was not affected (Figure 7C). Importantly, Dis1 protein levels were not affected by the production of antisense transcripts for all other loci tested in this study (Figure 7B), indicating that the reduction in Dis1 protein levels arising from either ectopic expression from an ectopic locus or the insertion of a marker in the dis1+ 3′UTR are the consequences of gene-specific expression rather than a non-specific impact such as simple titration of the RNAi machinery (see below).
Figure 8.
Transcription of a neighbouring marker towards, but not away from the dis1+ locus, perturbs Dis1 protein production and meiotic horsetail differentiation. (A) Cartoons depicting the architecture of the dis1:kanRAS and dis1:kanRs loci. (B) Immunoblots of samples taken at the indicated times from either dis1:kanR marker insertion alleles or dis1+ wild-type control diploid strains over the course of a pat1.114-induced sexual differentiation time course (upper panels). The lower panels show qPCR quantitation of the sense and antisense transcripts at the dis1 locus in each strain. (C) Tubulin and spindle pole staining (Sad1) at the indicated times as the dis1 marker insertion alleles are induced to undergo sexual differentiation by temperature-dependent inactivation of Pat1.114. The staining shown is indicated in labels in the left corner of each panel.
Involvement of Ago1, Dcr1, Clr4 and Rdp1 in antisense control
Given the widespread use of RNAi in fission yeast (Grewal, 2010), we asked whether the control of sexual differentiation by antisense RNA molecules used this system to modulate cell fate. The striking alteration of meiotic progression arising from production of antisense RNAs to the spk1AS, spo4AS and spo6AS loci was abolished in ago1.Δ, dcr1.Δ and rdp1.Δ backgrounds (Figure 6F–J; Supplementary Figure S11A–E and F–J, respectively; Supplementary Figures S6–S8). Similarly, dis1AS expression failed to reduce Dis1 protein levels or compromise horsetail movement upon deletion of any of these genes (Figure 7A–E; Supplementary Figure S9B). Removal of clr4+ also blocked the impact of dis1AS antisense production upon horsetail movement (Figure 7D and E), suggesting that this histone H3K9 methyltransferase contributes to the control of the dis1+ locus. However, at this stage is not possible to distinguish whether this impact arises as a consequence of heterochromatin formation (Volpe et al, 2002), possibly independently of H3K9 methylation (Gerace et al, 2010), or Mlo3 directed switching between TRAMP and siRNA pathways (Zhang et al, 2011).
This role for Clr4 in controlling dis1 could indicate that ARTs drive or facilitate heterochromatin formation. We therefore asked whether ARTs coincided with previously identified constitutive heterochromatic regions including subtelomeric, pericentromeric and mating type loci (Woolcock et al, 2011). Only 29/636 ARTs fell within these regions, of which only a single ART (complementary to SPAC186.09) was associated with Clr4-dependent enrichment of H3K9me, Swi6, Ago1, Rdp1 and Chp1 (Cam et al, 2005). The remaining 626 ARTs for which we were able to find a corresponding probe in the Cam et al data set (626/627) were enriched (>2-fold) for the euchromatin marker, H3K4me and showed little overlap with novel Swi6 (3), Rdp1 (0) and Dcr1 (5) associated regions, recently identified by Woolcock et al (2011). The significance of this apparent lack of correlation is tempered by the fact that these studies were conducted on log phase vegetative cultures, and the ARTs we define here are largely enriched in meiotic genes. A true appreciation of the interplay between heterochromatin formation and ART function awaits more in-depth analysis of chromatin structure and modifications during sexual differentiation.
If RNAi control makes a significant contribution to the fidelity of sexual differentiation, then we would anticipate that meiotic progression or the viability of spores produced from an RNAi-deficient meiosis should be compromised. While meiotic progression was not blocked by removal of any of these RITS-associated components, the number of cells undergoing meiosis in an h90 culture and the viability of the ensuing spores were diminished (Figure 5B; Hall et al, 2003). We conclude that RNAi control has a key role in modulating the fidelity of sexual differentiation in fission yeast. Our data suggest that significant component of the impact of this system upon meiotic progression arises from participation in antisense gene silencing at one or more meiotic stages.
The impact of antisense induction upon transcript levels
The striking correlations between the antisense induction and gene knockout phenotypes (Figure 6; Supplementary Figures S6–12) indicate that antisense production can silence gene function. It is possible that antisense production altered the overall sense/antisense ratio, the absolute level of sense transcripts or the translation of the sense transcripts. While the detailed mechanisms operating at each of these loci is beyond the scope of the current study, we performed strand-specific qPCR to monitor sense and antisense abundance during sexual differentiation in each of these contexts.
Induction of either spo4AS or spo6AS dramatically increased the levels of these ncRNAs (∼10-fold for spo6AS and 400-fold for spo4AS) and decreased the levels of the sense transcripts during a naturally induced sexual differentiation (Supplementary Figures S7B and S8B). Ago1 function was required for the stability of both antisense transcripts (Supplementary Figures S7G and S8D). The negative impact of antisense induction upon sense transcript abundance was independent of the method of meiotic induction, as spo6AS induction during a pat1.114-induced meiosis also increased spo6AS levels, and reduced spo6 sense transcript levels (Supplementary Figure S7J and K).
