Abstract
MicroRNAs (miRNAs) are short single-stranded non-coding molecules that function as negative regulators to silence or suppress gene expression. Aberrant miRNA expression has been implicated in a several cellular processes and pathogenic pathways of a number of diseases. Evidence is rapidly growing that miRNA regulation of gene expression may be affected by environmental chemicals. These environmental exposures include those that have frequently been associated with chronic diseases, such as heavy metals, air pollution, bisphenol A, and cigarette smoking. In this article, we review the published data on miRNAs in relation to the exposure to several environmental chemicals, and discuss the potential mechanisms that may link environmental chemicals to miRNA alterations. We further discuss the challenges in environmental-miRNA research and possible future directions. The cumulating evidence linking miRNAs to environmental chemicals, coupled with the unique regulatory role of miRNAs in gene expression, makes miRNAs potential biomarkers for better understanding the mechanisms of environmental diseases.
Keywords: MicroRNAs, Epigenetic, Environmental chemicals
1. Introduction
MicroRNAs (miRNAs) are short single-stranded RNAs of nearly 20–24 nucleotides in length that are transcribed from DNA but not translated into proteins. MiRNAs negatively regulate expression of target genes at the posttranscriptional level by binding to 3′ untranslated regions of target mRNAs [1]. Each mature miRNA is partially complementary to multiple target mRNAs and directs the RNA-induced silencing complex (RISC) to identify the target mRNAs for inactivation [2] (Figure 1). MiRNAs are initially transcribed as longer primary transcripts (pri-miRNAs) and processed first by the RNase enzyme complex, and then by Dicer, leading to incorporation of a single strand into the RISC. MiRNAs guide RISC to mRNAs that results in post-transcriptional repression. Recently, a number of studies have demonstrated that many miRNAs are involved in the regulation of gene expression through the targeting of mRNAs during cell proliferation, apoptosis, the control of stem cell self renewal, differentiation, metabolism, development, and tumor metastasis [3-4]. Compared with other mechanisms involved in gene expression, miRNAs act directly before protein synthesis and may be more directly involved in fine-tuning of gene expression or quantitative regulation [5-6]. Moreover, miRNAs also play key roles in modifying chromatin structure and participating in the maintenance of genome stability [7]. MiRNAs can regulate various physiological and pathological processes, such as cell growth, differentiation, proliferation, apoptosis, and metabolism [1, 8]. More than 10,000 miRNAs have been reported in animals, plants and viruses by using computational and experimental methods in miRNA-related public databases (Table 1). There are approximately 900 known human miRNAs [8]. The aberrant expression of miRNAs has been linked to various human diseases, such as Alzheimer's disease, cardiac hypertrophy, altered heart repolarization, lymphomas, leukemias and cancer at several sites [9-25].
Fig. 1.
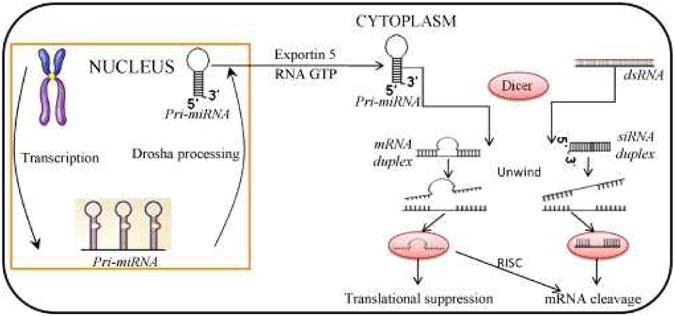
MiRNA biosynthesis and target mRNA inactivation. MiRNA genes encode long primary miRNA transcripts, which are initially transcribed from DNA and expressed as a part of pri-miRNAs. The miRNA portion of the pri-miRNA transcript usually forms a hairpin…
Table 1. Online Resources for miRNA Research.
