Abstract
Refractive error is the most common eye disorder worldwide, and a prominent cause of blindness. Myopia affects over 30% of Western populations, and up to 80% of Asians. The CREAM consortium conducted genome-wide meta-analyses including 37,382 individuals from 27 studies of European ancestry, and 8,376 from 5 Asian cohorts. We identified 16 new loci for refractive error in subjects of European ancestry, of which 8 were shared with Asians. Combined analysis revealed 8 additional loci. The new loci include genes with functions in neurotransmission (GRIA4), ion channels (KCNQ5), retinoic acid metabolism (RDH5), extracellular matrix remodeling (LAMA2, BMP2), and eye development (SIX6, PRSS56). We also confirmed previously reported associations with GJD2 and RASGRF1. Risk score analysis using associated SNPs showed a tenfold increased risk of myopia for subjects with the highest genetic load. Our results, accumulated across independent multi-ethnic studies, considerably advance understanding of mechanisms involved in refractive error and myopia.
Refractive error is the most important cause of visual impairment in the world1. Myopia, or nearsightedness, in particular is associated with structural changes of the eye, increasing the risk of severe complications such as macular degeneration, retinal detachment, and glaucoma. The prevalence of myopia has been rising dramatically over the past few decades2, and it is estimated that 2.5 billion people will be affected by myopia within a decade3. Although several genetic loci influencing refractive error have been identified4-10, their contribution to phenotypic variance is small, and many more loci are expected to explain its genetic architecture.
Here the Consortium for Refractive Error and Myopia (CREAM) presents results from the largest international genome-wide meta-analysis on refractive error with data from 32 studies from Europe, the United States, Australia, and Asia. The meta-analysis was performed in three stages: as a first step, we investigated genome-wide association study (GWAS) results of 37,382 individuals from 27 populations of European ancestry (Supplementary Note, Supplementary Table 1) using spherical equivalent as a continuous outcome; as a second step, we aimed to test cross-ethnic transferability of the statistical significant associations from the first stage in 8,376 individuals from 5 Asian cohorts (Supplementary Note, Supplementary Table 1). As a third step, we performed a GWAS meta-analysis on the combined populations (total n = 45,758). Subsequently, we examined the influence of associated alleles on the risk of myopia in a genetic risk score analysis, and lastly, we evaluated gene expression in ocular tissues and explored potential mechanisms by which newly found loci may exert their effect on refractive development.
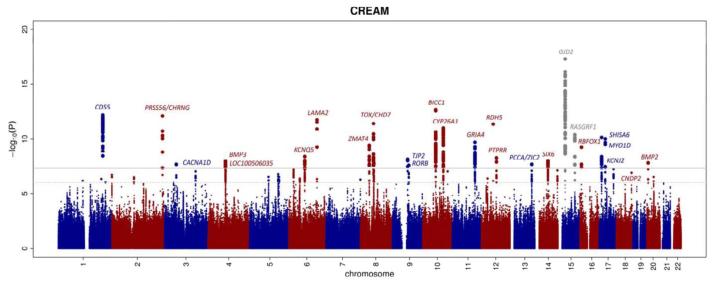
At step 1, we analyzed ~2.5 million autosomal single nucleotide polymorphisms (SNPs) which were obtained through whole-genome imputation of genotypes to HapMap 2. The inflation factors (γGC) of the test statistics in individual studies contributing to the meta-analysis ranged between 0.992 and 1.050, indicating excellent within-study control of population substructure (Supplementary Table 2). The overall lambda was 1.09, consistent with a polygenic inheritance model for refractive error (Q-Q plot, Supplementary Figure S1). We did not perform a lambda correction as Yang et al. have shown that in this situation substantial genomic inflation can be expected, even in the absence of population structure and technical artifacts11. We identified 309 SNPs that exceeding the conventional genome-wide significance threshold of P=5.0×10−8 in the European ancestry sample. These SNPs were clustered in 18 distinct genomic regions across 14 chromosomes (Figure 1, Table 1). At step 2, we investigated the 18 best associated SNPs in the Asian population: ten showed evidence of association (Table 1). The most significant association in both ancestry groups was at a previously identified locus on chromosome 15q14 in the proximity of the GJD2 gene (SNP rs524952; Pcombined=1.44×10−15)4,12. The locus near the RASGRF1 gene was also replicated in the meta-analysis (SNP rs4778879; Pcombined=4.25×10−11)9, the remaining 16 genome-wide significantly associated loci had not previously been reported in association with refractive error. Those loci that were not significant in the smaller sized Asian population mostly had a similar effect size and direction of effect as in the European ancestry sample. At step 3, we identified 8 additional loci which exceeded genome-wide significance in the combined analysis (Table 2). Regional and forest plots of the associated loci are provided in Supplementary Figures 2 and 3.
Figure 1. Manhattan plot of the GWAS meta-analysis for refractive error in the combined analysis (n = 45,758).
The plot shows −log10-transformed P values for all SNPs; the upper horizontal line represents the genome-wide significance threshold of P < 5.0 × 10−8; the lower line indicates P value of 10−5. Previously reported genes are depicted in grey. The A2BP1 gene is also known as RBFOX1.
Table 1. Genome-wide significant hits for refractive error in the European ancestry population with results in Asian population and combined analysis.
