Abstract
Malaria elimination strategies require surveillance of the parasite population for genetic changes that demand a public health response, such as new forms of drug resistance. 1,2 Here we describe methods for large-scale analysis of genetic variation in Plasmodium falciparum by deep sequencing of parasite DNA obtained from the blood of patients with malaria, either directly or after short term culture. Analysis of 86,158 exonic SNPs that passed genotyping quality control in 227 samples from Africa, Asia and Oceania provides genome-wide estimates of allele frequency distribution, population structure and linkage disequilibrium. By comparing the genetic diversity of individual infections with that of the local parasite population, we derive a metric of within-host diversity that is related to the level of inbreeding in the population. An open-access web application has been established for exploration of regional differences in allele frequency and of highly differentiated loci in the P. falciparum genome.
The genetic diversity and evolutionary plasticity of Plasmodium falciparum are major obstacles for malaria elimination. New forms of resistance against antimalarial drugs are continually emerging1,2 and new forms of antigenic variation represent a critical point of vulnerability for future malaria vaccines. Effective tools are needed to detect evolutionary changes in the parasite population and to monitor the spread of genetic variants that impact on malaria control.
Here we describe the use of deep sequencing to analyse P. falciparum diversity using blood samples from patients with malaria. The P. falciparum genome has several unusual features that greatly complicate sequence analysis, such as extreme AT bias, large tracts of non-unique sequence and several large families of intensely polymorphic genes3. Therefore our aim was not to determine the entire genome sequence of individual field samples – which would be prohibitively expensive with current technologies - but to define an initial set of SNPs distributed across the P. falciparum genome, whose genotype can be ascertained with confidence in parasitized blood samples by deep sequencing.
An additional complication for analysis of P. falciparum genome variation is that the billions of haploid parasites which infect a single individual can be a complex mixture of genetic types. Previous studies 4-8 have largely focused on laboratory-adapted parasite clones, but the intra-host diversity of natural infections is of fundamental biological interest. Parasites in the blood replicate asexually but, when they are taken up in the blood meal of an Anopheles mosquito, they undergo sexual mating. If the parasites in the blood are of diverse genetic types, this process of sexual mating can generate novel recombinant forms. Deep sequencing provides new ways of investigating within-host diversity and the role of sexual recombination in parasite evolution.
P. falciparum DNA was obtained from blood samples collected from 290 patients with malaria at clinics in Burkina Faso, Cambodia, Kenya, Mali, Papua New Guinea and Thailand (Supplementary Table S1). For 149 samples we used the conventional method of growing the parasites in short term blood culture before extracting the P. falciparum DNA. For 141 samples we used a new method by which P. falciparum DNA is extracted directly from venous blood samples after removing leucocytes 9. We refer to these as cultured and direct samples respectively.
Paired-end sequence reads were generated (median 0.7 × 109 bp per sample) using the Illumina Genome Analyser platform. Sequence analysis was divided into stages of SNP discovery, quality control filtering, genotyping and validation (see Methods and Supplementary Figure S1). After alignment to the 3D7 reference genome3, non-coding regions had much lower read depth than coding regions (Supplementary Figure S2): this can be ascribed to their high AT content (non-coding 87% AT, coding 70% AT). Read depth was also low in the highly polymorphic var, rifin and stevor coding regions. (Supplementary Figure S3). To reduce genotyping errors due to low coverage or copy number variation, for the purposes of this study we excluded all non-coding regions, as well as coding regions at the extremes of the read depth distribution. After these exclusions we were left with 70% of all exonic positions across the genome, with >50% of exonic positions for 71% of genes and >70% for 54% of genes (Supplementary Table S2).
Intra-host diversity complicates the process of excluding sequencing and alignment errors that manifest as false heterozygous genotypes. Two approaches were identified to address this problem (see Full Methods). We scored each position in the reference genome for its degree of uniqueness, and this was found to be a strong predictor of false heterozygous genotypes. We also observed a relationship between the population allele frequency of a SNP and its average level of within-sample heterozygosity, analogous to the Hardy-Weinberg relationship in diploid organisms. This enabled us to exclude SNPs displaying excessive levels of within-sample heterozygosity relative to their population frequency.
After applying the above filters, and excluding SNPs and samples with high levels of missing data, we obtained a final dataset of 86,158 SNPs genotyped in 227 samples (120 direct and 107 cultured) in which a median of 98% samples had valid genotyping data for each SNP, and a median of 98% SNPs had valid genotyping data for each sample (Supplementary Figure S4). This set of 86,158 SNPs (here referred to as the 86k SNP set) represents 10% of the SNPs discovered at the initial stage of sequence alignment. Comparison with the PlasmoDB 5.5 database indicates that 77,283 (89%) of these SNPs are novel, but it should be noted that previous genome-wide SNP discovery efforts have been largely based on low coverage capillary sequencing and the overall error rate is unknown 4-6.