| MiRNA databases |
|
MiRBase: the microRNA database miRBase DATABASE is a searchable database of published miRNA sequences and annotation; The miRBase REGISTRY is a naming service to provide investigators with unique names for novel miRNA genes prior to publication of results. http://www.mirbase.org/index.shtml |
|
MiRex. Infobase of microRNA gene expression A web resource for meta-analysis of microRNA gene expression. miRex collects, standardizes, and analyzes published data on microRNA gene expression. http://miracle.igib.res.in/mirex |
|
MiRNAMap A resource for experimentally verified microRNAs and experimentally verified miRNA target genes in human, mouse, rat, and other metazoan genomes. http://mirnamap.mbc.nctu.edu.tw/ |
| Target Prediction |
|
microRNA.org A comprehensive resource of microRNA target predictions and tissue-specific expression profiles. Target predictions are based on a development of the miRanda algorithm which incorporates current biological knowledge on target rules and on the use of an up-to-date compendium of mammalian microRNAs. http://www.microrna.org/microrna/home.do |
|
MicroCosm Targets A web resource, previously part of miRBase, containing computationally predicted targets for microRNAs across many species. http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/ |
|
TargetScan TargetScan predicts biological targets of miRNAs for 10 different vertebrate species, TargetScan Human considers matches to annotated human UTRs and their orthologs, as defined by UCSC whole-genome alignments. http://www.targetscan.org/ |
|
PicTar PicTar is an algorithm for the identification of microRNA targets on multiple vertebrate species, seven Drosophila species, and three nematode species. It is a searchable website that provides details on 3′ UTR alignments with predicted sites, and links to various public databases. http://pictar.mdc-berlin.de/ |
|
MicroInspector A scanning software for reverse target search. MicroInspector scans target sequences to detect miRNA binding sites. http://bioinfo.uni-plovdiv.bg/microinspector/ |
|
miRecords: An animal miRNA-target interaction database, including manually curated, experimentally validated miRNA targets based on literature, and predicted miRNA targets from multiple target prediction tools. http://mirecords.biolead.org/ |
|
RNAhybrid A tool for finding the minimum free energy hybridisation of a long and a short RNA that can be used for microRNA target prediction. http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/ |
| Other Resources |
|
miR2Disease A manually curated database providing microRNA information related to human diseases http://www.mir2disease.org/ |
|
PolymiRTS A database of naturally occurring DNA variation in putative miRNA target sites http://compbio.uthsc.edu/miRSNP/ |
|
SeqBuster A web-based bioinformatic tool offering a custom analysis of deep sequencing data at different levels, with special emphasis on the analysis of miRNA variants or isomiRs and the discovering of new small RNAs http://estivill_lab.crg.es/seqbuster/ |
Exposure to environmental chemicals is well known to increase risks for various diseases [26-27]. The etiological role of these chemical agents has been categorized according to their capability to alter the DNA sequence. Such information has been fundamental to determine environmental risks and our current regulatory efforts for exposure protection [28]. However, genetic mutations can only explain small portion of environmental diseases [29]. Recent evidence suggests that epigenetic factors, including DNA methylation, histone modification, and miRNAs, can regulate gene expression without involving DNA sequence changes [30-31]. MiRNAs are newly emerged as a gene expression regulatory factor that may link environmental chemicals and their related diseases.
In this article, we review the published data on miRNAs in relation to the exposure to several environmental chemicals, and discuss the potential mechanisms that may link environmental chemicals to miRNA alterations. We further discuss the challenges in environmental-miRNA research and possible future directions.
2. Environmental chemicals and miRNA
Gene expression can be changed as a response to exogenous stressors such as environmental chemicals [32-33]. Such changes may be regulated by specific miRNA(s). Growing evidence has demonstrated that environmental chemicals can induce changes in miRNA expression in experimental settings, and miRNA expression levels are associated with environmental exposures in observational studies [34-47]. A summary of these findings, together with their respective target genes and elicited potential biologic effects, is presented below and in Table 2.
Table 2. Environmental chemicals and microRNAs.
| Environmental chemicals | MiRNAs | Target genes or pathways | Function | Species or cell types | References |
|---|---|---|---|---|---|
| Sodium arsenite | ↑miR-222, ↓miR-210 | SAM, methyl-group donor | DNA methylation | TK6 cell line | [34] |
| Arsenic trioxide | ↓miR-19a | 143 genes, such as TMEM16A, PTEN, SNX17, and ATG16L1 | Cell growth arrest and apoptosis | T24 cell line | [35] |
| Cadmium | ↓miR-146a | BCL XL, CXCR4, EGFR, NF-κB, STAT, and TLR | Negative regulation of inflammation | Human | [36] |
| Aluminum-sulfate | ↑miR-146a | CFH | Inflammatory responses | HN cells | [37] |
| Aluminum-sulfate | ↑miR-9, -128, -125b | CSDA, PRDM6, DNAJC10, C1orf83, PAIP2, BAZ2B, CSNK2A1, FAM169B, and ZNF543 | Neurotoxicity | HN cells | [38] |
| Air pollution metal-rich PM | ↑miR-222 | MAPK and NGF | Increases leukocyte proliferation and induces inflammation | Human | [36] |
| Air pollution metal-rich PM | ↑miR-21 | PTEN, TGFβ, FAK, MAPK, P13K, and TP53 | Limit injuries from ROS | Human | [36] |
| Cigarette smoke | ↑miR-294 | Transcriptional repressor genes | Altered gene expression | Rat | [39] |
| Cigarette smoke | ↑miR-218 | Transcription factor and MAFG | Expression of MAFG | Human | [40] |
| Cigarette smoke | ↓let-7c, miR-34c, -222 | Genes involved in stress response and cell proliferation | Carcinogenesis | Rat | [41] |
| NNK | ↓miR-126 | Cytochrome P450 (CYP) 2A3 | Tumorigenesis | Rat | [42] |