While induction of spk1AS did not lead to the anticipated increase in the levels of this ncRNA, there was a marked decrease in the abundance of sense transcripts at the early stages of sexual differentiation (Supplementary Figure S6B). As with the spo4+ and spo6+ loci, this alteration in sense transcript levels relied upon Ago1 function (Supplementary Figure S6D).
dis1+ sense/antisense transcripts exhibited a third type of behaviour. While induction of antisense production phenocopied dis1.Δ in abolishing horsetail movement in a pat1.114-induced meiosis (Figure 7C), it did not have any impact upon the steady-state levels of either sense or antisense transcripts detected by qPCR (Supplementary Figure S10). In contrast, the insertion of the kanR marker 14 nucleotides upstream of the antisense start site increased dis1AS transcript levels throughout the sexual differentiation cycle in both the dis1:kanRAS and dis1:kanRs orientations (Figure 8B). Paradoxically, sense transcripts increased in dis1:kanRAS but decreased in the dis1:kanRs strain. Thus, while there is a strong correlation between the induction of antisense ncRNA transcripts and phenotype at this locus, the bulk levels of transcripts reveal a more complex pattern than at the spo4+, spo6+or spk1+ loci, suggesting that feedback controls may act at this dis1+ locus. Establishing the nature of such feedback controls is beyond the scope of this study as a range of possibilities exist, from alterations in histone occupancy through changes in chromatin modifications. However, two key points are pertinent for the current study: when the kanR marker transcribes in the AS orientation the phenotype mirrors that of the dis1.Δ null and the fact that the AS:s transcript ratio is elevated to around 2:1 at the phase of sexual differentiation during which Dis1-dependent horsetail migration occurs. In contrast, the AS:s ratio only reached around 1:1 when the marker was in the opposite orientation and there is no phenotype.
It is notable that abolition of Argonaute function decreased the ratio between the abundance of sense transcripts to atb2+ (α-tubulin) controls at the spo4+, spo6+ and spk1+ loci. This diminished ability to induce these meiotic genes may account for the reduction in the efficiency of meiotic progression and spore viability in Ago1 mutants (Figure 5B; Hall et al, 2003).
Discussion
We show that an extensive and elaborate array of ncRNA production accompanies sexual differentiation in the fission yeast S. pombe. Experimental manipulation suggests that these transcripts specifically regulate the function of the target genes. While previous studies have demonstrated the principle of gene control by antisense expression in fission yeast using exogenous genes (Arndt et al, 1995), the ubiquity, and importance of the endogenous genes subjected to ncRNA expression in our data sets, means that antisense interactions now move from a theoretical possibility to an intrinsic part of the regulatory machinery, with the same status and importance as other levels of control. Encouragingly, a similar model of ncRNA control of gene regulation has recently been derived from the analysis of stress responses in budding yeast (Xu et al, 2011).
The RNA sequencing experiments relied solely upon the pat.114 temperature sensitive mutation to relieve the block to meiotic differentiation imposed by Mei2. As graphically illustrated by the discrepancy in spo4/spo6 phenotypes described here and well documented in the literature, this approach to synchronising sexual differentiation, while enabling us to readily compare a range of genetic backgrounds with ease, is not representative of natural sexual differentiation, which combines pheromone signalling with nitrogen starvation signals (Chikashige et al, 2004; Arai et al, 2010; Asakawa et al, 2010; Cremona et al, 2011). It is therefore likely that the complexity of ncRNA controls during natural sexual differentiation will exceed those described here.
One phenomenon that emerged from our study is the extensive changes in the RNA profile between haploid and diploid strains. RNA levels differed at over 1000 loci. There are inherent differences between the haploid and diploid state. A recent report, that took a systematic approach to address the relationship between cell size and global transcription in haploid strains, revealed a tight coupling between transcription throughout the genome at different DNA/protein ratios (Zhurinsky et al, 2010). Thus, the changes seen in diploids are unlikely to arise from the simple doubling of genome content. Rather, diploid cells are longer, wider and less healthy than haploid cells, indicating significant differences in basic aspects of physiology. Perhaps, these changes reflect/invoke altered global transcriptional controls: an interesting topic for future molecular studies.
We found 321 genes that overlap to some extent with a gene on the opposite strand. About a quarter of these (77; 24%) are associated with known ncRNA genes corresponding to antisense loci that have been previously observed, but are yet to be assigned a function (Wilhelm et al, 2008). The extent of antisense transcripts is partly due to overlapping 3′ UTRs of convergent, adjacent, genes. If only coding sequences are considered, then the total number of overlapping gene pairs drops to 163. This serves to highlight the importance of using comprehensive genome annotations when designing gene disruption experiments, since care must be taken to avoid interfering with a regulatory antisense molecule on the opposite strand to the gene of interest. Furthermore, it is also vital to consider the possibility that the disruption of one gene can alter its ability to regulate a neighbouring gene via run-on transcription into the adjacent locus. The impact of marker insertion is exemplified by the orientation-dependent defect arising from marker insertion in the sequences 3′ of the dis1+ gene. Markers are routinely inserted immediately after stop codons when ‘PCR tagging’ approaches (Bähler et al, 1998) are used to fuse a protein tag to the 3′ end of a gene, to study protein function. Our data now show that it is dangerous to assume that a given gene is an independent entity, isolated from its neighbours, and that an extra set of controls may be required to ensure that the impact of a ‘deletion’ or protein tag is due entirely to the planned manipulation.
The UTRs of genes are typically more variable than their ORFs, making de-novo annotation of transcription start and end sites difficult (Brent, 2008). This holds both for protein-coding genes and ncRNAs. Consequently, at present, gene coordinates within GeneDB (Wood et al, 2002; Hertz-Fowler et al, 2004) represent only the extent of their coding sequences, although some annotation tables, determined by a visual inspection of array-derived transcription data (Dutrow et al, 2008), provide additional 5′ and/or 3′ UTR annotations for a subset of genes (Wilhelm et al, 2008; Lantermann et al, 2010). Re-annotating the genome using our RNA-Seq data and that of Rhind et al (2011), gives a more accurate definition of UTRs for the majority of genes in S. pombe; we expect this more detailed annotation to be a useful resource for the community and to provide an additional data source when interpreting global gene deletion data (Kim et al, 2010; http://annmap.picr.man.ac.uk). Importantly, we have found that the boundaries of both 5′ and 3′ UTRs in many loci changed during sexual differentiation, indicating that the definition of UTR boundaries in different genome data sets may apply only to the exact experimental conditions for each particular data set. The changes in UTR extent indicated by our studies suggest that a future, systematic, exploration of this phenomenon and the way UTR length is actively regulated in different contexts would be highly informative.
