Abstract
Cell-penetrating peptides (CPPs) are a promising tool to overcome cell membrane barriers. They have already been successfully applied as carriers for several problematic cargoes, like e.g. plasmid DNA and (si)RNA, opening doors for new therapeutics. Although several hundreds of CPPs are already described in the literature, only a few commercial applications of CPPs are currently available. Cellular uptake studies of these peptides suffer from inconsistencies in used techniques and other experimental conditions, leading to uncertainties about their uptake mechanisms and structural properties. To clarify the structural characteristics influencing the cell-penetrating properties of peptides, the chemical-functional space of peptides, already investigated for cellular uptake, was explored. For 186 peptides, a new cell-penetrating (CP)-response was proposed, based upon the scattered quantitative results for cellular influx available in the literature. Principal component analysis (PCA) and a quantitative structure-property relationship study (QSPR), using chemo-molecular descriptors and our newly defined CP-response, learned that besides typical well-known properties of CPPs, i.e. positive charge and amphipathicity, the shape, structure complexity and the 3D-pattern of constituting atoms influence the cellular uptake capacity of peptides.
Introduction
Since the discovery about 20 years ago by Frankel and Pabo that the Tat protein of the human immunodeficiency virus (HIV-1) can enter cells [1], cell-penetrating peptides (CPPs) are an increasingly growing part of fundamental and applied biomedical research. Throughout the literature, cell-penetrating peptides are traditionally defined as containing 5–30 amino acids, characterized by a net positive charge, which are able to cross cell barriers without causing significant membrane damage [2]. This property makes CPPs suitable to deliver hydrophilic macromolecules into the cell interior and to the different cellular compartments in vitro and in vivo [3]. They have already been successfully applied as carriers for problematic cargoes like plasmid DNA, oligonucleotides, short interfering RNA ((si)RNA), peptide-nucleic acids (PNA), proteins and other peptides, small molecules and liposome nanoparticles [4]. This implies that doors have been opened to new efficient peptide drugs [5].
During the last decade, several hundreds of CPPs have already been reported in the literature. In contrast to the traditional definition, CPPs actually present a chemically diverse group of peptides, showing a variety in constituent amino acids and 3D-structure. Three major classes can be distinguished: cationic, amphipathic and hydrophobic CPPs. This structural diversity accounts for the difference in uptake mechanism and level under different conditions between the groups of CPPs. Moreover, coupling the CPP to a cargo can also influence the level and mode of uptake into the cell [6]. Only a few structure-activity relationship (SAR) studies have tried to reveal which structural features are crucial for cellular uptake [7]–[16]. Hydrophobic alpha-helical structures seem to be important, as well as the positive charges from basic amino acids, with arginine favoured over lysine. Although equally contributing to the overall charge, the guanidinum group of arginine can donate two hydrogen bonds compared to one by lysine. Other factors apparently influencing cellular uptake are the peptide length and the conformation of the structure, which was demonstrated by the difference in cellular influx for pVEC and his scrambled analogue [2], [17]. The latter showed a reduced uptake into the cell, probably due to the loss of the N-terminal hydrophobic domain [7]. The influence of the peptide length was demonstrated for the SV40 T antigen, which showed an increase in cellular influx by adding a N-terminal sequence [17].
The available SAR studies only cover a limited set out of the diverse group of CPPs. Moreover, some publications show contradictory results [8], [9], possibly due to different experimental set ups. This impedes drawing general conclusions about the structural features important for cellular uptake. Furthermore, the uptake mechanism of the different CPP groups is still under debate. Today, endocytosis (energy dependent) and direct penetration (energy independent) are suggested to be the two major cellular uptake mechanisms. Depending on the experimental conditions, CPPs use two or more different mechanisms [2].
One approach for predicting CPPs is trial and error, which implies identifying sequences of a suitable length and rich in positive charges in a protein structure [18]. Another approach are the Sandberg expanded z-descriptors, used by Hällbrink et al. [19]. They calculated the bulk property values for a training set of known CPPs and known non-penetrating peptides and averaged over the total number of amino acids. The most relevant descriptors were Z1, Z2 and Z3, describing respectively lipophilicity, steric bulk properties and polarity, the latter having the most predictive power. Cell-penetrating properties of new sequences were predicted based on whether their bulk property values fall within preset intervals, derived from the values of the training set. Z-descriptors make it possible to predict cell-penetrating properties in silico, but a major disadvantage is that the sum of descriptors is calculated, hereby neglecting the order of the amino acids. Moreover, the Tat peptide was not considered as a CPP by their search criteria [19]. Another way to predict CPPs is data mining, which is based on finding similarity patterns in a large set of (experimental) data [18]. Artificial neural networks have already been used by Karelson and Dobchev to predict CPPs, based on quantitative structure-activity relationship (QSAR) derived features of a training set of about 100 known (non-)penetrating peptides [20]. Sanders et al. used support vector machine (SVM) classifiers, based on primary features derived from the biochemical properties of 111 known CPPs and 34 non-CPPs, to predict cell-penetrating properties [21]. The authors could experimentally confirm the cell-penetrating ability of the SVM classified CPPs. As primary biochemical properties of peptides were used, their classifiers provided insight in the structural requirements for cellular penetration, e.g. positional preference for certain amino acids, like positively charged and aromatic residues.
One can conclude that, although CPPs have been studied for over 20 years, a lot of structural and mechanistic properties still need to be unravelled. Furthermore, it is obvious that the variety of techniques and experimental conditions used to quantify the cellular uptake of CPPs, impedes to directly compare their extent of uptake. Together with the fact that the different CPPs differ structurally and mechanistically, controversies about the uptake mechanisms and artifactual results in the past [22], make it difficult to predict whether a peptide is cell-penetrating or not.
In this article, we explored the chemical space of a set of 186 peptides, for which quantitative data for cellular uptake are available, by use of chemo-molecular descriptors, which numerically express the peptide structure. In addition, we defined a new cell-penetrating (CP)-response, in order to compare the cell-penetrating properties of these peptides in a one-merit figure. This CP-response allows the use and comparison of experimental data obtained with a different experimental set up. By combining the chemical descriptors and the CP-responses, biomolecular modeling and clustering of peptides was performed. Our results confirm already described determining features for cellular uptake, but also provide new insights in structural requirements for cellular uptake of peptides.
Methods
Data
Articles describing the uptake of CPPs covering the last five years (2007– March 2012), were gathered using the search engines Web of Knowledge, Google and PubMed. The terms ‘cell penetrating peptides’, ‘uptake cell penetrating peptides’, ‘protein transduction domain’ each separately, as well as ‘cellular uptake’, ‘characterization’, ‘kinetics’, ‘quantification cellular uptake’ and ‘studying uptake’, using the Boolean operator ‘AND’ were used. Specific names of known cell-penetrating peptides (e.g. penetratin) were also included as search terms. More publications were obtained by searching in the reference list of suitable articles and reviews. This resulted in publications dating before 2007 (1998–2006). Only those were withheld, where the experimental set up was correct, i.e. use of non-fixed cells and removing or quenching of extracellular bound peptide [22]. Moreover, the publications should contain quantitative data or graphs expressing the cellular uptake of CPPs. When no quantitative data were explicitly mentioned in the text, these data were deduced from the available graphs.
Calculating Chemo-molecular Descriptors
Before the chemo-molecular descriptors of the 186 selected peptides could be calculated, the MM+ in vacuo optimized structure of the peptides (not amidated), representing the most fundamental peptide structure, was drawn and optimized using HyperChem 8.0 (Hypercube, Gainesville, FL, USA). The geometry optimization was obtained by the molecular mechanics force field method using the Polak–Ribière conjugate gradient algorithm with a root mean square (RMS) gradient of 0.1 kcal/(Å×mol) as stop criterion. Afterwards, these Cartesian coordinate matrices were used to calculate more than 3000 descriptors, using Dragon 5.5 (Talete, Milan, Italy), HyperChem 8.0 and MarvinSketch 5.10.3 (ChemAxon, Budapest, Hungary) software programs. The specific peptide descriptor LogSumAA, introduced by our research group, was also included in the descriptor set [23]. The non-discriminative descriptors, i.e. constant for all peptides, and one of two highly correlated descriptors, calculated using the Pearson correlation coefficient (absolute correlation >0.95), were eliminated, resulting in a final 186×454 data-matrix for the original descriptors. When all descriptors were divided by the molecular weight, a data matrix of 186×416 was obtained. Next, the data were transformed by z-scaling, ensuring equal contribution of each descriptor to the resulting model [24].
