Abstract
BACKGROUND & AIMS
Alterations in methylation of protein-coding genes are associated with Barrett’s esophagus (BE) and esophageal adenocarcinoma (EAC). Dys-regulation of noncoding RNAs occurs during carcinogen-esis but has never been studied in BE or EAC. We applied high-resolution methylome analysis to identify changes at genomic regions that encode noncoding RNAs in BE and EAC.
METHODS
We analyzed methylation of 1.8 million CpG sites using massively parallel sequencing-based HELP tagging in matched EAC, BE, and normal esophageal tissues. We also analyzed human EAC (OE33, SKGT4, and FLO-1) and normal (HEEpic) esophageal cells.
RESULTS
BE and EAC exhibited genome-wide hypomethylation, significantly affecting intragenic and repetitive genomic elements as well as noncoding regions. These methylation changes targeted small and long noncoding regions, discriminating normal from matched BE or EAC tissues. One long noncoding RNA, AFAP1-AS1, was extremely hypomethylated and overexpressed in BE and EAC tissues and EAC cells. Its silencing by small interfering RNA inhibited proliferation and colony-forming ability, induced apoptosis, and reduced EAC cell migration and invasion without altering the expression of its protein-coding counterpart, AFAP1.
CONCLUSIONS
BE and EAC exhibit reduced methylation that includes noncoding regions. Methylation of the long noncoding RNA AFAP1-AS1 is reduced in BE and EAC, and its expression inhibits cancer-related biologic functions of EAC cells.
Keywords: Esophageal Cancer Progression, Tumor Development, Gene Regulation, Noncoding RNA
Esophageal adenocarcinoma (EAC) is one of the fastest-growing cancers in the Western world. Ninety-five percent of EACs occur without any antecedent diagnosis of Barrett’s esophagus (BE), and the 5-year survival rate is only 15% in this group of patients.1 The rising incidence and poor prognosis of EAC have intensified research efforts into earlier methods to detect this disease.
Recently, increasing evidence has shown that eukaryotic transcriptomes and genomes are not the simple, wellordered substrates of gene transcription that they were once believed to be. It is now known that genomes are transcribed into a broad spectrum of RNA molecules, ranging from long protein-encoding messenger RNAs (mRNAs) to short noncoding transcripts, which frequently overlap or are interleaved on either strand.2 “Non-coding regions” refer to RNAs that are transcribed into RNA but not translated to protein. These noncoding regions are interspersed throughout genomic DNA. One subcategory of these transcripts, called long noncoding RNAs (lncRNAs), comprise noncoding RNA more than 200 nucleotides in length. lncRNAs are pervasively transcribed in the genome, but our understanding of the functions of these lncRNAs is limited. lncRNA transcription was previously believed to represent random transcriptional noise. However, expression levels of lncRNA transcripts have been observed to vary spatially, temporally, and in response to various stimuli.2,3 Moreover, several lncRNAs exhibit very precise expression patterns in different tissues. For example, Mercer et al observed exquisite patterning of lncRNA expression in the mouse brain, both in the tissue as a whole and in subcellular locations.4 Similarly, the expression of some lncRNAs has also been shown to be developmentally regulated.5
Despite this remarkable diversity in RNA species, only a few dysregulated lncRNAs have been implicated in cancer in humans.6,7 Examples include MALAT-1 in lung cancer,8 HULC in hepatocellular carcinoma,9 and PCGEM1 in prostate cancer,10 suggesting that lncRNAs may be involved in tumorigenesis or tumor progression. However, to our knowledge, studies of lncRNAs in EAC have not yet been reported.
In addition, emerging research has suggested mechanisms underlying the regulation of coding gene expression by lncRNAs. For example, lncRNAs can regulate chromosome structure in cis (XIST)11 or in trans (HOTAIR).12 Other lncRNAs modulate the activity of protein-binding partners.13,14 Many lncRNAs are antisense to protein-coding genes and may function by regulating splicing, editing, transport, translation, or degradation of their corresponding coding mRNA transcripts.15 In addition, lncRNAs may be posttranscriptionally processed into short non–protein-coding RNAs, which in turn regulate gene expression.16
In our previous study,17 unsupervised hierarchical clustering analyses showed that, at the level of the transcriptome, squamous mucosa clustered discretely from “glandular” epithelium (including gastric cardia as well as all stages of progression of BE); in contrast, at the level of the epigenome, “normal” mucosa (including both squamous and gastric cardia) clustered discretely from all “abnormal” (ie, BE) epithelia. These results showed similarity of epigenetic profiles between otherwise normal gastrointestinal tissues, despite obvious morphological differences. Having established this finding previously, our focus in the present study was to study epigenetic differences between normal esophagus (NE) and BE at a much higher resolution on the whole-genome level. Following this initial step, we sought to characterize lncRNAs that were both differentially methylated and differentially expressed in EAC versus NE. We found that one such differentially regulated and methylated lncRNA, AFAP1-AS1, was derived from the antisense strand of DNA at the AFAP1 coding gene locus and was hypomethylated and up-regulated in EAC tissues and cell lines. Inhibition of its expression in EAC cells resulted in diminished cell growth, migration, and invasion, as well as in increased apoptosis, thereby establishing, to our knowledge for the first time, a functional cancer-related consequence of epigenetic alteration at a lncRNA genomic locus. A schematic summary of experiments and a diagram of proposed AFAP1-AS1 mechanisms of action are shown in Supplementary Figure 1A–B, respectively.
Materials and Methods
Cell Culture
This study used 3 established human EAC cell lines (OE-33, SK-GT-4, and FLO-1) as well as human primary normal nonimmortalized esophageal epithelial cells (HEEpic; ScienCell Research Laboratories, Carlsbad, CA).
Tissue Specimens
Primary tissue samples were obtained at endoscopy performed for clinical diagnostic indications. All patients provided written informed consent under protocols approved by institutional review boards at The Johns Hopkins University School of Medicine, University of Maryland School of Medicine, or Baltimore Veterans Affairs Medical Center. All tissue samples were pathologically confirmed as NE, BE, or EAC. Specimens were stored in liquid nitrogen before RNA extraction. Three sets of NE/BE samples were studied by HELP-tagging analysis. Twelve pairs of NE/BE samples and 20 pairs of NE/EAC samples were also studied for differential expression of both AFAP1 and AFAP1-AS1.
HELP Tagging for Genome-Wide Methylation Analysis
The HELP-tagging assay applies massively parallel sequencing to analyze the status of 1.8 million CpGs distributed across the entire genome.18 To perform HELP-tagging assays,18 DNA samples were digested with Hpa II and ligated to customized Illumina (San Diego, CA) adapters with a complementary cohesive end. These adapters also contain an EcoP15 I site that cuts into the adjacent sequence 27 base pairs (bp) away, allowing us to polish that end and ligate the other Illumina adapter for library generation by polymerase chain reaction (PCR). The presence of the CCGG and EcoP15 I sequences at the ends of the reads allowed us to remove spurious sequences. We normalized the Hpa II signal with that of the deeply sequenced Msp I profiles, as performed previously.18 Results were generated using the WASP system and linked to a local mirror of the UCSC Genome Browser for visualization.