A disparate set of mechanisms generate antisense transcripts, including bidirectional transcription (Neil et al, 2009; Xu et al, 2009), simultaneous/asynchronous transcription from partially/completely overlapping transcripts and genes (Fahey et al, 2002; Munroe and Zhu, 2006), and transcription originating from independent antisense loci (Martens et al, 2004; Hongay et al, 2006). Expression patterns corresponding to each of these were apparent in our data (Figure 3A–D). Despite such diversity, some unifying patterns do emerge. First, in all loci tested by the production of antisense RNA, antisense function was entirely dependent upon the components of the RNAi machinery: Argonaute, Dicer and RNA-dependent polymerase. Second, the relative abundance of sense/antisense pairs is a significant marker of regulatory activity, irrespective either of the underlying mechanisms responsible for the biogenesis, or whether individual ARTs are operating pre- or post-transcriptionally. Thus, while our antisense expression experiments show that the control of key meiotic effectors by antisense production precisely regulates specific events, it is likely that similar studies of other pathways will identify similar control by antisense expression.
While the reliance of the specific phenotypes arising from ncRNA induction upon ago1, dcr1 and rdp1 indicates a key role for the RNAi machinery in implementing the control by ncRNA of gene function, it would be premature at this stage to draw precise conclusions about the level at which this control is executed at each particular locus. It will be important to disentangle genome-wide consequences of the removal of the RNAi machinery upon meiotic progression from targeted controls at each specific locus. The loss of the more generic RNA processing factor, Argonaute, in particular, clearly has a global impact upon both sense and antisense levels. Thus, any consideration of specific effects at a target locus must be considered in the context of genome-wide changes in the transcript profile. Until our understanding of the nature of such global changes improves, detailed interpretations of cause and effect, or even the relative contributions of one level of control over another, cannot be drawn. It is, however, clear that in every instance described here, the induction of ncRNA imposes a targeted attenuation of function that is both locus specific and RNAi dependent; it is simply not possible to infer the means by which the RNAi machinery exerts this control at present. Such detailed understanding can only arise from in-depth targeted analyses at key loci, such as dis1.
Antisense regulation of gene expression via heterochromatin formation raised an interesting ‘heterochromatin paradox’: How can you transcribe a gene that has been silenced in order to maintain silencing (Djupedal and Ekwall, 2008)? This was resolved by enhancing the temporal resolution of the study to the point where transcription and silencing were revealed to occur during discrete phases of the cell cycle, with antisense regulation in G1 resulting in the recruitment and accumulation of cohesin in S and G2 (Chen et al, 2008; Gullerova and Proudfoot, 2008). Thus, while the assessment of an asynchronous culture fails to resolve the phases of transcription and silencing during vegetative growth, analysis of successive cell-cycle phases in a synchronised culture revealed the underlying mechanism. We show that antisense regulation of protein expression in meiosis fluctuates at discrete phases of sexual differentiation, with antisense abundance exceeding sense abundance at specific stages of differentiation. Given that changes at a particular locus may oscillate more frequently than our sampling rates it is likely that an ‘ART paradox’ could obscure further ART loci.
The insertion of a marker 14 nucleotides upstream of the antisense start site of the dis1+ locus phenocopied both gene deletion and antisense production in reducing protein levels and abolishing horsetail movement. It did so only when its transcription ran in the same direction as the natural antisense transcript suggesting that persistent transcription extended from the inserted marker locus into the dis1+ locus. While this general conclusion is clear, and the insertion of the marker in the sense orientation had no impact upon the prevalence of horsetail nuclei, both Dis1 protein and RNA profiles were affected in this strain (Figure 8B). Rather than migrating as a single species, a more complex pattern of faster migrating Dis1 polypeptides accompanied the full-length molecule during sexual differentiation. Paradoxically, despite the persistence of Dis1 function in this strain, the sense/antisense ratios were also dramatically altered (Figure 8B). A detailed investigation of the mechanics of ncRNA production at the dis1+ locus and the means by which dis1AS alters protein levels is beyond the scope of this systems-based study. However, the isolation of the dis1+ locus from other protein-coding genes, the production of an independent ncRNA as its regulatory antisense transcript and an mRNA that encodes a protein-coding gene that generates a clear and well-defined phenotype suggest this locus may form the focus for future mechanistic studies.
The requirement for Dis1 function for horsetail movement has not been documented before. dis1+ antisense production or gene deletion abolished horsetail movement. This inhibitory affect would be cancelled out by simultaneous co-expression of sense transcript from an identical promoter (Supplementary Figure S9C). Significantly, increasing dis1+ sense transcript production in wild-type h90 cells actually increased the frequency of zygotes with horsetail movement (Supplementary Figure S9C), highlighting both the direct correlation between Dis1 levels and the induction of horsetail movement and the need to precisely control Dis1 levels (presumably by the ncRNA approach we uncover here) in order to regulate this important phase of meiotic recombination. The recent realisation that chTOG proteins act as microtubule polymerases (Al-Bassam and Chang, 2011) combine with earlier observations that Dis1 is enriched at SPBs (Nabeshima et al, 1995) to suggest that Dis1 promotes the extensive array of horsetail microtubules (Hagan and Yanagida, 1995; Svoboda et al, 1995) by accelerating microtubule polymerisation at the SPBs.
Control of the RNA profile appears to be a principal means by which sexual differentiation is regulated in S. pombe. It is regulated at the level of transcription, through stability (active stabilisation or destruction) and via at least two distinct types of splicing control (Shimoseki and Shimoda, 2001; Mata et al, 2002; Averbeck et al, 2005; Mata and Bähler, 2006; Xue-Franzen et al, 2006; Moldon et al, 2008; Djupedal et al, 2009; Amorim et al, 2010; Cremona et al, 2011). There appears to be significant overlap between these systems. For example, spo4+ and spo6+ are subject to both ncRNA and splicing controls (Averbeck et al, 2005). Examination of the ncRNA data set for overlap with the Mmi1/DSR system reveals low levels of antisense activity at seven DSR/Mmi1-regulated loci (e.g., ssm4+, mei4+), while a further three loci (meu1+, crs1+ and bqt1+) exhibited a statistically significant excess of antisense transcription in at least one data set. The antisense for crs1+ and bqt1+ originated from partially overlapping transcripts (∼200–300 bp) on the opposite strand, meu1+ antisense was considerably longer, and originated independently of neighbouring genes. Further overlap of controls at the crs1+ locus includes coordination between Mmi1-mediated control of both splicing and polyadenylation (McPheeters et al, 2009).