Multivariate Data-analysis
Multivariate data-analyses were performed using Principal Component Analysis (PCA) and Hierarchical Cluster Analysis (HCA) with SIMCA-P+12.0.0.0 (Umetrics AB, Umeå, Sweden) and SPSS Statistics 20.0.0 (IBM Corp., Armonk, NY, USA) software programs, respectively. Average-linkage HCA clustering was performed using the Euclidean distance as the dissimilarity criterion. After a first PCA-analysis of the dataset, feature selection was performed by selecting the descriptors having a predicted variation value of more than 0.30, resulting in a 186×248 data matrix. For the descriptor set divided by the molecular weight, a 186×210 matrix was obtained.
Multiple Linear Regression (MLR) analysis of the chemo-molecular descriptors, using SPSS Statistics 20.0.0, was performed to build a predictive model for cellular uptake of CPPs. The stepwise method was performed during the MLR process to identify the most significant descriptors using the following criteria: probability of F to enter ≤0.05 and probability of F to remove ≥0.10. After eliminating 12 outliers identified by the Grubbs outlier test (α = 0.05), the CP-responses of 174 peptides were used to build the model (information about the outliers see Table S2).
Statistics
All statistical analyses of the data were performed using SPSS Statistics 20.0.0 software. Throughout this article, the median of datasets was used as the best measure for central tendency for not normally distributed data.
Results
Data
Studies were selected when using protocols, including use of non-fixed cells and removing or quenching of extracellular bound peptide according to Richard et al. [22]. Only pure peptides, not coupled to cargoes or to fatty acid chains, were withheld for this study. At last, we selected only those peptides for which standardizing to the cellular influx of penetratin was possible, allowing to calculate the CP-response for cellular uptake. Finally, a dataset of 186 peptides was obtained, showing high to no or (very) low cellular uptake [7], [9], [11]–[13], [16], [17], [25]–[69] (see Table S1).
The different studies showed a remarkable variety in used techniques and operational parameters to test cellular uptake (Table 1). Inherent to the different techniques used, the protocols of the experiments varied between research groups. This may explain the inconsistent cellular uptake results for some CPPs in the literature, like Tat 48–60, which normally demonstrates a cellular uptake within the same range as penetratin and R9, but was not in reference [17]. The model amphipathic peptide (MAP) showed an unusual low uptake in the study of Wada et al., which is explained by the cell-specific uptake of this CPP [52].
Table 1. Experimental differences between studies for cellular uptake of peptides.
| Operational parameter | Examples | |||||
| Technique | Spectrofluorometry | MALDI-TOF MS | Confocal laser scanning microscopy (CLSM) | |||
| RP-HPLC | Flow cytometry (FACS) | Atomic Absorption Spectrometry | ||||
| Scintillometry | Splice correction assay | Quantitative image analysis of CLSM images | ||||
| Fluorescence microscopy | – | – | ||||
| Positive control | No | Tat 48–60 | Transportan 10 | |||
| Penetratin | Tat 47–57 | Transportan | ||||
| MAP | R9 | YGR6 | ||||
| pVEC | D-R9 | R8 | ||||
| Negative control | No | Dextran | Perforin | |||
| No peptide used | YDEGE | STRRSAMAPR | ||||
| Green fluorescent peptide | YDEEGGG | APRTPGGRR | ||||
| Units of quantitative data | µM or nM | pmol or nmol/mg cell protein | SI/mg cell protein | |||
| ng/mg cell protein | a.u. | Fold change in GeoMean fluorescence | ||||
| Mean fluorescence intensity | RLU/mg | Mean fluorescence intensity/mg cell protein | ||||
| Fold/basal fluorescence | Relative fluorescence intensity | Relative cellular uptake (to control) | ||||
| % of total peptide | % of added peptide | % cellular uptake | ||||
| Cellular fluorescence | Fold change in FITC medium | – | ||||
| Label | FITC | 5,6-carboxyfluorescein | 2-aminobenzoic acid | |||
| Biotin | Deuterium | Rhodamine | ||||
| NBD | TAMRA | Alexa 488 | ||||
| GaDOTA | Texas Red | 125I | ||||
| Cell line | AEC | BMC | HaCaT | HEK293 | MC57 | S. cerevisiae |
| HBCEC | CHO (−K1) | Caco-2 | HL60 | A549 | C. albicans | |
| bEnd | U2OS | Cos-7 | MDCK | A431 | E. coli | |
| MCF-7 | Jurkat | MOLT-4 | HeLa | Hela pLuc705 | B. megaterium | |
| NIH-3T3 | RAW264.7 | BA/F3 | K562 | BT-20 | N2a | |
| KB | RAW | U373 MG | Daudi | Sf9 | MDA-MB-231 | |
| HT-29 | SKMel37 | DAMI | A549 | U251 | KG1a | |
| TF-1 | ESC | NC | Sca-1+Lin− | HEK293 | L929 | |
| Calu-3 | MDA | HER | TM12 | CCRF-CEM | – | |
| Incubation concentration | 10 nM | 200 nM | 0.1 µM | 0.33 µM | 0.4 µM | 0.8 µM |
| 1 µM | 1.8 µM | 2 µM | 2.5 µM | 3 µM | 3.1 µM | |
| 3.5 µM | 4 µM | 4.5 µM | 5 µM | 6 µM | 6.3 µM | |
| 7.5 µM | 10 µM | 12.5 µM | 15 µM | 20 µM | 25 µM | |
| 30 µM | 40 µM | 50 µM | 100 µM | 110 µM | 200 µM | |
| 400 µM | 800 µM | 1.6 mM | – | – | – | |
Defining a Cell-penetrating Response
Because of the variety in experimental settings throughout the literature, the cellular uptake results of the available CPPs are difficult to directly compare and are expressed using different units, as listed in Table 1. Therefore, a cell-penetrating (CP)-response, a unified response expressing the cellular uptake efficiency of CPPs, would be of great help to obtain a clear overview over the cellular influx capacities of the CPPs described in the literature.
Penetratin, one of the first discovered CPPs and often described in the literature, is the most used positive control in uptake studies of other peptides. Therefore, penetratin was considered as a general positive control and used to normalize the responses for cellular uptake. Before a CP-response could be defined, several assumptions were made: (1) cell and label differences were neglected. As shown in Table 1, about 50 different cell lines and 12 different labels were used. The different nature of the labels was not considered when chemically defining the peptide structure. (2) The uptake of the negative control was considered to be negligible. (3) The maximal values of cellular uptake during an experiment were used to cope with a possible time effect. (4) If a positive control was used in a study, it was considered as an internal standard and could be used to average variations in operational parameters. Finally, (5) a linear correlation between the extracellular and intracellular peptide concentration was assumed, although it cannot be excluded that there is a specific concentration effect [37], [39], [41], [42], [60]. This last assumption was necessary, because to calculate the CP-response, the quantitative value for cellular uptake was first corrected for the incubation (extracellular) concentration resulting in a concentration normalized response. Then, the latter response was normalized to the positive control penetratin, according to the following equation:
| (1) |
where PCPP/CCPP and Ppen/Cpen are the concentration normalized influx responses for a CPP and penetratin respectively in the same study.
As already mentioned before, not all studies included penetratin as a positive control. When another positive control than penetratin was used, the median of all available ratios of that alternative positive control over penetratin was used to normalize the response to penetratin:
| (2) |
where PCPP/CCPP is the concentration normalized influx response for a CPP, PPC/CPC for a positive control in the same study different from penetratin and the response factor is the median of all ratios of the concentration normalized responses of the positive control over the concentration normalized responses of penetratin, as expressed in formula (1) (Table 2).
Table 2. Overview of the used positive controls in studies for cellular uptake of peptides and their CP-response.
| Positive control | CP-response |
| MAP | 2.05 |
| Penetratin | 1.00 |
| pVEC | 1.31 |
| R9 | 1.00 |
| Tat 47–57 | 0.31 |
| Tat 48–60 | 0.22 |
| Transportan 10 | 1.64 |
A third possibility was that no positive control was used in the cellular uptake study. Then, the CP- response was calculated using the following equation:
| (3) |
with PCPP/CCPP being the concentration normalized influx response for a CPP and 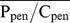 the median of all concentration normalized influx responses of penetratin, obtained using the same technique as the considered influx response (i.e. having the same unit).
the median of all concentration normalized influx responses of penetratin, obtained using the same technique as the considered influx response (i.e. having the same unit).