| NNK | ↓miR-34 | P53 | Damage response and apoptosis | Rat | [42] |
| RDX | ↑let-7, miR-15, -16, -26, -181; ↓miR-10b | Cell cycle regulators | Regulation of tumor pathogenesis | Mouse | [43] |
| RDX | ↑miR-206, -30, -195 | BDNFs | Neurotoxicity | Mouse | [43] |
| Carbon tetrachloride | ↓miR-298, -370 | Thioredoxinreductase 3 | Hepatotoxicity | Rat | [44] |
| Dioxin | ↑miR191 | MAPK | Apoptosis and proliferation | Rat | [45] |
| BPA | ↑miR-146a | DNA damage response | Cell proliferation and DNA damage | 3A placental cells | [46] |
Abbreviation: SAM, S-adenosyl methionine; TMEM16A, Transmembrane protein 16A; PTEN, Phosphatase and tensin homolog; SNX17, Sorting nexin 17; ATG16L1, ATG16 autophagy related 16-like 1; BCL XL, Bcl2 like 1 signaling; CXCR4, Chemokine c x c motif receptor 4 signaling; EGFR, Epidermal growth factor receptor signaling; NF-κB, Nuclear factor kappaB signaling; STAT, Signal transducer and activator of transcription signaling; TLR, Toll-like receptor signaling; CFH, Complement factor H; HN cells, Human neural cells; CSDA, Homo sapiens cold shock domain protein A mRNA; PRDM6, Homo sapiens PR domain containing 6 mRNA; DNAJC10, Homo sapiens DnaJ (Hsp40) homolog, subfamily C, member 10 mRNA; C1orf83, Homo sapiens chromosome 1 open reading frame 83 mRNA; PAIP2, Homo sapiens poly(A) binding protein interacting protein 2 mRNA; BAZ2B, Homo sapiens bromodomain adjacent to zinc finger domain, 2B mRNA; CSNK2A1, Homo sapiens casein kinase 2, alpha 1 polypeptide mRNA; FAM169B, Homo sapiens family with sequence similarity 169, member B mRNA; ZNF543, Homo sapiens zinc finger protein 543 mRNA; MAPK, Mitogen activated protein kinase signaling; NGF, Nerve growth factor signaling; TGFβ, transforming growth factor beta signaling; FAK, Focal adhesion kinase 1 signaling; MAPK, Mitogen activated protein kinase signaling; P13K, Phosphatidylinositol signaling; TP53, p53 signaling; ROS, reactive oxygen species; MAFG, A member of the small Maf protein family; NNK, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone; RDX, Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine; BDNFs, Brain-derived neurotrophic factors; BPA, Bisphenol A.
2.1. Metals
Heavy metals are widespread environmental contaminants. Several of them are essential in certain physiological processes and some of the environmental metals have been linked to a number of diseases, such as cancer, cardiovascular diseases, and neurological disorders [48]. In addition, exposure to certain metals may also play a role in the induction or exacerbation of several autoimmune diseases[49]. Metals have been shown to affect the immune system and increase oxidative stress in animal experiments. It has been accepted that the etiology roles of heavy metals are dependent on both the genetic background and amount and duration of exposures. In recent years, there has been a greater appreciation of the role of molecular factors, in the etiology of heavy metal-associated diseases [50-52]. Experimental and observational data have also linked altered miRNA expression with exposure to arsenic, cadmium, and aluminum, as detailed below.
2.1.1. Arsenic
Arsenic exposure is an established risk factor for cancers, cardiovascular diseases, and neurological disorders [53-57]. Marsit et al. have reported that human lymphoblast cell line TK-6 grown under arsenite exposure induced significant global increases in miRNA expression [34]. Arsenic trioxide (As2O3) has been used as a pharmacological treatment in acute promyelocytic leukemia (APL) [58]. Cao et al. also demonstrated that numerous miRNAs were up-regulated or down-regulated in T24 human bladder carcinoma cells exposed to arsenic trioxide [35]. In particular, miRNA-19a was significantly decreased, resulting in cell growth arrest and apoptosis. The arsenic-related changes in miRNA expression were shown to be reversible when the exposure was removed [35].
2.1.2. Cadmium
Epidemiology investigations have associated cadmium (Cd) exposure with increased risks of cancer and cardiovascular diseases [59-63]. Transcriptional and post-transcriptional gene regulation is critical in responses to Cd exposure, in which miRNAs may play important role [63-64]. Bollati et al. have recently demonstrated that increased expression of miR-146a in peripheral blood leukocytes was significant related to inhalation of Cd-rich air particles in steel workers [36]. MiRNA-146a expression is regulated by the transcription factor NF-κB (nuclear factor-kappa B), which has been implicated as an important causal link between inflammation and carcinogenesis [65].
2.1.3. Aluminum
Recent investigations have demonstrated a number of miRNAs that were altered in response to aluminum exposure. For example, miR-146a in human neural (HN) cells was significantly up-regulated after aluminum-sulfate treatment. Upregulation of miR-146a corresponded to the decreased expression of complement factor H (CFH), a repressor of inflammation [37]. In addition, a study on aluminum-sulfate-treated human neural cells in primary culture has demonstrated the increased expression of a set of miRNAs, including miR-9, miR-125b and miR-128. The same miRNAs were also found to be up-regulated in brain cells of Alzheimer patients, suggesting that aluminum exposure may induce genotoxicity via miRNA-related regulatory elements [66].
2.2. Air pollution
Exposure to the particulate component (particulate matter, PM) of ambient air pollution has been related to increased morbidity and mortality related to cardiovascular diseases, lung cancer, and adverse respiratory effects [67-68]. Recently, using microarray profiling, Jardim et al. have shown extensive alterations of miRNA expression profiles in human bronchial epithelial cells treated with diesel exhaust particles [68]. Out of 313 detected miRNAs, 197 were either up-regulated or down-regulated by at least 1.5-fold [68]. In workers in a steel plant nearby Milan, Italy, we have recently shown that exposure to metal-rich PM induced rapid changes in the expression of two inflammation-related miRNAs, i.e., miR-21 and miR-222, measured in peripheral blood leukocytes [69] .
