Summary
Bacterial pathogens can cause multiple plant diseases and plants rely on their innate immune system to recognize and actively respond to these microbes. The plant innate immune system is comprised of extracellular pattern recognition receptors that recognize conserved microbial patterns and intracellular nucleotide binding leucine-rich repeat (NLR) proteins that recognize specific bacterial effectors delivered into host cells. Plants lack the adaptive immune branch present in animals, but still afford flexibility to pathogen attack through systemic and transgenerational resistance. Here, we focus on current research in plant immune responses against bacterial pathogens. Recent studies shed light onto the activation and inactivation of pattern recognition receptors and systemic acquired resistance. New research has also uncovered additional layers of complexity surrounding NLR immune receptor activation, cooperation, and sub-cellular localizations. Taken together, these recent advances bring us closer to understanding the web of inter-molecular interactions responsible for coordinating defense responses and ultimately resistance.
Keywords: effector-triggered immunity (ETI), pattern-triggered immunity (PTI), plant innate immunity, resistance genes, systemic acquired resistance (SAR), transgenerational resistance
Introduction
Plants can be infected by a diverse array of microbial pathogens, resulting in significant damage to crop species and consequently food shortages and economic loss. In this review, we will focus on plant immune responses to foliar Gram-negative bacterial pathogens. These bacteria enter to their host through natural openings or wounds and proliferate in the apoplast. Examples are members of the Pseudomonads and Xanthomonads, which are responsible for several serious crop diseases such as bacterial spot and speck of tomato, black rot of crucifers, and bacterial blight on rice. The main virulence strategies of these gram-negative bacterial pathogens are toxins and effector proteins (Cui et al., 2009). They use the type three secretion system to deliver the effector proteins directly into host cells, where characterized effectors function as enzymes to inhibit plant immune perception and facilitate bacterial colonization (Cui et al., 2009).
To inhibit pathogen invasion and multiplication, plants rely on their innate immune system (Spoel & Dong, 2012). Surface localized immune receptors recognize conserved microbial features called Pathogen-Associated Molecular Patterns (PAMPs), such as bacterial flagellin, resulting in pattern-triggered immunity (PTI) (Spoel & Dong, 2012). In addition to microbial features, surface receptors can also recognize self-derived Damage-Associated Molecular Patterns (DAMPs) that are the result of wounding, initiating a similar immune response to PTI. Plants also employ intracellular immune receptors that can recognize specific bacterial effectors, leading to robust effector-triggered immunity (ETI). The plant immune system not only acts to limit current pathogen invaders, but can also prime the plant and its’ progeny for heightened resistance against subsequent attack. Localized ETI leads to subsequent transmission of mobile signals to distal plant tissue, priming defense responses resulting in systemic resistance against future attack. Pathogen infection can also induce epigenetic modifications conferring trans-generational immunity. These layers of the plant innate immune system function together and as a result, the vast majority of microbes are nonpathogenic on most plants. Here, we highlight some of the most recent findings in context with the current understanding of mechanisms governing plant immune responses informed by decades of research.
FLS2: the flagellin immune receptor as a model for PTI responses
Pattern-triggered immunity is widely considered to be effective in conferring resistance against most pathogens, and only specialized pathogens have evolved to overcome it. Several bacterial PAMPs have been identified, such as flagellin and elongation factor Tu, allowing for significant progress in the elucidation of the molecular mechanisms resulting in PTI. While most work on bacterial PAMP recognition has been performed in Arabidopsis, conserved flagellin PAMP receptors have also been found in other plant species. Furthermore, there are a set of PTI hallmarks that appear to be widely conserved independent of the PAMP that is recognized. These hallmarks include: MAP kinase (MAPK) and calcium-dependent protein kinase (CDPK) activation, generation of extracellular superoxide, defense gene activation and callose deposition (Boudsocq et al., 2010; Segonzac & Zipfel, 2011). These PTI markers have been well established, and can be broken into early and late events where calcium influx, MAPK and CDPK activation occurs within minutes, followed shortly by ROS production then defense gene activation. Finally, several hours later, callose deposition is observed (Fig. 1). It is also clear that these defense markers are not completely dependent on one another, as was demonstrated by the recent uncoupling of ROS production and MAPK activation (Segonzac et al., 2011).
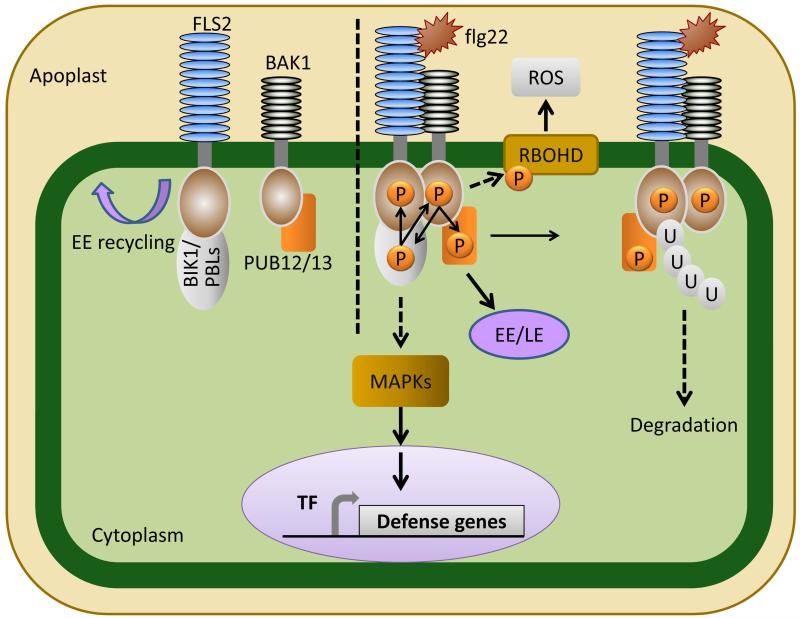
Fig. 1.
