Abstract
Amyloid has been traditionally viewed in the context of disease. However, the emerging concept of `functional amyloid' has taken a new direction into how we view amyloid. Recent studies have identified amyloid fibrils ranging from bacteria to humans that have a beneficial role, instead of being associated with a misfolded state that has been implicated in diseases such as Alzheimer's, Parkinson's and prion diseases. Here, we review our work on two human amyloidogenic polypeptides, one associated with Parkinson's disease, α-synuclein (α-syn), and the other important for melanin synthesis, the repeat domain (RPT) from Pmel17. Particularly, we focused our attention on spectroscopic studies of protein conformation and dynamics and their impact on α-syn amyloid formation and for RPT, we discussed the strict pH dependence of amyloid formation and its role in melanin biosynthesis.
1. Introduction
Since the term `amyloid' was first proposed, which nowadays reflects the mistaken identification of the substance as starch based on crude staining techniques;1 a more contemporary definition can now be used to define amyloid: `a highly-ordered protein aggregate with an unbranched filamentous morphology' These filaments are rich in β-sheet secondary structure that is described as a cross-β fold, where the β-strands are aligned perpendicular to the fibril axis.2 Both functional and pathological amyloids share this hallmark, yet recent advances in high resolution structural techniques show that fibrils do indeed have structural differences on the morphological and atomic level, even ones formed from the same polypeptide sequence.3
Because of their intimate and long time association with human diseases such as Alzheimer's and Parkinson's disease, a central question is then why are functional amyloids benign in the cellular millieux whereas the pathological ones are detrimental to cellular health. Recent studies have suggested that toxicity is attributable to oligomeric or annular intermediates that build-up during amyloid aggregation.4 Thus, one working hypothesis is that functional amyloidogenic polypeptides circumvent such misfolded fibrillar precursors during aggregation. Alternately, their amyloid formation and degradation are efficiently controlled by the cell and therefore, the fibrils are not cytotoxic.5
In this perspective, we begin with a brief historical background on the research efforts in structural determination of amyloid followed by detailed discussions on two specific polypeptides, α-synuclein (α-syn) and the repeat domain (RPT) from Pmel17 that form pathological and functional amyloid in Parkinson's disease and melanin biosynthesis, respectively. In the case of α-syn, we focused our attention on spectroscopic studies of protein conformation and dynamics and their impact on amyloid formation kinetics. For RPT, we discuss the strict pH dependence of amyloid formation and its functional role in the melanosome.
2. Historical background
The word `amyloid' (amylo (starch) + oid (like)) was first introduced in 1854 by Rudolph Virchow because of its reaction with iodine.6 For many years, it was thought that amyloid deposits were lipid or carbohydrate derived, until, in 1859, it was shown that proteins were present in these deposits. In 1875, the use of aniline dyes such as methyl violet was used in the recognition of amyloid (Figure 1). The aniline dye, Congo red was first shown as an amyloid dye by Bennhold in 1922, upon which its binding gave apple-green birefringence when viewed under polarized light.7 In 1959, Vassar and Culling used Thioflavin T (ThT) as a diagnostic tool to detect amyloid deposits.8 To this date, ThT has become a gold standard amongst amyloid fibril detection. Structural studies on amyloid began to emerge in the 1930's with the pioneering x-ray diffraction studies of Astbury and co-workers.9 In 1959, Cohen and Calkins showed that all amyloids gave a non-branching fibrillar structure when viewed under the electron microscope (EM).10 Eanes and Glenner reported in 1968 that x-ray diffraction of fibrils gave a cross-β x-ray pattern, which nowadays is a hallmark of amyloid.11 More recently, great strides have been made in amyloid structural determination on the residue level using solid state nuclear magnetic resonance (ssNMR) and electron paramagnetic resonance (EPR) spectroscopies (see recent reviews2,12–14). These techniques have shown that a parallel-in-register β-sheet structure is the most commonly observed feature for pathological amyloid.15
Fig. 1.
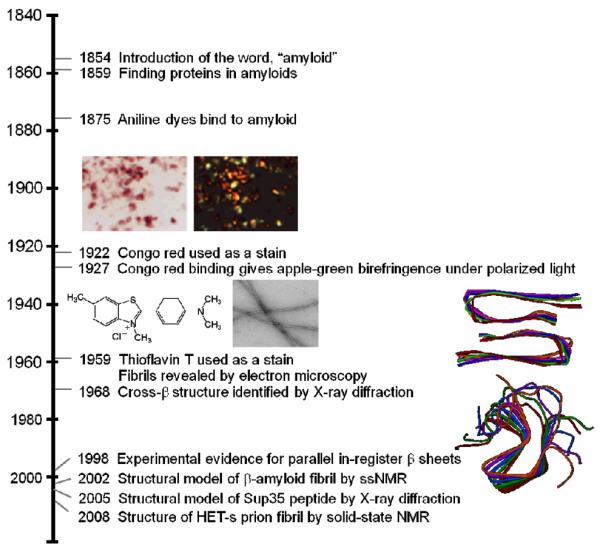
Time line showing important events in the history of the amyloid fibril.
Many of these disease-related amyloids have exhibited structural heterogeneity,3 otherwise known as fibril polymorphism, which has limited detailed atomic-level information. Nevertheless, for the amyloid-β, the peptide implicated in Alzheimer's disease, two distinct fibril structures have been determined by Tycko and coworkers by ssNMR.16,17 Figure 1 shows one variant which gives an in-register parallel β-sheet structure where each molecule has a hairpin conformation and two stacks of molecules are paired with a two-fold symmetry. For functional amyloids, the prion fungal protein, HET-s, gave high quality NMR data, which enabled Meir and coworkers to solve its three-dimensional structure with atomic resolution.18 Here, the β-strands wrap around the fibril axis in a helical arrangement (β-solenoid), and unlike pathological amyloids, parallel strands in helices have intra-molecular backbone hydrogen bonds.
While amyloids have yet to be crystallized and characterized by x-ray crystallography, short amyloidogenic segments from different amyloids have been crystallized and their atomic architecture determined.19–21 From these structures, the β-sheets have an interlocking of self-complementary side chains known as a steric zipper. Indeed, steric zippers could be one of the underlying motifs in stabilization of amyloid structure in certain types of amyloidogenic sequences.
3. α-Synuclein, a pathological amyloid
α-Synuclein and Parkinson's disease
α-Synuclein (α-syn), the protein encoded by the SNCA gene, has been associated with Parkinson's disease (PD) since 1997, when its missense mutation, A53T, was identified to cause early-onset autosomal dominant PD.22 Subsequent discovery of other mutations, A30P23 and E46K,24 as well as gene duplications and triplications25,26 has further bolstered its genetic connection to PD. Importantly, deposits of aggregated, fibrillar α-syn constitute as the major proteinacious material in Lewy bodies (LBs), classical pathological hallmarks of PD. While α-syn is widely implicated in PD, it is noteworthy that it was first identified in 1993 as the precursor protein for the non-β amyloid component (NAC) found in Alzheimer's disease plaques.27
The specific roles of α-syn and amyloid in PD remain controversial. Although some studies suggest LB formation is directly responsible for neurodegeneration,28–30 other reports support that LBs may actually serve a neuroprotective role.31–33 On the other hand, there is compelling evidence to suggest that oligomeric, smaller prefibrillar α-syn as the neurotoxic species (see recent reviews34–36). Nevertheless, these observations underscore the significance of biophysical studies for α-syn structure and dynamics both in solution and during amyloid formation process. Gaining insights on how a soluble, benign α-syn transitions to aggregates will allow elucidation of molecular events in amyloid pathology. In addition, because of α-syn is both cytoplasmic and membrane-associated in vivo, investigation of effects of solution conditions as well as other relevant biomolecules such as phospholipids are also necessary.