2.3. Cigarette smoking
Tobacco smoking has been estimated to account for 40% of all human cancers, and more than 5 million preventable deaths every year worldwide [40]. However, the exact mechanisms responsible for smoking-related diseases and deaths are still largely unknown. Schembri et al. have recently identified 28 miRNAs that were differentially expressed in bronchial airway epithelium in smokers when compared to non-smokers [40]. Most (82%) of these miRNAs were found to be down-regulated. Smoking-induced changes in miRNA expression were suggested to contribute to altered regulation of oncogenes, tumor suppressor genes, oxidative stress, xenobiotic metabolism, and inflammation. The miRNA changes were also found to be similar to those found in previous studies of miRNAs in lung cancer tissue [40].
Izzotti et al. have monitored the expression of 484 miRNAs in the lungs of mice exposed to cigarette smoking. In this study, downregulation of miRNA expression was observed in most of the miRNAs affected [70]. A more recent investigation by the same group has further shown that cigarette smoking induced down-regulation of 126 miRNAs by at least 2-fold, and of 24 miRNAs by more than 3-fold in lung tissue. These miRNA expression changes were associated with up-regulation of 107 genes (2.9% of the genes investigated) and 50 proteins (9.7% of the proteins investigated) [39]. The most remarkably down-regulated miRNAs belonged to several miRNA families, such as let-7, miR-10, miR-26, miR-30, miR-34, miR-99, miR-122, miR-123, miR-124, miR-125, miR-140, miR-145, miR-146, miR-191, miR-192, miR-219, miR-222, andmiR-223. These miRNAs regulate expression of genes involved in stress responses, apoptosis, proliferation, and angiogenesis [39]. Interestingly, for some miRNAs, cigarette smoking-induced down-regulation could be prevented by using chemopreventive agents, such as phenethyl isothiocyanate (PEITC) indole-3-carbinol (I3C), and N-acetyl-l-cysteine (NAC) [71-72], suggesting that miRNAs may represent potential new biomarkers for predicting the efficacy of cancer chemopreventive agents [71-72].
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a nitrosamine that can be found in a variety of tobacco products. NNK has been shown to promote the survival and growth of lung cancer cells [73]. Kalscheuer et al. have demonstrated that NKK exposure reduces the expression of several miRNAs, such as miR-101, miR-126, miR-199, and miR-34 in rat lungs [42]. The NKK-related miRNA expression profiles were similar to those observed in human lung cancer tissue [42, 74-75].
2.4. Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (also known as hexogen or cyclonite)
Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) is a common munitions constituent resulting from military and civil activities. Although most of this environmental pollutant is observed in soils, RDX and its metabolites are also found in water sources [76]. Exposure to RDX and its metabolites could cause neurotoxicity, immunotoxicity, and cancers [43]. Zhang et al. have recently evaluated the effects of RDX on miRNA expression in mouse brain and liver [43]. In this study, out of 113 miRNAs, 10 were significantly up-regulated and 3 were down-regulated. Most of the miRNAs that showed altered expression, including let-7, miR-17-92, miR-10b, miR-15, miR-16, miR-26, and miR-181, were related to toxicant-metabolizing enzymes, and genes related to carcinogenesis, and neurotoxicity [43].
2.5. Carbon tetrachloride
Carbon tetrachloride is a man-made chemical released in the environment through air emissions from industrial sites, as well as from landfill spillage and leaching. It has been associated with increased risk of liver cancer in epidemiology studies. Fukushima et al. have demonstrated that rat exposed to carbon tetrachloride showed down-regulation of miR298 and miR370 in the liver that was accompanied by hepatocyte necrosis and inflammation [77] .
2.6. Dioxin
As a known live carcinogen, the toxicity of dioxin is mediated by binding to the aryl hydrocarbon receptor [78-80]. In a xenograft mouse model of hepatocellular carcinoma, Elyakim et al. have found that miR-191 were upregulated by dioxin. MiR-191 has been shown to inhibit apoptosis and decreased cell proliferation [45], suggesting that dioxin may play its pathological role via miRNA-related mechanism.
2.7. Diethylstilbestrol
Diethylstilbestrol (DES) is a synthetic estrogen that was used to prevent miscarriages in pregnant women between the 1940s and the 1960s [81]. A moderate increase in breast cancer risk has been shown both in women who were treated with DES during pregnancy, and their daughters [82]. Hsu et al. have demonstrated that the expression of 82 miRNAs (9.1% of the 898 miRNAs evaluated) were altered in breast epithelial cells when exposed to DES [83]. In particular, the suppression of miR-9-3 expression was accompanied by promoter hypermethylation of the miR-9-3 coding gene in DES-treated epithelial cells [83].