We show that antisense expression is also critical for modulating gene function over the course of sexual differentiation. The wide variety of mechanisms for generating antisense (bidirectional transcription, nascent transcription from an independent locus, overlapping 3′ UTRs), coupled with the diversity of mechanisms by which these transcripts are exploited within cells (both pre- and post-transcriptional, in cis and in trans, with and without the establishment of heterochromatin), add further levels of complexity, such that each individual locus becomes the synthesis of a multitude of possible controls.
It is tempting to speculate that targeting of the individual transcripts by multiple pathways provides a robust, fail-safe approach to ensure the fidelity of sexual differentiation. In the data reported here, subjecting transcripts that escape Mmi1/Pab2-mediated destruction to antisense regulation imparts maximal control to the system. This is supported by a general loss of antisense expression to crs1+, bqt1+ and meu1+ synchronous with a dramatic increase in the sense strand (Supplementary Figure S13A–C). The complexity of these systems is also in keeping with the variety of expression patterns associated with antisense activity. Thus, while for each of the four loci described here, a strong characteristic phenotype is observed as a consequence of perturbations to their natural antisense profiles, different patterns of sense expression arise.
In a system comprising a set of interacting regulatory pathways, involving both positive and negative feedback loops, the lack of a simple relationship between sense and antisense expression is unsurprising, particularly given the number of alternate mechanisms by which antisense itself may be operating. Such complexity raises both challenges and opportunities, since a considerable degree of effort may be required not only to establish which combination of possible mechanisms are in operation at any given locus, but also to build sufficiently accurate models with which to represent these regulatory systems that span all levels of the central dogma. Extension of the systems-based approaches employed here will help identify model loci (such as dis1+) for detailed analysis by traditional, focused, molecular biology approaches.
A key debate over recent years has been the extent to which non-coding transcripts are simply ‘chatter’, occurring solely because evolutionary pressures against their expression have not been strong enough for them to be eliminated from the system. The data from our gene disruption experiments make it clear that phenomena such as improper termination and bidirectional transcription are not simply interesting artifacts arising from the complexities of transcription, but have an active and critical role in regulating gene expression. A final key issue to emerge from these data is a modification of our concept of a gene as a spatially distinct locus. This view is becoming increasingly untenable, since not only are the 5′ and 3′ ends of many genes indistinct, but that this lack of a hard and fast boundary is actively used by cells to control the transcription of adjacent and overlapping loci, and thus to regulate critical events in the life of a cell.
Materials and methods
RNA library preparation
RNA extraction
Total RNA was extracted from S. pombe cells as described previously (Lyne et al, 2003). Cells were treated with hot acidic phenol-chloroform, followed by phenol-chloroform extraction, RNA precipitation and column purification. RNA quality was determined using an Agilent 2100 Bioanalyser.
Ribosomal reduction
Total RNA (9 μg) was ‘ribosomally reduced’ using the Ribominus Eukaryote Kit for RNA-Seq (Invitrogen A10837-08) and the Ribominus Concentration Module (Invitrogen K155005). Samples had one or several rounds of ribosomal reduction until they showed a ribosomal reduction of 90% or above on an Agilent 2100 Bioanalyser.
Libraries were prepared with 500 ng of ribosomally reduced total RNA using the SOLiD Whole Transcriptome Analysis Kit for the first sequencing run; vegetative, IH5974 and IH2912 (before (time point 0) and 3, 5, and 10 h after temperature shift from 25 to 32°C) (Applied Biosystems 4425680) and the SOLiD Total RNA-Seq Kit for the second sequencing run that included additional 14 samples; vegetative, asynchronous IH5974 (wild type), and a vegetative, asynchronous IH3365 (wild-type diploid) alongside temperature shifted: IH2912 (pat1.114 diploid), IH8832 (atf21.Δ diploid) and IH8814 (atf31.Δ diploid) before, and 3, 5, and 10 h after a shift from 25 °C to 32°C to induce meiosis (Applied Biosystems 4445374) with the following changes: RNase III digestion time was increased to 25 min, size selection was carried using E-GEL Size Select 2% Agarose Gels (Invitrogen G661002) on the E-GEL iBase power system (Invitrogen G6400UK). Sample bar coding was carried out using the SOLiD Transcriptome Multiplexing Kit (Applied Biosystems 4427046). The libraries were measured on the Qubit Fluorometer (Invitrogen Q3287) using the Quant-IT DSDNA HS Assay Kit (Invitrogen Q32851) and the molarities assessed by a DNA1000 assay (Agilent Technologies) on the Agilent Bioanalyzer (G2938C) before pooling the libraries in equimolar quantities. Sequencing was performed on an AB SOLiD machine (first run 3.0; second run 3.0+).
Strand-specific quantitative reverse transcription PCR (ss-qRT–PCR)
Strand-specific reverse transcription was performed using TaqMan® Reverse Transcription Reagents (Applied Biosystems). ‘Reverse’ primers were used to reverse transcribe the sense transcript of any given gene. ‘Forward’ primers were used to reverse transcribe the antisense transcript. The reaction included 200 ng total RNA, 0.5 μM gene-specific primer, 1 × Taqman RT buffer, 5.5 mM Magnesium Chloride, 500 μM each dNTP, 0.4 U/μl RNase inhibitor, 1.25 U/μl Multiscribe Reverse Transcriptase and RNase-free water to a total volume of 10 μl. The mixture was incubated at 25°C for 10 min, 48°C for 30 min and 95°C for 5 min. ‘Minus RT’ and ‘minus primer’ controls were included within each time course. A minus primer control is a standard RT reaction minus primer and can generate cDNA products via the self priming of RNAs (Haddad et al, 2007). Sense tubulin alpha 2 (atb2+) was used as a housekeeping transcript because its low level of transcription is constitutive (Adachi et al, 1986; Mata et al, 2002; Rustici et al, 2004; Supplementary Table S12). Quantitative PCR was performed using the Universal ProbeLibrary (UPL, Roche Diagnostics) assay system. Primers used in the PCR were identical to the forward and reverse primers used in the reverse transcription. All UPL assays were validated using standard methods. PCR was performed using 1 × TaqMan® Gene Expression Mastermix (Applied Biosystems) containing 0.5 μM each primer, 5 ng cDNA and 0.25 μM UPL probe to a volume of 10 μl. Real-time PCR was performed as standard and expression normalised to atb2+.