α-Synuclein in its amyloid form
Though it has been linked to different biological roles (see recent review37), α-syn, a 140-residue presynaptic protein, is mostly studied for its propensity to form amyloid, the PD-associated conformational state. Like other pathological amyloids, α-syn fibrils also adopt a cross-β structure.38 Established by various experimental techniques,39–41 the fibril core has been identified to contain approximately the central polypeptide region from residues 30 to 100 (Figure 2). Further, NMR chemical shifts measurements suggest that the N-terminal portion is structurally heterogeneous whereas the C-terminal tail is characterized as flexible.42 As indicated from ssNMR,39,42 AFM,43 and TEM studies,44 α-syn fibrils are polymorphic and thus, a high resolution structure remains elusive. However, two independent models have been proposed based on site-directed spin labeling EPR spectroscopy as well as hydrogen/deuterium (H/D) exchange and ssNMR analyses.
Fig. 2.
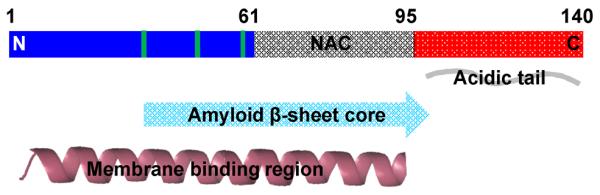
Schematic representation of the primary amino acid sequence of α-syn. Mutations linked to early-onset PD, A30P, E46K, and A53T, are shown in green. The N-terminal (residue 1–60), central region (residue 61–95), and C-terminal (residue 96–140) region has a net charge of positive (blue), neutral and hydrophobic (gray) and negative (red), respectively. Accordingly, α-syn adopts various conformations depending on the solution conditions. In its soluble state, α-syn is intrinsically disordered. Upon amyloid formation, the fibril core region is composed of residue 30–100, while the remaining N- and C-terminal regions are disordered. In the presence of membrane mimics such as acidic micelles and phospholipid vesicles, residues 1–95 bind and adopt α-helical structure.
Using site-directed spin labeling EPR spectroscopy, Langen and coworkers proposed that within the amyloid core region there are two antiparallel, β-strands with residues 62–67 forming the intervening β-turn.40,45 The individual β-strands are organized parallel-in-register and perpendicular to the fibril axis. Based on H/D exchange and ssNMR data, an alternative model was proposed by Riek and coworkers,39 identifying five β-strands comprised of the amyloid core residues, 35–96. The individual β-strands have approximate lengths of 6–8 and 9–13 residues.
Although α-syn fibril core is reasonably well characterized, the dynamics of polypeptide structural rearrangements during amyloid formation are ill-defined. In order to develop a mechanistic understanding of the aggregation process, studies that focus on solution polypeptide dynamics and provide information on the residue-level during aggregation are critical. In the following sections, we will discuss recent work on α-syn polypeptide conformation and dynamics in solution and research efforts in elucidating its aggregation mechanism.
Spectroscopic approaches to conformational mapping of α-synuclein
The α-syn amino acid sequence consists of three regions: the N-terminal region, constituting conserved, amphipathic hexamer motifs, KTKEGV, that can form helices in association with membranes; the central hydrophobic NAC region (residue 61–95), thought to be essential for aggregation; and the C-terminal tail, highly acidic and proline rich (Figure 2). At neutral pH, the N-terminal (residues 1–60), central (residues 61–95) and C-terminal (residues 96–140) region has a net charge of +4, −1, and −12, respectively. The distinct distribution of charges within the protein implies that its conformation is strongly dependent on solution conditions.
In the soluble state, α-syn lacks both secondary and tertiary structure and thus, has been classified as an intrinsically disordered protein (IDP).46 Like many other IDPs, the primary sequence of α-syn contains a high overall charge (−9) and low hydrophobic character.47 While traditional structural determination techniques such as x-ray crystallography and NMR spectroscopy provide little insight into the properties of dynamic and heterogeneous collection of conformations, other spectroscopic techniques such as time-resolved fluorescence energy transfer (trFET)48,49 and single molecule fluorescence spectroscopy,50–52 have been employed to gain residue-specific conformational changes under numerous solution conditions. Specifically, measurements of trFET have provided structural information on pseudo-WT48–52 and A30P α-syn49,51 because energy transfer rates or efficiencies depend on the inverse sixth power of the distance between a fluorescent donor (D) and energy acceptor (A).53 A DA pair is characterized by a critical length (ro) that defines the range of distances (r) that can be probed (0.1ro < r < 1.5ro). In heterogeneous systems, such as unstructured α-syn, analysis of FET kinetics allows the determination of DA distance distributions. Moreover, due to the high sensitivity of this technique it is possible to determine protein conformational changes down to a single polypeptide.50–52
Initial work using a naturally occurring DA pair, tryptophan and 3-nitro-tyrosine (3NY), probability distributions for 6 different pairs were obtained under various solution conditions.49 Indeed, these results revealed a heterogeneous protein ensemble composed of compact, intermediate, and extended conformations.
Based on these data, α-syn clearly behaves as a highly disordered polymer under all solutions examined. However, local conformational rearrangement and population redistribution were observed within the ensemble. For example, under acidic solution, the C-terminal region (94–136) adopts more collapsed structures and in the A30P mutant, pairs proximal to the Pro30 (4–19 and 19–39) indicate that the polypeptide is expanded in the vicinity of Pro30 (Figure 3). Subsequent work by numerous research groups also finds a rich conformational landscape for α-syn as in the case of solution pH52 and in particular, upon micelle50 and phospholipid vesicle binding.54
Fig. 3.
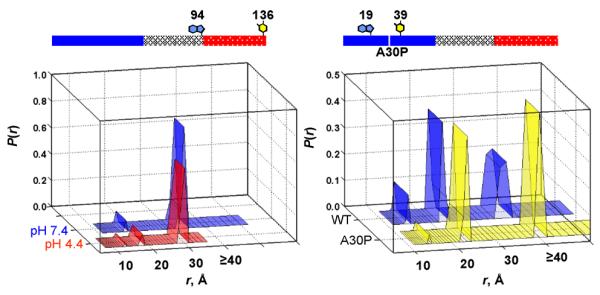
Probability distribution of Trp-to-3NY distances (P(r)) in α-syns extracted from trFET kinetics. Results are shown for the C-terminal pair (Trp94-3NY136) under neutral and acidic solutions (left panel) and a pair proximal (Trp19-3NY39) to the disease-related mutation, A30P at neutral pH (right panel). The experimental limit for distances is ~ 40 Å.