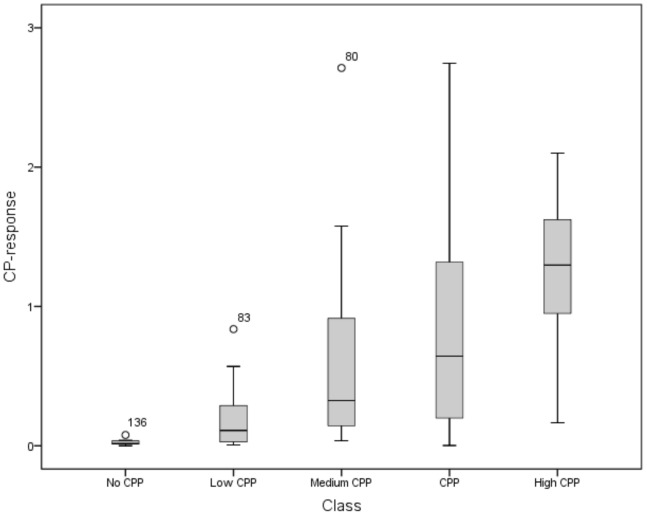
If more than one CP-response was available for a peptide, the median CP-response was calculated. Over all peptides, the CP-response ranged from 0.001378 to 2.744. The ranking of the peptides based on their CP-response, roughly corresponded with those found in the literature, e.g. the CP-response increased as follows: Tat 48–60< R9 ≈ penetratin <pVEC<transportan 10< MAP< transportan. This was in agreement with the overall study conclusions: Tat 48–60 mostly showed the lowest cellular influx [17], [26], [30], [31], [33], [34], [38], followed by R9 and penetratin [17], [25], [26], [28], [30]–[34], [38]. The peptides pVEC, transportan 10, MAP and transportan showed higher cellular influx than Tat 48–60, penetratin and R9. Transportan mostly showed a higher cellular influx than transportan 10 [10], [28]. Moreover, as a proof of concept, we investigated all manuscripts providing the quantitative data for cellular influx for the 186 peptides and compiled for each peptide how the authors estimated (subjectively) their cell-penetrating properties (see Table S3). We identified five classes: no CPP, low CPP (described as low CPP, low efficient, low effective, slow, nearly unmeasurable), medium CPP (described as medium CPP, efficient, effective) and high CPP (described as high CPP, highly, extremely effective, extremely efficient, rapid). When the authors only described the peptide as cell-penetrating, without any scaling or subjective ranking, these peptides were classified as CPP. Next, the distribution of the CP-responses in the five different classes was evaluated using Box-Whisker plots (see Figure 1). The median CP-response increased over the different classes from no CPP over low CPP, medium CPP and CPP to high CPP, indicating that peptides having a high or low calculated CP-response were also estimated in the same way by the researchers. This more detailed analysis thus demonstrated that the CP-response is indicative for the extent of cell-penetration of a peptide.
Figure 1. Distribution of the CP-responses in five different CPP classes as defined by the authors.
Exploration of the Chemical Space of CPPs
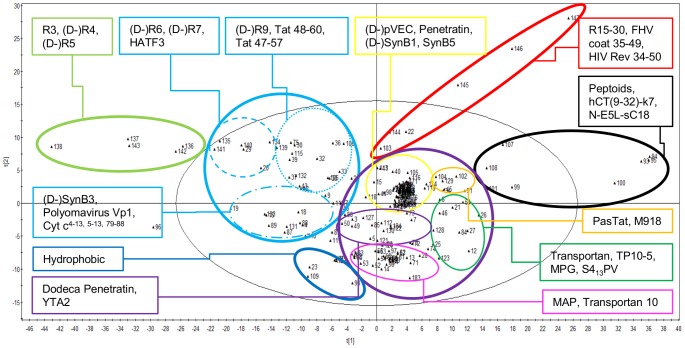
To determine the chemical space of a set of 186 peptides, which were investigated for cell-penetrating properties, a PCA and HCA-analysis of their calculated descriptors was performed. The first two principal components (PCs) of the calculated PCA-model explained already 62.6% of the total variability (Table 3). Based on the dendrogram of the HCA-analysis and the score plot of the first two PCs of the PCA-analysis, the 186 peptides could be categorized into six main clusters, which could be subdivided into eight subclusters (Figure 2).
Table 3. Summary of the PCA-analysis of the original descriptors, describing the eigenvalues of the covariance matrix, the total variance explained (cumulative R2) and the predictive ability (cumulative Q2).
| Principal Component | Eigenvalue | Cumulative R2 | Cumulative Q2 |
| 1 | 86.9 | 0.467 | 0.448 |
| 2 | 29.5 | 0.626 | 0.602 |
| 3 | 12.1 | 0.691 | 0.639 |
| 4 | 11.6 | 0.753 | 0.701 |
| 5 | 5.74 | 0.784 | 0.720 |
| 6 | 5.16 | 0.812 | 0.743 |
| 7 | 4.42 | 0.836 | 0.764 |
| 8 | 3.53 | 0.854 | 0.781 |
| 9 | 2.58 | 0.868 | 0.789 |
| 10 | 2.17 | 0.880 | 0.797 |
| 11 | 1.94 | 0.890 | 0.807 |
Figure 2. Score plot of the first versus the second principal component of the PCA-analysis of 186 peptides.
The six main clusters of peptides are indicated by a bold line (light green, light blue, red, purple, black and dark blue clusters), while the eight subclusters are encircled by a thin line (light blue dashed and/or dotted line, yellow, orange, dark green, purple and pink clusters). For each cluster, some examples of peptides are indicated.
The loading plot indicated that the first principle component (PC1) is mainly influenced by the mass, shape and connectivity of the peptides, while the second principle component (PC2) was determined by hydrophilicity and lipophilicity. In Figure 2, the peptides with high molecular weight (MW), surface area, molecular volume and number of hydrogen acceptor atoms were situated on the right along the horizontal axis and inherently these peptides had a higher number of peptide bonds (represented by the descriptors nRCONHR and C-040). The peptides on the right were also characterized by a more voluminous, complex and less compact structure. On the other side of the horizontal axis, the smaller, more symmetrical and compact peptides were located. On the PC2-axis, peptides mainly consisting of hydrophilic amino acids, like the basic arginine and lysine residues, represented by the high pI values of these peptides, were situated at the top. When descending to the bottom, the peptides turn more hydrophobic, indicated by higher log P values, hydration energy and BLI values (Kier benzene likeliness index), the latter describing the extent of molecular aromaticity.
The light green cluster at the left in the score plot represented short oligo-arginines (R3–R5), showing a very low median CP-response of 0.0769. The light blue subclusters contained cationic peptides, which differed in charge and peptide length (increasing from the left to the right). The light blue dashed-dotted subcluster (e.g. SynB3 and polyomavirus Vp1), showed a low median CP-response of 0.0392, while the dashed (e.g. R6, R7 and HATF3) and dotted (e.g. R9 and Tat 48–60) light blue subclusters had a mediocre cellular influx with median CP-responses of 0.323 and 0.464, respectively. The yellow and orange subclusters, which were centrally located in the PCA score plot, formed mixed clusters, as they contained both cationic and amphipathic peptides. The pink and purple amphipathic subclusters had median CP-responses of 0.181 and 0.302, respectively. The yellow subcluster (e.g. pVEC and penetratin), orange subcluster (e.g. PasTat and M918) and the dark green subcluster (e.g. transportan and MPG) had the highest values for the median CP-response, ranging from 0.511 to 0.729 and 0.798, respectively. These peptides were cationic and/or amphipathic and are composed of 15–27 amino acids. Remarkably, the group of peptides, showing a high CP-response could be subdivided in two groups: those having a positive PC2 value, which were mainly arginine rich (yellow and orange subcluster) and those having a negative PC2 value (dark green subcluster), which were mainly lysine rich. Although it was previously stated that arginine residues are favourable over lysine for cellular influx [2], our data did not confirm this statement. Peptides showing the highest CP-response had a high charge density or show amphipathicity. The latter peptides were centrally located in the score plot and were rich in sulfur-containing residues, especially methionine, as well as in aromatic amino acids.
The hydrophobic peptides, which are alanine, glycine, leucine, proline and valine rich, were located at the bottom of the score plot and showed a mediocre, but significant influx (median CP-response of 0.354). The peptides of the red cluster were highly charged and showed a high CP- response (median of 0.764). The cluster was mainly composed of oligoarginines of more than 15 residues, which are known for their cellular toxicity [12]. The black cluster consisted of voluminous, high molecular weight peptides, i.a. some peptoid structures, showing a very low cellular influx (median CP-response of 0.166).