FLS2 mediated flagellin perception and signaling. In the absence of flagellin perception, the FLS2 receptor associates with receptor-like cytoplasmic kinases (PBLs), including BIK1. FLS2’s co-receptor BAK1 associates with the E3 ligases PUB12/13. Upon perception of flagellin or the flg22 epitope, FLS2 heterodimerizes with BAK1, BAK1 phosphorylates BIK1 and PUB12/13, and BIK1 transphosphorylates both FLS2 and BAK1. BIK1 then disassociates from FLS2. Flagellin perception leads to the activation of MAP kinase signaling and defense gene expression as well as a burst of reactive oxygen species via the NADPH oxidase RBOHD. FLS2 activation is tightly controlled by PUB12/13, which after phosphorylation by BAK1, initiates ubiquitination and subsequent degradation of activated FLS2. The FLS2 receptor is also endocyosed in both activated and resting states. FLS2 is recycled in early endosomes (EE) in its resting state independently of BAK1. Activated FLS2 is endocytosed in a BAK1 dependent manner, first accumulating in early endosomes (EE), then in late endosomes (LE).
The best characterized PAMP receptor is the Arabidopsis flagellin receptor Flagellin Sensing 2 (FLS2) (Gomez-Gomez & Boller, 2000). FLS2 is a member of the receptor-like kinase (RLK) family, with an extracellular leucine-rich repeat (LRR) domain and an intracellular kinase domain. Perception of flagellin monomers or a 22-amino acid epitope of flagellin (flg22) induces dynamic changes in the FLS2 protein complex in addition to multiple post-translational modifications. One of the first of these dynamic changes is heterodimerization between FLS2 and its co-receptor BRI1-Associated Kinase (BAK1) after FLS2 binds flagellin (reviewed in (Segonzac & Zipfel, 2011)). BAK1 is also a receptor-like kinase and an important co-receptor for multiple RLKs involved in sensing PAMPs and DAMPs. BAK1 can function as a co-receptor for the EF-Tu Receptor EFR, the Pep Receptor 1 (PEPR1) and PEPR2, as well as the plant brassinosteriod receptor BRI1 (Yamaguchi et al., 2006; Segonzac & Zipfel, 2011). BAK1 is a member of the larger Somatic Embryogenesis Receptor-like Kinase (SERK) family, and other SERK members also interact with FLS2 and play a role in FLS2 signaling (Roux et al., 2011). Co-immunoprecipitation experiments between SERK members and either FLS2 or EFR indicate that both receptors heteromerize with multiple SERK members, but FLS2 strongly prefers to associate with BAK1 while EFR shows equal preference for multiple SERKs (Roux et al., 2011). Investigations into SERK co-receptors have revealed conserved mechanisms of receptor activation at the plasma membrane. In all cases, ligand binding by the receptor protein induces heteroligomerization, followed by transphosphorylation and activation of downstream signaling (Segonzac & Zipfel, 2011) (Fig. 1). Importantly, FLS2 function has also resulted in a set of PTI hallmarks, discussed above, that are conserved across activation of diverse RLKs in different plant species.
An essential kinase regulating transphosphorylation of the FLS2 complex is BIK1 (Botrytis Induced Kinase 1), a receptor-like cytoplasmic kinase (Lu et al., 2010; Zhang et al., 2010). Dynamics surrounding FLS2 activation include BAK1-FLS2 heteromerization, association and transphosphorylation between BIK1 and BAK1-FLS2, followed by release of phosphorylated BIK1 (Segonzac & Zipfel, 2011) (Fig. 1). Downstream substrates of the activated BAK1-FLS2 complex leading to activation of MAPKs have yet to be elucidated and will be key to a more complete understanding of PTI.
Data providing evidence for FLS2 regulation through both endocytic cycling as well as ubiquitination have inserted additional complexity in FLS2-triggered PTI (Robatzek et al., 2006; Beck et al., 2012). Resting-state FLS2 is recycled between the plasma membrane and early endosomes, while activated FLS2 accumulates in early then late endosomal compartments (Beck et al., 2012) (Fig. 1). Interestingly, BAK1 is only required for activated FLS2 accumulation in late endosomes (Beck et al., 2012). These data invoke some important questions about intermediary signaling complexes FLS2 may be involved in and the role of endocytosis in FLS2-triggered PTI. Is FLS2 forming signaling complexes within the endosomes, which could link to MAPK activation, or is it only sequestered for degradation? While much has been revealed about FLS2 activation, partners and its subcellular journey, the connection between these events and immediate downstream PTI signaling responses remain undiscovered. Furthermore, it is unknown if other PAMP or DAMP receptors also undergo endocytosis.