Measurements of α-synuclein tertiary contacts and long range interactions
From various biophysical measurements, it is evident that the α-syn ensemble is quite heterogeneous adopting a distribution of conformers and consequently, these subpopulations are likely dynamic and interchanging. While trFET studies provided ns snapshots of probability distance distributions, the short D excited state lifetime is insufficient for characterization of the time scales (tens of ns to μs) expected for intrachain diffusion and without considerations for dynamics only static distributions can be obtained.49 Therefore, experimental investigations of tertiary contact formation rates55–57 and long-range interactions58–62 have been aimed to elucidate conformational dynamics because large amplitude motions could play a crucial role during the aggregation process that is implicated in pathogenesis.
Determination of biopolymer contact times can be accomplished by exploiting measurements of rates of reactions with very small intrinsic barriers such as electron transfer (ET).55,56,63 By examining ET rates between a powerful electron donor, triplet excited state of W, and an acceptor, 3NY, ten different pairs have been examined in α-syn, probing loop sizes between 10 and 132 residues (Figure 4).55,56 The observed loop contact rates range between 105 and 107 s−1 (200 ns to 1 μs) which do not exhibit a monotonic correlation with loop size as would be expected if α-syn behaved as a random polymer. Interestingly, while rates measured for shorter loops (<20 residues) decrease with increasing size, larger loops level off in the range of 1–5 × 105 s−1 (2 to 10 μs), suggesting that the average distance between W and 3NY is not substantially changing for these loops.
Fig. 4.
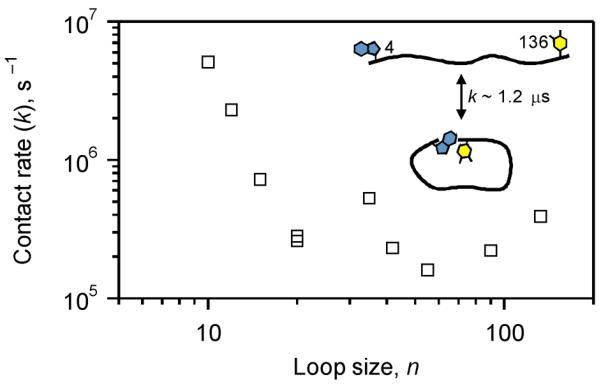
Contact rate constants as a function of loop size measured by 3NY quenching of the Trp triplet excited state in α-syn (20 mM sodium phosphate, pH 7.4, 22 °C).
Numerous NMR studies have provided additional insights into long-range and transient interactions in α-syn in solution.58–62 While inherent heterogeneity and dynamical nature of α-syn are limiting factors for conventional, high resolution structural characterization by NMR such as nuclear Overhauser effects, the introduction of paramagnetic relaxation enhancement (PRE) NMR measurements overcome these difficulties and allow characterization of distances up to 20 Å. Indeed, extremely good agreement was obtained for distance distributions extracted from trFET and ET studies55 and conformational ensembles generated by molecular dynamics simulations using experimental PRE constraints.61 Importantly, detailed structural analyses reveal that there are a number of significant long-range interactions that deviate from a random coil. Particularly, intramolecular contacts have been mapped between residues 120–140 of the C-terminus and central residues 30–100,58 residues 85–95 of the NAC region and residues 110–130 of the C-terminus,59 and N-terminal residues 1–20 and C-terminal residues 121–140.60
It is clear that there are nonrandom structural preferences in distinct regions of α-syn. The current consensus is that the C-terminal region is critical in modulating α-syn conformation due to its highly net negative charge and thus, it is proposed that it plays a role in the initiation and progression of aggregation. Indeed, C-terminally truncated α-syn has greater aggregation propensity and is capable of seeding fibril formation of the full length protein both in vivo and in vitro.64,65 Consistent with this idea, recent PRE data also point to the existence of a transient and weakly intermolecular contact between N- and C-terminus.62
Finally, in a biological context, α-syn is known to interact with more than 30 proteins and particularly, the C-terminal region has been suggested as a protein-protein interaction site, acting as a chaperone under certain physiological circumstances (see recent reviews37,66–68). These observations reiterate the need for continued research efforts in understanding how these interactions could affect the conformation and dynamics of α-syn as well as their potential physiological or pathological effects.
Residue-specific probes of amyloid formation kinetics
Characterization and evaluation of amyloid formation kinetics pose difficult technical challenges.69,70 But, through a combination of protein engineering, biophysical, and biochemical analyses including fluorescence spectroscopy and microscopy, studies of α-syn amyloid formation in a wide range of in vitro71–77 and in vivo78–81 systems are now possible. For example, research efforts in expansion of fluorophore diversity have yielded new amyloid-sensitive spectroscopic probes.74,76,82,83 Despite the high sensitivity and ability to monitor fibrils in a continuous assay, there are potential effects that an extrinsic or covalently-attached fluorophore can have on the aggregation process especially if larger, hydrophobic molecules are chosen. Moreover, while detection of early events (i.e. formation of oligomers) is a common aim as it is pertinent to disease, kinetic details at the residue level during amyloid formation are of equal importance. Therefore, an ideal fluorescent probe must report not just the average progress of aggregation but also able to provide residue-specific information without perturbing the kinetics.
To shed light on the α-syn aggregation mechanism, a recent study assessed and compared the involvement of N- and C-termini as well as residues proximal to amyloid core using site-specific labeling at 4 different sites and an environmentally sensitive fluorophore, dansyl (Dns).77 Because the fluorophore is relatively small and hydrophilic (two fused 6-member rings containing a charged sulfonate group), Dns-variants exhibited nearly identical conformational behavior as the WT protein in its soluble, micelle-binding, and aggregated states. Upon aggregation, spectroscopic changes indicate that all Dns sites are shielded from the aqueous solvent. The hydrophobic microenvironments for Dns7 and Dns136, sites outside the core, are likely due to their participation in interfilament interactions.
For the first time, direct kinetic comparisons showed that residues 7 and 136 exhibit local conformational changes prior to, whereas changes for residues 26 and 100 occur simultaneously with, macroscopic fibril formation. Notably, kinetics obtained for the disordered region (7 and 136) preceded proximal (26 and 100) amyloid core sites suggesting that the amyloid formation pathway may develop initially from the termini followed by residues toward the fibril center. Further evidence from seeded aggregation experiments support that this early event detection is occurring during the elongation process rather than during the nucleation step.
These results can be rationalized because the N- and C-terminal regions are involved in transient interactions in solution, as discussed earlier. In order for α-syn to form parallel-in-register β-strands and adopt a cross β-fold, local structural reorganization must occur at the disordered termini to break the conformational constraints arising from existing long-range intra/inter-molecular contacts (Figure 5). It is worth noting that, using two-dimensional Fourier transform infrared spectroscopy,84 Zanni and coworkers showed that for the human islet amyloid polypeptide implicated in type 2 diabetes, the fibril core exhibits earliest transitions, whereas the N- and C-termini occur later. For these two disease-related amyloids, each appears to have a distinct amyloid folding mechanism. Rather than having the final fold as the determinant for aggregation mechanism, the inherent differences in polypeptide properties such as charge distribution may ultimately dictate the pathway.