Summary of SNPs that showed genome-wide significant (P<5×10-8) association with spherical equivalent (SE) in subjects of European ancestry (stage 1), with results of replication in Asians (stage 2) and combined analysis (stage 3). Previously reported genes are marked in grey. We tested for heterogeneous effects between the Asian and the European ancestry sample, for which P values are shown. Abbreviations: SNP, single nucleotide polymorphism; Nearest gene, reference NCBI build 37; A1, reference allele; A2, other allele, MA, Minor Allele; MAF, average Minor Allele Frequency; Beta, effect size on spherical equivalent in diopters based on allele A1; SEM, standard error of the mean.
| Number of loci | SNP | Chromosome | Position | Nearest gene | A1/A2 | MAF | Beta | SEM | P value | MAF | Beta | SEM | P value | Beta | SEM | P value | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| Stage 1 (n = 37,382) | Stage 2 (n = 8,376) | Combined (n = 45,758) | Heterogeneity | ||||||||||||||
| 1 | rs1652333 | 1 | 203858855 | CD55 | G/A | 0.32 | −0.115 | 0.018 | 6.29E-11 | 0.42 | −0.099 | 0.035 | 5.00E-03 | −0.112 | 0.016 | 3.05E-12 | 0.94 |
| 2 | rs1656404 | 2 | 233205446 | PRSS56 | A/G | 0.21 | −0.151 | 0.025 | 2.38E-09 | 0.11 | −0.167 | 0.069 | 1.60E-02 | −0.153 | 0.024 | 7.86E-11 | 0.83 |
| rs1881492 | 2 | 233406997 | CHRNG | T/G | 0.22 | −0.145 | 0.022 | 1.28E-10 | 0.15 | −0.057 | 0.110 | 6.09E-01 | −0.139 | 0.021 | 5.15E-11 | 0.88 | |
| 3 | rs14165 | 3 | 53847407 | CACNA1D | A/G | 0.32 | 0.095 | 0.017 | 4.36E-08 | 0.12 | 0.120 | 0.100 | 2.29E-01 | 0.096 | 0.017 | 2.14E-08 | 0.25 |
| 4 | rs1960445 | 4 | 81930813 | BMP3 | C/T | 0.17 | −0.147 | 0.026 | 1.19E-08 | 0.11 | 0.034 | 0.055 | 5.32E-01 | −0.114 | 0.024 | 1.25E-06 | 0.31 |
| 5 | rs12205363 | 6 | 129834628 | LAMA2 | C/T | 0.10 | 0.228 | 0.034 | 1.13E-11 | 0.02 | 0.553 | 0.236 | 1.92E-02 | 0.235 | 0.033 | 1.79E-12 | 0.93 |
| 6 | rs4237036 | 8 | 61701056 | CHD7 | C/T | 0.35 | 0.097 | 0.017 | 1.52E-08 | 0.23 | 0.043 | 0.040 | 2.81E-01 | 0.089 | 0.016 | 1.82E-08 | 0.76 |
| rs7837791 | 8 | 60179085 | TOX | T/G | 0.49 | 0.106 | 0.017 | 9.22E-10 | 0.39 | 0.103 | 0.035 | 4.00E-03 | 0.106 | 0.015 | 3.99E-12 | 0.70 | |
| 7 | rs7829127 | 8 | 40726393 | ZMAT4 | G/A | 0.25 | 0.116 | 0.020 | 3.04E-09 | 0.11 | 0.112 | 0.055 | 4.23E-02 | 0.116 | 0.018 | 3.69E-10 | 0.66 |
| 8 | rs7042950 | 9 | 77149836 | RORB | G/A | 0.24 | −0.113 | 0.020 | 1.02E-08 | 0.42 | −0.040 | 0.037 | 2.72E-01 | −0.096 | 0.018 | 4.15E-08 | 0.83 |
| 9 | rs10882165 | 10 | 94924323 | CYP26A1 | T/A | 0.42 | −0.111 | 0.016 | 1.25E-11 | 0.20 | −0.060 | 0.056 | 2.84E-01 | −0.107 | 0.016 | 1.03E-11 | 0.90 |
| 10 | rs7084402 | 10 | 60265403 | BICC1 | G/A | 0.48 | −0.111 | 0.016 | 7.23E-12 | 0.50 | −0.094 | 0.035 | 7.34E-03 | −0.108 | 0.015 | 2.06E-13 | 0.71 |
| 11 | rs11601239 | 11 | 105061808 | GRIA4 | C/G | 0.46 | −0.092 | 0.017 | 3.45E-08 | 0.42 | −0.129 | 0.058 | 2.70E-02 | −0.095 | 0.016 | 5.92E-09 | 0.83 |
| 12 | rs3138144 | 12 | 56114768 | RDH5 | C/G | 0.48 | 0.113 | 0.018 | 4.28E-10 | 0.45 | 0.157 | 0.072 | 3.00E-02 | 0.119 | 0.017 | 4.44E-12 | 0.09 |
| 13 | rs2184971 | 13 | 100818091 | PCCA | G/A | 0.44 | 0.095 | 0.016 | 5.90E-09 | 0.22 | 0.022 | 0.040 | 5.84E-01 | 0.085 | 0.015 | 2.11E-08 | 0.96 |
| rs8000973 | 13 | 100691366 | ZIC2 | T/C | 0.47 | 0.089 | 0.016 | 4.24E-08 | 0.22 | 0.030 | 0.041 | 4.63E-01 | 0.081 | 0.015 | 5.10E-08 | 0.50 | |
| 14 | rs524952 | 15 | 35005885 | GJD2 | A/T | 0.48 | −0.154 | 0.021 | 1.11E-13 | 0.44 | −0.193 | 0.060 | 1.00E-03 | −0.158 | 0.020 | 1.44E-15 | 0.22 |
| 15 | rs4778879 | 15 | 79372874 | RASGRF1 | G/A | 0.44 | −0.103 | 0.017 | 1.27E-09 | 0.39 | −0.103 | 0.043 | 1.50E-02 | −0.102 | 0.015 | 4.25E-11 | 0.15 |