Methylation Analysis
HELP-tagging data were analyzed using an automated pipeline as described previously.18 Loci were defined in a continuous variable model, given the quantitative nature of this and comparable published assays.19 Methylation values were depicted from a range of 0 to 100, with 0 representing fully methylated to 100 representing fully hypomethylated loci. Mean methylation values for noncoding regions were obtained by averaging values over the whole transcript region.
Quantitative DNA Methylation Analysis by MassArray Epityping
Validation of HELP microarray findings was performed by matrix-assisted laser desorption/ionization time of flight mass spectrometry using EpiTyper by MassArray (Sequenom, San Diego, CA) on bisulfite-converted DNA as previously described.17,20,21 MassArray primers were designed to cover the flanking Hpa II sites for a given locus, as well as any other Hpa II sites found up to 2000 bp upstream of the downstream site and up to 2000 bp downstream of the upstream site, to cover all possible alternative sites of digestion.
Genomic Annotations
Genomic coordinates were obtained from HG18 build of the human genome from the UCSC browser using RefSeq annotations. Genomic regions 2 kilobases upstream and downstream of the transcription start sites were annotated as promoters. Two-kilobase flanking regions around the edges of CpG islands were annotated as CpG shores. RefSeq annotations with an NR prefix were categorized as noncoding transcripts. A size cutoff of 200 bp was used to distinguish between small and large noncoding transcripts.22
Small Interfering RNA Transfection and RNA Extraction
Two different small interfering RNAs (siRNAs) that targeted AFAP1-AS1 RNA (siRNA n262319 and n262320; Life Technologies, Grand Island, NY) and a scrambled siRNA control were used. The sequences of the 2 siRNAs were 5′-GGGCTTCAATTTA-CAAGCATT-3′ and 5′-CCTATCTGGTCAACACGTATT-3′.
Total RNA from tissue specimens and cells was extracted using TRIzol reagent (Invitrogen, Grand Island, NY). RNA concentration and integrity were determined by spectrophotometry and standard RNA gel electrophoresis.
The primer sequences for PCR are as follows: AFAP1-AS1, forward 5′-TCGCTCAATGGAGTGACGGCA-3′ and reverse 5′-CGGCTGAGACCGCTGAGAACTT-3′; AFAP1, forward 5′- CCGT-GCATCAACGGCTCGCTC-3′ and reverse 5′-TTCACAACA-GCCGCGGGATCC-3′. All PCRs were performed in triplicate. β-actin was used to normalize mRNA expression levels.
Cell Proliferation Assays
Cells were plated at a density of 1000 cells per well onto 96-well plates at day 0 (24 hours after siRNA transfection). Every other day until day 5, Cell Proliferation Reagent WST-1 (Roche, Mannheim, Germany) was added to each well and then incubated at 37°C for 1 hour. Optical density was measured at 660 nm (background) and 440 nm (signal) using a plate reader (Molecular Devices, Sunnyvale, CA).
Colony Formation Assays
Cells were trypsinized into a single-cell suspension. A total of 100 cells were plated in each well of a 6-well plate and maintained for 14 days to allow colony formation. Clones containing more than 50 cells were counted using a grid. Three independent experiments were performed. The formula for the colony formation ratio was as follows: Ratio = Numbers of Colony/Initiative Cells × 100%.
Cell Apoptosis Assays
After 48 hours of treatment with siRNA, OE33 cells were stained with Annexin V and PI using Annexin V-FITC/PI apoptosis detection kits (Vybrant Apoptosis Assay Kit, Grand Island, NY) and then examined by flow cytometry (BD FACSCalibur, Becton Dickinson, San Jose, CA). Cellular proteins were extracted 72 hours after siRNA transfection. Caspase-3 (Cell Signaling, Danvers, MA) expression was detected by Western blot.
Cell Cycle Analysis
After 48 hours of treatment with siRNA, OE33 cells were harvested, washed with ice-cold phosphate-buffered saline, fixed with 70% ethanol overnight, and pretreated with 5 mg/mL ribonuclease for 30 minutes at 37°C and then stained with propidium iodide (100 µg/mL). Cell cycle profile was determined by fl/m cytometric analysis of DNA content of cell nuclei (BD FACSCalibur).
Scratch Assays
Cells were grown to 90% confluence in 6-well culture plates. A p200 pipet tip was used to create a scratch on the cell monolayer. Images were captured immediately after wounding and 24 hours and 48 hours after wounding, and wound closure was monitored by microscopy. Wound sizes were verified with an ocular ruler to ensure that all wounds were the same width at the beginning of each experiment.
Cell Migration/Invasion Assays
Cell motility and invasiveness were measured on Transwell and Matrigel chamber plates, respectively (24-well format; BD Biosciences, St Louis, MO). Cells (5 × 104) were seeded onto Transwell or Matrigel insert membranes with 8-mm pores on day 2 following transfection. Growth medium containing 20% fetal bovine serum was used as a chemoattractant. After incubation at 37°C for 22 hours, cells that did not migrate or invade through the pores of the Transwell inserts were manually removed with a cotton swab. Cells present at the bottom of the membrane were fixed and stained using Diff-Quik Stain Set Kit (Dade Behring, Siemens, Schwalbach, Germany). After 10 minutes of incubation, the filters were washed thoroughly in water and suspended in 350 µL of 5% acetic acid and 5% methanol. Colorimetric readings were taken at an OD of 595 nm.
Statistical Analysis
Unsupervised clustering of HELP data by hierarchical clustering (1-Pearson correlation distance and Ward’s agglomeration method) was performed using the statistical software R, version 2.6.2. A 2-sample t test was used for each gene to summarize methylation differences between groups. Genes were ranked on the basis of this test statistic, and a set of top differentially methylated genes with an observed log fold change of >10 normalized angles between group means was identified. Genes were further grouped according to the direction of the methylation change (hypomethylated vs hypermethylated), and the relative frequencies of these changes were computed among the top candidates to explore global methylation patterns. We applied Significance Analysis of Microarrays for multiple testing based on 1000 permutations. This procedure allows control of the false discovery rate (FDR). The estimated FDR for each given “delta” was determined according to Tusher et al. The delta was chosen to result in an FDR ≤0.05, and all loci with P values less than .05 by t testing had FDR values <5%.23 Results of experiments are displayed as mean ±standard deviation. To evaluate statistical significance, Student t test was used unless otherwise noted. Differences were deemed statistically significant at P<.05.