The accuracy of genotype calls in the 86k SNP set was evaluated by five independent approaches (see Full Methods). We examined the evidence for 275 putative novel SNP using independent data from PCR-based capillary sequencing and Sequenom primer-extension mass spectrometry: the existence of the novel allele was confirmed for 270 of the 275 loci. The genotype concordance rate with Sequenom was 99.9% and with capillary sequencing it was 98.6%, excluding heterozygotes (Supplementary Tables S3 and S4). In the case of heterozygous genotypes, deep sequencing gives the allelic ratio whereas most other P. falciparum SNP typing methods give the majority allele or return a missing genotype. The observation of heterozygosity by deep sequencing correlated with Sequenom failing to call a majority allele, but when Sequenom made a majority allele call it agreed with deep sequencing data in 94.8% of cases (Supplementary Figure S5). Capillary sequencing data do not allow allelic ratios to be quantified precisely, but visual inspection of capillary sequence traces was consistent with heterozygous genotype calls in the deep sequencing data (Supplementary Figure S6). In a separate study to be reported elsewhere, we sequenced 90 laboratory-adapted parasite clones derived from three genetic crosses of P. falciparum and determined the rate of Mendelian errors in the 86k SNP set to be 0.05%.
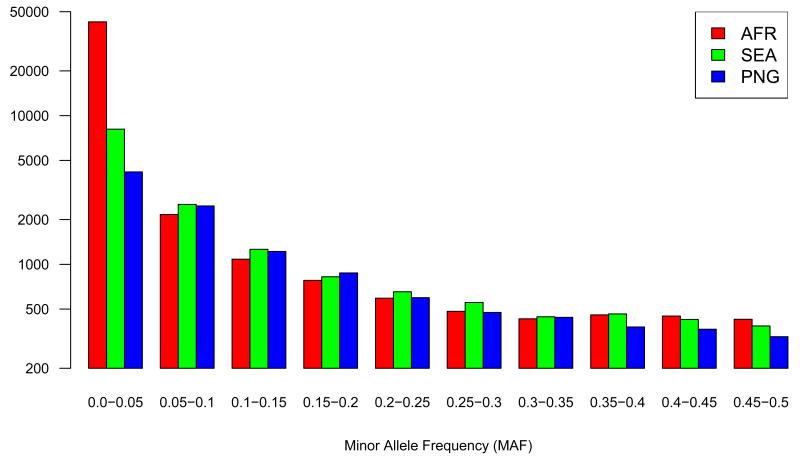
Population genetic analyses were carried out using the 86k SNP set typed in 227 samples as described above. The allele frequency spectrum is dominated by low frequency variants (Figure 1, Supplementary Figure S7) even when synonymous sites alone are considered, consistent with recent population expansion (Supplementary Table S5)10. Samples from Africa (AFR) had a greater number of low frequency variants than samples from Southeast Asia (SEA) or Papua New Guinea (PNG) with or without correction for sample size. Multiple lines of evidence indicate that P. falciparum originated in Africa, and loss of low frequency variation might have occurred as a result of population bottlenecks during migration out of Africa, as in human populations. 10,11
Figure 1.
(a) Minor allele frequency distribution of 86k SNPs set in samples from different continents (AFR, SEA and PNG). Vertical axis shows the number of SNPs in each category of allele frequency. Supplementary Figure S7 shows the data corrected for sample size (b) Considers SNPs that are private to either AFR, SEA or PNG, showing the ratio of nonsynonymous to synonymous substitutions (vertical axis) as a function of derived allele frequency (horizontal axis)
The most likely ancestral state of each SNP was determined from the P. reichenowi genome sequence but is difficult to estimate with confidence, since P. reichenowi might have diverged from P. falciparum relatively recently and its genome sequence has been determined for only one individual (refs 6,12 and Otto et al, manuscript in preparation). There appear to be more SNPs with low-frequency derived (non-ancestral) alleles in AFR than in SEA or PNG (Supplementary Figures S8 and S9). Focusing on SNPs that are private to one continent, those with high derived allele frequency show a considerable excess of non-synonymous substitutions, suggesting that these are largely the result of directional selection (Figure 1b, Supplementary Figure S10).
Many SNPs (64%) were observed in only one continent, but most were low-frequency variants and larger sample sizes are needed to determine how many of these are truly private. Corrected for sample size, the number of private SNPs was greatest in East Africa (EAF) and least in SEA, both of which comprised cultured samples (Supplementary Figure S11). Intermediate numbers were observed in West Africa (WAF) and PNG, both of which comprised direct samples. Thus the effect of culturing on SNP ascertainment appears to be relatively small compared to the effect of geographical location.