As PC1 was mainly dominated by the molecular weight, the same PCA-analysis was performed, but using all descriptors divided by the molecular weight in order to neutralize its MW size-effect, although some descriptors were already corrected for the MW. However, this modification of the descriptors did not deliver extra information. The calculated PCA-model resulted in similar clusters of CPPs (see Table S4 and Figure S1).
Functional Diversity of CPPs
Using our newly defined CP-response and the calculated chemo-molecular descriptors of the peptides, a stepwise MLR-model was constructed to predict the cell-penetrating ability of new peptides. Variability in the CP-response, due to the experimental variations as well as to the assumptions made, was also taken into consideration by introducing random response noise ranging between 0.90 and 1.10. With those in silico noised responses, covering thus 20% of variability, new datasets were created (MLR1 to MLR10). By performing the MLR-analysis of these datasets (Table 4), the descriptors most robustly influencing the CP-response, i.e. descriptors which were withheld in more than half of the MLR-models, were selected. In Table 5, the meaning of these robust descriptors influencing the cell-penetrating properties are listed.
Table 4. Overview of the most robust descriptors influencing the CP-responses in the 11 MLR-models.
| MLR | MLR1 | MLR2 | MLR3 | MLR4 | MLR5 | MLR6 | MLR7 | MLR8 | MLR9 | MLR10 | Mean | |
| R2 | 0.621 | 0.589 | 0.493 | 0.515 | 0.619 | 0.617 | 0.508 | 0.587 | 0.525 | 0.572 | 0.615 | 0.569 |
| Adjusted R2 | 0.577 | 0.545 | 0.458 | 0.478 | 0.578 | 0.572 | 0.471 | 0.542 | 0.487 | 0.532 | 0.567 | 0.528 |
| Descriptor | Coefficients 1 | # | ||||||||||
| B04[N-N] | 0.175 | 0.285 | 0.287 | 0.298 | 0.228 | 0.154 | 0.183 | 0.251 | 0.203 | 0.305 | 0.187 | 11 |
| GATS5m | 0.401 | 0.573 | 0.321 | 0.298 | 0.541 | 0.435 | 0.389 | 0.443 | 0.396 | 0.612 | 0.670 | 11 |
| G2e | −0.184 | −0.141 | −0.186 | −0.221 | −0.205 | −0.186 | −0.218 | −0.226 | −0.215 | −0.177 | −0.181 | 11 |
| nCt | 0.465 | 0.482 | 0.570 | 0.547 | – | 0.453 | 0.491 | 0.588 | 0.555 | 0.215 | – | 9 |
| nROR | 0.244 | 0.198 | – | – | 0.322 | 0.231 | – | – | – | 0.300 | 0.320 | 6 |
| T(N.S) | 0.912 | 0.461 | – | – | 0.897 | 0.940 | – | – | – | 0.799 | 0.607 | 6 |
| G3u | −0.184 | −0.137 | – | – | −0.224 | −0.183 | – | – | – | −0.190 | – | 5 |
| Mp | 0.548 | 0.352 | – | – | 0.525 | 0.553 | – | 0.307 | – | – | – | 5 |
| Mor15p | −0.656 | – | – | – | −0.791 | −0.673 | – | – | – | −0.366 | −0.275 | 5 |
| Mor26m | −0.319 | – | −0.202 | −0.209 | −0.361 | −0.318 | – | – | −0.191 | −0.305 | – | 7 |
| GATS7e | – | – | 0.922 | 1.066 | – | – | 1.127 | 1.233 | 1.143 | – | – | 5 |
| GATS7p | – | – | −0.682 | −0.761 | – | – | −0.798 | −0.944 | −0.806 | – | – | 5 |
| Mor16p | – | −0.316 | −0.385 | −0.419 | – | – | −0.478 | −0.482 | −0.391 | – | −0.301 | 7 |
| Mor27m | – | – | −0.410 | −0.404 | – | – | −0.327 | −0.298 | −0.387 | – | – | 5 |
| Mor27e | – | – | 0.248 | 0.291 | – | – | 0.202 | 0.217 | 0.274 | – | – | 5 |
For each model, the coefficients of the significant descriptors are indicated.
Table 5. Meanings of the robust descriptors influencing significantly the CP-response of peptides.
| Descriptor | Meaning | Class |
| B04[N-N] | Presence/absence of N-N at topological distance 4 | 2D binary fingerprints |
| GATS5m | Geary autocorrelation - lag 5/weighted by atomic masses | 2D autocorrelations |
| G2e | 2st component symmetry directional WHIM index/weighted by atomic Sanderson electronegativities | WHIM1 descriptors |
| nCt | Number of total tertiary C(sp3) | Functional group counts |
| nROR | Number of ethers (aliphatic) | Functional group counts |
| T(N.S) | Sum of topological distances between N.S | Topological descriptors |
| G3u | 3st component symmetry directional WHIM index/unweighted | WHIM descriptors |
| Mp | Mean atomic polarizability (scaled on Carbon atom) | Constitutional descriptors |
| Mor15p | 3D-MoRSE - signal 15/weighted by atomic polarizabilities | 3D-MoRSE2 descriptors |
| Mor26m | 3D-MoRSE - signal 26/weighted by atomic masses | 3D-MoRSE2 descriptors |
| GATS7e | Geary autocorrelation - lag 7/weighted by atomic Sanderson electronegativities | 2D autocorrelations |
| GATS7p | Geary autocorrelation - lag 7/weighted by atomic polarizabilities | 2D autocorrelations |
| Mor16p | 3D-MoRSE - signal 16/weighted by atomic polarizabilities | 3D-MoRSE2 descriptors |
| Mor27m | 3D-MoRSE - signal 27/weighted by atomic masses | 3D-MoRSE2 descriptors |
| Mor27e | 3D-MoRSE - signal 27/weighted by atomic Sanderson electronegativities | 3D-MoRSE2 descriptors |
Weighted Holistic Invariant Molecular descriptors.
23D-Molecular Representation of Structures based on Electron diffraction.
The descriptor B04[N-N] is a 2D-binary fingerprint descriptor, representing the presence or absence of the specific atom pair N-N at a topological distance of four bonds. Our models indicated that the presence of such a N-N pair has a positive influence on the cell-penetrating response. When looking at the amino acid structures, this N-N bond at topological distance four is found in asparagine and histidine residues. The latter is a weak alpha-helix former and thus may be important to establish the secondary amphipathic structure of peptides [70]. The GATS5m, GATS7p and GATS7e descriptors are Geary 2D-autocorrelation descriptors, which describe the topology of the peptide in association with atomic masses (m), polarizabilities (p) and Sanderson electronegativities (e). At specific path length (lag) five, the atomic masses have a high positive contribution to the cell-penetrating properties, while at lag seven, a positive (weighted by atomic Sanderson electronegativities) or negative (weighted by atomic polarizabilities) influence on our CP-response was observed. GATS7e shows the dispersion of electronegative atoms at a topological distance equal to seven bonds in a peptide, while the value of GATS7p shows the importance of atomic polarizabilities over the same topological distance. Peptides having high (GATS5m and GATS7e) or low (GATS7p) values of these descriptors, were rich in basic amino acids, arginine and lysine, as well as the aromatic amino acid tryptophan.
3D-Molecule Representation of Structures based on Electron diffraction (3D-MoRSE) descriptors are 3D-molecular descriptors derived from scattering transform functions, reflecting various physicochemical properties, like atomic polarizability (signals 15 and 16), atomic masses (signals 26 and 27) and atomic electronegativity (signal 27) [71]. From these 3D-MoRSE descriptors could be derived that the position of these physicochemical properties in the 3D-space is crucial for cell-penetrating properties. Based on these descriptors, a favourable cellular influx was predicted for the amphipathic and/or cationic subclusters of the PCA-analysis, i.e. the dark green, pink, purple and yellow subclusters. Moreover, the peptides belonging to the dark green and yellow subclusters showed the highest median CP-response, which was also predicted based on their values of the robust 3D-MoRSE descriptors. 3D-descriptors characterizing the symmetry of the peptides also robustly influenced the CP-response: the symmetry-directional WHIM descriptors G2e (weighted by atomic Sanderson electronegativities) and G3u (unweighted) negatively influenced the cell-penetrating properties, indicating that the cellular influx of peptides increased with decreasing peptide symmetry [71]. Peptides containing branched and hydrophobic amino acids, e.g. valine, leucine and isoleucine, as indicated by the descriptor nCt, accounting for the number of tertiary carbon atoms showed a higher CP-response. Also the T(N.S) descriptor referring to the presence of sulfur-containing amino acids, and the mean atomic polarizability (Mp) contributed positively to the cellular penetration. Methionine as well as the hydrophobic amino acids are also (strong) alpha-helix formers and thus important for establishing a secondary amphipathic structure. Finally, the nROR descriptor, which was an unexpected robust descriptor, also positively influenced the CP-response. The cationic amphiphilic polyproline helices (CAPHs) contain such ether functions to link the hydrophobic and hydrophilic residues. Although the MLR-analysis did not directly point to the importance of a positive charge for cellular uptake, the information contained in the robust descriptors indicated its influence as well as of a secondary amphipathic structure.