2.8. Bisphenol A
Bisphenol A (BPA) is a chemical with estrogenic properties that has been widely used as an industrial plasticizer in epoxy resins used in food and beverage containers, baby bottles, and dental composites [84]. BPA is considered an endocrine disruptor with potential reproductive effects and a weak carcinogen associated with increased cancer risk in adult life through fetal exposures [85-86]. Using microarray analyses, Avissar-Whiting et al. have observed an elevated overexpression of miR-146a in BPA-treated placental cell lines [46]. MiR-146a expression was associated with slower cell proliferation and higher sensitivity bleomycin-induced DNA damage [46].
3. Hypothesized mechanisms linking environmental chemicals and miRNA alterations
The exact mechanisms by which environmental factors alter miRNA expression are not fully understood. In Figure 2, we present a hypothesized conceptual model that may help explain the mechanisms by which environmental chemicals induce miRNA-expression alterations. Our model is centered upon inflammation and oxidative stress, which have been shown to directly affect miRNA expression [87-92]. As such, this model might be limited to those environmental chemicals for which inflammation and oxidative stress are primary mediators of toxicity. We hypothesize that environmental chemicals may cause miRNA alterations via increasing oxidative stress and/or triggering inflammatory responses. Both oxidative stress and inflammation have been implicated and play important pathological roles in various diseases.
Fig. 2.

Hypothesized conceptual model for environmental chemicals and microRNAs. Environmental chemicals may induce alterations of miRNA expression through inflammation and oxidative stress pathways. Altered miRNA expression may cause target gene expression…
This hypothesis is supported by our recent investigation in which we have observed that exposure to ambient PM2.5 and its metal components altered the expression of two inflammation-related miRNAs, i.e., miR-21 and miR-222 [69]. These results set the stage for further analyses that can expand upon and further delineate specific mechanisms related to environmental exposure-induced miRNA changes.
Similar to protein-coding genes, miRNA expression can also be controlled or regulated by their encoding gene DNA methylation status. Whether environmental exposures affect DNA methylation of miRNA non-coding genes remains to be determined.
4. Challenges and future directions of environment-miRNA research
Our understanding of miRNA biology has advanced greatly in recent years. We are now at the forefront of the exciting, yet challenging, task of translating our understanding of the effects of environmental factors on miRNAs in relation to disease outcomes and to their use as biomarkers in disease prevention and treatment. However, there are a variety of questions that remain to be solved in environmental miRNA research.
4.1. Limitations of analytical technologies for miRNA expression analysis
A major challenge is represented by technological limitations of miRNA detection and discovery to achieve genome-wide, high throughput, sensitive, and accurate analysis. There is still the need for a considerable amount of work to validate miRNA profiling by either microarray or deep-sequencing profiling. The need, particularly in human environmental health studies, to analyze large numbers of samples is constrained by the relative high costs of currently available technologies. The standardization of the methods and cross-platform reproducibility are critical issues that will need to be addressed in the near future. The continuous technological advances in accurate and cost-effective miRNA detection prospect a very promising role for miRNAs as novel biomarkers of environmental chemical exposure-related diseases.
4.2. Prediction and identification of mRNA targets
A major challenge in miRNA research is the complete identification of both mRNA targets and cellular functions of miRNAs. RISC-induced mRNA cleavage or inactivation requires only partial complementarity between miRNA-mRNA pairs [2]. Therefore, based on sequence pairings, each miRNA is expected to regulate hundreds of target mRNAs. The level of complementarity is different between target site–miRNA pairs, and finding target genes has proved to be a steeper challenge than expected. Identifying and validating more targets will inevitably help to improve the predictions and our understanding of the biological role of miRNAs. There are several approaches available for target hunters, including bioinformatic predictions and experimental techniques. Thomas et al. have recently reviewed the different approaches available to the identification of miRNA-regulated genes [94], including bioinformatic prediction, genetic approaches, mRNA microarray analysis and proteomics, miRNA overexpression or knockdown, and identification of miRISC-associated mRNAs. Computational methods have been widely used to predict miRNA targets, and to identify mRNA and proteins for further experimental validation. A list of online free-access tools for target prediction is reported in Table 1. To date, however, the different computational methods are not in agreement with each other and cannot predict all miRNA targets as defined using high-throughput experimental methods [95].
4.3. Identify chemical-specific miRNA(s)
The interactions of miRNAs with multiple putative mRNA targets and cellular functions puts ahead of us the opportunity to identify complex interconnected signatures of environmental exposures at different molecular substrates, including but not limited to miRNAs, mRNAs, and proteins. Identifying chemical-specific miRNA(s) will not only help our understanding of environmental disease, but may open the way to novel biomonitoring and preventive strategies. Therefore, it is critically important to be able to identify and validate miRNAs that can be induced by specific environmental chemicals and regulate gene expression.
4.4. MiRNA-environment associations: Causal mechanisms or epiphenomena?
Because the environment-related miRNA changes in healthy subjects, unlike in miRNA changes in diseased tissues, are often small and may cumulate over time, it is difficult to establish the precise cause-effect relationships among environmental chemicals, miRNA alterations, and diseases. The induced miRNA alterations are also often reversible upon the removal or mitigation of the environmental exposure, suggesting that chronic exposure may be necessary to permanently alter the expression of miRNAs [34]. A longitudinal study approach with multiple measures of exposures and miRNA expression at multiple times would be ideal to study miRNA dynamics in relation to environmental chemical exposures. Future studies will need to demonstrate the contribution of environment-miRNA interaction to environmental human disease.