Results
High-Resolution Methylome Analysis Reveals Genome-Wide Hypomethylation in BE
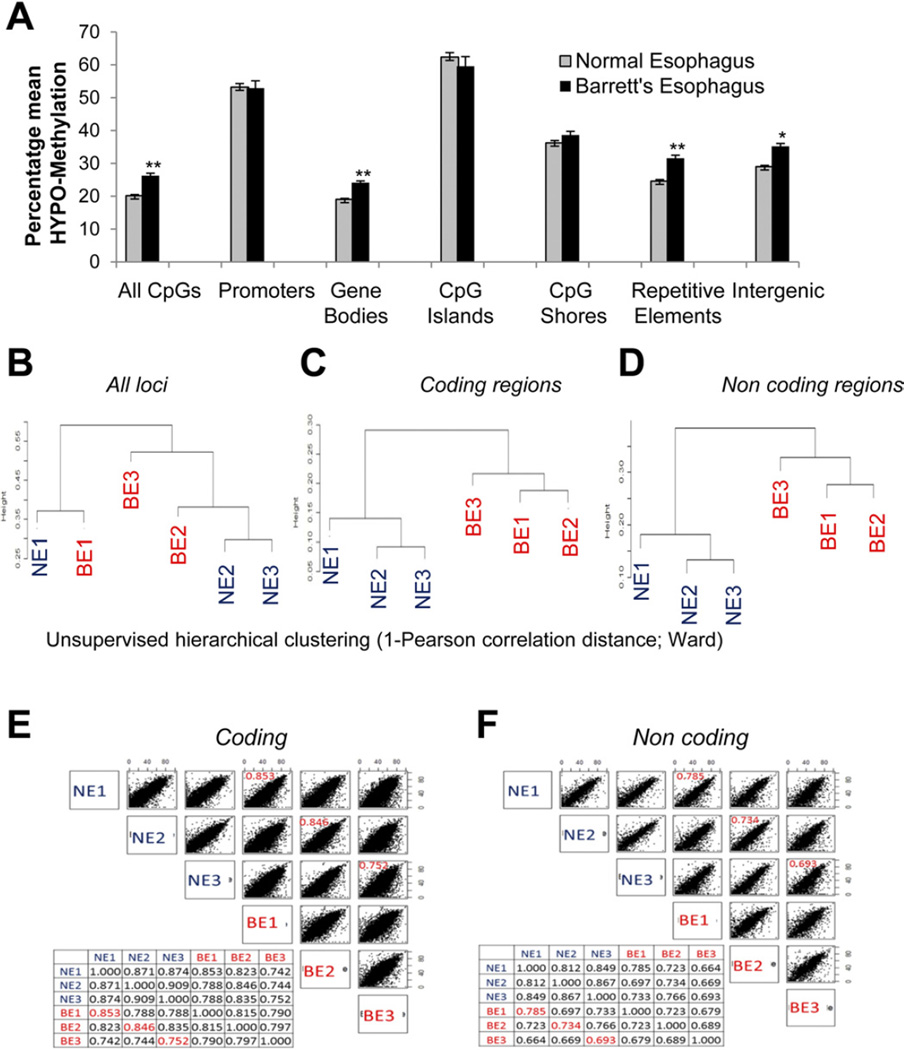
Although various studies have reported epigenetic alterations in BE, these studies have so far been restricted to promoter CpG methylation.17,24 We sought to elucidate the methylome of BE using a high-resolution assay (HELP tagging) with massively parallel sequencing to determine the CpG methylation status of 1.8 million loci distributed throughout the genome.18 Three sets of histologically validated endoscopic mucosal biopsy specimens, representing matched normal esophageal squamous mucosa and BE metaplasia, were obtained. Methylome profiling of these samples showed that hypomethylation was the predominant change in BE (Figure 1A). The magnitude of hypomethylation was most striking in gene bodies and at repetitive elements of the genome. Interestingly, promoters and CpG islands did not exhibit significant differential methylation. Because intragenic regions showed significant differential methylation and included both coding and non-coding parts of the genome, we next determined the discriminatory power of these epigenetic changes. Unsupervised clustering based on CpG methylation of all probes was unable to distinguish between NE and BE (Figure 1B). Unsupervised clustering based on methylation of all coding and noncoding regions, on the other hand, strikingly discriminated between NE and BE, even in matched patient sets (Figure 1C and D), establishing the importance of these novel changes. Furthermore, a comparison of epigenetic alterations at coding versus noncoding sites revealed that noncoding regions had a larger magnitude of methylation change in BE, as evident from the lower correlation coefficients between these samples. Less correlation was observed in the methylation status of noncoding loci between matched samples of NE and BE (marked in red), revealing a greater magnitude of change at these loci (Figure 1E and F). In fact, there was even less correlation among the BE samples for noncoding methylation changes, suggesting that these loci represent active areas of epigenetic change. These data suggest that novel noncoding epigenetic changes occur during evolution of NE to BE.
Figure 1.
Genome-wide hypomethylation in BE. (A) Three sets of endoscopic biopsy specimens representing NE and BE were examined using HELP tagging. Hypomethylation was seen in BE (t test, **P< .005; *P< .05). Hierarchical clustering was based on CpG methylation for all (B) probes, (C) coding regions, and (D) noncoding regions. Correlation plots were created based on methylation of (E) coding and (F) noncoding regions.