| 16 | rs17183295 | 17 | 31078271 | MYO1D | T/C | 0.23 | −0.132 | 0.021 | 3.04E-10 | 0.16 | −0.166 | 0.144 | 2.49E-01 | −0.131 | 0.020 | 9.66E-11 | 0.34 |
| 17 | rs4793501 | 17 | 68718733 | KCNJ2 | C/T | 0.42 | 0.096 | 0.016 | 3.21E-09 | 0.44 | 0.010 | 0.034 | 7.64E-01 | 0.080 | 0.014 | 2.79E-08 | 0.04 |
| 18 | rs12971120 | 18 | 72174022 | CNDP2 | G/A | 0.23 | 0.108 | 0.020 | 4.39E-08 | 0.30 | 0.014 | 0.063 | 8.27E-01 | 0.099 | 0.019 | 1.85E-07 | 0.49 |
Table 2. Additional genome-wide significant hits from the combined meta-analysis (n = 45,758).
Summary of SNPs that showed genome-wide significant (P<5×10-8) association with spherical equivalent (SE) in the combined analysis (stage 3), with results in subjects of European ancestry (stage 1) and Asians (stage 2). We tested for heterogeneous effects between the two ancestries, for which P values are shown. Abbreviations: SNP, single nucleotide polymorphism; Nearest gene, reference NCBI build 37; A1, reference allele; A2, other allele, MA, Minor Allele; MAF, average Minor Allele Frequency; Beta, effect size on spherical equivalent in diopters based on allele A1; SEM, standard error of the mean. The A2BP1 gene is also known as RBFOX1.
| Number of loci | SNP | Chromosome | Position | Nearest gene | A1/A2 | Beta | SEM | P value | MAF | Beta | SEM | P value | MAF | Beta | SEM | P value | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| Combined (n = 45,758) | Stage 1 (n = 37,382) | Stage 2 (n = 8,376) | Heterogeneity | ||||||||||||||
| 1 | rs9307551 | 4 | 80530670 | LOC100506035 | A/C | −0.099 | 0.017 | 1.09E-08 | 0.25 | −0.097 | 0.020 | 1.37E-06 | 0.50 | −0.105 | 0.035 | 3.06E-03 | 0.70 |
| 2 | rs7744813 | 6 | 73643288 | KCNQ5 | C/A | 0.112 | 0.019 | 4.18E-09 | 0.41 | 0.114 | 0.021 | 6.80E-08 | 0.33 | 0.094 | 0.046 | 4.30E-02 | 0.14 |
| 3 | rs11145465 | 9 | 71766592 | TJP2 | A/C | −0.124 | 0.021 | 7.26E-09 | 0.25 | −0.125 | 0.023 | 6.92E-08 | 0.07 | −0.136 | 0.091 | 1.35E-01 | 0.14 |
| 4 | rs12229663 | 12 | 71249995 | PTPRR | G/A | 0.099 | 0.017 | 5.47E-09 | 0.27 | 0.104 | 0.019 | 5.46E-08 | 0.36 | 0.080 | 0.052 | 1.23E-01 | 0.74 |
| 5 | rs1254319 | 14 | 60903756 | SIX6 | A/G | −0.088 | 0.015 | 1.00E-08 | 0.32 | −0.088 | 0.017 | 2.03E-07 | 0.34 | −0.087 | 0.036 | 1.57E-02 | 0.59 |
| 6 | rs17648524 | 16 | 7459682 | A2BP1 | C/G | −0.118 | 0.019 | 5.64E-10 | 0.36 | −0.116 | 0.022 | 7.48E-08 | 0.14 | −0.140 | 0.058 | 1.60E-02 | 0.24 |
| 7 | rs2969180 | 17 | 11407900 | SHISA6 | A/G | −0.101 | 0.015 | 7.29E-11 | 0.36 | −0.101 | 0.019 | 7.51E-08 | 0.45 | −0.097 | 0.034 | 4.00E-03 | 0.41 |
| 8 | rs235770 | 20 | 6761764 | BMP2 | T/C | −0.089 | 0.016 | 1.57E-08 | 0.39 | −0.088 | 0.017 | 1.34E-07 | 0.33 | −0.087 | 0.050 | 8.20E-02 | 0.78 |
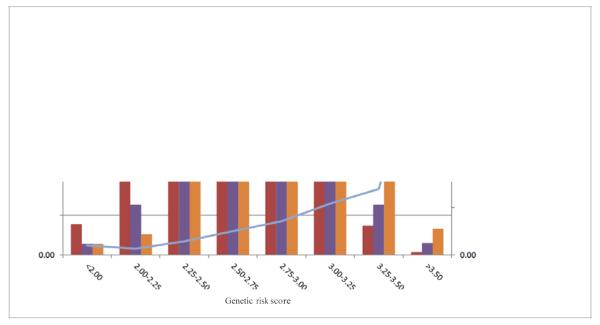
Genotype distributions of the risk alleles were evaluated in Rotterdam Studies 1-3 (n=9,307). The clinical utility for the prediction of risk of myopia was evaluated by a weighted genetic risk score analysis based on the aggregate of effects (regression coefficients betas) of individual SNPs derived from the meta-analysis, using the middle risk category as a reference. Risk scores ranged from a mean risk score of 1.88 (95% CI 1.86 - 1.89) in the lowest risk score category to 3.63 (95% CI 3.61-3.65) in the highest risk score category. Having the lowest or the highest genetic risk score was associated with an odds ratio of 0.38 (95% CI 0.18-0.77), and an odds ratio of 10.97 (95% CI 3.727-31.251) of myopia, respectively (Figure 2). The predictive value (area under receiver operating characteristic curve, AUC) of myopia versus hyperopia was 0.67 (95% CI 0.65-0.69), a relatively high value for genetic factors in a complex trait13,14. The genetic variants explain 3.4% of the phenotypic variation in refractive error in the Rotterdam Study.