5. Conclusion
The rapidly growing evidence linking miRNAs and environmental chemical, coupled with the unique regulatory role of miRNAs in gene expression, makes miRNAs potential biomarkers for elucidating the mechanisms and developing more effective prevention strategies for environmental diseases. To achieve a better understanding of miRNA biology and its pathological role, large longitudinal population studies of miRNAs in relation to exposure to environmental chemicals are warranted. Future studies will also need to integrate analyses of down-stream target mRNAs and investigate the interplays with genetic and other epigenetic factors, such as DNA methylation of miRNA encoding genes.
Acknowledgments
We apologize to colleagues whose work was not cited due to space constraints. Our work is partially supported by grants from the HSPH-NIEHS Center for Environmental Health New Investigator Fund (P30ES000002)
References
- 1.Singh SK, Pal Bhadra M, Girschick HJ, Bhadra U. MicroRNAs--micro in size but macro in function. FEBS J. 2008;275:4929–4944. doi: 10.1111/j.1742-4658.2008.06624.x. [DOI] [PubMed] [Google Scholar]
- 2.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA Sequencing for Context-Specific Identification of In Vivo MicroRNA Targets. Circ Res. 2010 doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2010 doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 5.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257–268. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 7.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Backes C, Meese E, Lenhof HP, Keller A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010;38:4476–4486. doi: 10.1093/nar/gkq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho L, Fivecoat H, Wang J, Pasinetti GM. Alzheimer's disease biomarker discovery in symptomatic and asymptomatic patients: experimental approaches and future clinical applications. Exp Gerontol. 2010;45:15–22. doi: 10.1016/j.exger.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Provost P. Interpretation and applicability of microRNA data to the context of Alzheimer's and age-related diseases. Aging (Albany NY) 2010;2:166–169. doi: 10.18632/aging.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer's and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3:251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montgomery RL, van Rooij E. MicroRNA regulation as a therapeutic strategy for cardiovascular disease. Curr Drug Targets. 2010;11:936–942. doi: 10.2174/138945010791591368. [DOI] [PubMed] [Google Scholar]
- 14.Shen E, Diao X, Wei C, Wu Z, Zhang L, Hu B. MicroRNAs target gene and signaling pathway by bioinformatics analysis in the cardiac hypertrophy. Biochem Biophys Res Commun. 2010;397:380–385. doi: 10.1016/j.bbrc.2010.05.116. [DOI] [PubMed] [Google Scholar]
- 15.Swynghedauw B, Delcayre C, Samuel JL, Mebazaa A, Cohen-Solal A. Molecular mechanisms in evolutionary cardiology failure. Ann N Y Acad Sci. 2010;1188:58–67. doi: 10.1111/j.1749-6632.2009.05084.x. [DOI] [PubMed] [Google Scholar]
- 16.Fabbri M, Croce CM, Calin GA. MicroRNAs in the ontogeny of leukemias and lymphomas. Leuk Lymphoma. 2009;50:160–170. doi: 10.1080/10428190802535114. [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 18.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcucci G, Radmacher MD, Mrozek K, Bloomfield CD. MicroRNA expression in acute myeloid leukemia. Curr Hematol Malig Rep. 2009;4:83–88. doi: 10.1007/s11899-009-0012-7. [DOI] [PubMed] [Google Scholar]
- 20.Motyckova G, Stone RM. The role of molecular tests in acute myelogenous leukemia treatment decisions. Curr Hematol Malig Rep. 2010;5:109–117. doi: 10.1007/s11899-010-0049-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: tiny molecule, great function. Crit Rev Oncol Hematol. 2010;74:149–155. doi: 10.1016/j.critrevonc.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Xu X. Diet, epigenetic, and cancer prevention. Adv Genet. 2010;71:237–255. doi: 10.1016/B978-0-12-380864-6.00008-0. [DOI] [PubMed] [Google Scholar]
- 23.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–1148. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 26.Crinnion WJ. The CDC fourth national report on human exposure to environmental chemicals: what it tells us about our toxic burden and how it assist environmental medicine physicians. Altern Med Rev. 2010;15:101–109. [PubMed] [Google Scholar]
- 27.Newbold RR. Impact of environmental endocrine disrupting chemicals on the development of obesity. Hormones (Athens) 2010;9:206–217. doi: 10.14310/horm.2002.1271. [DOI] [PubMed] [Google Scholar]
- 28.Weisburger JH, Williams GM. The distinct health risk analyses required for genotoxic carcinogens and promoting agents. Environ Health Perspect. 1983;50:233–245. doi: 10.1289/ehp.8350233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foley DL, Craig JM, Morley R, Olsson CA, Dwyer T, Smith K, Saffery R. Prospects for epigenetic epidemiology. Am J Epidemiol. 2009;169:389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reamon-Buettner SM, Mutschler V, Borlak J. The next innovation cycle in toxicogenomics: environmental epigenetics. Mutat Res. 2008;659:158–165. doi: 10.1016/j.mrrev.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda K. Effect of environmental chemicals on the genes and the gene expression. Yakugaku Zasshi. 2009;129:1501–1506. doi: 10.1248/yakushi.129.1501. [DOI] [PubMed] [Google Scholar]