Fig. 5.
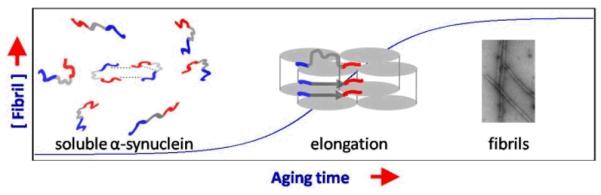
Schematic representation for a proposed aggregation mechanism of α-syn. Soluble α-syn ensemble is consisted of conformers that are heterogeneous, dynamic, and interchanging. Various long range and transient inter/intra-molecular interactions in α-syn have been characterized. The highly acidic C-terminal tail has been suggested to play a key role in modulating α-syn solution conformation. Due to constraints arising from existing long-range interactions in the soluble α-syn, local structural rearrangement at the N-and C-terminus is expected to occur in order to favor α-syn forming a cross β-fold in its amyloid state. Hence, the N- and C-termini experience early environmental changes preceding residues near the amyloid core during the elongation phase.
4. Pmel17, a functional amyloid
Pmel17 and its role in melanin synthesis
Pmel17 is a structural protein important for melanin deposition that forms intralumenal fibrous striations in melanosomes, organelles where melanin is formed and stored.85 Melanin synthesis involves a series of oxidation steps initiated by action of tyrosinase on the substrate tyrosine. Specifically, tyrosine is converted to l-DOPA (L-3,4-dihydroxyphenylalanine), which is readily oxidized to the melanin precursor, indole-5,6-quinone. The subsequent polymerization of these precursors to form melanin is proposed to occur in the presence of the structural protein Pmel17, which acts as a scaffold for depositing melanin.
Pmel17 was first identified genetically in 1930, as the `silver gene' in mice, whose mutation resulted in hypopigmentation.86 It was not until 1991 that the gene was mapped to the silver locus and cloned.87 Pmel17 is a transmembrane protein processed by a series of proteolytic and glycosylation steps.88 Expression of Pmel17 alone in a nonmelanocytic cell line induces filament formation, reminiscent to those seen in melanosomes.89
Melanosome maturation consists of four morphological stages at the ultrastructural level (Figure 6).90 Stages I and II are defined by formation of filaments which intiate during stage I and completed by stage II. Ellipsoidal stage II melanosomes harbor mature filaments that are found in parallel arrays, spanning the length of the organelle. These filaments are approximately 6–10 nm in diameter. Once mature filaments are formed, melanin synthesis begins with enrichment of the enzyme, tyrosinase, catalyzing melanin formation during stages III/IV.
Fig. 6.
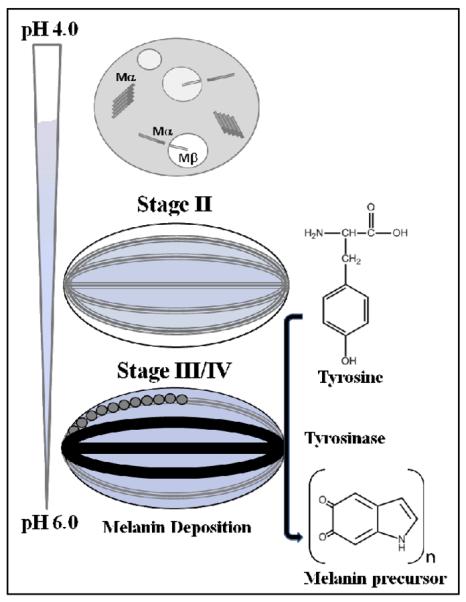
Schematic representation showing the four morphological stages of melanosome maturation. Stage I contains full-length Pmel17, which undergoes membrane cleavage, liberating the Mα from the Mβ transmembrane fragment. Further proteolytic processing of Mα generates pre-fibrillar aggregates. Stage II consists of long fibrillar striations that run in parallel arrays spanning the length of the melanosome. At least two domains, including RPT are proposed to contribute to the fibrillar matrix. Stages III and IV contain melanins, synthesized from tyrosine by the enzyme, tyrosinase. The melanin precursors are depicted as circles and melanin as black polymer deposits on the fibrils. Maturation of the melanosome is accompanied with a pH change, with a gradient ranging from 4.0–6.0.
During melanin synthesis, these fibrous striations act as scaffolds for pigment deposition; EM images reveal that these fibrils grow 2-fold in diameter reaching 14–20 nm. In highly pigmented cells, the internal structure of the melanosome is masked with melanin. Recently, Kelly and coworkers showed that these filaments are composed of amyloid, based on positive staining of melanosome extracts with thioflavin S, a known amyloidogenic dye.91 Moreover, a Pmel17-specific antibody demonstrated colocalization with these amyloid-positive extracts.
It is suggested that at least two domains contribute to the fibrillar matrix, PKD (polycystic kidney disease domain)92 and RPT (repeat domain, Figure 7 top).93 Pmel17 antibodies specific for these two domains decorate the fibrils in vivo. Deletion of the RPT domain abolished its capacity to form the melanosomal matrix fibrils and therefore arrested melanosome maturation.93 A closely related homolog to Pmel17 named GPNMB (glycoprotein nonmetastatic melanoma protein b) lacks an RPT domain and is incapable of forming a fibrillar matrix, suggesting a structural role for RPT.94 In the following sections, we will focus on recent work on the repeat domain of Pmel17 (Pmel17:RPT), highlighting the intimate relationship between solution pH and RPT amyloid formation.
Fig. 7.
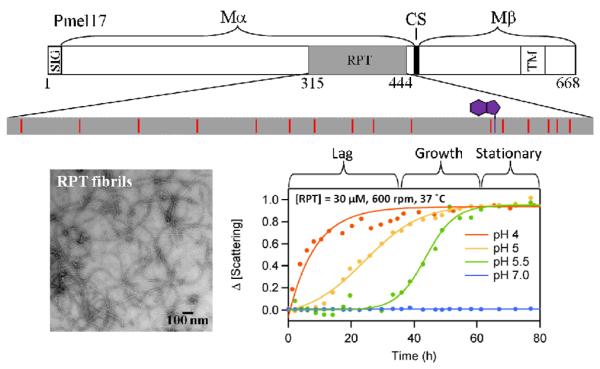
Schematic representation of the primary amino acid sequence of Pmel17 (top). The Mα (residues 1–467) and Mβ (residues 468–669) fragments along with cleavage site (CS) are shown. Processing of Mα forms the fibrillar matrix, which includes RPT. The ionizable side chains, 14 Glu and 1 Asp, (red) in RPT are highlighted. The native tryptophan is shown in purple. EM image of RPT fibrils formed at pH 5.0 (bottom left). pH dependent aggregation of RPT monitored by light scattering is shown (bottom right).