Figure 2. Genetic risk score for myopia.
Distribution of subjects from Rotterdam Study 1-3 (n= 9,307) with myopia (SE ≤ −3 D), emmetropia (SE ≥ −1.5 D & ≤ 1.5 D) and hyperopia (SE ≥ 3 D) as a function of genetic risk score. This score is based on the regression coefficients and allele dosages of the associated SNPs for all 26 loci identified in the meta-analysis. The mean OR of myopia was calculated per risk category, using the middle risk score category (risk score 2.50; 2.75) as a reference.
We examined the expression of genes harboring a genetic association signal by measuring levels of RNA in various eye tissues, and found most of these genes expressed in the eye (Supplementary Table 3). The genes PRSS56, LOC100506035, and SHISA6 were not available in the expression data set; all other genes were expressed in the retina. Subsequently, we assessed the areas where our SNP hits reside for H3K27ac modification marks15, and HaploReg16 annotations for marks of active regulatory elements (Supplementary Table 4, Supplementary Figure 4). We found that many hits contain these elements, and alteration of regulatory function is therefore a suggestive mechanism.
The widely-accepted model for myopia development is that eye growth is triggered by a visually-evoked signaling cascade, which originates from the sensory retina, traverses the retinal pigment epithelium and choroid, and terminates in the sclera, where active extracellular matrix (ECM) remodeling results in a relative elongation of the eye17. Many of the genes in or near the identified loci can be linked to biological processes that drive this cascade. Neurotransmission in the retina is a necessary mechanism for eye growth regulation; the most significantly associated gene GJD2 plays a role herein. This gene forms a gap junction between neuronal cells in the retina, enabling intracellular exchange of small molecules and ions. The other previously-reported gene RASGRF1 is a nuclear exchange factor that promotes GDP/GTP exchange on Ras-family GTPases, and is involved in synaptic transmission of photoreceptor responses18,19. Both GJD2 and RASGRF1 knockout mice show retinal photoreception defects18,20. One of the newly identified genes, GRIA4 (SNP rs11601239; Pcombined=5.92×10−9) also has a potential function in this pathway. This gene is a glutamate-gated ion channel that mediates fast synaptic excitatory neurotransmission21, is present in various retinal cells22, has been shown to be critical for light signaling in the retina23 and emmetropization24. Another gene involved in synaptic transmission is A2BP1 (also known as RBFOX1; SNP rs17648524; Pcombined=5.64×10−10, an RNA-binding splicing regulator which modulates membrane excitability25.
We identified for the first time a number of genes involved in ion transport, channel activity and maintenance of membrane potential. KCNQ5, a potassium channel regulator (SNP rs7744813; Pcombined=4.18×10−9), participates in the transport of K+ from the retina to the choroid, and may contribute to voltage-gated K+ channels in photoreceptors and retinal neurons associated with myopia26,27. CD55 (SNP rs1652333; Pcombined=3.05×10−12) is known to elevate cytosolic calcium ion concentration. Other ion channel genes include CACNA1D, a voltage-sensitive calcium channel regulator; KCNJ2, a regulator of potassium ion transport; CHRNG, a nicotinic cholinergic receptor; and MYO1D, a putative binder of calmodulin, which mediates Ca+ sensitivity to KCNQ5 ion channels.
Retinoic acid is synthesized in the retina and highly expressed in the choroid and has been implicated in eye growth in experimental myopia models28-30. Retinol dehydrogenase 5 (RDH5), a novel refractive error susceptibility gene (SNP rs3138144; Pcombined=4.44×10−12), is involved in the recycling of 11-cis-retinal in the visual cycle31. Mutations in RDH5 cause congenital stationary night blindness (OMIM# 136880), a disease associated with myopia. Other genes involved in retinoic acid metabolism are RORB (RAR-related orphan receptor), and CYP26A1, genes that were significant in the European ancestry studies. Notably, retinoic acid contributes to ECM remodeling by regulating cell differentiation.