- 33.Patel CJ, Butte AJ. Predicting environmental chemical factors associated with disease-related gene expression data. BMC Med Genomics. 2010;3:17. doi: 10.1186/1755-8794-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 35.Cao Y, Yu SL, Wang Y, Guo GY, Ding Q, An RH. MicroRNA-dependent regulation of PTEN after arsenic trioxide treatment in bladder cancer cell line T24. Tumour Biol. 2010 doi: 10.1007/s13277-010-0111-z. [DOI] [PubMed] [Google Scholar]
- 36.Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, Baccarelli A. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect. 2010;118:763–768. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TP, Percy ME, Tarr MA, Lukiw WJ. Characterization of an NF-kappaB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009;103:1591–1595. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Bowers J, Vaziri C, Ott K, Sensinger K, Collins JJ, Brody JS, Getts R, Lenburg ME, Spira A. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4- (methylnitrosamino)-1- (3-pyridyl)-1-butanone. Carcinogenesis. 2008;29:2394–2399. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B, Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ Health Perspect. 2009;117:231–240. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukushima T, Hamada Y, Yamada H, Horii I. Changes of micro-RNA expression in rat liver treated by acetaminophen or carbon tetrachloride--regulating role of micro-RNA for RNA expression. Journal of Toxicological Sciences. 2007;32:401–409. doi: 10.2131/jts.32.401. [DOI] [PubMed] [Google Scholar]
- 45.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, Cohen D, Yerushalmi N. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010;70:8077–8087. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 46.Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol. 2010;29:401–406. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lema C, Cunningham MJ. MicroRNAs and their implications in toxicological research. Toxicology Letters. 2010;198:100–105. doi: 10.1016/j.toxlet.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 48.Howard Hu. Human Health and Heavy Metals Exposure. In: McCally M, editor. Life Support: The Environment and Human Health. Massachusetts Institute of Technology; Boston: 2002. pp. 65–81. [Google Scholar]
- 49.Hemdan NY, Emmrich F, Faber S, Lehmann J, Sack U. Alterations of TH1/TH2 reactivity by heavy metals: possible consequences include induction of autoimmune diseases. Ann N Y Acad Sci. 2007;1109:129–137. doi: 10.1196/annals.1398.015. [DOI] [PubMed] [Google Scholar]
- 50.Waalkes MP. Cadmium carcinogenesis. Mutat Res. 2003;533:107–120. doi: 10.1016/j.mrfmmm.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C, Ke Q, Costa M, Shi X. Molecular mechanisms of arsenic carcinogenesis. Mol Cell Biochem. 2004;255:57–66. doi: 10.1023/b:mcbi.0000007261.04684.78. [DOI] [PubMed] [Google Scholar]
- 53.Roman D, Pizarro I, Rivera L, Camara C, Palacios MA, Gomez MM, Solar C. An approach to the arsenic status in cardiovascular tissues of patients with coronary heart disease. Hum Exp Toxicol. 2010 doi: 10.1177/0960327110389835. [DOI] [PubMed] [Google Scholar]
- 54.Tchounwou PB, Centeno JA, Patlolla AK. Arsenic toxicity, mutagenesis, and carcinogenesis--a health risk assessment and management approach. Mol Cell Biochem. 2004;255:47–55. doi: 10.1023/b:mcbi.0000007260.32981.b9. [DOI] [PubMed] [Google Scholar]
- 55.Ali N, Hoque MA, Haque A, Salam KA, Karim MR, Rahman A, Islam K, Saud ZA, Khalek MA, Akhand AA, Hossain M, Mandal A, Miyataka H, Himeno S, Hossain K. Association between arsenic exposure and plasma cholinesterase activity: a population based study in Bangladesh. Environ Health. 2010;9:36. doi: 10.1186/1476-069X-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma A, Sharma MK, Kumar M. Modulatory role of Emblica officinalis fruit extract against arsenic induced oxidative stress in Swiss albino mice. Chem Biol Interact. 2009;180:20–30. doi: 10.1016/j.cbi.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 57.Roy M, Sinha D, Mukherjee S, Paul S, Bhattacharya RK. Protective effect of dietary phytochemicals against arsenite induced genotoxicity in mammalian V79 cells, Indian. J Exp Biol. 2008;46:690–697. [PubMed] [Google Scholar]
- 58.Binet F, Antoine F, Girard D. Interaction between arsenic trioxide and human primary cells: emphasis on human cells of myeloid origin. Inflamm Allergy Drug Targets. 2009;8:21–27. doi: 10.2174/187152809787582516. [DOI] [PubMed] [Google Scholar]
- 59.Jarup L, Akesson A. Current status of cadmium as an environmental health problem. Toxicol Appl Pharm. 2009;238:201–208. doi: 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 60.Ishido M. The modification of biocellular chemical reactions by environmental physicochemicals. Prog Theor Phys Supp. 2008:124–133. [Google Scholar]
- 61.Akesson A, Julin B, Wolk A. Long-term dietary cadmium intake and postmenopausal endometrial cancer incidence: A population-based prospective cohort study. Cancer Res. 2008;68:6435–6441. doi: 10.1158/0008-5472.CAN-08-0329. [DOI] [PubMed] [Google Scholar]
- 62.McElroy JA, Shafer MM, Trentham-Dietz A, Hampton JM, Newcomb PA. Cadmium exposure and breast cancer risk. J Natl Cancer I. 2006;98:869–873. doi: 10.1093/jnci/djj233. [DOI] [PubMed] [Google Scholar]