The RPT domain of Pmel17 forms amyloid in vitro and promotes melanin synthesis
RPT, named due to the 10-imperfect repeats rich in proline, serine, threonine and glutamic acid residues,93 is capable of forming amyloid in vitro whereas Pmel17 fragments encompassing other domains failed,95 supporting that this domain comprises the amyloid core in vivo. Importantly, RPT amyloid formation occurs only under mildly acidic conditions (pH 5.0), typical of the melanosomal pH, with no amyloid formation detected at pH 6 or above. RPT aggregates were shown to be filamentous by EM, rich in β-sheet by circular dichroism (CD) spectroscopy, gave enhanced ThT fluorescence, and birefringence upon staining with Congo red, meeting many of the criteria to be classified as amyloid. Moreover, electron diffraction with unoriented fibrils showed a band at 4.5±0.2 Å, indicative of the presence of ordered β-sheets. Preliminary solid-state NMR data also indicated chemical shifts characteristic of a β-sheet rich structure.95
To recapitulate the function of RPT as a scaffold for melanin deposition, an in vitro assay was used that monitored melanin formation. In the presence of RPT fibrils, the activity of tyrosinase and its substrate l-DOPA exhibited a three-fold increase in melanin formation; soluble protein had no detectable effect on activity. Interestingly, other amyloids such as HET-s18 increased the efficiency of melanin synthesis at rates comparable to RPT, suggesting that the common cross-β structure may be sufficient to template the synthesis of melanin in vitro.
pH dependence of RPT amyloid formation
It is known that maturation of the melanosome is associated with a pH change, where stage I is highly acidic (pH ~4.0) and stage III/IV reaching ~pH 6.0 (Figure 6).96,97 To shed light on the pH sensitivity of RPT fibril formation, a combination of techniques that included ThT fluorescence, EM and CD were used to monitor morphological and secondary structural changes as a function of pH.98 Fibril formation showed a sigmoidal kinetics behavior, consisting of a lag, growth, and stationary phase, typically observed in amyloid aggregation (Figure 7 bottom right). Specifically, aggregation occurred from pH 4.0 to 5.5, accelerating under more acidic conditions. Fibrils formed only below 6.0 with no aggregation observed at higher pH. At pH 4.0, RPT rapidly aggregates forming small pre-fibrillar structures that have an amorphous appearance. However, in slightly more alkaline conditions (pH 5.0 – 5.5), long unbranched fibrils are seen with associated differences in kinetics and fibril morphology (Figure 7 bottom left). At pH 5.5, aged RPT fibrils are more homogeneous and reminiscent to those present in stage II melanosomes.
To examine the effect of pH in mediating RPT amyloid formation during melanosome maturation, an in vitro pH titration was performed. Upon titration of preformed RPT aggregates at pH 4.0 to 5.0, prefibrillar structures changed into long striated fibrils. This morphological change is reminiscent of the transition observed in stage I to II melanosomes. Interestingly, further titration to pH 7.0 resulted in almost complete disassembly of the fibrils, supporting a highly reversible aggregation/disaggregation process.
Role of the C-terminus in RPT amyloid formation
To examine residue level environmental changes upon aggregation, fluorescence measurements of the only intrinsic tryptophan (W423) was used. The single native tryptophan (in the second to last repeat Figure 7 top) is an exquisitely sensitive, fluorescent probe of RPT aggregation kinetics and fibril morphology.98 For all acidic conditions examined (pH < 6), distinct spectral and temporal changes were observed and can be correlated to macroscopic differences in fibril morphology. For example, between pH 5.0 – 5.5, significant intensity decreases are observed along with spectral blue-shift, indicating a more hydrophobic local environment and potential quenching due to Trp-Trp stacking interactions, which would be consistent with a parallel in-register β-sheet conformation. The high responsiveness of W423 during RPT aggregation points towards a key role for the C-terminal region in fibril assembly.
Other lines of evidence also support this hypothesis. Firstly, in vivo, the repeat domain is O-glycosylated with the N-terminal region containing sialylated glycans located between repeats 2 and 3.88 Addition of these modifications could cause a steric constraint around this region preventing aggregation. Secondly, proline residues are primarily located between repeats 1–7 and 10 would destabilize any amyloid structure and so these regions would less likely contribute to the fibril core. Lastly, computational methods for predicting amyloid-forming segments such as WALTZ, TANGO, PASTA and Zyggregator predict that residues in the C-terminus will participate in the fibril core.
The RPT domain of Pmel17 orthologs forms amyloid
To evaluate the primary amino acid sequence dependence of RPT aggregation propensities, very recent work investigated the Pmel17:RPT domains from mouse and zebrafish as well as a splice variant of human RPT.99,100 While these Pmel17 orthologs share moderate sequence similarity to one another, there is little sequence conservation within the RPT domain with mouse and no similarity with zebrafish. The RPT splice variant contains a 42 amino acid truncation between repeats 5 and 8. Upon aggregation at pH 5.0, all RPT domains were capable of forming amyloid. Similar to what was shown for human RPT, no aggregation could be detected at neutral pH. Fibrils formed at pH 5.0 were sensitive to neutral pH, showing disassembly of the fibrils when monitored by ThT.
Using ssNMR, both mouse and zebrafish RPT adopt the in-register parallel β-sheet architecture that is often observed in human pathogenic amyloids (Figure 8 top). By labelling at least one or a few residues with 13C- labelled amino acid in the carbonyl position, the nearest intra/intermolecular 13C distance can be measured.101 The distance between β-strands in a β-sheet is about 0.5 nm for parallel in-register β-sheets and a distance of ~0.5 nm consistent with this model was measured (Figure 8 bottom).
Fig. 8.
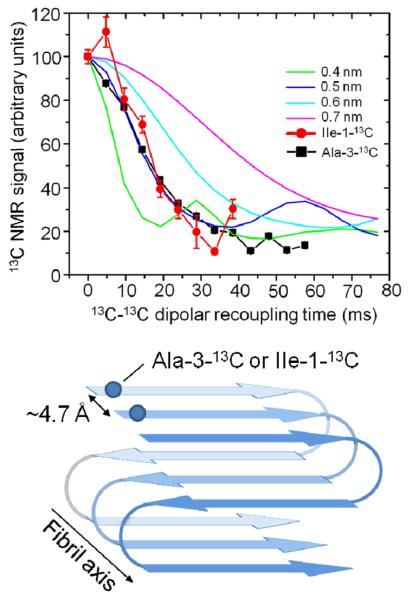
Solid state NMR 13C-C13 dipolar recoupling experiments of Ala-3-13C or Ile-1-13C labeled mouse RPT fibrils using PITHIRDSCT101 pulse sequence (top). Schematic representation of RPT amyloid showing a parallel in-register structure (bottom).