ECM remodeling of the sclera is the pathological hallmark in myopia development. LAMA2 (laminin α2, SNP rs12205363; Pcombined=1.79×10−12) is the most prominent gene in this respect. LAMA2 forms a subunit of the heterotrimer laminins which are essential components of basement membranes, stabilizing cellular structures and facilitating cell migration32. The two bone morphogenic genes (BMP2, SNP rs235770; Pcombined=1.57×10−8; BMP3) can also be placed within the ECM architecture. They are members of the transforming growth factor-β (TGFβ) super-family, regulate growth and differentiation of mesenchymal cells, and may orchestrate the organization of other connective tissues than bone such as sclera. Remarkably, BMP2 shows bidirectional expression in retinal pigment epithelium in myopia animal models33.
Genes involved in eye development appeared as a separate entity among the gene functions. SIX6 (SNP rs1254319; Pcombined=1.00×10−8 has been linked to anophthalmia and glaucoma34,35, PRSS56 (protease serine 56, SNP rs1656404; Pcombined=7.86×10−11) to microphthalmia36-38, CHD7 to CHARGE syndrome, a congenital condition with severe eye structural defects, and ZIC2 to brain development including visual perception. For the remaining novel gene associations, a mechanism in the pathogenesis of myopia is not immediately clear. Results from Ingenuity and the Protein Link Evaluator39 (Supplementary Figure 5) visualize the subcellular location of all associated genes, and illustrates their interrelationships. Direct connections between genes were surprisingly infrequent, suggesting molecular disease heterogeneity or functional redundancy in the pathobiological events involved in development of refractive error and myopia.
In summary, we identified 24 new chromosomal loci associated with refractive error through a large-scale meta-analysis of GWAS from international multi-ethnic studies. The significant overlap in genetic loci for refractive error between subjects of European ancestry and Asians provides evidence for shared genetic risk factors between the populations. The tenfold increased risk of myopia for those carrying the highest number of risk alleles depicts the clinical significance of our findings. Further elucidation of the mechanisms by which these loci affect eye growth carries the potential to improve the visual outcome of this common trait.
Online methods
Study design
We performed a meta-analysis on directly genotyped and imputed SNPs from individuals of European ancestry in 27 studies, with a total of 37,382 individuals. Subsequently, we evaluated significant SNPs in 8,376 subjects of Asian origin from 5 different studies, and performed a meta-analysis on all studies combined.
Subjects and phenotyping
All studies participating in this meta-analysis are part of the Consortium for Refractive Error and Myopia (CREAM). All studies had a population-based design and had a similar protocol for phenotyping (Supplementary Table 1). Eligible participants underwent a complete ophthalmologic examination including a non-dilated measurement of refractive error of both eyes. Exclusion criteria were all conditions that could alter refraction, such as cataract surgery, laser refractive procedures, retinal detachment surgery, keratoconus, or ocular or systemic syndromes. Inclusion criteria were persons aged 25 years and over who had data on refractive error and genotype.
The meta-analysis of step 1 was based on 27 studies of European ancestry: 1958 British Birth Cohort, ALSPAC, ANZRAG, AREDS1a1b, AREDS1c, CROATIA-Korcula, CROATIA-Split, CROATIA-Vis, EGCUT, FECD, TEST/BATS, FITSA, Framingham, GHS 1, GHS 2, KORA, ORCADES, TwinsUK, WESDR, YFS, ERF, DCCT, BMES, RS1, RS2, RS3, and OGP Talana. The second step was formed by 5 Asian studies: Beijing Eye Study, SCES, SIMES, SINDI, and SP2.
General methods, demographics and phenotyping and genotyping methods of the study cohorts can be found in the Supplementary Note and Supplementary Table 1. All studies were performed with the approval of their local Medical Ethics Committee, and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
Genotyping and imputation
Particulars of genotyping in each cohort, particular platforms used to generate genotyping and methods of imputation can be found in more detail in the Supplementary Table 5. To produce consistent datasets and enable meta-analysis of studies across different genotyping platforms, the cohorts performed genomic imputation on the HapMap Phase 2 available genotypes with MACH40 or IMPUTE41, using the appropriate ancestry as templates.
Each cohort applied stringent quality control procedures prior to the imputation, including minor allele frequency cutoffs, Hardy-Weinberg equilibrium (P > 10−7), genotypic success rate (>95%), Mendelian inconsistencies, exclusion of individuals with more than 5% shared ancestry (exception made for family-based cohorts in which due adjustment for family relationship was made) and removal of all individuals whose ancestry as determined through genetic analysis did not match the prevailing ancestry group of the own cohort. SNPs with low imputation quality were filtered using metrics specific to the imputation method and thresholds used in their previous GWAS analyses. Hence, imputation quality criteria varied slightly among studies, and low-confidence imputed SNPs were omitted in the meta-analysis for individual studies.
Statistical analysis
Spherical equivalent was calculated according to the standard formula (SE=sphere + ½ cylinder), and the mean of two eyes was used for analysis. When data from only one eye was available, the SE of this eye was used.
Each cohort performed association analyses in which the spherical equivalent (determined as described above) was the dependent variable and genotypes (number of alleles in each of the HapMap2 loci) as the independent variables. Analyses in all cases also adjusted for sex and age at the time of phenotype measurement. In family-based cohorts score-test based association test was used to adjust for within-family relatedness (see Supplementary Note)42,43. Study-specific lambda estimates are shown in Supplementary Table 2.