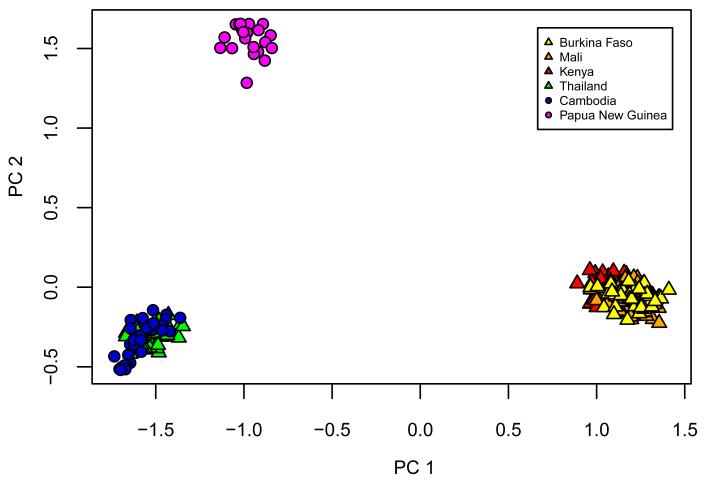
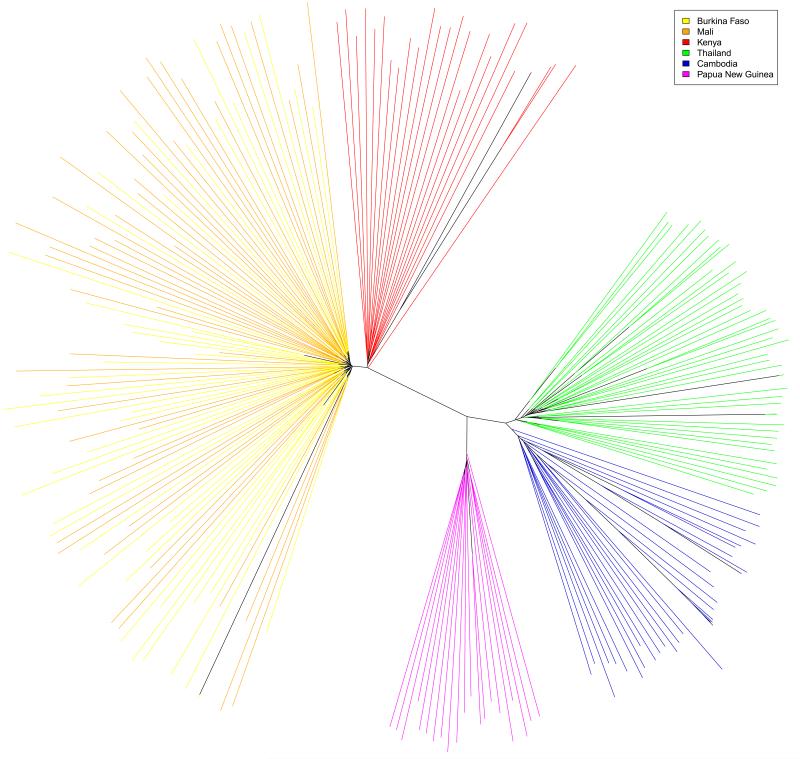
The global population structure of P. falciparum shows a clear division by continent (Figure 2a). Mean FST values between continents ranged from 0.19 to 0.28 (Supplementary Table S6). Population structure within continents is evident from FST values, principal components analysis (Supplementary Figure S12), and a neighbour-joining tree (Figure 2b). All of these methods show greater degree of population structure in Southeast Asia than West Africa, i.e. samples from Cambodia and Thailand form separate clusters, while samples from Mali and Burkina Faso are intermixed. These data are consistent with previous evidence that parasite population structure tends to be increased in regions of low or patchy malaria transmission. 13
Figure 2.
Representations of a pairwise distance matrix between the 227 samples analyzed. (a) Principal components analysis (b) Unrooted neighbour-joining tree. Leaf branches are coloured according to the country of origin of the sample.
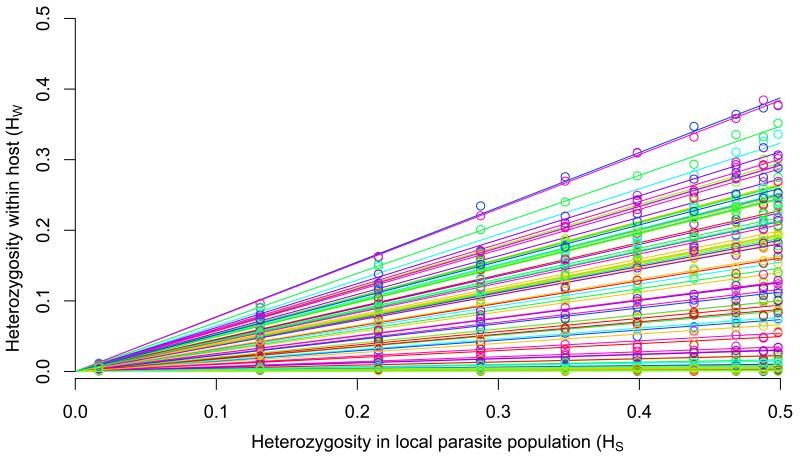
To understand the hierarchical population structure of P. falciparum, methods are needed to quantify the genetic diversity of individual infections relative to the genetic diversity of the parasite population as a whole. With deep sequencing data, we can estimate levels of heterozygosity both within an individual sample (HW) and within the local parasite population (HS). For a biallelic SNP, we define HW as 2pWqW where pW and qW denote the proportions of the two alleles in the sequence reads of an individual sample; and HS as 2pSqS where pS and qS denote the corresponding population allele frequencies at that geographical location. We observe a strong linear relationship between HW and HS when data for all 86k SNPs are aggregated for an individual sample (Figure 3a, Supplementary Figure S13). More specifically, each sample shows a linear relationship between HW and HS but the gradient of the line varies considerably between samples. This gradient is essentially a genome-wide estimate of HW/HS for the sample in question. Thus for each sample we can derive the metric FWS where
This is closely related to Wright’s inbreeding coefficient FIS which can be formulated as
where HI is the heterozygosity of the individual and HS is that of the local population. 14 Estimation of FIS is of practical relevance for malaria control since high rates of inbreeding are thought to favour the emergence of multigenic drug resistance. 15,16 FIS is conventionally measured at the oocyst stage of infection, i.e. after the parasites have undergone sexual mating within the mosquito and before they develop into separate haploid forms, but this is technically demanding and difficult to implement on a large scale 15,17. Since parasites undergo sexual mating shortly after the mosquito has ingested blood from an infected person, the level of within-host diversity determines the potential for inbreeding or outcrossing in the next generation. Thus FWS values observed in blood samples provide a proxy indicator of inbreeding rates in the population. The precise relationship to inbreeding rates quantified in oocysts merits further investigation. We report elsewhere a study of how FWS relates to standard methods of estimating multiplicity of infection 18.