Inappropriate or prolonged activation of immune responses is deleterious for plant health and productivity. Thus, immune receptor activation must be tightly regulated. Recently, the E3 ligases PUB12 and PUB13 were shown to rapidly ubiquitinate activated FLS2, highlighting the tight regulation of immune responses (Lu et al., 2011). PUB12 and PUB13 associate with BAK1 at the plasma membrane and are rapidly phosphorylated by BAK1 upon FLS2 activation. Phosphorylated PUB12/13 ubiquitinate activated FLS2 and target it for degradation (Lu et al., 2011). FLS2 can associate with PUB12 and PUB13 within 30 s post FLS2 activation, underscoring the rapidity with which activated FLS2 responds to ligand perception and forms a complex (Lu et al., 2011). Evidence suggests that FLS2 ubiquitination targets the activated receptor for proteasomal degradation in order to finely tune defense signaling (Fig. 1). Consistent with this hypothesis, pub12/13 mutants have an enhanced disease resistance phenotype when inoculated with virulent bacteria (Lu et al., 2011). Future research investigating the duration of PUB12/13’s association with activated FLS2 and if these proteins are coordinately endocytosed will shed light onto the dynamics of FLS2 signaling.
Recently, two manuscripts suggested broad-spectrum ligand activation for FLS2, where it was shown to respond to CLAVATA 3 peptides (Lee et al., 2011) and sulfonated Ax21 (Danna et al., 2011), which are unrelated to the flg22 epitope. Independent confirmation of this binding is so far unsuccessful for both CLV3p and Ax21 (Mueller et al., 2012; Segonzac et al., 2012). Future investigations will be necessary to determine if FLS2 can bind and be activated by diverse peptides, or if issues with contamination during synthetic peptide synthesis can explain these disparate results.
Effector triggered immunity (ETI): domain architecture, subcellular localization and downstream genetic loci
Intracellular plant immune receptors recognize pathogen effectors inside host cells, leading to ETI and robust disease resistance. Although PTI and ETI both elicit reactive oxygen species (ROS) production and defense gene induction, there are major differences in their strength, duration and timing of activation (Thomma et al., 2011). Definitive ETI markers are less established than those for PTI, but the programmed cell death hypersensitive response (HR) is regarded as a hallmark of ETI.
During an incompatible interaction with a bacterial pathogen, a combination of PTI and ETI will be induced, resulting in arrested pathogen growth. Microarray analyses have highlighted significant overlap in defense gene induction after perception of PAMPs or effectors (Navarro et al., 2004). It is possible that ETI is simply an amplification of PTI responses. Alternatively, some network overlap as well as distinct signaling events may occur in each system. The extent of overlap between PTI and ETI signaling overlap remains a major unanswered research question.
Most characterized intracellular ETI receptors belong to a group of structurally similar Resistance proteins called NLRs with Nucleotide-binding and Leucine-Rich repeat domains, which recognize effectors from diverse microbial pathogens. NLR receptors can recognize corresponding pathogen effectors directly or indirectly through effector-mediated perturbations of NLR interacting proteins (Elmore et al., 2011). Furthermore, these NLRs belong to the larger STAND (signal transduction ATPases with numerous domains) superfamily, whose members also include animal NLRs and pro-apoptotic proteins APAF-1 and CED4 (Leipe et al., 2004). NLR domain architecture is conserved between plants and animals, although animal NLR receptors have greater domain diversity at their N-termini. Plant NLRs are subdivided into two main groups based on their N-terminus, which contains either a Toll and interleukin-1-like receptor (TIR) domain or a coiled-coil (CC) domain (Meyers et al., 2003). Genetic evidence in plants and animals highlights the importance of an intact P-loop motif that coordinates ATP binding. ATP binding and hydrolysis has only been demonstrated for two recombinant tomato NLR receptors: I2 recognizing the fungus Fusarium oxysporum f. sp. lycopersici and Mi recognizing the root knot nematode, potato aphid, and whitefly (Tameling et al., 2002). Plant NLR immune receptors with mutations that should inhibit ATP hydrolysis are typically autoactive; whereas NLR P-loop (GxxxxGKT/S) mutants unable to bind ATP/ADP are inactive (Takken & Goverse, 2012). NLRs RPM1 and RPS2, recognizing bacterial effectors from Pseudomonas syringae are consistent with this model, and mutations in the P-loop of either protein render it inactive (Takken et al., 2006). Thus, the current model of NLR activation indicates that they switch from an ADP-bound ‘off’ state to an ATP-bound ‘on’ state, resulting in intra- and inter-molecular changes facilitating downstream signaling (Takken & Goverse, 2012).