Discussion
Studies of the cellular uptake of cell-penetrating peptides demonstrate a great variety in experimental conditions, as illustrated in Table 1. These differences in used techniques and operational parameters, are at least partly responsible for discrepancies in conclusions about the cellular uptake of certain CPPs, like e.g. the uptake mechanism. In Table S3, the available information on the mechanism of cellular uptake of our selected peptides is listed. There are three main mechanisms of cellular entry: (1) direct penetration, wich can be subdivided into (a) inverted micelle formation, (b) pore formation, (c) carpet-like model, (d) membrane thinning and (e) nucleation zones. The second mechanism is (2) endocytosis, with subcategories (a) micropinocytosis, (b) dependent on coat proteins and (c) independent on coat proteins. Some publications also define a third mechanism: (3) energy-dependent, but not endocytosis [2], [12], [72], [73]. From Table S3 can be derived that the different studies on the uptake mechanism of CPPs show an inconsistency in cellular uptake mechanism. Cell-penetrating peptides use different mechanisms of entry, either simultaneously or as function of experimental factors, like the extracellular concentration, cell line, presence of a cargo, incubation time and temperature [2], [42], [44].
Clearly, there is an urgent need for harmonization of the experimental conditions in the investigations of cellular uptake of peptides, like other authors have already suggested in the past [18], [20]. Especially, the use of a standard positive control or controls, e.g. penetratin, is recommended, as it allows to neutralize to some extent the differences in experimental conditions. Therefore, we defined a CP-response, a unified response which allows the comparison of experimental data of the cellular influx of peptides. Several assumptions were made, which cause, together with the existing experimental variations, some variability in our CP-response. Nevertheless, the hitherto described cell-penetrating peptides can be compared using this CP-response and new conclusions about the structure-activity modeling of these peptides can be drawn.
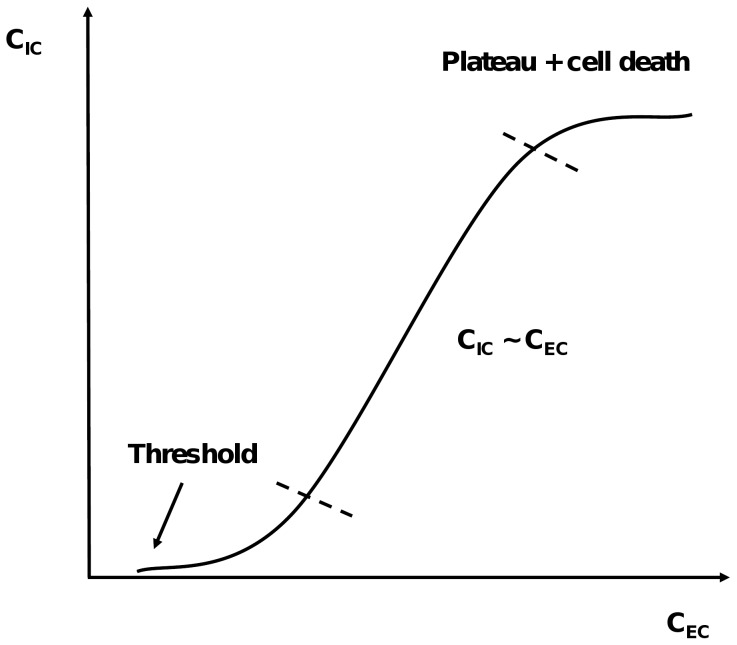
As a first assumption, cell and label differences were neglected, as a wide range of cell lines and detection labels are used throughout the literature. It is clear that different cell lines have different membrane characteristics, which influence the cell-penetrating properties [17], [22], [25], [27], [30], [32]–[34], [37], [38], [40], [41], [43]–[45], [53], [55], [56], [58], [64]–[67]. We also assumed penetratin as a general positive control, because it is quite often used and is well characterized, being one of the first described CPPs. It was also necessary to correct the uptake responses for the incubation concentration, as there exists a clear relationship between the extracellular and intracellular concentration of CPPs. Therefore, we assumed a simple linear relationship, justified by the fact that only a few studies have already investigated the internalization dependence on the extracellular peptide concentration, not allowing more complex models to be used. For most CPPs, there is indeed a correlation between the intracellular and the extracellular concentration [37], [39], [41], [42], [60]. On the other hand, some peptides, like R9, hLF and Tat 47–57, show a sudden sharp increase in intracellular concentration, when a certain extracellular concentration is reached [41], [42]. Still for other peptides, the extracellular concentration needs to exceed a threshold concentration before cellular uptake takes place. Some authors explain this phenomenon by the fact that the uptake mechanism of CPPs depends on the extracellular concentration [42]. Moreover, Hällbrink et al. [74] showed that the uptake of CPPs may also be dependent on the peptide-to-cell ratio, as demonstrated for MAP and penetratin. Besides, some CPPs show toxic effects starting from a certain extracellular concentration [37], [39]. Taking the above findings in consideration, we visualized the intracellular versus extracellular concentration curve for CPPs as a sigmoid (see Figure 3), characterized by a threshold value for influx, which was for all available peptide data about 1 µM. When the threshold is reached, the intracellular concentration increases in function of the extracellular concentration, followed by flattening of the curve until a plateau value for intracellular concentration is reached, possibly due to cell death. The threshold value for influx is CPP and cell line dependent. For most CPPs however, only one extracellular concentration is investigated, which makes it impossible to reconstruct the full sigmoid curve dependence. We applied a linear model, realizing that this approach is an over-simplification, leading to increased variability and bias. It is clear that studying the correlations between intracellular and extracellular concentration, would give more insights into the uptake mechanisms of the peptides, as well as into the toxicity profile.
Figure 3. Supposed dependence of the intracellular CPP concentration on the extracellular concentration when performing cellular influx studies.
Our dataset contained peptides showing very low to high cellular influx (CP-response of 0.001378 to 2.744), indicating that our dataset covered a sufficiently wide range of cell-penetrating responses. Moreover, the ranking of the peptides based on the CP-responses, corresponds roughly with those found in the literature, when considering the most studied and compared CPPs. This indicates that our approach is a valuable quantitative way to assess CPP properties, which was also demonstrated by the evaluation of the distribution of the CP-responses in the five different classes of CPPs as defined by the authors. From Figure 1 can be derived that the medium CP-response increases over the different classes from no CPP to high CPP. Still there exists a clear overlap in CP-responses between the different classes. The lower whiskers of the distribution of the medium CPP, CPP and high CPP classes are extended to almost zero response, indicating that they also contain non- or low-penetrating peptides, according to our proposed CP-response. We evaluated the peptides composing these lowest values and concluded that they can often be explained by an incorrect descriptive conclusion of the authors. Possible reasons are that the classification was based on experiments without trypsinization, while also experiments with trypsinization of the cells were performed, or that much higher incubation concentrations than normally applied are used in order to reach cell-penetration, leading to low CP-responses as they are concentration corrected [17], [51]. Nevertheless, this observed consistency strengthens the value of our CP-response.
The exploration of the chemical space of the 186 peptides, investigated for cell-penetrating properties, confirmed some known features about CPPs, thus supporting our approach, but also revealed some new insights in the structural diversity of these peptides. The molecular weight, surface area, molecular volume, the number of hydrogen bond acceptors, hydrophobicity and charge determined the main clusters in the PCA-analysis. These characteristics join with previous findings about important properties for cellular influx, i.a. z-scales used by Hällbrink et al. [19]. However, our PCA-analysis indicated that also the shape and complexity of the structure differ within the group of CPPs. In the score plot of the PCA-analysis (Figure 2), there was a clear trend in symmetry, complexity and compactness of the structure: extremes for these descriptors give low CP-responses for the peptides. From this exploration of the chemical space of CPPs, it can be derived that not only the constituent amino acids determine cell-penetrating properties but also their position. This contrasts the current general opinion that the 3D-structure is not significantly influencing the cellular uptake, except for the secondary amphipathic CPPs [6]. Moreover, our 3D-structures are calculated based on a theoretical phase, i.e. MM+ in vacuo optimized structures according to Hyperchem molecular mechanics, which is independent from its biological medium and interactions.