Figure 3.
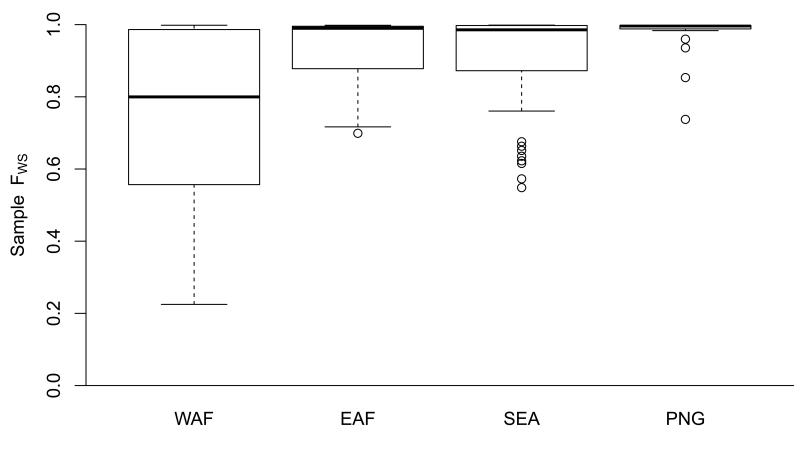
(a) Relationship between heterozygosity in the local parasite population (HS, horizontal axis) and within-host heterozygosity (HW, vertical axis) for all samples in the WAF population. Each line represents a different sample, whose within-host heterozygosity values were averages across all SNPs, categorised according to their heterozygosity in the local parasite population. Separate plots for each population are shown in Supplementary Figure S17). (b) Boxplot showing the distribution of FWS estimates in samples from each of the four populations.
We observe marked differences in FWS between locations (Figure 3b). High levels of FWS (≥0.95) were much more common in PNG (89% of samples) than in WAF (38%), with intermediate rates in SEA (67%) and EAF (63%). Culturing might affect FWS estimation, but the samples from PNG and WAF were not cultured. In general, high levels of inbreeding tend to be associated with low transmission intensity 13 and these data are therefore somewhat surprising since the entomological inoculation rate (EIR) has been estimated to be in the range of 45-293 in Madang in Papua New Guinea 19 where the PNG samples were collected, compared to 140-389 in Burkina Faso 19, ~6 in rural areas of Cambodia 20 and ~1 on the Thai-Burmese border21. Acknowledging that EIR can be highly variable within a locality and that these estimates are indicative, it appears unlikely that the high levels of FWS in PNG are primarily due to low transmission intensity. An alternative explanation is that, in this geographical region, people tend to live in small isolated communities, which might reduce the likelihood of infection with parasites of different genetic types. The small size of the PNG sample provides limited information about local parasite population structure (Supplementary Figure S14) but previous studies indicate that this is very high in some villages within this area of Papua New Guinea22.
These data allow linkage disequilibrium (LD) in the P. falciparum genome to be estimated with greater precision than has previously been possible. In particular, we can begin to distinguish LD due to haplotype structure, which decays with distance in the genome, from LD due to population structure, which is independent of distance in the genome (see Methods, Supplementary Tables S8-S9 and Supplementary Figures S15-S17). Averaged across the genome, after correcting for population structure and other confounders, we find that r2 decays to <0.1 within 1kb in all populations studied here, while and D’ decays to <0.1 within approximately 1kb in WAF and EAF, and with 50kb in SEA and PNG (Supplementary Figure S18). These findings imply that high levels of haplotypic diversity exist at all of these locations, despite low transmission intensity and high rates of inbreeding at some locations. This might be partly due to the high rate of meiotic recombination in P. falciparum, estimated to be 17kb/cM. 23 It is also possible that much of the haplotypic diversity seen in contemporary P. falciparum populations has ancient origins, and arose in Africa before P. falciparum was spread around the world by human migration. This would be analogous to the situation that is seen in human populations, where migration out of Africa was associated with a series of population bottlenecks, which have led to reduction in haplotypic diversity in descendant populations around the world 11. The higher levels of LD observed in SEA and PNG than in WAF and EAF are consistent with both of these possibilities
A web application is provided for browsing, querying and downloading information about all of the SNPs genotyped in this study and their allele frequencies in different geographical regions (http://www.malariagen.net/data/pfalciparum). It can be used, for example, to view regional patterns of variation in known antimalarial drug resistance genes: from these data it is immediately apparent that the pfcrt K76T allele has markedly different haplotypic backgrounds in Southeast Asia and Papua New Guinea, consistent with previous evidence that chloroquine resistance has evolved independently in multiple locations (Supplementary Table S9)1,24. It can also be used to search for genes that are highly differentiated between geographical regions (Supplementary Tables S10 and S11). For example, two genes that affect the fertility of gametocytes, Pfs230 and Pf47, are among the most highly differentiated loci in this dataset .25 Two SNPs in Pfs230 codon 1566 result in three amino acid variants: N (widespread), T (private to SEA, frequency 0.87) and K (private to AFR, frequency 0.79). Codon variant T236I of Pf47 has a fixed difference between AFR and other populations. These data lend weight to previous reports of extreme differentiation in Pf47 and the related gene Pfs48/4526, which is suggested to be due to evolutionary selection of gamete recognition and compatibility. Another example is codon variant F368S of the putative transporter gene PFA0245w27 which has a fixed difference between PNG and other populations, raising the question of whether this plays a role in drug resistance; it is also noteworthy that the P. berghei orthologue of this gene is critical for sexual development of the parasite28.