All study effect estimates were corrected using genomic control and were oriented to the positive strand of the NCBI build 36 reference sequence of the human genome, which was the genomic build on which most available genotyping platforms were based. The coordinates and further annotations of the SNPs were further converted into build 37, the most recent of the available builds at the time of writing.
Meta-analyses used effect size estimations (beta regression coefficients) and standard errors from individual cohorts’ summary statistics. Random-effects were assumed for all the meta-analyses which were performed using GWAMA44. We tested for heterogeneous effects between the two ancestries using METAL45 for Linux. For the purpose of these analyses, we defined significance as equal to or better than the conventional multiple testing genome-wide thresholds of association (P<5.0 × 10−8) for stage 1 and nominally significant probabilities (P<0.05) for stage 2. Manhattan, regional plots and forest plots were made using R (see URLs) and Locuszoom (see URLs)46.
For the Rotterdam Study 1-3, a weighted genetic risk score per individual was calculated using the regression coefficients from the GWAS meta-analysis model for the association of SNPs within the associated 26 loci (Table 1, Table 2; per locus only one SNP was included in the analysis) and the individual allele dosages per genotype to evaluate the relationships between myopia (SE ≤ − 3 D), emmetropia (−1.5 D ≤ SE ≤ 1.5 D) and hyperopia (SE ≥ +3 SD). The weighted risk scores were categorized and mean odds ratios per risk score category were calculated for subjects with myopia versus hyperopia, using the middle risk score category as a reference. Subsequently, the area under the receiver curve (AUC) was calculated for myopia versus emmetropia and myopia versus hyperopia. Lastly, the proportion of variance of spherical equivalent explained by the identified SNPs was calculated. For these analyses, we used SPSS version 20.0.0 (SPSS Inc.).
Gene expression data in human eye tissue
Independently designed, collected, and reported human ocular tissue array data from two different sources, as well as literature reviews were used to verify evidence of expression of the candidate genes.
RPE, photoreceptors and choroid
Human gene expression data of RPE, photoreceptors and choroid were obtained essentially as described47 and the dataset has been deposited in NCBI’s Gene Expression Omnibus48 (GEO series accession number GSE20191; see URLs). In short, postmortem eye bulbs (retinal pigment epithelium was obtained from six donor eyes, choroid was obtained from three donor eyes and photoreceptors were obtained from three donor eyes), provided by the Corneabank Amsterdam, were rapidly frozen using liquid nitrogen. Donors were between 63 and 78 years old and had no known history of eye pathology. Cryosections were cut from the macula, and histology was used to confirm a normal histological appearance. Retinal pigment epithelium, photoreceptor and choroidal cells were isolated from macular sections using a Laser Microdissection System (PALM). Total RNA was isolated and the mRNA component was amplified, labeled and hybridized to a 44K microarray (Agilent Technologies)49. At least three to six microarrays were performed per tissue. Sample isolation, procedures and expression microarray analysis were carried out according to MIAMI guidelines. As a measure of the level of expression, we sorted all the genes represented on the 44K microarray by increasing their expression, and we calculated the corresponding percentiles (Supplementary Table 3a).
Sclera, cornea and optic nerve
We assessed expression of the associated genes in sclera, cornea and optic nerve tissue in an additional dataset (unpublished data). Adult eyes were obtained from the North Carolina Eye Bank (Winston-Salem, North Carolina). All whole globes were immersed in RNALater (Quiagen, Hilden, Germany) within 6.5 hours of collection, shipped overnight on ice, and dissected on the day of arrival. The retina, choroid and scleral tissues were isolated at the posterior pole using a circular, double embedded technique using round 7 mm and 5 mm biopsy punches. To reduce contamination of retina to the other ocular tissues samples, the second biopsy punch of 5 mm was used in the center of the 7 mm punch after retinal removal. RNA samples (quality control of RNA concentration and 260/280 nm ratios using Nanodrop®) (Invitrogen, Carlsbad, California, USA) were hybridized to whole genome microaray Illumina® HumanHT-12 v4 Expression BeadChips (over 25,000 genes and 48,000 probes) in two batches. The first batch was hybridized to adult RPE, choroid, and sclera RNA samples (n=6). The second batch of newer chips with additional probes was hybridized to adult optic nerve and cornea samples (n=6). The data were exported from Illumina® GenomeStudio and log2 transformed. Sample outliers were determined by principle component analyses using the Hoteling’s T2 test50 (at 95% confidence interval) and removed from further analyses. The data intensity was normalized by Quantile normalization followed by Multichip Averaging51 to reduce chip effects. For each tissue type, the probes with signal intensities below background levels and those with the lowest (5%) signal intensities (detection P<0.10) were excluded. Evidence of expression in the remaining probes was defined by detection P<0.05. Probes with detection P values < 0.10 and > 0.05 required additional tissue expression support from EyeSAGE or literature reports (Supplementary Table 3b).
Search for regulatory elements
We used the ‘Integrated Regulation from ENCODE’ track in the UCSC genome browser to look at H3K27ac modificiation marks as a mark of active regulatory elements. Numbers of H3K27ac modification marks were counted between the associated topSNP from a locus and the nearest gene and within (the nearest) gene itself. We also used HaploReg16 annotations to look for other signs of regulatory activity at the site of the associated SNP itself, such as enhancer histone marks, DNAse hypersensitivity sites, binding proteins and motifs changed.