The light green cluster in Figure 2 consists of oligo-arginines of up to five arginines and shows a very low to negligible CP-response, consistent with the conclusions of Mitchell et al. [12]. On the other hand, based on the characteristics of the clusters with the highest unified response, high density of positive charges and amphipathicity favour cellular uptake. The amphipathic peptides were located centrally in the score plot of the PCA-analysis and were characterized by a high extent of sulfur-containing residues, as well as aromatic amino acids. These features are indeed important for establishing a secondary amphipathic structure. According to Chou and Fasman, methionine and the aromatic amino acids, phenylalanine and tryptophan, are (strong) alpha-helix formers and as hydrophobic amino acids they contribute to hydrophobic interactions when establishing the secondary structure [70].
Although MLR only captures a linear correlation between descriptors [21], it gives us valuable information about which descriptors influence cellular uptake. By adding 20% noise around our calculated CP-response, we included the expected variability of the CP-responses, caused by experimental variations as well as by our assumptions. We evaluated the most robust descriptors, i.e. those descriptors which were incorporated in more than half of the obtained MLR-models. This MLR-analysis revealed that a positive charge, represented by the basic amino acids arginine and lysine, and an amphipathic structure are discriminating properties for cellular influx of peptides. We also identified the symmetry and the compactness of the peptide structure as determining. Furthermore, the 3D-MoRSE descriptors indicate that certain patterns in the molecular structure influence whether a peptide is efficiently cell-penetrating or not. This refers to an amphipathic structure or more in general to recurrent functional groups, like e.g. the guanidinium group of arginine. Indeed, based on the 3D-MoRSE descriptors, a favoured cellular influx is predicted for the amphipathic peptides. The results of the MLR-analysis correspond well with the identified important features for cellular uptake during the exploration of the chemical space of the 186 peptides.
Cell-penetrating peptides form a chemically diverse group of peptides, as we demonstrated during the PCA-analysis, and can be classified in three chemically different groups according to Milletti [6]: (1) cationic CPPs (C), which contain a stretch of positive charges and their 3D-structure is not an amphipathic helix. (2) Amphipathic CPPs (A), which are characterized by a hydrophobic and hydrophilic part by adapting a helix structure. Amphipathic peptides may have a cationic nature (AC) or their hydrophilic part can be neutral, anionic or polar (A). The (3) hydrophobic CPPs (H) are peptides containing only apolar residues, with low net charge or have hydrophobic amino acid groups that are crucial for cellular uptake. Hydrophobic CPPs may also have a cationic (CH) or amphipathic nature (AC). In Table S3, the chemical classes of the individual peptides of our dataset are listed and are schematically visualized in Figure 4. Using this chemical classification method, there is a clear overlap demonstrated for the different classes, especially for the amphipathic-cationic peptides.
Figure 4. Schematic representation of the main CPP chemical classes from our dataset.
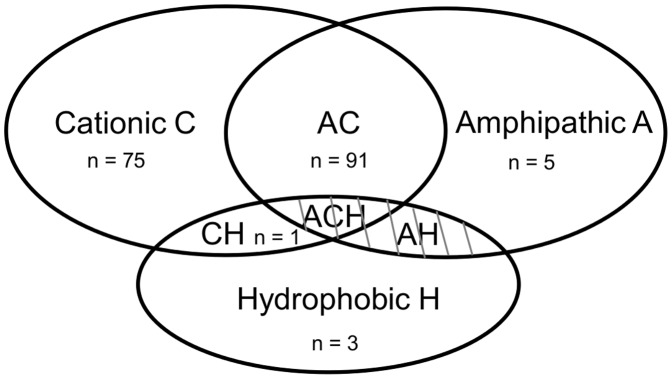
We believe that our CP-response, as a more objective and quantitative measure for cellular penetration, will foster the discussion of the cellular uptake mechanisms, as well as the definition and the classification of the CPPs.
Conclusion
When gathering quantitative data for cellular influx of peptides, it was clear that harmonization of these studies is highly needed. By defining a cell-penetrating response, the quantitative evaluation of the cellular influx characteristics of 186 peptides was possible. This CP-response, together with chemo-molecular descriptors of the peptides, was used to explore the chemical-functional space of CPPs. Our study indicated that besides already reported CPP-determing features, like i.a. positive charge and amphipathicity, also the shape, complexity and compactness of the structures, play an import role for influx into the cell. As our CP-response is a more objective and quantitative measure for cellular penetration of peptides, it will help to classify these peptides, to unravel the different uptake mechanisms, as well as to establish a common evaluation tool.
Supporting Information
Score plot of the first versus the second principal component of the PCA-analysis of 186 peptides after dividing their descriptors by the molecular weight. The colors of the clusters correspond with the clusters found in the score plot of the PCA-analysis using the original descriptors (Figure 2). For each cluster, some examples of peptides are indicated.
(PDF)
Overview of the 186 (non-) CPPs, including their CP-response.
(PDF)
List of cell-penetrating peptides whose CP-response is an outlier.
(PDF)
Classification of (non-) CPPs based on chemical class, literature data and their uptake mechanisms.
(PDF)
Summary of the PCA-analysis of the descriptors divided by the molecular weight, describing the eigenvalues of the covariance matrix, the total variance explained (cumulative R2) and the predictive ability (cumulative Q2).
(PDF)
Funding Statement
This work was supported by the Special Research Fund (BOF) of Ghent University (http://www.ugent.be/en/research/funding/phd/bof/fjd/jointdoctorate.htm) to SS (01D38811) and EW (01J22510); Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen: http://www.iwt.be/english/funding) to BG (121512) and MD (101529). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Frankel AD, Pabo CO (1988) Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55: 1189–1193. [DOI] [PubMed] [Google Scholar]
- 2.Madani F, Lindberg S, Langel Ü, Futaki S, Gräslund A (2011) Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. J Biophys DOI: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed]
- 3. Jarver P, Langel Ü (2006) Cell-penetrating peptides – A brief introduction. BBA-Biomembranes 1758: 260–263. [DOI] [PubMed] [Google Scholar]
- 4.Lindgren M, Langel Ü (2011) Classes and Prediction of Cell-Penetrating Peptides. In: Langel Ü, editor. Cell-Penetrating Peptides: Methods and Materials. New York: Humana Press. 3–19. [DOI] [PubMed]
- 5. Vergote V, Burvenich C, Van de Wiele C, De Spiegeleer B (2009) Quality specifications for peptide drugs: a regulatory-pharmaceutical approach. J Pept Sci 15: 697–710. [DOI] [PubMed] [Google Scholar]
- 6. Milletti F (2012) Cell-penetrating peptides: classes, origin, and current land scape. Drug Discov Today 17: 850–860. [DOI] [PubMed] [Google Scholar]
- 7. Elmquist A, Hansen M, Langel Ü (2006) Structure-activity relationship study of the cell-penetrating peptide pVEC. BBA-Biomembranes 1758: 721–729. [DOI] [PubMed] [Google Scholar]
- 8. Fischer PM, Zhelev NZ, Wang S, Melville JE, Fåhraeus R, et al. (2000) Structure-activity relationship of truncated and substituted analogues of the intracellular delivery vector Penetratin. J Peptide Res 55: 163–172. [DOI] [PubMed] [Google Scholar]
- 9. Drin G, Mazel M, Clair P, Mathieu D, Kaczorek M, et al. (2001) Physico-chemical requirements for cellular uptake of pAntp peptide: Role of lipid-binding affinity. Eur J Biochem 268: 1304–1314. [DOI] [PubMed] [Google Scholar]