Pmel17: a highly regulated functional amyloid
Pmel17 is highly regulated in vivo with the full-length polypeptide incapable of forming amyloid fibrils. Upon transmembrane cleavage, the lumenal Mα fragment is released,89 which upon further proteolysis and glycosylation forms the melanosomal matrix fibrils. The timing and sequence of these events prevent amyloid formation happening too early, therefore allowing correct processing. For example, cellular compartments such as ER and Golgi, which lie upstream of melanosome development, would be protected from aberrant Pmel17 fibril assembly. Cellular factors such as chaperones may be recruited to regulate the process of propagation, degradation, and toxic intermediate build up. Indeed, the protein MART-1 (melanoma antigen recognized by T cells) has been shown to regulate Pmel17 folding and amyloid formation.102
Clearly, in vitro RPT only forms amyloid at the acidic melanosomal pH and these fibrils quickly dissolve when subjected to near neutral pH. During stage I, protein aggregation is initiated with fibril growth arrested until stage II, at which point the melanosomal pH reaches ~5.0 and the fibrils then begin to elongate. This pH sensitivity and stability can easily be understood in light of the many glutamic acid residues present in the repeat domains of Pmel17 orthologs.
The striking observation that the RPT fibrils dissolve at neutral pH suggests that upon deprotonation of specific carboxylates, the electrostatic charge repulsion increases with these glutamate residues leading to the dissolution of fibrils. Such an observation with disease-related amyloids has not been noted due to their extreme difficulty in disassembly e.g. by proteolysis and denaturation. From a biological perspective, if RPT was to escape from the melanosome, it would be exposed to a more neutral environment, and eventually dissolve and maintain a soluble benign form. This reversible mechanism ensures that RPT fibril formation is sequestered and aggregated only in the melanosome.
5. Concluding Remarks
The emerging concept of `functional amyloid' has forced us to rethink the notion that amyloids are only associated with diseases. It is becoming evident that there is a `yin' and `yang' side to amyloids and likely many other facets to them as more amyloidgenic proteins are discovered. By reviewing our work on both α-syn and RPT, we can begin to postulate about how they are biologically different from one another. For α-syn as well as other amyloidogenic polypeptides such as amyloid-β, the current thought is that toxicity is associated with pre-fibrillar or oligomeric species that form during early stages of aggregation. Notably, pre-fibrillar aggregates have been shown to permeabilize membranes,103 suggesting a specific molecular mechanism for pathogenesis. By utilizing biophysical techniques that allow characterization of heterogeneity in the polypeptide ensemble, we are beginning to elucidate the conformational dynamics of α-syn in solution. Moreover, site-specific fluorescent probes have been particularly useful for studies of the amyloid formation process by capturing early events during the aggregation process. With these spectroscopic tools in hand, we are now poised to obtain mechanistic insight into the role of intermediates and their interactions with other biomolecules such as membranes.
On the other hand for functional amyloids, in particular the RPT domain, if oligomeric species are generated, they could rupture melanosome membranes causing leakage of pre-melanin intermediates that are known to be cytotoxic; but clearly, this does not occur. One step to circumvent this is to mitigate or limit the build-up of these potentially membrane disruptive intermediates during amyloid formation.91 In addition, its pH dependent amyloid forming behavior offers important insights into Pmel17 function, a scaffold for melanin deposition. Firstly, the amyloid filament allows for the acceleration of melanin formation by increasing the concentration and orientation of melanin precursors. The positioning of these intermediates on the fibrils restricts the environment for melanin polymerization. Secondly, melanin precursors are known to be highly cytotoxic and could easily diffuse through the melanosome membrane and cause detriment to other cellular compartments. Sequestering these intermediates along the fibrillar axis would mitigate such effects. Lastly, melanin deposition could protect the fibrils if the melanosome pH becomes too alkaline. Alternatively, dissassembly of RPT could be a way for melanosomes to recycle amyloid fibrils.
It is clear from our work as well as others that while functional and pathological amyloids share similarities at the structural level, there are distinct differences as to how they form. For RPT, the strict pH requirement for aggregation suggests a highly regulated process that allows amyloid to be formed during melanin synthesis, at which point it acts as a scaffold for melanin deposition. In contrast, α-syn aggregation occurs in the absence of a controlled cellular system, which could permit the build-up of intermediates leading to cytotoxicity and eventual cell death.
Acknowledgements
This work was supported by the Intramural Research Program at the National Institutes of Health, National Heart, Lung, and Blood Institute. We thank Rob Tycko in providing the Aβ amyloid structure and Candace Pfefferkorn for discussion.
References
- 1.Kyle RA. Amyloidosis: A convoluted story. Br. J. Haematol. 2001;114:529. doi: 10.1046/j.1365-2141.2001.02999.x. [DOI] [PubMed] [Google Scholar]
- 2.Greenwald J, Riek R. Biology of amyloid: Structure, function, and regulation. Structure. 2010;18:1244. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Petkova AT, Leapman RD, Guo ZH, et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 4.Glabe CG. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler DM, Koulov AV, Balch WE, et al. Functional amyloid - from bacteria to humans. Trends Biochem. Sci. 2007;32:217. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Sipe JD, Cohen AS. Review: History of the amyloid fibril. J. Struct. Biol. 2000;130:88. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 7.Divry P, Florkin M. The optic properties of amyloid. Comptes Rendus Seances Soc. Biol. Fil. 1927;97:1808. [Google Scholar]
- 8.Vassar PS, Culling CFA, Taylor HE. A fluorescent staining method for demonstrating amyloid. Am. J. Pathol. 1959;35:718. [Google Scholar]
- 9.Astbury WT, Dickinson S. The x-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 1935;29:2351. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen AS, Calkins E. Electron microscopic observations on a fibrous component in amyloid of diverse origins. Nature. 1959;183:1202. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- 11.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968;16:673. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 12.Bockmann A, Meier BH. Prions En route from structural models to structures. Prion. 2010;4:8. doi: 10.4161/pri.4.2.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: Molecular insights from electron paramagnetic resonance spectroscopy. Q. Rev. Biophys. 2008;41:265. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 14.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 2011;62:279. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benzinger TLS, Gregory DM, Burkoth TS, et al. Propagating structure of Alzheimer's β-amyloid((10-35)) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13407. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paravastu AK, Leapman RD, Yau WM, et al. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18349. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasmer C, Lange A, Van Melckebeke H, et al. Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 19.Nelson R, Sawaya MR, Balbirnie M, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawaya MR, Sambashivan S, Nelson R, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 21.Apostol MI, Sawaya MR, Cascio D, et al. Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease. J. Biol. Chem. 2010;285:29671. doi: 10.1074/jbc.C110.158303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 23.Kruger R, Kuhn W, Muller T, et al. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 1998;18:106. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 24.Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 25.Singleton A, Gwinn-Hardy K, Sharabi Y, et al. Association between cardiac denervation and parkinsonism caused by α-synuclein gene triplication. Brain. 2004;127:768. doi: 10.1093/brain/awh081. [DOI] [PubMed] [Google Scholar]