Primers are as follows: Dis1 (Fwd: accccagaattccataccg, Rev: caacgcttcctttcgatctt, Probe: 7); Spo4 (Fwd: tccatttgagaaaacgatagca, Rev: ggccttggatcatgtagca, Probe: 65); Spo6 (Fwd: accaagaaccagcattcgag, Rev: aagttgggaaacgccaatc, Probe: 140); Spk1 (Fwd: atcaaattttgcgcgctct, Rev: tgatggcttcaggtctctatga, Probe: 67); Tubulin (Fwd: atggcaacgtgtttgctgta, Rev: tggtcgttacagcagcttga, Probe: 7).
Fission yeast techniques
Standard fission yeast and molecular biology methods were used throughout (Moreno et al, 1991). PCR deletion was, according to Bähler et al (1998), using pFA6kanRMX6, or pFA6natMX6 (Hentges et al, 2005) as templates. The strains listed in Supplementary Table S11 were used. Cells were grown in EMM2 media at 25°C. For pat1.114-induced meioses, the appropriate pat1.114 diploid strains were transferred to 32°C and samples taken before (0) and 3, 5 and 10 h after the shift. To assess the impact of antisense expression upon meiotic progression, cells were grown to mid-log phase in EMM2 media containing 10 μM thiamine at 25°C, washed three times in thiamine-free EMM2 and then maintained in mid-log phase at 25°C in thiamine-free EMM2 medium to induce production of antisense RNA. Cells were then diluted to maintain them in mid-log phase of the cell cycle during induction (72 h for integrated strains and 48 h for pREP multicopy plasmid-based experiments in Figure 6K–P and Supplementary Figure S9). Sexual differentiation was then induced by filtering the culture and washing once in minimal sporulating medium (MSL—which lacks a nitrogen source) before re-suspending the cultures at a density equivalent to early log phase in MSL. Twelve hours later, cells were fixed with 70% EtOH and stained with DAPI (Moreno et al, 1991). Immunostaining with TAT1 antibodies to reveal microtubules and AP9.5 antibodies against the spindle pole marker Sad1 were conducted as described previously (Hagan and Yanagida, 1995). For live-cell imaging, diploid cells homozygous for the pat1.114 mutation harboured an atb2.GFP fusion gene whose expression was under the control of the native atb2+ promoter integrated at the leu1 locus. Cells were grown to early log phase overnight in EMM2 at 25°C. Cells were stained with 10 μg/μl Hoescht 33342 for 10 min before filtering and returning to EMM2 and mounting for imaging in a Bioptechs chamber. One hour before observation, the temperature was shifted to 32°C. Images were captured every 6 min on a Deltavision platform. Twenty sections, separated by 0.4 μm were captured for every time point and maximum projections generated to create the images in the montages in Supplementary Figure S1.
Protein extracts were prepared according to Grallert et al (2004) run on SDS–PAGE, transferred onto PVDF and probed with the ECF (Amersham RPN578) before detection on a Bio-Rad pharosFXplus molecular imager.
Antisense ectopic expression and disruption
To assess the impact of antisense expression, it was necessary to place a precise copy of the desired antisense transcript under the control of an inducible promoter. The scheme is summarised below and in cartoon form in Figure 4. Assessment of the nmt1+/thi3+ transcript levels through a pat1.114-induced sexual differentiation revealed no changes from the high level of transcription detected in vegetative growth (Supplementary Figure S3C), indicating that this repressible promoter represented an ideal candidate. We therefore used a version of the pREP1 plasmid (Maundrell, 1990) in which the LEU2 marker had been replaced with the natMX6 marker, pREP1N (Grallert and Hagan, in preparation). The rpl42+ ‘suicide’ marker (Roguev et al, 2007) was inserted between the Nde1 and BamH1 sites of pREP1N to generate pREP1N.death. We then determined as accurately as possible from the RNA sequencing data, the initiation and termination sites of nmt1 RNA. Genomic DNA was used as a template for PCR amplification of the transcripts as follows. One end of the primer corresponded to the 80 nucleotides upstream or downstream of the nmt1+ transcript start and stop sites. The other end of the primer corresponded to the sequences at the beginning or end of the relevant antisense transcripts defined as follows: 3 826 785–3 828 889 on chromosome 2 (spo4AS), 3 102 749–3 104 936 on chromosome 2 (spo6AS), 2 999 626–3 001 223 on chromosome 1 (spk1AS), 2 999 088–3 000 939 on chromosome 1 (ups1sense); 342 774–346 100 on chromosome 3 (dis1AS-short) and 342 879–346 451 on chromosome 3 (dis1AS-long). The resulting PCR fragments contained a precise copy of the relevant antisense RNA transcript flanked by sequences directly upstream and downstream of the nmt1+ transcript. The PCR transcript was co-transformed with the pREP1N.death plasmid into an rpl42.cyhR strain. rpl42.cyhR is a recessive mutation that confers resistance to cyclohexamide (Roguev et al, 2007). The presence of a wild-type rpl42+ allele in this background kills cells in the presence of cyclohexamide. Elimination of the rpl42+ sequences by recombination between the PCR fragment and the vector enables the rpl42.cyhR allele to confer resistance to cyclohexamide such that cells survive on YES plates containing nourseothricin, cyclohexamide and thiamine. Through this approach, we derived new vectors in which each desired antisense transcript precisely replaced the nmt1+ transcript so that it was regulated by the nmt1+ promoter (whose transcription does not fluctuate during sexual differentiation). After plasmid recovery and sequencing to check the fidelity of the recombination and the veracity of the antisense transcript sequence, the plasmids were either transformed into IH347 host to assess the biological impact of antisense production, or the entire cassette encompassing the nmt1+ regulatory regions was cloned into the Pst1/Sac1 sites of pINTA (Petersen and Hagan, 2003).