These data represent the first stage in development of methods for population-based genome sequencing of P. falciparum. Work is ongoing to increase the number of SNPs that can be reliably genotyped, and to develop accurate methods for typing indels, copy number polymorphisms and large structural variations. Future studies will benefit from new methods to reduce the effects of AT bias on sequencing library preparation 29,30 and the increasing length and accuracy of sequencing reads will allow greater access to highly polymorphic regions of the genome. Such technical advances will enable an expanding range of applications, e.g. high-resolution analyses of local population structure to explore models of space-time clustering and immunological strain selection.
Genome sequencing of parasites in clinical blood samples is an important step towards translation to public health applications, e.g. developing effective genetic markers to track the spread of antimalarial drug resistance, and to monitor evolutionary changes in the parasite population 7,8. There is a need to develop protocols, tools and resources and to enable researchers in malaria endemic countries to integrate parasite genome sequencing into clinical and epidemiological investigations, and to facilitate open-access sharing of large-scale population genomic data.
FULL METHODS (TO BE INCLUDED IN ONLINE VERSION)
For further details, see the Supplementary Methods section of the Supplementary Materials
Sample Sequencing
All samples from patients were collected with informed consent from the patient, or from a parent or guardian in the case of minors. Blood collection was approved by local ethics committees (details in Supplementary Methods section). At each location, sample collection was approved by the appropriate local ethics committee. For 141 samples, parasitized erythrocytes were obtained directly from the blood samples after leukocyte-depletion to remove the majority of human DNA31. For the remaining 149 samples, parasites were established in culture in vitro prior to DNA extraction. After genomic DNA was extracted from erythrocytes, total DNA and level of human DNA contamination were determined for each sample31. Samples with >1 μg DNA and <60 % human DNA contamination were deemed suitable for sequencing. Standard Illumina sequencing libraries were prepared following the manufacturer’s recommended protocol 32, avoiding PCR amplification if sufficient quantity of sample DNA (> 1 ug) was available33 Samples were sequenced with between 37 to 76 cycles of paired-end sequencing per read, depending on available technology at time of sampling.
Prior to P. falciparum genome alignment, we removed reads that map to the human genome. This is done for ethical reasons, to limit open access to sequencing data originating from parasite DNA. From an analytical perspective, we found that the presence of human DNA reads made negligible difference to our genotyping (see Supplementary Methods)
Discovery of potential SNPs
To discover an initial list of potential SNPs, short sequence reads were aligned against the P. falciparum 3D7 reference sequence V2.1.5 using the bwa program. To maximise the list of potential SNPs, we included read data from 139 additional samples belonging to other studies, including field samples from Mali, Kenya, Gambia, Ghana, Tanzania, Peru, Cambodia, Vietnam and Thailand; from UK travellers; and from laboratory strains and their experimental crosses. The alignments were processed by samtools to generate a read pileup consensus, and a list of potential SNPs. By merging lists from all samples, we found a total of 1,313,570 potential SNPs. These were subjected to filtering based on quality measures produced by samtools. The quality criteria (CQ >= 36, SQ >= 36, MMQ >= 26) were determined from analyses of the SNP distributions, as was the effect of applying the filters (see Supplementary Methods for details). To reduce false positives, we realigned each sample using the stringent SNP-o-matic algorithm 34, applying a base quality score threshold of 27 and only allowing variations listed in the potential SNPs catalogue. The catalogue was thus reduced to 975,935 potential SNPs.
Quality Filtering
We subjected potential SNPs to a series of filtering steps, to eliminate various classes of artefacts.
To minimize suspected alignment errors, we discarded potential SNPs, unless a minor allele either occurred in at least 1% of all reads across all samples, or was represented by at least 10 reads in at least one sample. We also restricted the catalogue to biallelic SNPS containing a reference allele and an alternate allele (third alleles supported by single spurious read were ignored without discarding the SNP). For the remaining 868,117 potential SNP, we plotted the distribution of coverage (total read counts across all samples), separating coding and non-coding SNPs (Supplementary Figure S2). Because of lower coverage in non-coding regions (possibly a result of problematic alignments due to higher A-T content and low-complexity regions), we restricted our catalogue to positions in the 15%-85% coverage range of coding regions of nuclear chromosomes, totalling 142,779 biallelic potential SNPs.