Despite their structural conservation, a variety of pre- and post- activation modes exist for different NLR receptors. Some NLRs dramatically change their oligomerization status in response to effector perception. Two TIR-NLRs, N from tobacco recognizing tobacco mosaic virus, and L6 from flax recognizing the fungus Melampsora lini, exist as monomers pre-activation, but form homo-dimers and higher-order complexes post-activation based on co-immunoprecipitation analyses (Mestre & Baulcombe, 2006; Bernoux et al., 2011). However, some NLRs do not change their self-association in response to effector perception. For example, the Arabidopsis CC-NLR RPS5, recognizing the P. syringae effector AvrPphB, and the barley MLA10 CC-NLR, recognizing the fungus Blumeria graminis f. sp. hordei, dimerize both pre- and post-activation (Ade et al., 2007; Maekawa et al., 2011). Most oligomerization experiments were conducted on NLRs recognizing viral or fungal pathogens. However, work with the NLR RPS5 suggests that diverse pre- and post-activation modes may be broadly applicable. Therefore, the oligomerization status of activated and resting NLRs cannot be generalized across this family or even by microbial pathogen type, but rather a diversity of associations can exist. Since the cloning of the first NLR plant immune receptors in the 1990’s (Salmeron et al., 1996; Jones & Jones, 1997), some of the most pressing questions in the field are focused on how pathogen components result in intra- and intermolecular changes in immune receptors, ultimately leading to disease resistance. Technical advancements enabling purification of full-length NLR receptors is required in order to decipher their conformational changes between active and inactive states. Ultimately, structure-based knowledge is likely necessary in order to efficiently engineer NLRs for broad-spectrum pathogen resistance.
An emerging aspect of NLR function in plant innate immune responses is their ability to act in concert with other NLRs or as helpers to other NLRs. The Arabidopsis immune receptors RRS1 and RPS4 are genetically located within a large cluster of NLRs in the Arabidopsis Ws-0 ecotype (Botella et al., 1997). Both RRS1 and RPS4 cooperate genetically to regulate defense responses to R. solanacearum carrying the PopP2 effector and the P. syringae pv. tomato effector AvrRps4 (Narusaka et al., 2009). It is not yet known how this cooperation works and if there is physical interaction between RRS1 and RPS4. Bacterial titers in rps4/rrs1 are not as high as those attained in Col-0 with virulent P. syringae, indicating that there may still be additional NLR(s) or NLR pairs that act to regulate resistance in Ws-0, adding another layer of complexity. The RRS1/RPS4 dual resistance was also shown to be required for resistance to the fungal pathogen Colletotrichum higginsianum, demonstrating that specific NLRs can function generally against many microbial pathogens (Narusaka et al., 2009). Additionally, multiple members of a subgroup of NLRs lacking the canonical EDVID motif present in classical CC-NLRs are required for partial or full resistance imparted by some plant immune receptors. They are termed ‘helper’ NLRs as they are positioned genetically downstream of activation of the primary immune receptor (Bonardi et al., 2011). ADR1 is an example of a ‘helper’ NLR which is required for full resistance mediated through the CC-NLR RPS2, and TIR-NLRs RPP4 and RPP2 in Arabidopsis (Bonardi et al., 2011) (Fig. 2). Interestingly, they must have a different activation mechanism than previously characterized NLRs as an intact P-loop has been shown to be dispensable for function in ADR1 (Bonardi et al., 2011). Helper NLRs like ADR1 could function via intermolecular interactions with other NLRs or downstream signaling components. ADR1 functions partially through regulation of salicylic acid accumulation and also plays a role in execution of basal defenses (Bonardi et al., 2011). These examples illustrate that NLRs are involved in multiple levels of defense signaling that are not rigidly pathogen-specific. A greater understanding of NLR cooperative activity will provide a more complete picture of how defense responses are coordinated downstream of receptor activation.
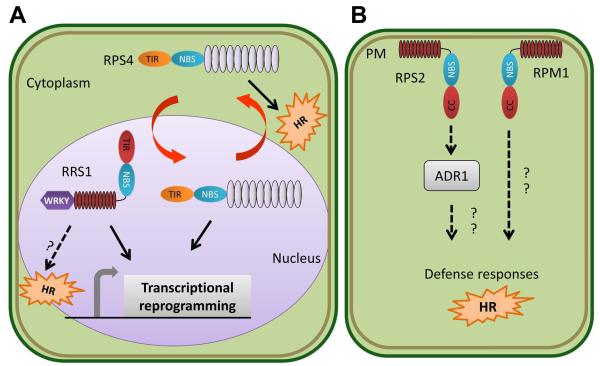
Fig. 2.