To modify the regulation of the dis1+ antisense transcript, the kanRMX6 marker was inserted in either orientation relative to the ORF at position 346 465 on chromosome 3 (14 nucleotides upstream of the detected antisense transcript) to create the dis1 alleles; dis1:kanRAS (reading antisense to the dis1+ORF)and dis1:kanRs (reading sense to the dis1+ORF).
Genome level alignments and annotation
Reads of length 50 bases originating from each sample were aligned using Bowtie (Langmead et al, 2009) to the S. pombe genome sequence (Ensembl S. pombe, Build EF1, version 58.1a) as well as to its corresponding exon–exon junctions database. We performed two alignments per sample, one in which no mismatches were tolerated and a second in which up to three mismatches within the seed alignment (seed length=28) were allowed. Reads that matched multiple loci were removed from further analysis and the resultant alignment files pre-processed to generate ‘pile-ups’ against each chromosome. The numbers of mappable reads without mismatches in each sample are given in Supplementary Table S8.
Exon–exon junctions
We anticipated that some of the reads that did not map contiguously to the S. pombe genome would align to exon–exon junctions, therefore, searches were performed against the genome sequence combined with a data set of 4820 known exon–exon junctions as defined by Ensembl S. pombe release-5. To ensure that a 50 base read mapped to a splice junction, only the last 44 bases of the first exon and the first 44 bases of the second exon were considered (if the exon exceeded length 44). In this way, reads that overlapped a junction by <6 nucleotides were excluded. Reads that matched to more than one junction or elsewhere on the genome were also discarded.
Defining TBlocks, gene extensions and data partitioning
Each residue on the forward and reverse strands of each chromosome was replaced with the number of sequence reads starting at that location.
The algorithm was set to identify the longest stretch of contiguous transcription across the different samples, defining TBlocks (76 315 TBlocks) boundaries.
TBlocks were positioned relative to the known genome annotation, using Perl and the implementation of Ensembl S. pombe annotation database (version 5) (Kersey et al, 2009) in the xmapcore Bioconductor/R package (Gentleman et al, 2004; Yates et al, 2008) (available from http://annmap.picr.man.ac.uk). TBlocks that were found to extend genes (Supplementary Table S1; 7160) were used to define the TSS and TTS of the genes they extended. Ambiguous gene extensions that overlapped with more than one gene were ignored. Known genome annotation was then combined with our 5′ and/or 3′ gene extensions (7287 extensions associated with 4414 protein-coding genes, 26 pseudogenes and 275 ncRNAs, Supplementary Table S2). Using the expanded annotation, a set of 10 677 exonic regions (5857 protein-coding known/novel and non-coding RNA genes) was then used to annotate each residue in the genome (Perl), to derive intronic (4825) and intergenic (5834) coordinate sets.
TBlocks were then re-positioned relative to the expanded genome annotation (Supplementary Table S1). TBlocks that were located completely within annotation in both orientations (within genes/introns/UTRs) were discarded. Only intergenic TBlocks (19 931) and those found to extend genes were considered (35 and 1446, in the sense or antisense orientation, respectively). The latter group was partitioned in both orientations accordingly, while only the intergenic portions left following the split were considered. Using our coordinate system, we then retrieved expression data for 5857 genes and their 5857 antisense counterparts, 4825 introns and 21 199 intergenic TBlocks. Regions that were not expressed were removed, leaving a total of 32 891 expressed regions (Supplementary Table S3). The same annotation was used to retrieve expression data generated following the second sequencing run. In total, 29 837 were found to be expressed in at least 1 of the 14 conditions interrogated (5547 genes, 4990 antisense loci, 3173 intronic and 16 127 intergenic).
RNA-seq expression level
Normalised expression levels (E) for individual exons, UTRs, introns, antisense loci and TBlocks were calculated using the formula [1] as described in Mortazavi et al (2008) and Bradford et al (2010) (RPKM measure). Shortly, the number of reads (R) detected across a given region at a given time point (i) was multiplied by a constant (C=1 × 1010) and divided by the total number of reads at that time point (Ti) multiplied by the regions length (L).
 |
A small constant was added (10−5) to all expression values to avoid taking logs of zero. Gene level and antisense expression values were summarised using exons and UTRs data in both orientations, respectively. Sample-specific expression levels for all loci interrogated in this study are provided in Supplementary Table S12. Raw expression data files are available from Gene Expression Omnibus (GEO accession: GSE28113; http://www.ncbi.nlm.nih.gov/geo/).
Statistical differential expression
Initially differential expressed regions were defined using read counts, and G-test of independence. The aim was to identify region changing in at least one of the five samples interrogated (vegetative asynchronous IH5974 (wild type) and IH2912 before (time point 0) and 3, 5 and 10 h after temperature shift from 25 to 32°C). The number of read counts for a given region at a given sample was normalised by the total number of reads at that sample. In all, 32 891 expressed regions were analysed using a G-test function (with Williams' correction) implemented in Bioconductor/R. P-values were then processed using the qvalue package (Storey, 2004), and a cutoff q-value <0.05 was applied, leaving 6599 regions flagged as significantly changing between samples (Supplementary Figure S2). The same approach was employed to define differentially expressed regions across all 14 samples in the second sequencing run.
To derive lists of differentially expressed loci between vegetative IH5974 (haploid) and vegetative IH2912 (pat1.114 diploid); vegetative IH5974 (haploid) and vegetative IH3365 (pat1+ diploid) and between the vegetative IH2912 (pat1.114 diploid) and the vegetative IH3365 (pat1+ diploid). A series of pair-wise comparisons were conducted using count data and Fisher's exact test as described in Bloom et al (2009) implemented in Bioconductor/R (Gentleman et al, 2004). Briefly, 2 × 2 contingency tables containing sample and region read counts were used for calculating P-values for each region (29 837). These were then adjusted by the qvalue package (Storey, 2004), as before, to correct for multiple testing. Thereafter, the resultant lists of differentially expressed loci were compared (Figure 2; Supplementary Table S5).