- 26.Singleton AB, Farrer M, Johnson J, et al. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 27.Ueda K, Fukushima H, Masliah E, et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11282. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waxman EA, Giasson BI. Molecular mechanisms of α-synuclein neurodegeneration. Biochim. Biophys. Acta-Mol. Basis Dis. 2009;1792:616. doi: 10.1016/j.bbadis.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greffard S, Verny M, Bonnet AM, et al. A stable proportion of Lewy body bearing neurons in the substantia nigra suggests a model in which the Lewy body causes neuronal death. Neurobiol. Aging. 2010;31:99. doi: 10.1016/j.neurobiolaging.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 31.Hindle JV. Ageing, neurodegeneration and Parkinson's disease. Age Ageing. 2010;39:156. doi: 10.1093/ageing/afp223. [DOI] [PubMed] [Google Scholar]
- 32.Olanow CW, Perl DP, DeMartino GN, et al. Lewy-body formation is an aggresome-related process: a hypothesis. Lancet Neurol. 2004;3:496. doi: 10.1016/S1474-4422(04)00827-0. [DOI] [PubMed] [Google Scholar]
- 33.Gertz HJ, Siegers A, Kuchinke J. Stability of cell-size and nucleolar size in Lewy body containing neurons of substantia-nigra in Parkinson's disease. Brain Res. 1994;637:339. doi: 10.1016/0006-8993(94)91257-2. [DOI] [PubMed] [Google Scholar]
- 34.van Rooijen BD, Claessens MM, Subramaniam V. Membrane interactions of oligomeric α-synuclein: Potential role in Parkinson's disease. Curr. Protein. Pept. Sci. 2010;11:334. doi: 10.2174/138920310791330659. [DOI] [PubMed] [Google Scholar]
- 35.Cookson MR, van der Brug M. Cell systems and the toxic mechanism(s) of α-synuclein. Exp. Neurol. 2008;209:5. doi: 10.1016/j.expneurol.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cookson MR. α-Synuclein and neuronal cell death. Mol. Neurodegener. 2009;4 doi: 10.1186/1750-1326-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dev KK, Hofele K, Barbieri S, et al. Part II: α-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacology. 2003;45:14. doi: 10.1016/s0028-3908(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 38.Serpell LC, Berriman J, Jakes R, et al. Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-β conformation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4897. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilar M, Chou HT, Luhrs T, et al. The fold of α-synuclein fibrils. Proc. Natl. Acad. ScI. U S A. 2008;105:8637. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M, Margittai M, Chen J, et al. Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 2007;282:24970. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 41.Qin Z, Hu D, Han S, et al. Role of different regions of α-synuclein in the assembly of fibrils. Biochemistry. 2007;46:13322. doi: 10.1021/bi7014053. [DOI] [PubMed] [Google Scholar]
- 42.Heise H, Hoyer W, Becker S, et al. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15871. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conway KA, Harper JD, Lansbury PT. Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry. 2000;39:2552. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 44.Hoyer W, Antony T, Cherny D, et al. Dependence of α-synuclein aggregate morphology on solution conditions. J. Mol. Biol. 2002;322:383. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 45.Der-Sarkissian A, Jao CC, Chen J, et al. Structural organization of α-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 2003;278:37530. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 46.Weinreb PH, Zhen WG, Poon AW, et al. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35:13709. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 47.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 48.Grupi A, Haas E. Segmental conformational disorder and dynamics in the intrinsically disordered protein α-synuclein and its chain length dependence. J. Mol. Biol. 2011;405:1267. doi: 10.1016/j.jmb.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 49.Lee JC, Langen R, Hummel PA, et al. α-Synuclein structures from fluorescence energy-transfer kinetics: Implications for the role of the protein in Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16466. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreon AC, Gambin Y, Lemke EA, et al. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5645. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreon AC, Moran CR, Ferreon JC, et al. Alteration of the α-synuclein folding landscape by a mutation related to Parkinson's disease. Angew. Chem. 2010;49:3469. doi: 10.1002/anie.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trexler AJ, Rhoades E. Single molecule characterization of α-synuclein in aggregation-prone states. Biophys. J. 2010;99:3048. doi: 10.1016/j.bpj.2010.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakowicz JR. Principles of fluorescence spectroscopy. Springer; New York: 2006. [Google Scholar]
- 54.Trexler AJ, Rhoades E. α-Synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009;48:2304. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JC, Gray HB, Winkler JR. Tertiary contact formation in α-synuclein probed by electron transfer. J. Am. Chem. Soc. 2005;127:16388. doi: 10.1021/ja0561901. [DOI] [PubMed] [Google Scholar]
- 56.Lee JC, Lai BT, Kozak JJ, et al. α-Synuclein tertiary contact dynamics. J. Phys. Chem. B. 2007;111:2107. doi: 10.1021/jp068604y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urie KG, Angulo D, Lee JC, et al. Synchronous vs asynchronous chain motion in α-synuclein contact dynamics. J. Phys. Chem. B. 2009;113:522. doi: 10.1021/jp806727e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dedmon MM, Lindorff-Larsen K, Christodoulou J, et al. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 59.Bertoncini CW, Jung YS, Fernandez CO, et al. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1430. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernado P, Bertoncini CW, Griesinger C, et al. Defining long-range order and local disorder in native α-synuclein using residual dipolar couplings. J. Am. Chem. Soc. 2005;127:17968. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- 61.Allison JR, Varnai P, Dobson CM, et al. Determination of the free energy landscape of α-synuclein using spin label nuclear magnetic resonance measurements. J. Am. Chem. Soc. 2009;131:18314. doi: 10.1021/ja904716h. [DOI] [PubMed] [Google Scholar]
- 62.Wu KP, Baum J. Detection of transient interchain interactions in the intrinsically disordered protein α-synuclein by NMR paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2010;132:5546. doi: 10.1021/ja9105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang IJ, Lee JC, Winkler JR, et al. The protein-folding speed limit: Intrachain diffusion times set by electron-transfer rates in denatured Ru(NH3)5(His-33)-Zn-cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3838. doi: 10.1073/pnas.0637283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li WX, West N, Colla E, et al. Aggregation promoting C-terminal truncation of α-synuclein is a normal cellular process and is enhanced by the familial Parkinson's disease-linked mutations. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2162. doi: 10.1073/pnas.0406976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoyer W, Cherny D, Subramaniam V, et al. Impact of the acidic C-terminal region comprising amino acids 109-140 on α-synuclein aggregation in vitro. Biochemistry. 2004;43:16233. doi: 10.1021/bi048453u. [DOI] [PubMed] [Google Scholar]