Hypomethylation of Noncoding Regions Occurs in BE
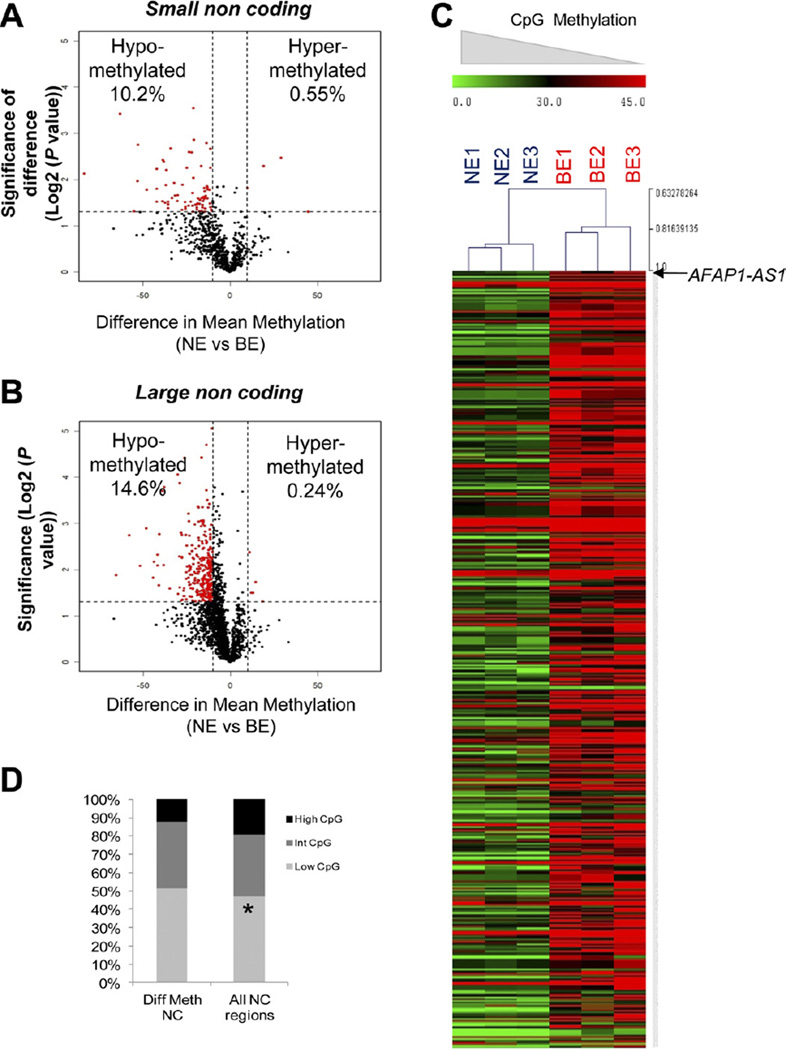
Because little was known about epigenetic regulation of noncoding regions during disease, we decided to focus on CpG methylation changes in noncoding regions. We observed that both small (<200 bp) and large (>200 bp) noncoding regions were characterized by hypomethylation (Figure 2A and B). In fact, a greater proportion of large noncoding regions were affected by aberrant hypomethylation (92/901 differentially methylated small vs 367/2501 differentially methylated large noncoding regions, P= .001, proportions test). We used Significance Analysis of Microarrays for multiple testing based on 1000 permutations to calculate the FDR. All differentially methylated loci with P values less than .05 by t testing were found to have an FDR of <5%.23 Furthermore, hierarchical clustering revealed a signature of 470 differentially methylated noncoding regions, which included numerous novel transcript regions that have not been studied previously in cancer. The top 20 most-altered transcripts (coding and noncoding) are shown in Supplementary Tables 1 and 2.
Figure 2.
Hypomethylation affects noncoding regions. (A and B) Volcano plots display differences in mean methylation between NE and BE samples (x-axis) and logs of P values between means (y-axis). Differentially methylated loci (red) reveal hypomethylation in both large and small noncoding regions. (C) Heat map displaying differential CpG methylation at all noncoding regions shows predominant hypomethylation. (D) Bar graph illustrating distribution of CpG densities according to differential methylation.
Because CpG island regions have previously been considered a principal target of epigenetic dysregulation in cancer, we next sought to determine whether noncoding regions affected by aberrant methylation were disproportionately associated with a higher density of CpGs. We annotated the genome into regions of low, intermediate, and high CpG density and then determined the correlation of differentially methylated noncoding loci with CpG density. We found that the majority of noncoding loci exhibiting differential methylation during progression of BE lay, paradoxically, outside of CpG-dense regions. These novel data support the hypothesis that epigenetic changes are not restricted to CpG-dense regions, such as CpG islands.
Finally, we decided to focus on a particular large noncoding transcript, AFAP1-AS1, to study functional consequences of epigenetic changes at noncoding loci. AFAP1-AS1 was chosen because it was significantly aberrantly hypomethylated in BE; it was a very large lncRNA (6810 bp); and its coding counterpart, the AFAP1 protein, is known to be involved in human cancers.25 We could find no published studies of this lncRNA in any human illness or disease model.
AFAP1-AS1 Is Hypomethylated and Overexpressed in BE
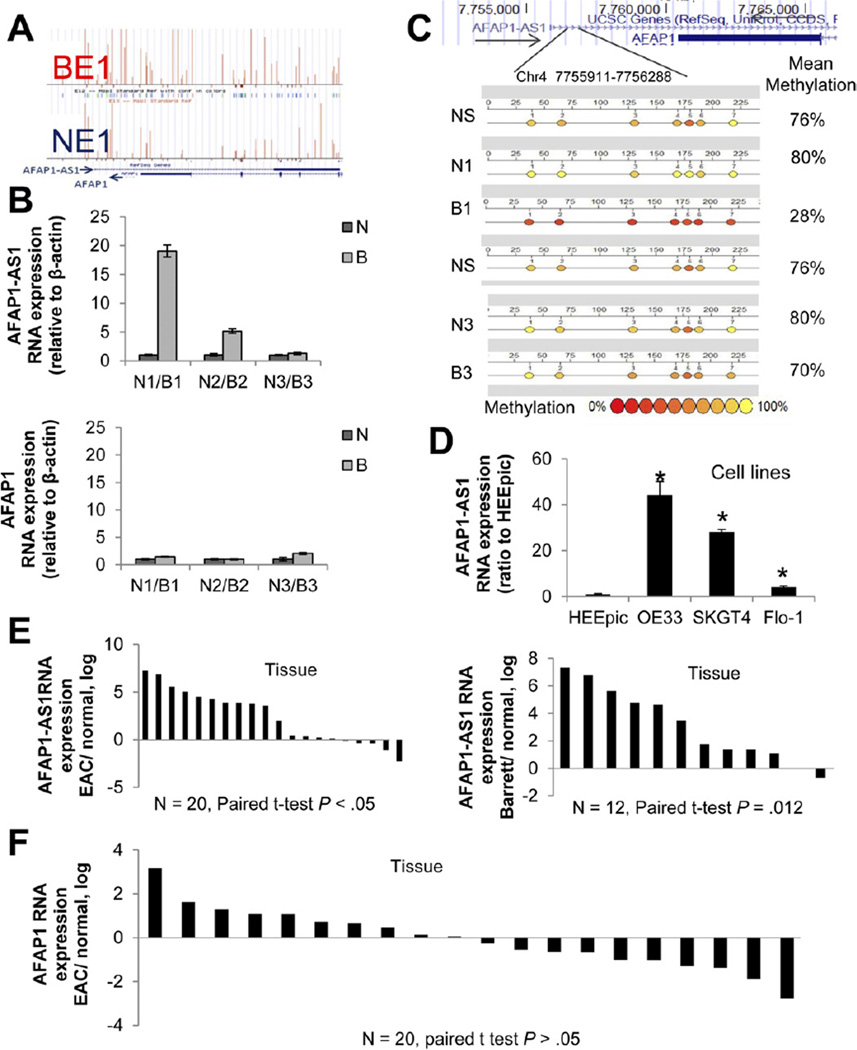
The AFAP1-AS1 locus was characterized by striking hy-pomethylation in BE in all 3 matched NE-BE tissue pairs. Hypomethylation occurred near the AFAP1-AS1 transcription start site and throughout its intragenic regions (Figure 3A), as depicted by the taller and more numerous vertical bars (proportional to percent hypomethylation) in a representative BE sample in this figure. These samples also exhibited increased expression of AFAP1-AS1 (Figure 3B, upper panel). Interestingly, the start site and promoter of the AFAP1 protein-coding gene were not differentially methylated in these BE samples, and expression of AFAP1 was significantly lower than that of AFAP1-AS1 (Figure 3B, lower panel). Bisulfite MassArray analysis of methylation of the AFAP1-AS1 locus revealed hypomethylation in the B1 (BE) sample when compared with the matched N1 (NE) sample. Normal stomach (NS) was also methylated similarly to sample N1. Sample B3 was not hypomethylated when compared with N3; methylation values correlated with expression values for paired sets N1/B1 and N3/B3 (Figure 3C). Next, we measured expression of AFAP1-AS1 in esophageal cell lines, finding overexpression in 3 EAC cell lines