Each location was assigned a uniqueness score, which is the smallest n such that all n-mer sequences overlapping the position have no identical match across the reference genome. To reduce the impact of misalignments in low complexity regions, we discarded all potential SNPs with uniqueness score ≥26 (Supplementary Methods Figure M6) at which at least one sample presented reads for multiple alleles. 104,156 potential SNPs were retained after this filtering step.
For the present analysis it was important to remove SNPs and samples with high levels of missingness (insufficient read data to establish a genotype). SNPs were ordered by the proportion of samples covered for each SNP, and samples by the proportion of SNPs covered for each sample. Plots of coverage (Supplementary Figure S4) suggested suitable cutoff levels: we discarded SNPs with <220 samples covered at least 5×, and samples with <83,000 SNPs at the same coverage level. As a result, 89,324 potential SNPs and 227 samples were retained.
Heterozygosity (the probability of observing multiple alleles in the same sample) is expected to be related to allele frequency in the population, and we sought to identify positions (termed “hyperheterozygous SNP”) which significantly diverge from this relationship, i.e. where an unusually high percentage of samples present within-sample variation (Figure M7). To identify hyperheterozygous SNPs, we computed a pseudo-likelihood score λ for each SNP, which is a measure of the likelihood that the observed levels of heterozygosity (estimated by the proportion of samples that present multi-allele genotypes) are consistent with the average levels of heterozygosity in a population, for SNPs with similar allele frequency (details in the Supplementary Methods). A higher-than-normal score signifies that a SNP is likely to be hyperheterozygous. SNPs were ordered by λ values to identify suitable cut-off values for each population (Figure M8). Potential SNPs with λ above the cut-off score in at least one population were discarded, resulting in a catalogue of 86,089 typable SNPs. There were supplemented by 79 manually inspected SNPs in four genes (Pfcrt, Pfdhfr, Pfdhps and Pfmdr1) confirmed to be involved in drug resistance, bringing the SNP catalogue to a total of 86,158 typable SNPs, and 227 typable samples.
Genotyping and Validation
All typable samples were genotyped at each typable SNP by a single allele. At positions with fewer than 5 reads, the genotype was undetermined; at all other positions, the genotype was chosen to be the allele with the most reads. We used several independent approaches to evaluate genotyping accuracy in our 86k SNP set and to confirm novel allele calls, combining different technologies, approaches and prior knowledge to confirm calls for various classes of SNPs.
The Sequenom® mass spectrometry platform was used to validate genotype calls for 102 novel SNPs (not included in the PlasmoDB 5.5 list of known SNPs) in the majority (195/227) of samples in our final dataset. A high proportion exhibited of tested SNPs non-reference alleles at low frequency in our dataset (79 with non-reference allele frequency <0.05, 24 with NRAF ≥0.05). Details of SNP selection, multiplex design, sample preparation, assay screening and genotyping are given in the Supplementary Methods. Of the initial 5 multiplexes, each with 39 P. falciparum assays (195 assays), a total of 142 assays (Supplementary Methods Table M3) were taken forward. Of these, three failed to produce usable results from field isolates, and eight had no Illumina calls for non-reference alleles in the subset of tested samples. Finally, 29 assays were disregarded because Sequenom could not call a genotype in samples where Illumina identified non-reference alleles, leaving 102 assays that were informative for confirming novel alleles. The presence of the novel allele was confirmed in all of these assays. For calls where Illumina genotype was a single allele, genotype concordance rate was 99.9% overall, and 98.8% where Illumina called a novel allele. Concordance did not vary significantly with allele frequency: it was 99.9% for NRAF <0.05 and 99.9% for NRAF ≥0.05 (Supplementary Table S3). We observed that Illumina heterozygous calls correlated with high levels of missingness in Sequenom data, reflecting the difficulty of assigning a majority allele (Supplementary Figure S5). When Illumina yielded a heterozygous genotype and Sequenom a valid call, the two methods agreed on the majority allele in 94.8% of cases.
PCR-based capillary sequencing was used to validate genotype calls for a total of 173 novel SNPs, selected with representation across the allele frequency spectrum, in 53 field isolates obtained directly from clinical blood samples, i.e. without culturing. Details of SNP selection, sample preparation and sequencing are given in the Supplementary Methods. All capillary reads were aligned to the 3D7 reference sequence, discarding fragments <30. The novel allele was confirmed in 168 of the 173 SNPs. (Supplementary Methods Table M4). These included 55 SNPs with NRAF <0.1 and 118 with NRAF ≥0.1. Excluding Illumina heterozygote calls, the genotype concordance rate between the two methods was 99.1% overall, and 96.6% where Illumina called a novel allele (Supplementary Table S4). Concordance did not vary significantly with allele frequency: it was 98.7% for SNPs of NRAF<0.1 and 98.6% for NRAF ≥0.1.
A number of samples were genotyped using an Illumina BeadArray assay, described elsewhere.35 The array assayed 384 previously reported SNPs, 91 of which overlapped with our 86k SNP set. Details are given in the Supplementary Methods. A total of 103 samples analysed in the present study were genotyped by this method, using the same starting DNA but independent sample processing and amplification steps. An overall concordance of 98% was estimated based on majority allele calls.