- 10. Soomets U, Lindgren M, Gallet X, Hällbrink M, Elmquist A, et al. (2000) Deletion analogues of transportan. BBA-Biomembranes 1467: 165–176. [DOI] [PubMed] [Google Scholar]
- 11. Song J, Kai M, Zhang W, Zhang J, Liu L, et al. (2011) Cellular uptake of transportan 10 and its analogs in live cells: Selectivity and structure-activity relationship studies. Peptides 32: 1934–1941. [DOI] [PubMed] [Google Scholar]
- 12. Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB (2000) Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res 56: 318–325. [DOI] [PubMed] [Google Scholar]
- 13. Scheller A, Oehlke J, Wiesner B, Dathe M, Krause E, et al. (1999) Structural Requirements for Cellular Uptake of α-Helical Amphipathic Peptides. J Pept Sci 5: 185–194. [DOI] [PubMed] [Google Scholar]
- 14. Vivès E, Brodin P, Lebleu B (1997) A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J Biol Chem 272: 16010–16017. [DOI] [PubMed] [Google Scholar]
- 15. Vivès E, Granier C, Prevot P, Lebleu B (1997) Structure-activity relationship study of the plasma membrane translocating potential of a short peptide from HIV-1 Tat protein. Lett Pept Sci 4: 429–436. [Google Scholar]
- 16. Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, et al. (2000) The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. P Natl Acad Sci USA 97: 13003–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mueller J, Kretzschmar I, Volkmer R, Boisguerin P (2008) Comparison of Cellular Uptake Using 22 CPPs in 4 Different Cell Lines. Bioconjugate Chem 19: 2363–2374. [DOI] [PubMed] [Google Scholar]
- 18. Hansen M, Kilk K, Langel Ü (2008) Predicting cell-penetrating peptides. Adv Drug Deliver Rev 60: 572–579. [DOI] [PubMed] [Google Scholar]
- 19. Hällbrink M, Kilk K, Elmquist A, Lundberg P, Lindgren M, et al. (2005) Prediction of Cell-Penetrating Peptides. Int J Pept Res Ther 11: 249–259. [Google Scholar]
- 20. Karelson M, Dobchev D (2011) Using artificial neural networks to predict cell-penetrating compounds. Expert Opin Drug Dis 6: 783–796. [DOI] [PubMed] [Google Scholar]
- 21. Sanders WS, Johnston CI, Bridges SM, Burgess SC, Willeford KO (2011) Prediction of Cell-Penetrating Peptides by Support Vector Machines. PLOS Comput Biol 7: e1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richard JP, Melikov K, Vivès E, Ramos C, Verbeure B, et al. (2003) Cell-penetrating Peptides: A reevaluation of the mechanism of cellular uptake. J Biol Chem 278: 585–590. [DOI] [PubMed] [Google Scholar]
- 23. D’Hondt M, Gevaert B, Stalmans S, Van Dorpe S, Wynendaele E, et al. (2013) Reversed-phase fused-core HPLC modeling of peptides. J Pharm Anal 3: 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, et al.. (2006) Multi- and Megavariate Data Analysis: Part I – Basic Principles and Applications, Second revised and enlarged edition. Umeå: Umetrics AB. 39–101.
- 25. Elmquist A, Lindgren M, Bartfai T, Langel Ü (2001) VE-Cadherin-Derived Cell-Penetrating Peptide, pVEC, with Carrier Functions. Exp Cell Res 269: 237–244. [DOI] [PubMed] [Google Scholar]
- 26. Lundberg P, Langel Ü (2006) Uptake Mechanisms of Cell-Penetrating Peptides Derived from Alzheimer's Disease Associated Gamma-Secretase Complex. Int J Pept Res Ther 12: 105–114. [Google Scholar]
- 27. Holm T, Netzereab S, Hansen M, Langel Ü, Hällbrink M (2005) Uptake of cell-penetrating peptides in yeasts. FEBS Lett 579: 5217–5222. [DOI] [PubMed] [Google Scholar]
- 28. Lindgren ME, Hällbrink MM, Elmquist AM, Langel Ü (2004) Passage of cell-penetrating peptides across a human epithelial cell layer in vitro . Biochem J 377: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palm C, Jayamanne M, Kjellander M, Hällbrink M (2007) Peptide degradation is a critical determinant for cell-penetrating peptide uptake. BBA-Biomembranes 1768: 1769–1776. [DOI] [PubMed] [Google Scholar]
- 30. El-Andaloussi S, Järver P, Johansson HJ, Langel Ü (2007) Cargo-dependent cytotoxicity and delivery efficacy of cell-penetrating peptides: a comparative study. Bioch J 407: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burlina F, Sagan S, Bolbach G, Chassaing G (2005) Quantification of the Cellular Uptake of Cell-Penetrating Peptides by MALDI-TOF Mass Spectrometry. Angew Chem Int Edit 44: 4244–4247. [DOI] [PubMed] [Google Scholar]
- 32. El-Andaloussi S, Johansson HJ, Holm T, Langel Ü (2007) A Novel Cell-Penetrating Peptide, M918, for Efficient Delivery of Proteins and Peptide Nucleic Acids. Mol Ther 15: 1820–1826. [DOI] [PubMed] [Google Scholar]
- 33. Fischer R, Köhler K, Fotin-Mleczek M, Brock R (2004) A Stepwise Dissection of the Intracellular Fate of Cationic Cell-penetrating Peptides. J Biol Chem 279: 12625–12635. [DOI] [PubMed] [Google Scholar]
- 34. Jones SW, Christison R, Bundell K, Voyce CJ, Brockbank SMV, et al. (2005) Characterisation of cell-penetrating peptide-mediated peptide delivery. Brit J Pharmacol 145: 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walrant A, Correia I, Jiao CY, Lequin O, Bent EH, et al. (2011) Different membrane behaviour and cellular uptake of three basic arginine-rich peptides. Biochim Biophys Acta 1808: 382–393. [DOI] [PubMed] [Google Scholar]
- 36. Maiolo JR, Ferrer M, Ottinger EA (2005) Effects of cargo molecules on the cellular uptake of arginine-rich cell-penetrating peptides. BBA-Biomembranes 1712: 161–172. [DOI] [PubMed] [Google Scholar]
- 37. Oehlke J, Scheller A, Wiesner B, Krause E, Beyermann M, et al. (1998) Cellular uptake of an α-helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior non-endocytically. BBA- Biomembranes 1414: 127–139. [DOI] [PubMed] [Google Scholar]
- 38. Sugita T, Yoshikawa T, Mukai Y, Yamanada N, Imai S, et al. (2008) Comparative study on transduction and toxicity of protein transduction domains. Brit J Pharmacol 153: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Drin G, Cottin S, Blanc E, Rees AR, Temsamani J (2003) Studies on the Internalization Mechanism of Cationic Cell-Penetrating Peptides. J Biol Chem 278: 31192–31201. [DOI] [PubMed] [Google Scholar]
- 40. Johansson HJ, El-Andaloussi S, Holm T, Mäe M, Jänes J, et al. (2008) Characterization of a Novel Cytotoxic Cell-Penetrating Peptide Derived From p14ARF Protein. Mol Ther 16: 115–123. [DOI] [PubMed] [Google Scholar]
- 41. Duchardt F, Ruttekolk IR, Verdurmen WPR, Lortat-Jacob H, Bürck J, et al. (2009) A Cell-penetrating Peptide Derived from Human Lactoferrin with Conformation-dependent Uptake Efficiency. J Biol Chem 284: 36099–36108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R (2007) A Comprehensive Model for the Cellular Uptake of Cationic Cell-penetrating Peptides. Traffic 8: 848–866. [DOI] [PubMed] [Google Scholar]
- 43. Verdurmen WPR, Bovee-Geurts PH, Wadhwani P, Ulrich AS, Hällbrink M, et al. (2011) Preferential Uptake of L- versus D-Amino Acid Cell-Penetrating Peptides in a Cell Type-Dependent Manner. Chem Biol 18: 1000–1010. [DOI] [PubMed] [Google Scholar]
- 44. Alves ID, Bechara C, Walrant A, Zaltsman Y, Jiao CY, et al. (2011) Relationships between Membrane Binding, Affinity and Cell Internalization Efficacy of a Cell-Penetrating Peptide: Penetratin as a Case Study. Plos One 6: e24096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jones S, Howl J (2011) Enantiomer-Specific Bioactivities of Peptidomimetic Analogues of Mastoparan and Mitoparan: Characterization of Inverso Mastoparan as a Highly Efficient Cell Penetrating Peptide. Bioconjugate Chem 23: 47–56. [DOI] [PubMed] [Google Scholar]