The same approach was also used to derive the lists of differentially expressed genes/loci between IH2912 (pat1.114 diploid) and IH8832 (atf21.Δ diploid) or IH8814 (atf31.Δdiploid) strains. However, a four times more stringent q-value (FDR <0.0125) was adopted to account for all time points interrogated in these comparisons, that is, time points 0, 3, 5 and 10 h were systematically compared between each mutant and wild-type strains (pair-wise comparison at a time).
Fold changes and identification of ARTs
Antisense/sense fold changes were calculated for all unambiguous protein-coding loci (4966), that is, excluding genes flagged as dubious or pseudogenes by GeneDB. Fold changes were calculated simply by subtraction of mRNA-sense log2 expression values at all samples from the corresponding antisense expression values. Only antisense loci that displayed a fold change (log2 ratio) >0 in at least one sample and that were found to be differentially expressed in at least one sample in either orientation (G-test FDR <0.05) were considered to be ARTs. To allocate ARTs to different samples, the significance of sense/antisense ratio was evaluated using Fisher's exact test, as before (FDR <0.05). Significant ARTs were then allocated to different samples according to the sample in which they displayed the highest fold change (Supplementary Table S6).
GO analysis
The 354 accessions of known protein-coding genes found opposite of the newly identified ARTs were processed using the ‘GO Term Finder’ algorithm implemented in Perl (Ashburner et al, 2000; Boyle et al, 2004) (cutoff of P-value <5 × 10−5, with Bonferroni correction). Analysis was repeated with permuted data (1000 simulations), in which randomly selected sets of accessions of the same size as the original group (354) were independently processed. Random gene sets were taken from a pool of unambiguous protein-coding genes that were found to be differentially expressed in this study (4011).
The recent web version of the Amigo algorithm (http://amigo.geneontology.org) was also used to find enriched terms (Supplementary Table S5). All other GO analyses were performed once under the same settings and once at a more relaxed cutoff (cutoff of P-value <0.01, with Bonferroni correction), using ‘Go Term Finder’ and Amigo algorithms, albeit without any additional simulations (Supplementary Tables S7 and S9).
Data availability
RNA-Seq data sets are available from the GEO under accession number GSE28113.
Supplementary Material
Supplementary Figures S1–13 and Supplementary Tables S1, S3, S8 and S11
Acknowledgments
This work was supported by Cancer Research UK (CRUK) grant number C147/A6058. We thank Profs M Yanagida and K Gull for αDis1 and TAT1 antibodies, respectively; R Allshire, N Krogan, V Simanis and T Toda for strains; A Carr, A Patel and C Wilkinson for molecular biology reagents; and S Bagley and the Advanced Imaging Facility for technical assistance. We thank C Wirth for assistance with R scripting.
Author contributions: DB, AG, IH and CJM designed the experiments. AG performed the bench experiments. PS performed RNA extractions and qPCR experiments. SP and YH performed the RNA sequencing. JB and DB aligned sequence reads to the genome. YL, CJM and DB designed the statistical approach. DB conducted the bioinformatics analysis. TY implemented the genome database and associated visualisation tools. DB, IH and CJM wrote the manuscript. AG, JB, PS and YH also contributed to the final version of the paper. CJM, DB and IH conceived the study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi Y, Toda T, Niwa O, Yanagida M (1986) Differential expressions of essential and nonessential alpha-tubulin genes in Schizosaccharomyces pombe. Mol Cell Biol 6: 2168–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F (2011) Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol 21: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R (2011) Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23: 258–265 [DOI] [PubMed] [Google Scholar]
- Amorim MJ, Cotobal C, Duncan C, Mata J (2010) Global coordination of transcriptional control and mRNA decay during cellular differentiation. Mol Syst Biol 6: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K, Sato M, Tanaka K, Yamamoto M (2010) Nuclear compartmentalization is abolished during fission yeast meiosis. Curr Biol 20: 1913–1918 [DOI] [PubMed] [Google Scholar]
- Arndt GM, Atkins D, Patrikakis M, Izant JG (1995) Gene regulation by antisense RNA in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 248: 293–300 [DOI] [PubMed] [Google Scholar]
- Asakawa H, Kojidani T, Mori C, Osakada H, Sato M, Ding DQ, Hiraoka Y, Haraguchi T (2010) Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr Biol 20: 1919–1925 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J (2005) Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell 18: 491–498 [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bitton DA, Wood V, Scutt PJ, Grallert A, Yates T, Smith DL, Hagan IM, Miller CJ (2011) Augmented annotation of the Schizosaccharomyces pombe genome reveals additional genes required for growth and viability. Genetics 187: 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA (2009) Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics 10: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO∷TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, Miller CJ (2010) A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics 11: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MR (2008) Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat Rev Genet 9: 62–73 [DOI] [PubMed] [Google Scholar]
- Bühler M, Spies N, Bartel DP, Moazed D (2008) TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kurokawa R, Haraguchi T, Hiraoka Y (2004) Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells 9: 671–684 [DOI] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Bühler M, Dlakic M, Moazed D (2007) Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Cremona N, Potter K, Wise JA (2011) A meiotic gene regulatory cascade driven by alternative fates for newly synthesized transcripts. Mol Biol Cell 22: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Ekwall K (2008) Molecular biology. The paradox of silent heterochromatin. Science 320: 624–625 [DOI] [PubMed] [Google Scholar]
- Djupedal I, Kos-Braun IC, Mosher RA, Soderholm N, Simmer F, Hardcastle TJ, Fender A, Heidrich N, Kagansky A, Bayne E, Wagner EG, Baulcombe DC, Allshire RC, Ekwall K (2009) Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J 28: 3832–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP (2009) RNAi in budding yeast. Science 326: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR (2008) Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet 40: 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R (2004) Fission yeast in general genetics. In The Molecular Biology of Schizosaccharomyces pombe: Genetics, Genomics and Beyond, Egel R (ed), pp 1–12. Heidelberg: Springer-Verlag [Google Scholar]
- Ender C, Meister G (2010) Argonaute proteins at a glance. J Cell Sci 123: 1819–1823 [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey ME, Moore TF, Higgins DG (2002) Overlapping antisense transcription in the human genome. Comp Funct Genomics 3: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Vardy L, Koonrugsa N, Toda T (2001) Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J 20: 3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace EL, Halic M, Moazed D (2010) The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell 39: 360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM (2004) Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev 18: 1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K (2005) Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol 15: 1663–1669 [DOI] [PubMed] [Google Scholar]
- Grewal SI (2010) RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Moazed D, Proudfoot NJ (2011) Autoregulation of convergent RNAi genes in fission yeast. Genes Dev 25: 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ (2008) Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995 [DOI] [PubMed] [Google Scholar]