but not in normal esophageal epithelial cells (HEEpic; Figure 3D). Finally, we sought to determine whether AFAP1-AS1 was overexpressed in a larger cohort of primary human esophageal tissues. Using quantitative reverse-transcription PCR, we assessed expression levels of AFAP1-AS1 in 20 matched pairs of human EAC and adjacent NE as well as in 12 matched pairs of human benign BE and adjacent NE. AFAP1-AS1 expression was elevated relative to NE in the majority of EACs (15/20) and BEs (11/12) (Figure 3E). These data suggest that AFAP1-AS1 expression is up-regulated in both EAC cell lines and primary EAC tissues, consistent with the DNA hypomethylation observed in these same samples. We also measured the expression of the protein-coding gene AFAP1 in the same matched NE-EAC pairs, and the results revealed no significant change in levels of AFAP1 (Figure 3F). Expression levels of both AFAP1-AS1 (RNA) and AFAP1 (RNA) in NE, BE, and EAC tissues were measured in 3 patients (Supplementary Figure 2A). Two of these showed higher RNA levels of both AFAP1-AS1 and AFAP1 in Barrett’s and tumor tissues, while the third showed no significant change in either RNA. Protein levels of AFAP1 were in accordance with RNA levels in patient 1 (Supplementary Figure 2B). In addition, HELP-tag-ging data showed that the methylation profile at the start site of the AFAP1 gene was very similar between matched NE and BE (Supplementary Figure 3). These data suggest that noncoding RNA AFAP1-AS1 is hypomethylated and up-regulated in BE and EAC but that this dysregulation appears to have no effect on the expression of its coding counterpart, AFAP1.
Figure 3.
Aberrant methylation and expression ofAFAP1-AS1 in human EAC. (A) HELP-tagging results for matched BE and NE samples. Upward vertical bars (proportional to percent hypom-ethylation) are taller and more numerous in BE1 than in NE1, showing relative hypomethyla-tion at the AFAP1-AS1 locus in BE. (B) Expression of AFAP1-AS1 (upper panel) and AFAP1 (lower panel) in tissues studied for methylation. AFAP1-AS1 expression was significantly higher in BE than NEin2 of the 3 tissue pairs. AFAP1 expression was lower than AFAP1-AS1 expression but not significantly different between BE and NE. (C) Bisulfite MassArray analysis of methylation of the AFAP1-AS1 locus reveals hypomethylation in sample B1 (BE) relative to its matched N1 (NE) sample. Normal stomach (NS) is methylated similarly to NE. Sample B3 is not hypomethylated compared with N3, in agreement with expression levels for this paired set. (D) AFAP1-AS1 expression was elevated in all EAC cell lines tested (OE33, SKGT4, FLO-1). HEEpic, nonimmortalized normal primary esophageal epithelial cells. * P<.05 compared with HEEpic. (E) In 20 EAC cases, AFAP1-AS1 expression was elevated in 75% of EACs (15/20) versus NE tissues. AFAP1-AS1 expression was elevated in 91% of BEs (11/ 12) relative to NE tissues. (F) Expression levels of AFAP1 in NE and EAC tissues from 20 patients. There was no significant difference between NE and EAC.
Specific Inhibition of AFAP1-AS1 Is Achieved With siRNAs, Without Effects on AFAP1 Expression
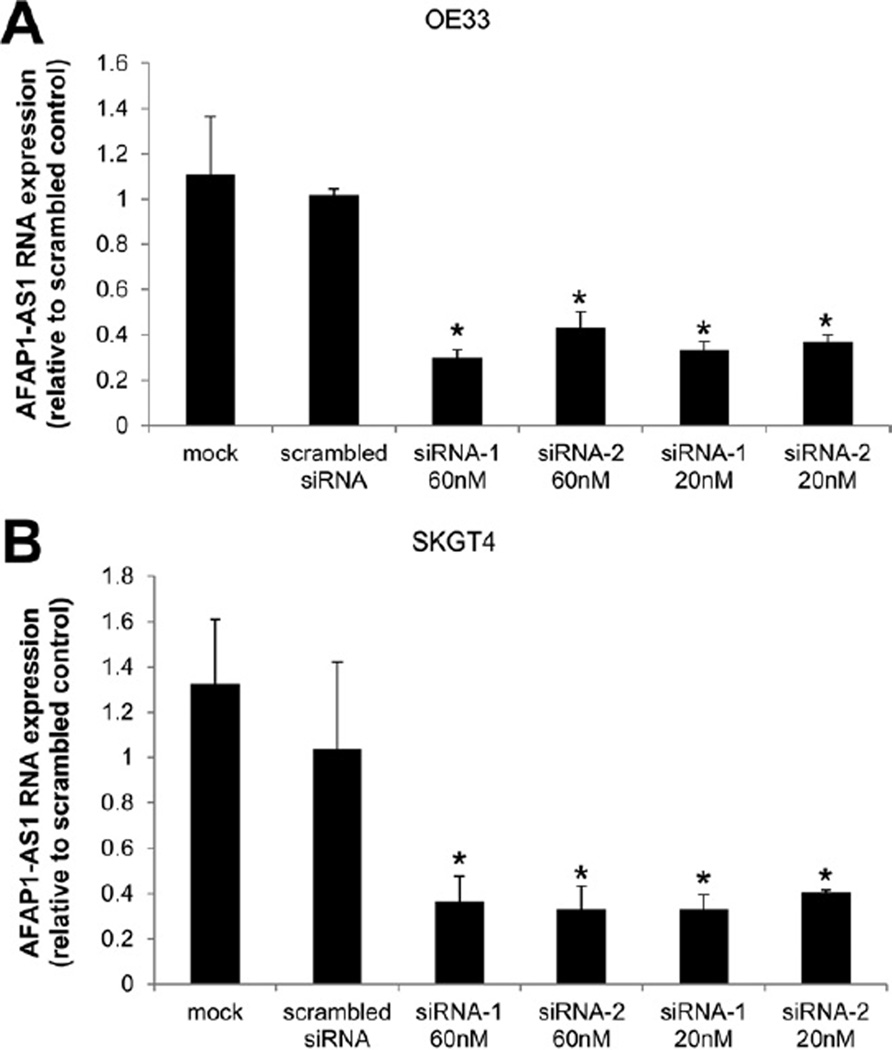
To investigate the functional involvement of AFAP1-AS1 in human EAC, we used the siRNA knockdown strategy to inhibit AFAP1-AS1 expression in EAC cells. Two different siRNAs were tested for knockdown efficiency, and both caused >60% reduction of AFAP1-AS1 levels in 2 EAC cell lines (OE33 and SKGT4) (Figure 4A and B). To determine the effect of AFAP1-AS1 inhibition on AFAP1 expression in these 2 cell lines, we used quantitative reverse-transcription PCR and Western blot to examine the expression of AFAP1 following siRNA-mediated knockdown of AFAP1-AS1. The level of AFAP1 expression was not significantly altered following AFAP1-AS1 knockdown relative to a scrambled siRNA control (Supplementary Figure 4A and B). These results confirm that these siRNAs did not affect the expression level of AFAP1, suggesting that phenotypic effects observed following knockdown of AFAP1-AS1 were driven directly by AFAP1-AS1, rather than indirectly via AFAP1.