Pathway analyses
We used two different programs for pathway analsysis; Ingenuity (see URLs), version August 2012, application build 172788, content version 14197757) and the Disease Association Protein-Protein Link Evaluator (DAPPLE)39.
Subcellular localization assignment and functional annotation of myopia associated disease genes as well as molecular pathway analysis was carried out using the Ingenuity knowledge database. The candidate myopia disease genes discovered in this study were entered into the Ingenuity knowledge database (IPA). We used the “IPA toggle subcellular layout” function to show the subcellular location (extracellular, plasma membrane, cytoplasm, nucleus, unknown) of the proteins corresponding to these genes, which yield a first glance which signaling molecules and pathways are involved in myopia. Subsequently, we used the IPA “connect” function to discover potential direct or indirect functional relationships or molecular pathways in between these entries. This yielded surprisingly little hits, which suggest molecular disease heterogeneity and/or functional redundancy in the pathobiological events leading to myopia. Next, we used the IPA “overlay” function to annotate the myopia candidate disease genes with (their involvement in) “functions and diseases”, “canonical pathways” and a range of custom made gene lists from previous studies, including photoreceptor, RPE, and choroidal specific transcripts (partly published52). Lastly, we used the Disease Association Protein-Protein Link Evaluator (DAPPLE)39 to look for physical connections between proteins encoded from disease-genes associated regions.
Supplementary Material
Acknowledgements
We gratefully thank the invaluable contributions of all study participants, their relatives and staff at the recruitment centers. Complete funding information and acknowledgements per study can be found in the Supplementary Note.
Footnotes
Author contributions V.J.M.V, P.G.H., R.W., C.J.H., C.C.W.K, A.W.H., D.A.M., T.L.Y., and C.M.v.D. performed analyses and drafted the manuscript. C.C.W.K, D.S., C.J.H., J.E.B.W., S.M.S., C.M.v.D., A.H., D.A.M., S.M., A.P., V.V, C.W., P.N.B., T.Y.W., J.R., T.L.Y., K.O., O.P., S.P.Y., J.G., A.M., M.P., S.K.I., and N.P. jointly conceived the project and supervised the work. J.E.B.W., S.M.S., D.A.M., T.L.Y., C.J.H., C.C.W.K, D.S., J.E.B.W., C.M.v.D., R.W., P.G.H., V.J.M.V., K.O., Y.Y.T., T.Y.W., P.N.B., V.V., N.A., B.A.O., A.H., J.R.V., F.R., A.G.U., N.P., C.M., A.M., T.Z., B.F., J.F.W., Z.V., O.P., A.F.W., C.H., I.R., S.K.I., E.C., J.H.L., R.P.I.J., S.J., M.S., J.J.W., P.M., I.C., J.S.R., P.M.C., C.E.P., G.W.M., A.M., W.A., F.M., M.P., L.C.K., T.D.S., E.Y.D., A.N., O.R., C.C.K., T.M., A.D., R.T.O., Y.Z., J.L., R.L., P.C., V.A.B., W.T.T., E.V., T.A., E.S.T., C.C.W.K., A.M., T.H., R.K., B.E.K.K., J.E.C., K.P.B., L.J.C., C.P.P., D.W.H.H., S.P.Y., J.W., O.P., J.B.J., L.X., H.S.W., S.M.H., A.D.P., M.K., T.L., K.M.M., C.L.S., C.W., N.J.T., D.M.E., B.S.P., J.P.K., G.M., G.H.S., M.K.I., X.Z., C.Y.C., A.W.H., S.M., R.H., J.A.G., and Q.F. were responsible for study-specific data. G.H.S., Q.F., and J.A.G. were involved in the genetic risk score analysis. T.L.Y., A.A.B.B., T.G.M.F.G., and F.H. performed the data expression experiments. A.A.B.B., T.G.M.F.G., A.M., and S.M. were involved in pathway analyses. J.E.B.W., S.M.S., D.A.M., T.L.Y., K.O., T.Y.W., P.N.B., T.G.M.F.G., S.K.I., E.C., J.J.W., A.J.M.H.V., C.C.K., B.E.K.K., S.P.Y., C.W., N.J.T., G.H.S., M.K.I., A.W.H., and J.A.G. critically reviewed the manuscript.
Competing financial interests The authors declare no competing financial interests.