Subcellular partitioning of NLR-mediated plant immune responses. (a) RPS4 and RRS1 mediate activation of plant immune responses against Pseudomonas and Ralstonia bacterial pathogens carrying the cognate effectors AvrRps4 and PopP2. RPS4 is a TIR-NLR which predominately localizes to endomembranes in a resting state and upon activation undergoes shuttling between the cytoplasm and nucleus. Nuclear localization is required for RPS4-mediated transcriptional reprogramming, while recognition in the cytoplasm is required for the HR. However, however robust HR and sustained transcriptional reprogramming requires coordination between nuclear and cytoplasmic pools of activated RPS4. RRS1 exhibits novel domain architecture, with a TIR-NLR fused to a C-terminal WRKY domain. WRKY domains are found in multiple plant transcription factors required for innate immune reprogramming. RRS1 is nuclear localized, presumably where its WRKY domain acts as a transcriptional regulator upon recognition of PopP2, leading to the transcription of defense related genes. (b) RPS2 and RPM1, two CC-NLRs that recognize Pseudomonas effectors AvrRpt2, and AvrRpm1/ AvrB respectively, reside at the plasma membrane. RPM1’s subcellular localization does not change in response to activation. Thus, dynamic re-localization of plant immune receptors between cellular compartments is not necessary for all NLRs. RPS2 downstream signaling also requires the ‘helper’ NLR ADR1, which may interface with components relaying the activation of defense responses in other subcellular compartments.
There are also a few cases of cloned intracellular plant Resistance genes that do not exhibit NLR domain architecture. For example, pepper BS3 encodes a flavin monooxygenase that is sufficient to confer robust disease resistance against the bacterial pathogen Xanthomonas campestris pv. vesicatoria carrying the effector AvrBs3 (Romer et al., 2007). Another example of non-NLR resistance is SOBER1, an Arabidopsis phospholipase. Resistance to the X. campestris effector AvrBsT is characterized by the presence of an HR, and is due to the sober1 recessive allele (Kirik & Mudgett, 2009). SOBER1 is thought to suppress Phosphatidic Acid (PA) production by phospholipase D. Increased PA levels have been correlated with the HR in other NLR-mediated systems, but it is unknown how the PA signal may result in HR and resistance. Therefore, it is possible that these novel Resistance genes represent activation of downstream defense related genes, by-passing initial NLR signaling in a way that is still sufficient for robust resistance.
Location is everything: sub-cellular partitioning of plant immune responses
Plant immune receptors have diverse subcellular localizations, potentially enabling surveillance of diverse effector targets. Intracellular NLR receptors can localize to the plasma membrane, endoplasmic reticulum, chloroplast, nucleus, or cytoplasm. Additionally, a subset of NLR receptors recognizing viral, fungal, and bacterial pathogens shuttle between the cytoplasm and nucleus (Fig. 2). Examples of receptors exhibiting nuclear-cytoplasmic localizations are RPS4, RRS1, Rx, N, and MLA10. RRS1 was the first confirmed NLR to be described as nuclear localized (Deslandes et al., 2003). As discussed above, RRS1 functions in concert with RPS4 (Narusaka et al., 2009). RPS4 localizes predominately to the endomembranes before activation (Wirthmueller et al., 2007), but nuclear localization is required for immunity (Heidrich et al., 2011). The Enhanced Disease Susceptibility-1 (EDS1) protein is required for TIR-NLR function, including RPS4-mediated defense responses to AvrRps4. EDS1 exhibits nuclear-cytoplasmic localization, has been shown to co-immunoprecipitate with both AvrRps4 and RPS4, and it is postulated that RPS4 recognizes the EDS1-AvrRps4 complex (Bhattacharjee et al., 2011; Heidrich et al., 2011). Using P. syringae carrying AvrRps4 fused to nuclear localization or nuclear export signals, the role of specific cellular compartments for RPS4-mediated ETI was investigated (Heidrich et al., 2011). Recognition of nuclear-localized AvrRps4 was sufficient to suppress bacterial growth, but recognition in the cytoplasm was necessary for the HR (Fig. 2). Furthermore, the coordination of nuclear and cytoplasmic pools was necessary for robust HR and sustained transcriptional reprogramming (Heidrich et al., 2011) (Fig. 2).
Nuclear-cytoplasmic shuttling has also been demonstrated for NLRs recognizing other pathogen types. MLA10’s cytoplasmic pool is required for the HR, whereas its nuclear pool allows association with WRKY transcription factors leading cellular reprogramming for defense (Shen et al., 2007; Bai et al., 2012). Similarly, the potato CC-NLR Rx requires both nuclear and cytoplasmic localization for establishment of both HR and transcriptional reprogramming (Slootweg et al., 2010; Tameling et al., 2010). Thus, nuclear-cytoplasmic partitioning is often important for defense signaling, with different defense outputs segregated in different subcellular compartments (Fig. 2). Future research investigating the composition of these signaling platforms, their relative contributions to disease resistance, and if the partitioning threshold also acts as a trigger for defenses will provide a more complete understanding of immune signaling.
Not all NLRs undergo dynamic re-localization in response to pathogen perception. RPM1, a CC-NLR that recognizes the P. syringae effectors AvrB and AvrRpm1 (Grant et al., 1995), localizes to the plasma membrane (Boyes et al., 1998). Furthermore, RPM1 engineered to be constitutively tethered to the plasma membrane was found to still elicit an HR, indicating that re-localization to the nucleus is likely not required for RPM1-mediated defense responses (Gao et al., 2011) (Fig. 2). Undoubtedly, future investigations will focus on identifying signaling components that link NLRs at the membrane to activation of WRKY transcription factors or transcriptional machinery in the nucleus.