- 63.Bhatnagar A. Environmental cardiology - Studying mechanistic links between pollution and heart disease. Circ Res. 2006;99:692–705. doi: 10.1161/01.RES.0000243586.99701.cf. [DOI] [PubMed] [Google Scholar]
- 64.Tellez-Plaza M, Navas-Acien A, Crainiceanu CM, Guallar E. Cadmium exposure and hypertension in the 1999-2004 National Health and Nutrition Examination Survey (NHANES) Environ Health Persp. 2008;116:51–56. doi: 10.1289/ehp.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. 2008;36:1211–1215. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- 66.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. Journal of Inorganic Biochemistry. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baccarelli A, Cassano PA, Litonjua A, Park SK, Suh H, Sparrow D, Vokonas P, Schwartz J. Cardiac autonomic dysfunction - Effects from particulate air pollution and protection by dietary methyl nutrients and metabolic Polymorphisms. Circulation. 2008;117:1802–1809. doi: 10.1161/CIRCULATIONAHA.107.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect. 2009;117:1745–1751. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, Baccarelli A. Exposure to Metal-rich Particulate Matter Modifies the Expression of Candidate MicroRNAs in Peripheral Blood Leukocytes. Environ Health Perspect. 2010 doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM, De Flora S. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila) 2010;3:62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Izzotti A, Larghero P, Cartiglia C, Longobardi M, Pfeffer U, Steele VE, De Flora S. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010;31:894–901. doi: 10.1093/carcin/bgq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang RY, Li MY, Hsin MK, Underwood MJ, Ma LT, Mok TS, Warner TD, Chen GG. 4-Methylnitrosamino-1-3-pyridyl-1-butanone (NNK) promotes lung cancer cell survival by stimulating thromboxane A(2) and its receptor. Oncogene. 2010 doi: 10.1038/onc.2010.390. [DOI] [PubMed] [Google Scholar]
- 74.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. P Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Colin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 76.Beller HR, Tiemeier K. Use of liquid chromatography/tandem mass spectrometry to detect distinctive indicators of in situ RDX transformation in contaminated groundwater. Environ Sci Technol. 2002;36:2060–2066. doi: 10.1021/es0157696. [DOI] [PubMed] [Google Scholar]
- 77.Fukushima T, Hamada Y, Yamada H, Horii I. Changes of micro-RNA expression in rat liver treated by acetaminophen or carbon tetrachloride--regulating role of micro-RNA for RNA expression. J Toxicol Sci. 2007;32:401–409. doi: 10.2131/jts.32.401. [DOI] [PubMed] [Google Scholar]
- 78.Okey AB. Special contribution - An aryl hydrocarbon receptor odyssey to the shores of toxicology: The deichmann lecture, international congress of toxicology-XI. Toxicol Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- 79.Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 80.Baccarelli A, Pesatori AC, Masten SA, Patterson DG, Needham LL, Mocarelli P, Caporaso NE, Consonni D, Grassman JA, Bertazzi PA, Landi MT. Aryl-hydrocarbon receptor-dependent pathway and toxic effects of TCDD in humans: a population-based study in Seveso, Italy. Toxicology Letters. 2004;149:287–293. doi: 10.1016/j.toxlet.2003.12.062. [DOI] [PubMed] [Google Scholar]
- 81.Laitman CJ. DES exposure and the aging woman: mothers and daughters. Curr Womens Health Rep. 2002;2:390–393. [PubMed] [Google Scholar]
- 82.Verloop J, van Leeuwen FE, Helmerhorst TJ, van Boven HH, Rookus MA. Cancer risk in DES daughters. Cancer Causes Control. 2010;21:999–1007. doi: 10.1007/s10552-010-9526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng AS, Huang TH. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–5945. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.vom Saal FS, Hughes C. An extensive new literature concerning low-dose effects of bisphenol A shows the need for a new risk assessment. Environmental Health Perspectives. 2005;113:926–933. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keri RA, Ho SM, Hunt PA, Knudsen KE, Soto AM, Prins GS. An evaluation of evidence for the carcinogenic activity of bisphenol A. Reproductive Toxicology. 2007;24:240–252. doi: 10.1016/j.reprotox.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho SM, Tang WY, de Frausto JB, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Research. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 88.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 89.Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O (2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J Vasc Res. 2009;46:527–540. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 93.Thomas M. Desperately seeking miRNA targets 2010 doi: 10.1038/nsmb.1921. ? ed. [DOI] [PubMed] [Google Scholar]
- 94.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 95.Nelson PT, Kiriakidou M, Mourelatos Z, Tan GS, Jennings MH, Xie K, Wang WX. High-throughput experimental studies to identify miRNA targets directly, with special focus on the mammalian brain. Brain Res. 2010;1338:122–130. doi: 10.1016/j.brainres.2010.03.108. [DOI] [PMC free article] [PubMed] [Google Scholar]