- 46. Jones S, Holm T, Mäger I, Langel Ü, Howl J (2010) Characterization of Bioactive Cell Penetrating Peptides from Human Cytochrome c: Protein Mimicry and the Development of a Novel Apoptogenic Agent. Chem Biol 17: 735–744. [DOI] [PubMed] [Google Scholar]
- 47. Paramelle D, Subra G, Vezenkov LL, Maynadier M, André C, et al. (2010) A Straightforward Approach for Cellular-Uptake Quantification. Angew Chem Int Edit 49: 8240–8243. [DOI] [PubMed] [Google Scholar]
- 48. Fischer R, Waizenegger T, Köhler K, Brock R (2002) A quantitative validation of fluorophore-labelled cell-permeable peptide conjugates: fluorophore and cargo dependence of import. BBA- Biomembranes 1564: 365–374. [DOI] [PubMed] [Google Scholar]
- 49. Kalafut D, Anderson TN, Chmielewski J (2012) Mitochondrial targeting of a cationic amphiphilic polyproline helix. Bioorg Med Chem Lett 22: 561–563. [DOI] [PubMed] [Google Scholar]
- 50. Geisler IM, Chmielewski J (2011) Dimeric Cationic Amphiphilic Polyproline Helices for Mitochondrial Targeting. Pharm Res 28: 2797–2807. [DOI] [PubMed] [Google Scholar]
- 51. Gomez JA, Chen J, Ngo J, Hajkova D, Yeh IJ, et al. (2010) Cell-Penetrating Penta-Peptides (CPP5s): Measurement of Cell Entry and Protein-Transduction Activity. Pharmaceuticals 3: 3594–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wada S-i, Tsuda H, Okada T, Urata H (2011) Cellular uptake of Aib-containing amphipathic helix peptide. Bioorg Med Chem Lett 21: 5688–5691. [DOI] [PubMed] [Google Scholar]
- 53. Palm C, Netzereab S, Hällbrink M (2006) Quantitatively determined uptake of cell-penetrating peptides in non-mammalian cells with an evaluation of degradation and antimicrobial effects. Peptides 27: 1710–1716. [DOI] [PubMed] [Google Scholar]
- 54. Neundorf I, Rennert R, Hoyer J, Schramm F, Löbner K, et al. (2009) Fusion of a Short HA2-Derived Peptide Sequence to Cell-Penetrating Peptides Improves Cytosolic Uptake, but Enhances Cytotoxic Activity. Pharmaceuticals 2: 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takayama K, Nakase I, Michiue H, Takeuchi T, Tomizawa K, et al. (2009) Enhanced intracellular delivery using arginine-rich peptides by the addition of penetration accelerating sequences (Pas). J Control Release 138: 128–133. [DOI] [PubMed] [Google Scholar]
- 56. Nakase I, Hirose H, Tanaka G, Tadokoro A, Kobayashi S, et al. (2009) Cell-surface Accumulation of Flock House Virus-derived Peptide leads to Efficient Internalization via Micropinocytosis. Mol Ther 17: 1868–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Walther C, Ott I, Gust R, Neundorf I (2009) Specific Labeling With Potent Radiolabels Alters the Uptake of Cell-Penetrating Peptides. Biopolymers 92: 445–451. [DOI] [PubMed] [Google Scholar]
- 58. Manceur A, Wu A, Audet J (2007) Flow cytometric screening of cell-penetrating peptides for their uptake into embryonic and adult stem cells. Anal Biochem 364: 51–59. [DOI] [PubMed] [Google Scholar]
- 59. Foged C, Franzyk H, Bahrami S, Frokjaer S, Jaroszewski JW, et al. (2008) Cellular uptake and membrane-destabilising properties of α-peptide/β-peptoid chimeras: lessons for the design of new cell-penetrating peptides. Biochim Biophys Acta 1778: 2487–2495. [DOI] [PubMed] [Google Scholar]
- 60. Åmand HL, Fant K, Nordén B, Esbjörner EK (2008) Stimulated endocytosis in penetratin uptake: Effect of arginine and lysine. Bioch Bioph Res Com 371: 621–625. [DOI] [PubMed] [Google Scholar]
- 61. Bodor N, Tóth-Sarudy E, Holm T, Pallagi I, Vass E, et al. (2007) Novel, cell-penetrating molecular transporters with flexible backbones and permanently charged side-chains. J Pharm Pharmacol 59: 1065–1076. [DOI] [PubMed] [Google Scholar]
- 62. Östlund P, Kilk K, Lindgren M, Hällbrink M, Jiang Y, et al. (2005) Cell-Penetrating Mimics of Agonist-Activated G-Protein Coupled Receptors. Int J Pept Res Ther 11: 237–247. [Google Scholar]
- 63. Aubry S, Aussedat B, Delaroche D, Jiao CY, Bolbach G, et al. (2010) MALDI-TOF mass spectrometry: A powerful tool to study the internalization of cell-penetrating peptides. BBA- Biomembranes 1798: 2182–2189. [DOI] [PubMed] [Google Scholar]
- 64. Letoha T, Gaál S, Somlai C, Venkei Z, Glavinas H, et al. (2005) Investigation of penetratin peptides. Part 2. In vitro uptake of penetratin and two of its derivatives. J Pept Sci 11: 805–811. [DOI] [PubMed] [Google Scholar]
- 65. Takeshima K, Chikushi A, Lee KK, Yonehara S, Matsuzaki K (2003) Translocation of Analogues of the Antimicrobial Peptides Magainin and Buforin across Human Cell Membranes. J Biol Chem 278: 1310–1315. [DOI] [PubMed] [Google Scholar]
- 66. Myrberg H, Lindgren M, Langel Ü (2007) Protein Delivery by the Cell-Penetrating Peptide YTA2. Bioconjugate Chem 18: 170–174. [DOI] [PubMed] [Google Scholar]
- 67. Martin I, Teixidó M, Giralt E (2011) Design, Synthesis and Characterization of a New Anionic Cell-Penetrating Peptide: SAP(E). ChemBioChem 12: 896–903. [DOI] [PubMed] [Google Scholar]
- 68. Lindgren M, Gallet X, Soomets U, Hällbrink M, Bråkenhielm E, et al. (2000) Translocation Properties of Novel Cell Penetrating Transportan and Penetratin Analogues. Bioconjugate Chem 11: 619–626. [DOI] [PubMed] [Google Scholar]
- 69. Balayssac S, Burlina F, Convert O, Bolbach G, Chassaing G, et al. (2006) Comparison of Penetratin and Other Homeodomain-Derived Cell-Penetrating Peptides: Interaction in a Membrane-Mimicking Environment and Cellular Uptake Efficiency. Biochemistry 45: 1408–1420. [DOI] [PubMed] [Google Scholar]
- 70. Chou PY, Fasman GD (1977) Secondary structural prediction of proteins from their amino acid sequence. Trends Biochem Sci 2: 128–131. [Google Scholar]
- 71.Todeschini R, Consonni V (2003) Descriptors from Molecular Geometry. In: Gasteiger J, editor. Handbook of Chemoinformatics: From Data to Knowledge in 4 Volumes, First edition. Weinheim: Wiley-VCH Verlag GmbH & Co. 1004–1033.
- 72. Trabulo S, Cardoso AL, Mano M, Pedroso de Lima MC (2010) Cell-Penetrating Peptides – Mechanisms of Cellular Uptake and Generation of Delivery Systems. Pharmaceuticals 3: 961–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alves ID, Walrant A, Bechara C, Sagan S (2012) Is There Anybody in There? On The Mechanism of Wall Crossing of Cell Penetrating Peptides. Curr Protein Pept Sci 13: 658–571. [DOI] [PubMed] [Google Scholar]
- 74. Hällbrink M, Oehlke J, Papsdorf G, Bienert M (2004) Uptake of cell-penetrating peptides is dependent on peptide-to-cell ratio rather than on peptide concentration. BBA-Biomembranes 1667: 222–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Score plot of the first versus the second principal component of the PCA-analysis of 186 peptides after dividing their descriptors by the molecular weight. The colors of the clusters correspond with the clusters found in the score plot of the PCA-analysis using the original descriptors (Figure 2). For each cluster, some examples of peptides are indicated.
(PDF)
Overview of the 186 (non-) CPPs, including their CP-response.
(PDF)
List of cell-penetrating peptides whose CP-response is an outlier.
(PDF)
Classification of (non-) CPPs based on chemical class, literature data and their uptake mechanisms.
(PDF)
Summary of the PCA-analysis of the descriptors divided by the molecular weight, describing the eigenvalues of the covariance matrix, the total variance explained (cumulative R2) and the predictive ability (cumulative Q2).
(PDF)