A NimbleGen microarray platform with optimized probe design comprising 45,524 SNPs, described previously36, was used to genotype 5 samples from this study. Details are given in the Supplementary Methods. A total of 9,658 of the SNPs assayed by the microarray platform overlapped with our 86k SNP set. In line with the array’s genotyping capabilities, heterozygote Illumina calls were excluded from concordance calculations. Concordance rate for each sample ranged between 93% and 99%, with a mean of 96% of genotype calls in agreement between the two methods (Supplementary Methods Table M5).
Finally, we estimated the error rate of our genotyping methods using an approach that does not depend on comparison with other platforms. In study to be reported elsewhere, we applied the same methods of sequencing and genotyping described in this paper to 90 clonal lines of P. falciparum derived from the parents and F1 progeny of three experimental genetic crosses that were previously carried out at the National Institutes of Health, Bethesda, MD, USA37-39. The sample comprised both parents and 20 progeny of the 3D7 × HB3 cross; both parents and 32 progeny of the HB3 × DD2 cross; and both parents and 34 progeny of the 7G8 × GB4 cross. We compared genotypes observed in the progeny and the parents to detect inconsistencies (referred to as Mendel errors), e.g. where the progeny has an allele seen in neither of the parents, which we considered as potential genotyping errors. We found a Mendel error rate of 1.3% at the stage of sequence alignment, which drops to 0.05% after applying the various QC filters described, i.e. in our final genotyping set of 86,158 SNPs we find a mean of 43 Mendel errors per sample.
Determination of Allele Frequencies
Allele frequencies in each population were determined for all SNPs, by analysing all genotyped samples. The non-reference allele frequency (NRAF) is as the proportion of genotyped samples whose genotype was not the reference allele. The minor allele frequency (MAF) within a population is the proportion of genotyped samples carrying the least common genotype for that population. We classified a SNP as private if one of the alleles (the private allele) was at non-zero frequency only in a single population, while all other populations exhibit only the other allele without variation.
Allele Status Determination
We determined the putative ancestral state of SNPs is by comparison with outgroup homologous sequences in P reichenowi (Pr), a parasite with recent common ancestry. At SNPs where the homologous Pr allele is one of the two alleles observed in our Pf dataset, we defined the alternative Pf allele to be the putative derived allele as; otherwise it was undefined. To reduce incorrect inferences of ancestral state (Supplementary Figure S8), we reasoned that if a putative derived allele is private to one continental population, this provides additional circumstantial evidence that it is truly the derived allele; and that private alleles in SEA and PNG are very likely to be derived, whereas it is less certain that private alleles in AFR are derived, assuming that P. falciparum originated in Africa. Hence we retained the putative derived allele inferred from Pr, discarding those private to non-African samples where the putative derived allele was not the private allele. The three approaches show only marginal differences in allele frequency spectrum, affecting high-frequency more than low-frequency alleles, but greatly reducing the proportion of putative derived alleles observed to be at fixation (Supplementary Figure S9). The derived allele frequency (DAF) is the frequency of the derived allele in a population.
All typable SNPs defined in this study are in gene coding regions and were classified as synonymous or nonsynonymous, according to whether an amino acid change occurs when substituting the reference allele with the non-reference allele at that SNP in the 3D7 reference genome sequence, without any other changes. The reading frame and exon boundaries were determined from the PlasmoDB 5.5 annotation of the 3D7 genome40.
Analysis of relatedness between samples
Principal component analysis (PCA) of pairwise distance matrices was performed using the Classical Multidimensional Scaling (CMS) method 41. For each PCA analysis of a subset of N samples, all typable SNPs were used to build a pairwise distance matrix. Pairwise distance was calculated as the proportion of SNPs at which the two samples are genotyped with different alleles, excluding those SNPs where at least on e of the two genotypes was undetermined. The CMS algorithm was applied using the R language cmdscale() implementation. The same pairwise distance matrix was used to produce a neighbour-joining tree 42 using the nj() implementation in the R ape package.
Heterozygosity and Inbreeding Coefficient Analysis
For heterozygosity analysis, allele frequencies at SNPs were estimated by using allele read counts, rather than by genotyping each sample with a single allele. For each sample s, we computed allele frequencies (f1 and f2) at a given SNP from the sample’s read counts for the two alleles. The sample’s heterozygosity at the SNP was thus derived: Hs,x=1−(f12 + f22). To measure population-wide MAF at a given SNP, we deriving the MAF from total read counts for the two alleles across all samples in the population. The population-wide heterozygosity was thus derived: HP,x=1−(fMAF2 + (1−fMAF)2). SNPs were binned into ten equally-sized MAF intervals ([0.0-0.05], [0.05-0.1] … [0.45-0.5]), and for each bin we computed the mean within-population heterozygosity HP,f. Similarly, for each sample in the population, we computed the mean within-sample heterozygosity Hs,f at each bin. We plotted Hs,f against Hp,f for each sample and fitted a linear regression to estimate FWS=1− (Hs,f / Hp,f).