Genetic loci required for effector-triggered immunity (ETI)
Many genetic screens have been conducted to identify loci important for ETI as well as PTI. However, relatively few genes have been identified that are absolutely required for immune recognition aside from the receptors themselves or loci that are required for receptor biogenesis and/or subcellular localization. Of the few genes that have been identified, those having a strong effect on ETI include the conserved chaperone complex controlling aspects of receptor folding, stability, and activation. This chaperone complex consists of Heat Shock Protein 90 (HSP90), the Suppressor of the G2 allele of SKIP1 (SGT1) and required for MLA12 Resistance (RAR1) (Takken & Goverse, 2012). Genetic screens have also identified genes differentially required for CC or TIR NLR-mediated responses. As mentioned above, EDS1 is required for TIR-NLR defenses (Parker et al., 1996), while Non-Race Specific Disease Resistance-1 (NDR1) is required for defense mediated by multiple, but not all, CC-NLRs (Century et al., 1997). Other genes that have been uncovered primarily exhibit intermediate phenotypes, indicating that ETI requires the action of many downstream genes with minor effects, elicits a relatively short signaling network, or additional core components of immune signaling are essential for plant survival.
Systemic acquired resistance: a long-lasting broad spectrum immune response
A crucial aspect of plant immune responses is the ability to ward off subsequent pathogen attack. Activation of ETI induces Systemic Acquired Resistance (SAR) in leaves distal to the inoculation site, conferring broad spectrum resistance against biotrophic pathogens. SAR relies on the local accumulation of the plant hormone salicylic acid, which is regulated by small diffusible signals originating from the point of infection.
The search for pathogen-inducible mobile signals is ongoing, with several different signals identified to date. Methyl salicylate was the first mobile signal to be fully described, and is essential in triggering SAR in tobacco (reviewed in (Spoel & Dong, 2012)). However, several other mobile signals have been uncovered in Arabidopsis. Recently, the non-protein amino acid pipecolic acid, was shown to be involved in defense amplification and priming during SAR (Návarová et al., 2012). Additional SAR inducers include lipid-based signals derived from the apoplastic lipid-transfer protein DIR1 (Defective in Induced Resistance 1), azelaic acid, and glycerol-3-phosphate (Spoel & Dong, 2012). Interestingly, DIR1 is only required for systemic resistance and cooperatively functions with other mobile signals. For example, DIR1 activity is required for resistance induced by exogenously applied azelaic acid, and glycerol-3-phosphate requires DIR1 for mobile signal transduction (Chanda et al., 2011). One way to explain these findings is that different hormone and lipid-based mobile SAR signals may function cooperatively, potentially allowing for more specialized responses.
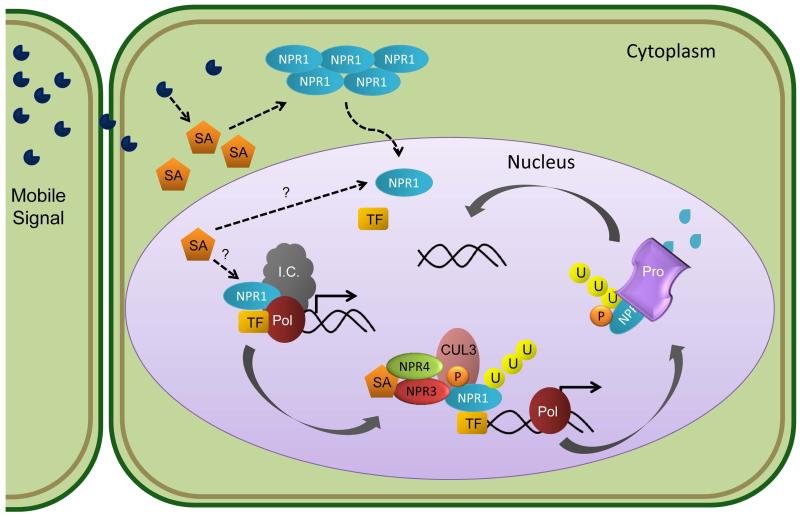
The presence of SAR signals in distal tissues triggers the accumulation of SA resulting in defense gene expression. SA-induced transcriptional reprogramming requires the transcription co-factor NPR1 (non-expressor of pathogenesis-related gene 1) in distally activated tissues to regulate the differential expression of over 2,000 genes, including pathogenesis related genes (reviewed in Spoel & Dong, 2012). NPR1 is a redox-sensitive protein, existing as an oligomer in the absence of SAR. In a reducing environment, NPR1’s disulfide bonds are reduced, enabling NPR1 monomers to enter into the nucleus. NPR1 is also post-transcriptionally regulated by the proteasome. Upon immune activation, NPR1’s phosphodegredon motif is phosphorylated, resulting in ubiquitination through association with Cullin 3, a scaffold for the E3 ligase complex. Ubiquitinated NPR1 is subsequently degraded by the proteasome (Spoel et al., 2009). Based on these results, the current model suggests that NPR1 may interact with transcription factors to recruit transcriptional machinery to the promoters of target genes. NPR1 phosphorylation could occur after transcriptional initiation, resulting in degradation and enabling fresh active NPR1 to recruit another round of gene expression (Fig. 3).