Inhibition of AFAP1-AS1 in EAC Cells Leads to Reduced Proliferation and Anchorage-Dependent Growth
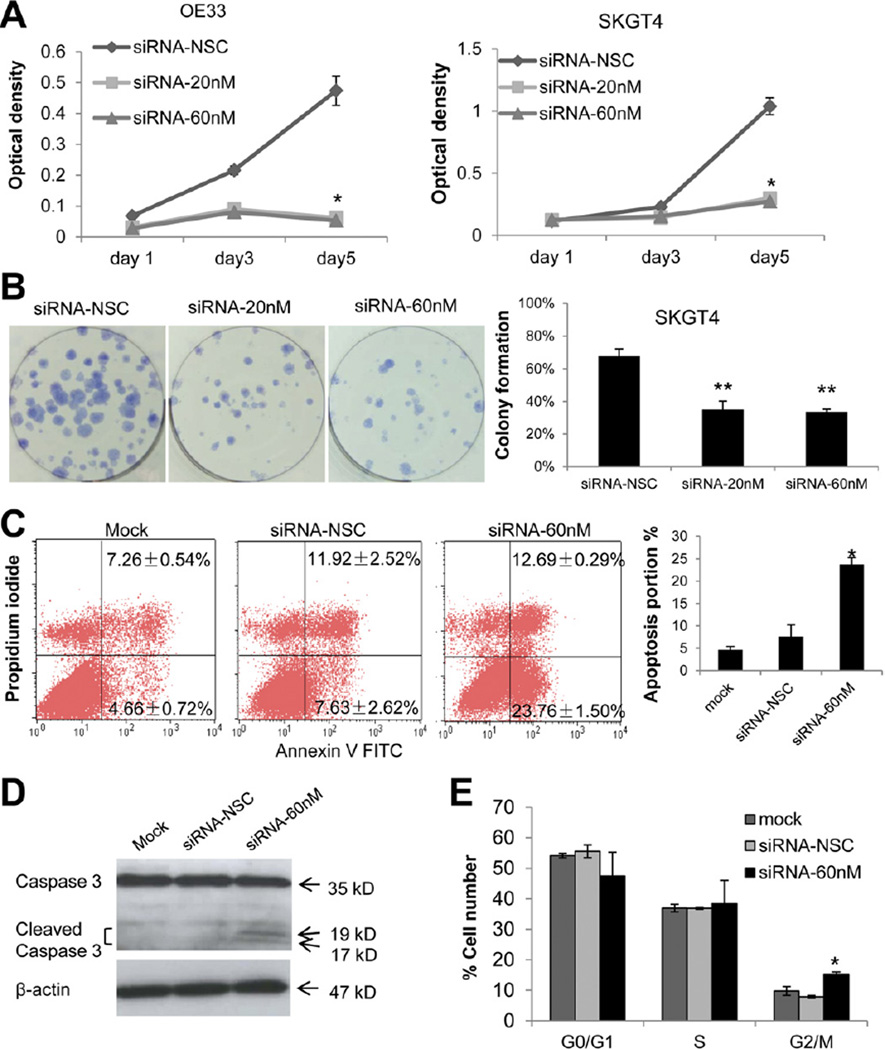
To determine the functional consequences of deregulated AFAP1-AS1 expression, several in vitro assays were performed. In comparison with cells transfected with a scrambled control siRNA, transfection with specific siRNAs significantly decreased growth at day 5 in both SKGT4 and OE33 EAC cells (Figure 5A). In addition, siRNA-treated cells exhibited significantly decreased anchorage-dependent growth versus a scrambled siRNA control. The capacity of specific siRNA-treated cells to form colonies was reduced by 50% in SKGT4 cells (Figure 5B).
Figure 5.
Inhibition of AFAP1-AS1 significantly reduces proliferation and colony formation in EAC cells. (A) WST-1 cell proliferation assays in OE33 and SKGT4 cells. Both the 20 nmol/L and 60 nmol/L concentrations induced significantly lower growth rates *(P< .01 and *P< .01, respectively). (B) Colony formation assays.(Left panel) AFAP1-AS1-specific siRNA-treated cells (2 wells at right) versus scrambled control siRNA-treated cells (leftmost well) (original magnification 10x). (Right panel) Mean ± standard deviation of 3 independent experiments. **P< .05. (C) Relative to treatment with a scrambled siRNA, transfection with a specific AFAP1-AS1 siRNA induced significant apoptosis (lower right quadrant, 23.76% ± 1.50% vs 7.63% ± 2.62%; *P< .05) in OE33 cells. (D) Western blotting revealed cleaved caspase-3 only in AFAP1-AS1 siRNA-treated cells. (E) Cell cycle results in OE33 EAC cells. Relative to treatment with a scrambled siRNA, transfection with a specific AFAP1-AS1 siRNA induced significant G2/M-phase cell cycle arrest. *P<.05. NSC, nonspecific control siRNA.