Linkage Disequilibrium (LD) Analysis
We analyzed the decay of LD with genomic distance for each population separately. LD was measured by computing two commonly used measures (D’ and r2) for pairs of SNPs of varying distance 43,44. After categorizing SNPs into equally spaced MAF intervals, LD calculations were conducted separately for each frequency bin, and later combined (Supplementary Figures S15-S17). We accounted for offsets due to population structure by a sample rotation method, and by measuring “random” LD between SNPs on different chromosomes. Complete details are given in the Supplementary Methods (Supplementary Tables S7 and S8).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Gordon Dougan and Nick Day for support, and to Tim Anderson and Margaret Mackinnon for helpful comments. The sequencing and analysis components of this study were supported by the Wellcome Trust through Sanger Institute core funding (077012/Z/05/Z; 098051) and a Strategic Award (090770/Z/09/Z); the Medical Research Council through the MRC Centre for Genomics and Global Health (G0600230) and an MRC Professorship to Dominic Kwiatkowski (G19/9). Other parts of this study were partly supported by the Wellcome Trust; the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health; and a Howard Hughes Medical Institute International Scholarship (55005502) to Abdoulaye Djimde.
REFERENCES
- 1.Wootton JC, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 5.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 6.Jeffares DC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neafsey DE, et al. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 2008;9:R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu J, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet. 2010;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auburn S, et al. An effective method to purify Plasmodium falciparum DNA directly from clinical blood samples for whole genome high-throughput sequencing. PLoS One. 2011;6:e22213. doi: 10.1371/journal.pone.0022213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joy DA, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 11.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 12.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci U S A. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson TJ, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 14.Hartl D, Clark AG. Principles of population genetics. 4th edn Sinauer Associates; 2007. [Google Scholar]
- 15.Paul RE, et al. Mating patterns in malaria parasite populations of Papua New Guinea. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 16.Dye C, Williams BG. Multigenic drug resistance among inbred malaria parasites. Proc Biol Sci. 1997;264:61–67. doi: 10.1098/rspb.1997.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill WG, Babiker HA, Ranford-Cartwright LC, Walliker D. Estimation of inbreeding coefficients from genotypic data on multiple alleles, and application to estimation of clonality in malaria parasites. Genet Res. 1995;65:53–61. doi: 10.1017/s0016672300033000. [DOI] [PubMed] [Google Scholar]
- 18.Auburn S, et al. Characterization of Within-Host Plasmodium falciparum Diversity using Next-Generation Sequence Data. PLoS ONE. doi: 10.1371/journal.pone.0032891. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DL, Drakeley CJ, Chiyaka C, Hay SI. A quantitative analysis of transmission efficiency versus intensity for malaria. Nature Communications. 2010;1:108. doi: 10.1038/ncomms1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trung HD, et al. Malaria transmission and major malaria vectors in different geographical areas of Southeast Asia. Tropical Medicine & International Health: TM & IH. 2004;9:230–237. doi: 10.1046/j.1365-3156.2003.01179.x. [DOI] [PubMed] [Google Scholar]
- 21.Paul RE, et al. Genetic analysis of Plasmodium falciparum infections on the north-western border of Thailand. Trans R Soc Trop Med Hyg. 1999;93:587–593. doi: 10.1016/s0035-9203(99)90057-3. [DOI] [PubMed] [Google Scholar]
- 22.Schultz L, et al. Multilocus haplotypes reveal variable levels of diversity and population structure of Plasmodium falciparum in Papua New Guinea, a region of intense perennial transmission. Malaria journal. 2010;9:336. doi: 10.1186/1475-2875-9-336. doi:10.1186/1475-2875-9-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su X, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 24.Mehlotra RK, et al. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci U S A. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dijk MR, et al. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 2010;6:e1000853. doi: 10.1371/journal.ppat.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anthony TG, Polley SD, Vogler AP, Conway DJ. Evidence of non-neutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol Biochem Parasitol. 2007;156:117–123. doi: 10.1016/j.molbiopara.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Martin RE, Henry RI, Abbey JL, Clements JD, Kirk K. The ‘permeome’ of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:R26. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boisson B, et al. The novel putative transporter NPT1 plays a critical role in early stages of Plasmodium berghei sexual development. Molecular Microbiology. 2011 doi: 10.1111/j.1365-2958.2011.07767.x. [DOI] [PubMed] [Google Scholar]
- 29.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oyola SO, et al. Optimizing Illumina Next-Generation Sequencing library preparation for extremely AT-biased genomes. BMC genomics. 2012;13:1. doi: 10.1186/1471-2164-13-1. doi:10.1186/1471-2164-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES FOR ONLINE METHODS
- 31.Auburn S, et al. An effective method to purify Plasmodium falciparum DNA directly from clinical blood samples for whole genome high-throughput sequencing. PLoS One. 2011;6:e22213. doi: 10.1371/journal.pone.0022213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manske HM, Kwiatkowski DP. SNP-o-matic. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campino S, et al. Population Genetic Analysis of Plasmodium falciparum Parasites Using a Customized Illumina GoldenGate Genotyping Assay. PLoS One. 2011;6:e20251. doi: 10.1371/journal.pone.0020251. doi:10.1371/journal.pone.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan JC, et al. An optimized microarray platform for assaying genomic variation in Plasmodium falciparum field populations. Genome Biology. 2011;12:R35. doi: 10.1186/gb-2011-12-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walliker D, et al. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 38.Wellems TE, et al. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 39.Hayton K, et al. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe. 2008;4:40–51. doi: 10.1016/j.chom.2008.06.001. doi:10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–328. [Google Scholar]
- 42.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 43.Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theoret Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.