- Haddad F, Qin AX, Giger JM, Guo H, Baldwin KM (2007) Potential pitfalls in the accuracy of analysis of natural sense-antisense RNA pairs by reverse transcription-PCR. BMC Biotechnol 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M (1995) The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 129: 1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI (2003) RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA 100: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Yamamoto M (2007) Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res 15: 523–537 [DOI] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005) Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, Parkhill J, Ivens AC, Rajandream MA, Barrell B (2004) GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res 32: D339–D343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR (2006) Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745 [DOI] [PubMed] [Google Scholar]
- Iino Y, Yamamoto M (1985) Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet 198: 416–421 [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, Kinsella RJ, Kulesha E, Maheswari U, Megy K, Nuhn M, Proctor G, Staines D, Valentin F, Vilella AJ, Yates A (2009) Ensembl genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res 38: D563–D569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR et al. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, Liu Y (2010) Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell 38: 803–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC (2011) Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23: 258–265 [DOI] [PubMed] [Google Scholar]
- Li P, McLeod M (1996) Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell 87: 869–880 [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett C, Rustici G, Chen D, Langford C, Vetrie D, Bähler J (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, Smith GR, Moreno S (2005) A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Bähler J (2006) Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci USA 103: 15517–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147 [DOI] [PubMed] [Google Scholar]
- Mata J, Wilbrey A, Bähler J (2007) Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol 8: R217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K (1990) nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem 265: 10857–10864 [PubMed] [Google Scholar]
- McLeod M, Beach D (1988) A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature 332: 509–514 [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA (2009) A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol 16: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J (2008) Promoter-driven splicing regulation in fission yeast. Nature 455: 997–1000 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Morita T, Yamada T, Yamada S, Matsumoto K, Ohta K (2011) Fission yeast ATF/CREB family protein Atf21 plays important roles in production of normal spores. Genes Cells 16: 217–230 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Munroe SH, Zhu J (2006) Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol Life Sci 63: 2102–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M (1995) p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev 9: 1572–1585 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kishida M, Shimoda C (2000) The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells 5: 463–479 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Nakamura-Kubo M, Nakamura T, Shimoda C (2002) Novel fission yeast Cdc7-Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol Cell Biol 22: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M (2001) M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol 11: 537–549 [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d′Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Ni T, Tu K, Wang Z, Song S, Wu H, Xie B, Scott KC, Grewal SI, Gao Y, Zhu J (2010) The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLoS One 5: e15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O (2004) Mating-type control and differentiation. In The Molecular Biology of Schizosaccharomyces pombe: Genetics, Genomics and Beyond, Egel R (ed), pp 281–296. Heidelberg: Springer-Verlag [Google Scholar]
- Nurse P (1985) Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet 198: 497–502 [Google Scholar]
- Petersen J, Hagan IM (2003) S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol 13: 590–597 [DOI] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ (2002) Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA 99: 8796–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintales L, Sanchez M, Antequera F (2010) Analysis of DNA strand-specific differential expression with high density tiling microarrays. BMC Bioinformatics 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol 18: 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM et al. (2011) Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ (2007) High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods 4: 861–866 [DOI] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J (2004) Periodic gene expression program of the fission yeast cell cycle. Nat Genet 36: 809–817 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamashita A, Yamamoto M (2003) The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell 14: 2461–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoseki M, Shimoda C (2001) The 5′ terminal region of the Schizosaccharomyces pombe mes1 mRNA is crucial for its meiosis-specific splicing. Mol Genet Genomics 265: 673–682 [DOI] [PubMed] [Google Scholar]
- Sigova A, Rhind N, Zamore PD (2004) A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev 18: 2359–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Buscaino A, Kos-Braun IC, Kagansky A, Boukaba A, Urano T, Kerr AR, Allshire RC (2010) Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep 11: 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD (2004) Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc A Stat Soc 66: 187 [Google Scholar]
- Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M (1991) Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev 5: 1990–1999 [DOI] [PubMed] [Google Scholar]
- Svoboda A, Bähler J, Kohli J (1995) Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma 104: 203–214 [DOI] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ (2007) A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA 104: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Shinozaki-Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M (1997) Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386: 187–190 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M (1994) S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78: 487–498 [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O (1995) Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol Cell Biol 15: 4964–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Bühler M (2011) Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol 18: 94–99 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Münster S, Smolik M, Huber W, Steinmetz LM (2011) Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue-Franzen Y, Kjaerulff S, Holmberg C, Wright A, Nielsen O (2006) Genomewide identification of pheromone-targeted transcription in fission yeast. BMC Genomics 7: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol 145: 1233–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M (2010) Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J 29: 2173–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T, Okoniewski MJ, Miller CJ (2008) X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res 36: D780–D786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI (2011) Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331: 1624–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Leonhard K, Watt S, Marguerat S, Bähler J, Nurse P (2010) A coordinated global control over cellular transcription. Curr Biol 20: 2010–2015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–13 and Supplementary Tables S1, S3, S8 and S11
Data Availability Statement
RNA-Seq data sets are available from the GEO under accession number GSE28113.