URLs R, http://www.r-project.org; Locuszoom, http://csg.sph.umich.edu/locuszoom; Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20191; Ingenuity, http://www.ingenuity.com
References
- 1.Wojciechowski R. Nature and nurture: the complex genetics of myopia and refractive error. Clin Genet. 2011;79:301–20. doi: 10.1111/j.1399-0004.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutti DO, et al. Axial growth and changes in lenticular and corneal power during emmetropization in infants. Invest Ophthalmol Vis Sci. 2005;46:3074–80. doi: 10.1167/iovs.04-1040. [DOI] [PubMed] [Google Scholar]
- 3.Smith TS, Frick KD, Holden BA, Fricke TR, Naidoo KS. Potential lost productivity resulting from the global burden of uncorrected refractive error. Bull World Health Organ. 2009;87:431–7. doi: 10.2471/BLT.08.055673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solouki AM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y, et al. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet. 2011;88:805–13. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakanishi H, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20:2861–8. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 8.Li YJ, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–75. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hysi PG, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Q, et al. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 2012;8:e1002753. doi: 10.1371/journal.pgen.1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–12. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verhoeven VJ, et al. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012;131:1467–80. doi: 10.1007/s00439-012-1176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrada K, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rymer J, Wildsoet CF. The role of the retinal pigment epithelium in eye growth regulation and myopia: a review. Vis Neurosci. 2005;22:251–61. doi: 10.1017/S0952523805223015. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Medarde A, et al. RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J Neurochem. 2009;110:641–52. doi: 10.1111/j.1471-4159.2009.06162.x. [DOI] [PubMed] [Google Scholar]
- 19.Tonini R, et al. Expression of Ras-GRF in the SK-N-BE neuroblastoma accelerates retinoic-acid-induced neuronal differentiation and increases the functional expression of the IRK1 potassium channel. Eur J Neurosci. 1999;11:959–66. doi: 10.1046/j.1460-9568.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 20.Abd-El-Barr MM, et al. Genetic dissection of rod and cone pathways in the dark-adapted mouse retina. J Neurophysiol. 2009;102:1945–55. doi: 10.1152/jn.00142.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyer B, et al. Absence seizures in C3H/HeJ and knockout mice caused by mutation of the AMPA receptor subunit Gria4. Hum Mol Genet. 2008;17:1738–49. doi: 10.1093/hmg/ddn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connaughton V. Glutamate and Glutamate Receptors in the Vertebrate Retina. In: Kolb H, Fernandez E, Nelson R, editors. Webvision: The Organization of the Retina and Visual System. Salt Lake City (UT): 1995. [Google Scholar]
- 23.Yang J, Nemargut JP, Wang GY. The roles of ionotropic glutamate receptors along the On and Off signaling pathways in the light-adapted mouse retina. Brain Res. 2011;1390:70–9. doi: 10.1016/j.brainres.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Smith EL, 3rd, Fox DA, Duncan GC. Refractive-error changes in kitten eyes produced by chronic on-channel blockade. Vision Res. 1991;31:833–44. doi: 10.1016/0042-6989(91)90150-4. [DOI] [PubMed] [Google Scholar]
- 25.Fogel BL, et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–86. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Yang D, Hughes BA. KCNQ5/K(v)7.5 potassium channel expression and subcellular localization in primate retinal pigment epithelium and neural retina. Am J Physiol Cell Physiol. 2011;301:C1017–26. doi: 10.1152/ajpcell.00185.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pattnaik BR, Hughes BA. Effects of KCNQ channel modulators on the M-type potassium current in primate retinal pigment epithelium. Am J Physiol Cell Physiol. 2012;302:C821–33. doi: 10.1152/ajpcell.00269.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troilo D, Nickla DL, Mertz JR, Summers Rada JA. Change in the synthesis rates of ocular retinoic acid and scleral glycosaminoglycan during experimentally altered eye growth in marmosets. Invest Ophthalmol Vis Sci. 2006;47:1768–77. doi: 10.1167/iovs.05-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mertz JR, Wallman J. Choroidal retinoic acid synthesis: a possible mediator between refractive error and compensatory eye growth. Exp Eye Res. 2000;70:519–27. doi: 10.1006/exer.1999.0813. [DOI] [PubMed] [Google Scholar]
- 30.McFadden SA, Howlett MH, Mertz JR. Retinoic acid signals the direction of ocular elongation in the guinea pig eye. Vision Res. 2004;44:643–53. doi: 10.1016/j.visres.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Parker RO, Crouch RK. Retinol dehydrogenases (RDHs) in the visual cycle. Exp Eye Res. 2010;91:788–92. doi: 10.1016/j.exer.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheele S, et al. Laminin isoforms in development and disease. J Mol Med (Berl) 2007;85:825–36. doi: 10.1007/s00109-007-0182-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu Y, Wildsoet CF. Bidirectional, Optical Sign-Dependent Regulation of BMP2 Gene Expression in Chick Retinal Pigment Epithelium. Invest Ophthalmol Vis Sci. 2012;53:6072–80. doi: 10.1167/iovs.12-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramdas WD, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallardo ME, et al. Analysis of the developmental SIX6 homeobox gene in patients with anophthalmia/microphthalmia. Am J Med Genet A. 2004;129A:92–4. doi: 10.1002/ajmg.a.30126. [DOI] [PubMed] [Google Scholar]
- 36.Gal A, et al. Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease. Am J Hum Genet. 2011;88:382–90. doi: 10.1016/j.ajhg.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orr A, et al. Mutations in a novel serine protease PRSS56 in families with nanophthalmos. Mol Vis. 2011;17:1850–61. [PMC free article] [PubMed] [Google Scholar]
- 38.Nair KS, et al. Alteration of the serine protease PRSS56 causes angle-closure glaucoma in mice and posterior microphthalmia in humans and mice. Nat Genet. 2011;43:579–84. doi: 10.1038/ng.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 42.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Booij JC, et al. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics. 2009;10:164. doi: 10.1186/1471-2164-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Soest SS, et al. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch’s membrane. Mol Vis. 2007;13:1608–17. [PubMed] [Google Scholar]
- 50.Hotelling H. The Generalization of Student’s Ratio. The Annals of Mathematical Statistics. 1931;2:360–378. [Google Scholar]
- 51.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 52.Booij JC, et al. A new strategy to identify and annotate human RPE-specific gene expression. PLoS One. 2010;5:e9341. doi: 10.1371/journal.pone.0009341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.