Fig. 3.
Model of salicylic acid perception and subsequent transcriptional reprogramming. Mobile Systemic Acquired Resistance (SAR) signals trigger the accumulation of Salicylic Acid (SA) in systemic leaves distal to the inoculation site. NPR1 is a master regulator of SAR. NPR1 is sequestered in the cytoplasm as an oligomeric complex in unchallenged plants. SA accumulation leads to an increase in cellular reduction potential, enabling NPR1 reduction from oligomeric to monomeric status. Monomeric NPR1 then enters the nucleus where it acts as a transcriptional co-factor, facilitating differential expression of thousands of genes. NPR1 may interact with transcription factors to help recruit the initiation complex (I.C.) and RNA Polymerase II (Pol) to the promoters of target genes. NPR1 is phosphorylated at its phosphodegron motif, resulting in ubiquitination through its association with Cullin 3 (CUL3), a scaffold for the E3 ligase complex, and subsequent degradation by the proteasome (Pro). The SA receptors NPR3 and NPR4 help facilitate NPR1 degradation. NPR3 interacts with NPR1 and CUL3, while NPR4 interacts with NPR3, CUL3 and SA. These interactions lead to CUL3 mediated ubiquitination of NPR1, targeting it for degradation by the proteasome (Pro). Proteasome degradation of NPR1 may then free promoters once again for fresh NPR1 to recruit transcriptional machinery.
The link between NPR1 activation and SA accumulation has long remained elusive. Recently, progress in understanding NPR1-mediated transcriptional reprogramming has been made via the identification of multiple SA receptors (Fu et al., 2012; Wu et al., 2012). Recombinant NPR3 and NPR4, paralogs of NPR1, strongly bind SA (Fu et al., 2012). Genetic and biochemical analyses provide evidence that NPR3 and 4 act as adapter proteins for Cullin 3 to mediate the degradation of NPR1 (Fig. 3). Interestingly, in contrast to the npr1 mutant which is compromised in basal resistance, the npr3/npr4 mutant exhibits enhanced disease resistance to virulent bacteria. However, the npr3/npr4 mutant is not able to elicit SAR in response to inoculation with the avirulent pathogen P. syringae pv. maculicola expressing AvrRpt2 and is also partially compromised in ETI (Fu et al., 2012). This suggests that while NPR 3 and 4 are required for SA-defense pathways, other NPR1-mediated defense responses are still functional in the npr3/npr4 double mutant. Evidence supporting NPR1 as the SA receptor has also recently been published (Wu et al., 2012). Using a metal ion affinity column, the transition metal copper was required for NPR1 binding to SA (Wu et al., 2012). Side-by-side comparisons of binding affinities for all NPR proteins in the presence of a transition metal will shed light to their relative affinities for SA. Future research investigating how NPR1 and its paralogs dynamically associate in response to SA perception in planta is important for a clearer picture of SAR induction.
Epigenetics and transgenerational resistance
Not only can plants achieve immunity within their own lifetime, but pathogen recognition results in epigenetic modifications leading to immune priming in subsequent generations. Treatment of Arabidopsis with the SAR inducer β–amino-butyric acid (BABA), as well as virulent or avirulent strains of P. syringae pv. tomato, led to their progeny displaying enhanced disease resistance, with faster and stronger expression of SA-dependent defense-related genes (Luna et al., 2012; Slaughter et al., 2012). Progeny from primed plants also exhibited enhanced resistance to the biotrophic oomycete Hyaloperonospora arabidopsidis (Slaughter et al., 2012). This phenotype lasted for one stress-free generation with enhanced penetrance occurring after pathogen treatments in subsequent generations (Slaughter et al., 2012). Transgenerational resistance was blocked in the npr1 mutant (Luna et al., 2012), highlighting the importance of NPR1 in this process.
Epigenetic modifications such as DNA methylation and chromatin remodeling are implicated in the regulation of transgenerational resistance. Somatic homologous recombination was also reported to be involved in regulation of transgenerational stress memory (Molinier et al., 2006). Here, flg22 or ultraviolet-C treated Arabidopsis plants displayed increased somatic homologous recombination both in parental lines and in up to four subsequent generations (Molinier et al., 2006). Future studies focusing on elucidating molecular details of how plants pass their immune memories or experiences to subsequent generations will result in important mechanistic discoveries that could be exploited for disease control.
Conclusions and future directions
The field of plant microbe biology has made dramatic progress in understanding plant immune function since the cloning of the first immune receptors in the 1990’s. Multiple PTI receptors have been identified from different plant species. A detailed understanding of how a subset of these PTI receptors are activated as well as the induction of key immune markers has emerged, highlighting conservation in immune signaling upon perception of microbial patterns from diverse pathogens. Furthermore, many NLR immune receptors and their corresponding pathogen effectors have been identified. Additional layers of complexity upon NLR activation have emerged, highlighting diverse subcellular localizations, cooperativity, and their complexities in downstream defense signaling. Despite this progress, many questions still remain. What are the signaling proteins immediately downstream of immune receptor activation? How can NLRs with disparate subcellular localizations elicit ETI and the hypersensitive response? What are the biochemical and structural changes in immune receptors that occur upon pathogen perception? Answers to these and other pressing questions will undoubtedly keep the field busy for years to come.