- 66.Burgoyne RD, Morgan A. Chaperoning the SNAREs: A role in preventing neurodegeneration? Nat. Cell. Biol. 2011;13:8. doi: 10.1038/ncb0111-8. [DOI] [PubMed] [Google Scholar]
- 67.Witt SN. Hsp70 molecular chaperones and Parkinson's disease. Biopolymers. 2010;93:218. doi: 10.1002/bip.21302. [DOI] [PubMed] [Google Scholar]
- 68.Cuervo AM, Wong ES, Martinez-Vicente M. Protein degradation, aggregation, and misfolding. Mov. Disord. 2010;25(Suppl 1):S49. doi: 10.1002/mds.22718. [DOI] [PubMed] [Google Scholar]
- 69.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 70.Giehm L, Otzen DE. Strategies to increase the reproducibility of protein fibrillization in plate reader assays. Anal. Biochem. 2010;400:270. doi: 10.1016/j.ab.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 71.Dusa A, Kaylor J, Edridge S, et al. Characterization of oligomers during α-synuclein aggregation using intrinsic tryptophan fluorescence. Biochemistry. 2006;45:2752. doi: 10.1021/bi051426z. [DOI] [PubMed] [Google Scholar]
- 72.Kaylor J, Bodner N, Edridge S, et al. Characterization of oligomeric intermediates in α-synuclein fibrillation: FRET studies of Y125W/Y133F/Y136F α-synuclein. J. Mol. Biol. 2005;353:357. doi: 10.1016/j.jmb.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 73.Nath S, Meuvis J, Hendrix J, et al. Early aggregation steps in α-synuclein as measured by FCS and FRET: Evidence for a contagious conformational change. Biophy. J. 2010;98:1302. doi: 10.1016/j.bpj.2009.12.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thirunavukkuarasu S, Jares-Erijman EA, Jovin TM. Multiparametric fluorescence detection of early stages in the amyloid protein aggregation of pyrene-labeled α-synuclein. J. Mol. Biol. 2008;378:1064. doi: 10.1016/j.jmb.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 75.van Rooijen BD, van Leijenhorst-Groener KA, Claessens MMAE, et al. Tryptophan fluorescence reveals structural features of α-synuclein oligomers. J. Mol. Biol. 2009;394:826. doi: 10.1016/j.jmb.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 76.Yushchenko DA, Fauerbach JA, Thirunavukkuarasu S, et al. Fluorescent ratiometric MFC probe sensitive to early stages of α-synuclein aggregation. J. Am. Chem. Soc. 2010;132:7860. doi: 10.1021/ja102838n. [DOI] [PubMed] [Google Scholar]
- 77.Yap TL, Pfefferkorn CM, Lee JC. Residue-specific fluorescent probes of α-synuclein: Detection of early events at the N- and C-termini during fibril assembly. Biochemistry. 2011;50:1963. doi: 10.1021/bi2000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klucken J, Outeiro TF, Nguyen P, et al. Detection of novel intracellular α-synuclein oligomeric species by fluorescence lifetime imaging. FASEB J. 2006;20:2050. doi: 10.1096/fj.05-5422com. [DOI] [PubMed] [Google Scholar]
- 79.Outeiro TF, Putcha P, Tetzlaff JE, et al. Formation of toxic oligomeric α-synuclein species in living cells. PLoS One. 2008;3:e1867. doi: 10.1371/journal.pone.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Opazo F, Krenz A, Heermann S, et al. Accumulation and clearance of α-synuclein aggregates demonstrated by time-lapse imaging. J. Neurochem. 2008;106:529. doi: 10.1111/j.1471-4159.2008.05407.x. [DOI] [PubMed] [Google Scholar]
- 81.Roberti MJ, Bertoncini CW, Klement R, et al. Fluorescence imaging of amyloid formation in living cells by a functional, tetracysteine-tagged α-synuclein. Nat. Methods. 2007;4:345. doi: 10.1038/nmeth1026. [DOI] [PubMed] [Google Scholar]
- 82.Celej MS, Caarls W, Demchenko AP, et al. A triple-emission fluorescent probe reveals distinctive amyloid fibrillar polymorphism of wild-type α-synuclein and its familial Parkinson's disease mutants. Biochemistry. 2009;48:7465. doi: 10.1021/bi9003843. [DOI] [PubMed] [Google Scholar]
- 83.Celej MS, Jares-Erijman EA, Jovin TM. Fluorescent N-arylaminonaphthalene sulfonate probes for amyloid aggregation of α-synuclein. Biophys. J. 2008;94:4867. doi: 10.1529/biophysj.107.125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shim SH, Gupta R, Ling YL, et al. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6614. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berson JF, Theos AC, Harper DC, et al. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 2003;161:521. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dunn LC, Thigpen LW. The silver mouse - A recessive color variation. J. Hered. 1930;21:495. [Google Scholar]
- 87.Kwon BS, Chintamaneni C, Kozak CA, et al. A melanocyte-specific gene, Pmel-17, maps near the silver coat color locus on mouse chromosome-10 and is in a syntenic region on human chromosome-12. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9228. doi: 10.1073/pnas.88.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valencia JC, Rouzaud F, Julien S, et al. Sialylated core 1 O-glycans influence the sorting of Pmel17/gp100 and determine its capacity to form fibrils. J. Biol. Chem. 2007;282:11266. doi: 10.1074/jbc.M608449200. [DOI] [PubMed] [Google Scholar]
- 89.Berson JF, Harper DC, Tenza D, et al. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol. Biol. Cell. 2001;12:3451. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hurbain I, Geerts WJC, Boudier T, et al. Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19726. doi: 10.1073/pnas.0803488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fowler DM, Koulov AV, Alory-Jost C, et al. Functional amyloid formation within mammalian tissue. PLoS. Biol. 2006;4:100. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Watt B, van Niel G, Fowler DM, et al. N-terminal domains elicit formation of functional Pmel17 amyloid fibrils. J. Biol. Chem. 2009;284:35543. doi: 10.1074/jbc.M109.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoashi T, Muller J, Vieira WD, et al. The repeat domain of the melanosomal matrix protein Pmel17/gp100 is required for the formation of organellar fibers. J. Biol. Chem. 2006;281:21198. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 94.Hoashi T, Sato S, Yamaguchi Y, et al. Glycoprotein nonmetastatic melanoma protein b, a melanocytic cell marker, is a melanosome-specific and proteolytically released protein. FASEB J. 2010;24:1616. doi: 10.1096/fj.09-151019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McGlinchey RP, Shewmaker F, McPhie P, et al. The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13731. doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Puri N, Gardner JM, Brilliant MH. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J. Invest. Dermatol. 2000;115:607. doi: 10.1046/j.1523-1747.2000.00108.x. [DOI] [PubMed] [Google Scholar]
- 97.Brilliant MH. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigm. Cell. Res. 2001;14:86. doi: 10.1034/j.1600-0749.2001.140203.x. [DOI] [PubMed] [Google Scholar]
- 98.Pfefferkorn CM, McGlinchey RP, Lee JC. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21447. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nichols SE, Harper DC, Berson JF, et al. A novel splice variant of Pmel17 expressed by human melanocytes and melanoma cells lacking some of the internal repeats. J. Invest. Dermatol. 2003;121:821. doi: 10.1046/j.1523-1747.2003.12474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McGlinchey RP, Shewmaker F, Hu KN, et al. Repeat domains of melanosome matrix protein Pmel17 orthologs form amyloid fibrils at the acidic melanosomal pH. J. Biol. Chem. 2011;286:8385. doi: 10.1074/jbc.M110.197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J. Chem. Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 102.Hoashi T, Watabe H, Muller J, et al. Mart-1 is required for the function of the melanosomal matrix protein Pmel17/gp100 and the maturation of melanosomes. J. Biol. Chem. 2005;280:14006. doi: 10.1074/jbc.M413692200. [DOI] [PubMed] [Google Scholar]
- 103.Uversky VN, Eliezer D. Biophysics of Parkinson's disease: Structure and aggregation of α-synuclein. Curr. Protein Pept. Sci. 2009;10:483. doi: 10.2174/138920309789351921. [DOI] [PMC free article] [PubMed] [Google Scholar]