We next performed experiments to assess the mechanism of growth inhibition induced by AFAP1-AS1 inhibition (Figure 5C). The induction of apoptosis following 48-hour treatment with AFAP1-AS1 or scrambled control siRNAs in OE33 cells was examined using flow cytometry. Knockdown of AFAP1-AS1 significantly increased apoptosis in EAC cells (23.76% ±1.5% vs 7.63% ± 2.62%; t test P < .05, Figure 5C). Moreover, we measured caspase-3 protein levels in siRNA-treated versus untreated OE33 cells. Cleavage of caspase-3 into smaller bands (17 and 19 kilodaltons; Figure 5D) occurred only after AFAP1-AS1 siRNA treatment, suggesting that inhibition of AFAP1-AS1 induces apoptosis. We also performed cell cycle assays after siRNA treatment using flow cytometry (Figure 5E). Knockdown of AFAP1-AS1 significantly induced G2/M-phase arrest (15.22%± 0.79% vs 7.89% ± 0.43%; t test P< .05). Taken together, these findings suggest that the ln-cRNA AFAP1-AS1 modulates both proliferation and programmed cell death in esophageal cancer cells.
Inhibition of AFAP1-AS1 in EAC Cells Leads to Reduced Invasion
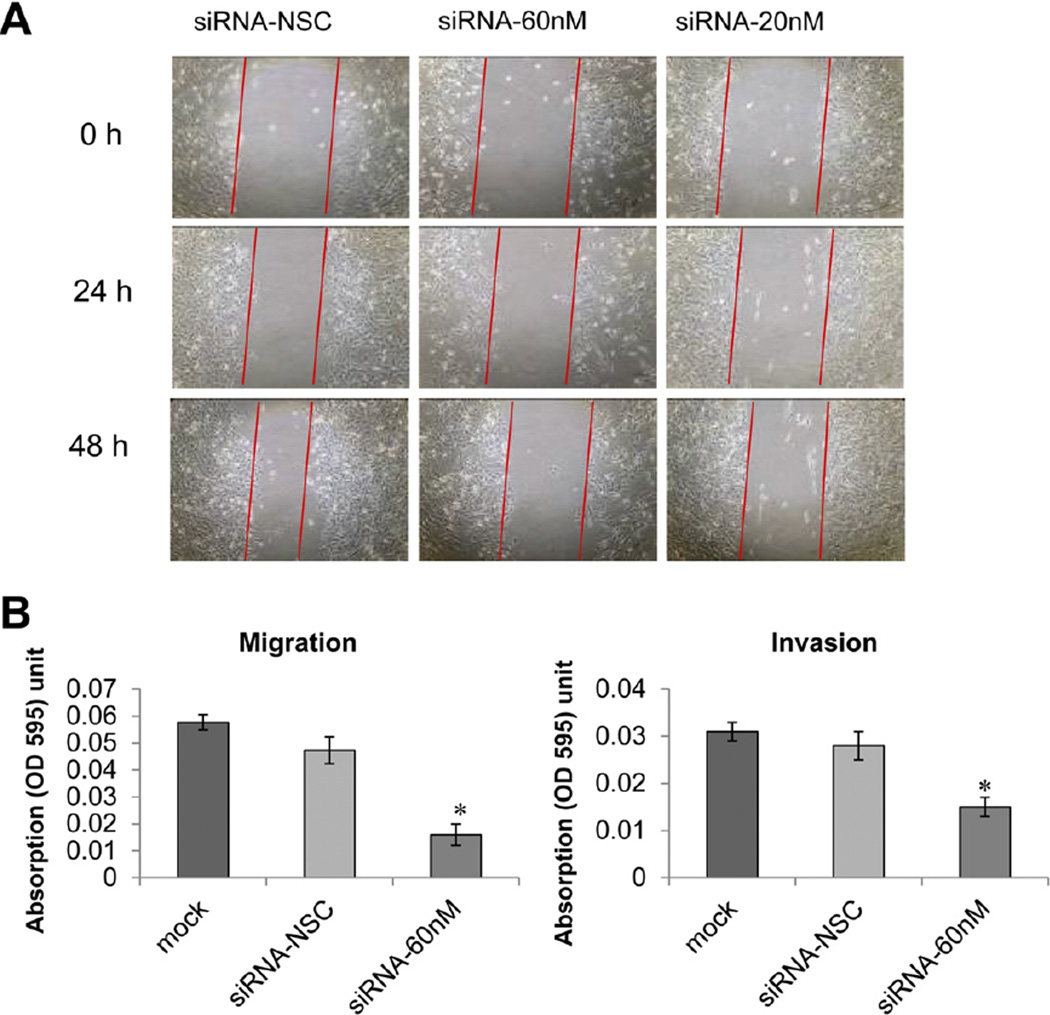
Invasiveness is a hallmark of all cancer cells. Therefore, wound healing assays were performed to gauge the effect of AFAP1-AS1 suppression on cell motility. AFAP1-AS1 knockdown resulted in attenuated motility of SKGT4 and OE33 cells. Specifically, compared with the scrambled siRNA control-treated cells, wound recovery was significantly delayed in AFAP1-AS1-specific siRNA-treated SKGT4 (Figure 6A)and OE33 cells (Supplementary Figure 5).
Figure 6.
AFAP1-AS1 suppression inhibits cell motility. (A) Wound healing assays. AFAP1-AS1 suppression by siRNA decreased SKGT4 cell motility (original magnification 10x). (B) In vitro migration and invasion assays. *P< .05. NSC, nonspecific control siRNA.
Furthermore, the migration and invasiveness of EAC cells were assessed using the migration and invasion assays as described in Materials and Methods. As shown in Figure 6B, SKGT4 cell migration and invasion were reduced by 36.0% (P < .05) and 75.9% (P < .05), respectively, following AFAP1-AS1 inhibition. Thus, these data suggest that suppression of AFAP1-AS1 expression reduces the migration and invasiveness of EAC cells.
Discussion
The prognosis of EAC is quite poor because most patients present at late stages, when treatment regimens are less effective. By the time symptoms of dysphagia become manifest, this disease is usually advanced, and most patients die within the first year following diagnosis.1 Thus, a better understanding of this disease could lead to earlier detection and improved treatment outcomes.
By using high-resolution deep sequencing–based HELP tagging, we found that hypomethylation, rather than hypermethylation, is the predominant epigenetic alteration during early progression of BE. Interestingly, we also found that this genome-wide early hypomethylation seems to affect both coding and noncoding regions of the genome. Although global hypomethylation has been reported in many epigenetic studies of cancer,26 the functional consequences of this change have not been completely elucidated. It has been hypothesized that loss of methylation leads to carcinogenesis by encouraging genomic instability27 and aberrantly activating oncogenes.28 Our data establish that hypomethylation is associated with the overexpression of lncRNA transcripts, which exert functional pro-cancerous effects in esophageal cells.