Acknowledgements
We thank DongHyuk Lee, Zuh Jyh D. Lin, Jun Liu, and James M. Elmore for critical reading of the manuscript. GC is supported by grants from the National Science Foundation MCB-1054298, the National Institutes of Health RO1GM092772, and the USDA National Institute of Food and Agriculture 2010-65108-20527. K.A.Y. is supported by a National Science Foundation Grant MCB-1054298 (awarded to G.C.). E.H. is supported by a National Institutes of Health Grant RO1GM092772 (awarded to G.C.).
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104(7):2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FL, et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012;8(6):e1002752. doi: 10.1371/journal.ppat.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S. Spatio-temporal cellular d namics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell. 2012;24(10):4205–4219. doi: 10.1105/tpc.112.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9(3):200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science. 2011;334(6061):1405–1408. doi: 10.1126/science.1211592. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proceedings of the National Academy of Sciences. 2011;108(39):16463–16468. doi: 10.1073/pnas.1113726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Coleman MJ, Hughes DE, Nishimura MT, Jones JD, Somerville SC. Map positions of 47 Arabidopsis sequences with sequence similarity to disease resistance genes. Plant J. 1997;12(5):1197–1211. doi: 10.1046/j.1365-313x.1997.12051197.x. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 2010;464(7287):418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci U S A. 1998;95(26):15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278(5345):1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- Chanda B, Xia Y, Mandal MK, Yu K, Sekine KT, Gao QM, Selote D, Hu Y, Stromberg A, Navarre D, et al. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat Genet. 2011;43(5):421–427. doi: 10.1038/ng.798. [DOI] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol. 2009;11(10):1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- Danna CH, Millet YA, Koller T, Han SW, Bent AF, Ronald PC, Ausubel FM. The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc Natl Acad Sci U S A. 2011;108(22):9286–9291. doi: 10.1073/pnas.1106366108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100(13):8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJ, Coaker G. Plant NB-LRR signaling: upstreams and downstreams. Curr Opin Plant Biol. 2011;14(4):365–371. doi: 10.1016/j.pbi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486(7402):228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chung EH, Eitas TK, Dangl JL. Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci U S A. 2011;108(18):7619–7624. doi: 10.1073/pnas.1104410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269(5225):843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. Arabidopsis EDS1 connects pathogen effector recognition to cell compartmentspecific immune responses. Science. 2011;334(6061):1401–1404. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- Jones DA, Jones JDG. The role of leucine-rich repeat proteins in plant defences. Advances in Botanical Research. 1997;24:89–167. [Google Scholar]
- Kirik A, Mudgett MB. SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc Natl Acad Sci U S A. 2009;106(48):20532–20537. doi: 10.1073/pnas.0903859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chah OK, Sheen J. Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature. 2011;473(7347):376–379. doi: 10.1038/nature09958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343(1):1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332(6036):1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Bruce TJ, Roberts MR, Flors V, Ton J. Next-generation systemic acquired resistance. Plant Physiol. 2012;158(2):844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Toller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9(3):187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18(2):491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15(4):809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442(7106):1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Mueller K, Chinchilla D, Albert M, Jehle AK, Kalbacher H, Boller T, Felix G. Contamination risks in work with synthetic peptides: flg22 as an example of a pirate in commercial peptide preparations. The Plant Cell. 2012;24(8):3193–3197. doi: 10.1105/tpc.111.093815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60(2):218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring A-C, Zeier J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. The Plant Cell. 2012;24(12):5123–41. doi: 10.1105/tpc.112.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135(2):1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. The Plant Cell. 1996;8(11):2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20(5):537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318(5850):645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. The Arabidopsis leucine-rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. The Plant Cell. 2011;23(6):2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86(1):123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, Rathjen JP. Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 2011;156(2):687–699. doi: 10.1104/pp.110.171249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Nimchuk ZL, Beck M, Tarr PT, Robatzek S, Meyerowitz EM, Zipfel C. The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. The Plant Cell. 2012;24(8):3186–92. doi: 10.1105/tpc.111.091264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Current Opinion in Microbiology. 2011;14(1):54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Shen Q-H, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ülker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315(5815):1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012;158(2):835–843. doi: 10.1104/pp.111.191593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 2010;22(12):4195–4215. doi: 10.1105/tpc.110.077537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12(2):89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137(5):860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI. Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol. 2006;9(4):383–390. doi: 10.1016/j.pbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Takken FL, Goverse A. How to build a pathogen detector: structural basis of NBLRR function. Curr Opin Plant Biol. 2012;15(4):375–384. doi: 10.1016/j.pbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14(11):2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell. 2010;22(12):4176–4194. doi: 10.1105/tpc.110.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Nurnberger T, Joosten MH. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell. 2011;23(1):4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17(23):2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Despres C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports. 2012;1(6):639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences. 2006;103(26):10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]