Although approximately 90% of the human genome is transcribed,29 the ENCODE project has shown that a surprisingly small amount of this RNA (approximately 2%) actually encodes proteins; thus, most transcripts are non–protein-coding RNAs. lncRNAs (longer than 200 nucleotides) are emerging as a novel class of non-coding RNAs. Several lncRNAs have been identified as being linked to human disease and exerting specific functions.30 Our data show that the AFAP1-AS1 lncRNA is overexpressed in primary BE and EAC tissues as well as in EAC cell lines. In vitro functional analyses support an oncogenic role for this lncRNA in the esophagus. Proliferation assays showed that inhibition of AFAP1-AS1 by siRNA diminished cell proliferation. Furthermore, treatment with siRNA inhibited colony formation and reduced migration and invasion. Moreover, inhibition of AFAP1-AS1 increased apoptosis and G2/ M-phase arrest. Taken together, these findings suggest that AFAP1-AS1 is a functional lncRNA in human EAC cells and suggest the potential utility of AFAP1-AS1 as a biomarker of EAC.
The AFAP1-AS1 transcript is derived from the antisense strand of AFAP1 genomic DNA, the opposite strand of which encodes AFAP1. AFAP1 modulates actin filament integrity and serves as an adaptor protein linking Src family members and other signaling proteins to actin filaments.31 AFAP1 is involved in cancer cell pathophysiology; it is required for actin stress fiber formation and cell adhesion in breast cancer cells.32 Similarly, AFAP1 is overexpressed in prostate cancer and contributes to tumorigenic growth by regulating focal cell contacts.25 We hypothesized that the antisense RNA AFAP1-AS1 might regulate expression of its cognate sense gene, AFAP1. However, we did not observe an inverse correlation between expression levels of AFAP1-AS1 and AFAP1 in primary tumors or tumor cells (Figure 3B, E, and F and Supplementary Figure 2). Thus, AFAP1-AS1 may not bind to its sense cognate gene; its effects may involve an AFAP1-independent mechanism during development or progression of EAC. Nevertheless, it is noteworthy that AFAP1-AS1 localizes to the antisense genomic DNA strand near the C-terminus of AFAP1, at the actin-binding domain of AFAP1. Thus, it will be of great interest in the future to investigate whether and how AFAP1-AS1 is involved in actin stress fiber formation.
The evolutionary conservation of sense mRNAs with their corresponding antisense cognate noncoding RNAs, in conjunction with the vast number of lncRNAs that exist, suggests a role for these RNAs in organismal complexity.33 Non–protein-coding RNAs have recently been shown to exert control over gene transcription via several different pathways: transcriptional gene silencing through the targeted recruitment of epigenetic silencing complexes to particular loci34,35; posttranscriptional gene silencing; degradation of transcriptionally active mRNAs, as exhibited for RNA interference, siRNA, and microRNA; and STAU-1–mediated RNA decay.36
Moreover, evolutionary retention of miRNAs may be linked to the regulation of antisense lncRNAs. Specifically, miRNAs can control both sense and antisense transcripts, based on their relative abundance.34 An example of this ability of microRNAs to regulate bidirectional transcription can be found within miR-373, which binds to the antisense noncoding RNA for Ecadherin.37
In summary, we have shown that AFAP1-AS1 expression is substantially increased in EAC versus NE tissues as well as in EAC cell lines. This elevated expression of AFAP1-AS1, its role in cell proliferation and apoptosis, and its effect on cell migration and invasion suggest that dysregulated expression of AFAP1-AS1 is involved in development or progression of EAC and that AFAP1-AS1 represents a functional lncRNA in esophageal carcinogenesis.
Supplementary Material
Figure 4.
Knockdown efficiency of AFAP1-AS1–specific siRNAs in EAC cells. (A and B) Two siRNA molecules were tested. Both siRNAs at 20 nmol/L and 60 nmol/L yielded ≥60% reduced expression of AFAP1-AS1 in OE33 and SKGT4 EAC cells (vs scrambled siRNAs). * P<.05.
Acknowledgments
Funding
Supported by the American Cancer Society, Leukemia & Lymphoma Society, and US Public Health Service grants DK087454, CA146799, and CA133012. S.J.M. is the Harry and Betty Myerberg/ Thomas R. Hendrix Professor of Gastroenterology. W.W. was supported by an Exchange Scholarship from the China Scholarship Council.
Abbreviations used in this paper
- bp
base pairs
- BE
Barrett’s esophagus
- EAC
esophageal adenocarcinoma
- FDR
false discovery rate
- lncRNA
long noncoding RNA
- mRNA
messenger RNA
- NE
normal esophagus
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2013.01.019.
References
- 1.Polednak AP. Trends in survival for both histologic types of esoph-ageal cancer in US surveillance, epidemiology and end results areas. Int J Cancer. 2003;105:98–100. doi: 10.1002/ijc.11029. [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Sunkin SM, et al. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 6.Perez DS, Hoage TR, Pritchett JR, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 7.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 9.Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 10.Fu X, Ravindranath L, Tran N, et al. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 11.Jonkers I, Monkhorst K, Rentmeester E, et al. Xist RNA is confined to the nuclear territory of the silenced X chromosome throughout the cell cycle. Mol Cell Biol. 2008;28:5583–5594. doi: 10.1128/MCB.02269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinoza CA, Allen TA, Hieb AR, et al. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 14.Mariner PD, Walters RD, Espinoza CA, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez H, Opalinska J, Zhou L, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet. 2011;7:e1001356. doi: 10.1371/journal.pgen.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki M, Jing Q, Lia D, et al. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol. 2010;11:R36. doi: 10.1186/gb-2010-11-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa ME, Reimers M, Thompson RF, et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS One. 2008;3:e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa ME, Wouters BJ, Skrabanek L, et al. Genome-wide epigenetic analysis delineates a biologically distinct immature acute leukemia with myeloid/T-lymphoid features. Blood. 2009;113:2795–2804. doi: 10.1182/blood-2008-08-172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaral PP, Clark MB, Gascoigne DK, et al. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microar-rays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Z, Cheng Y, Gu W, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 2009;69:4112–4115. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Park SI, Artime MC, et al. AFAP-110 is overexpressed in prostate cancer and contributes to tumorigenic growth by regulating focal contacts. J Clin Invest. 2007;117:2962–2973. doi: 10.1172/JCI30710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 28.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 29.Wilhelm BT, Marguerat S, Watt S, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 30.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 31.Baisden JM, Qian Y, Zot HM, et al. The actin filament-associated protein AFAP-110 is an adaptor protein that modulates changes in actin filament integrity. Oncogene. 2001;20:6435–6447. doi: 10.1038/sj.onc.1204784. [DOI] [PubMed] [Google Scholar]
- 32.Dorfleutner A, Stehlik C, Zhang J, et al. AFAP-110 is required for actin stress fiber formation and cell adhesion in MDA-MB-231 breast cancer cells. J Cell Physiol. 2007;213:740–749. doi: 10.1002/jcp.21143. [DOI] [PubMed] [Google Scholar]
- 33.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 34.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris KV. Non-coding RNAs, epigenetic memory and the passage of information to progeny. RNA Biol. 2009;6:242–247. doi: 10.4161/rna.6.3.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris KV, Santoso S, Turner AM, et al. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.