Abstract
An rpn11-1 temperature-sensitive mutant shows defect in proteolysis, mitochondrial function and proteasome assembly. The Rpn11 protein is a proteasome subunit that deubiquitinates proteolytic substrates. Multiubiquitinated proteins interact with proteasome receptors, such as Rpn10, which intriguingly is also required for promoting proteasome stability. We report here that Rpn10 binds Rpn11, and genetic studies revealed synthetic lethality of an rpn11-1 rpn10Δ double mutant. The carboxy-terminus of Rpn11 is critical for function, as deletion of 7 C-terminal residues prevented suppression of rpn11-1 rpn10Δ. Native gel electrophoresis showed increased levels of the proteasome 20S catalytic particle in rpn11-1 rpn10Δ, and altered assembly. The inviability of rpn11-1 rpn10Δ was suppressed by rpn10uim, a mutant that can bind the proteasome, but not multiubiquitin chains. rpn10uim reduced the levels of free 20S, and increased formation of intact proteasomes. In contrast, rpn10vwa, which binds multiubiquitin chains but not the proteasome, failed to suppress rpn11-1 rpn10Δ. Moreover, high levels of multiubiquitinated proteins were bound to rpn10vwa, but were not delivered to the proteasome. Based on these findings, we propose that the lethality of rpn11-1 rpn10Δ results primarily from altered proteasome integrity. It is conceivable that Rpn10/Rpn11 interaction couples proteasome assembly to substrate binding.
Keywords: Proteasome, Rpn11, Rpn10, Ubiquitin, Proteolysis
Introduction
The turnover of intracellular proteins occurs primarily through the ubiquitin/proteasome system (UPS), and involves highly conserved targeting factors termed ubiquitin activating (E1), ubiquitin conjugating (E2) and ubiquitin ligase (E3) enzymes (Glickman and Ciechanover 2002; Pickart and Cohen 2004). The majority of proteins that are degraded by the proteasome are covalently attached to a multiubiquitin (multiUb) chain that comprises at least four ubiquitins (Chau et al. 1989; Thrower et al. 2000). Linkage of the K48 lysine residue in a multiUb chain creates an extended hydrophobic surface that binds receptors in the proteasome (Beal et al. 1998; Pickart 1997). Recent studies indicate that alternate lysine residues in ubiquitin can also support the assembly of multiubiquitin chains that target substrates to the proteasome for degradation (Xu et al. 2009). The conjugation of ubiquitin to a substrate is dynamic, since multiUb chain processivity (E4) factors (Koegl et al. 1999), and multiUb chain disassembly (DUB) proteins can influence protein stability by either accelerating entry into the degradation pathway, or by rapidly withdrawing a protein from a proteolytic fate (Chen et al. 2002). The complete removal of ubiquitin from substrates that are correctly targeted to the proteasomes is also required for recycling Ub and initiating degradation (Hanna et al. 2006; Verma et al. 2002; Yao and Cohen 2002). Strikingly, substrate deubiquitination is required for successful proteolysis (Verma et al. 2002; Yao and Cohen 2002), and complementary roles of two deubiquitinating enzymes (Rpn11 and Ubp6) in yeast proteasomes has been described (Guterman and Glickman 2004). Additional deubiquitinating enzymes have been detected in proteasomes characterized in higher eukaryotes (Koulich et al. 2008). Rpn11 is a metalloprotease, and mutations in residues that contribute to its active site abolish function and cause lethality (Verma et al. 2002; Yao and Cohen 2002). Ubp6 was reported to function non-catalytically to delay substrate degradation, and prevent inadvertent entry of ubiquitin into the catalytic 20S particle (Hanna et al. 2006). An rpn11-1 temperature-sensitive mutant is defective in proteolysis, mitochondrial function and shows proteasome instability (Rinaldi et al. 1998, 2002; Verma et al. 2002). We recently reported that the loss of 31 carboxy-terminal residues in rpn11-1 contributed to a proteasome assembly defect, and a failure of proteasomes to bind multiubiquitinated substrates (Chandra et al. 2010). Strikingly, expression of this C-terminal domain in trans could partially overcome the defects of rpn11-1.
To understand the mechanism of substrate translocation to the proteasome we have characterized the Rad23 shuttle factor (Chen and Madura 2002), and Rpn10, a proteasome receptor that binds multiUb chains (Deveraux et al. 1994; van Nocker et al. 1996). We reported previously that the rad23Δ rpn10Δ double mutant shows poor growth, accumulates high molecular weight multiUb proteins, and displays sensitivity to drugs that cause protein unfolding (Lambertson et al. 1999). We proposed that these pleiotropic defects of rad23Δ rpn10Δ were caused by a failure to degrade specific cellular proteins, since bulk protein turnover was not affected significantly. In agreement, the degradation of the Cdk inhibitor Sic1 is prevented by loss of either Rad23 (rad23Δ) or Rpn10 (rpn10Δ), due to a failure to target this ubiquitinated substrate to the proteasome (Verma et al. 2004). This might explain why deubiquitination of Sic1 is defective in both rad23Δ and rpn10Δ (Verma et al. 2004). Two other shuttle factors in Saccharomyces cerevisiae (Dsk2; Ddi1), were reported to play overlapping, but non-redundant roles with Rad23 (Kim et al. 2004). Despite these diverse genetic interactions by Rad23 and Rpn10, it is clear that the lethality caused specifically by the loss of both Rad23 and Rpn10 indicates a significant role for these two proteins in intracellular proteolysis.
While the link between Rad23 and Rpn10 has been characterized extensively (Elsasser et al. 2004; Lambertson et al. 1999; Verma et al. 2004), the events that occur after substrate delivery to the proteasome have remained uncertain. We surmised that the transfer of multiUb substrates to the proteasome must occur in a manner that facilitates deubiquitination by the resident deubiquitinating enzymes, such as Rpn11 and Ubp6. To test this idea we examined interactions between Rpn11 and multiple proteasome subunits and identified Rpn10 as a candidate binding partner. This report describes in biochemical detail the interactions between Rpn11/Rpn10. A critical role for these two proteins is underscored by the synthetic lethality of a double mutant rpn11-1 rpn10Δ; we note that neither single mutant has a significant growth defect at the permissive temperature (Fu et al. 1998; Madura and Prakash 1990; van Nocker et al. 1996). Further study of the critical carboxy terminus of Rpn11 (Chandra et al. 2010; Rinaldi et al. 2004, 2008; Verma et al. 2002) showed that a mutant lacking just seven amino acid residues was unable to suppress the inviability of rpn11-1 rpn10Δ.
We investigated the key structural determinants in Rpn10 (VWA and UIM) to identify their contribution to the suppression of rpn11-1 rpn10Δ. An rpn10uim mutant that can bind the proteasome, but not multiUb chains suppressed rpn11-1 rpn10Δ. However, an rpn10vwa mutant that binds multiUb proteins (Elsasser et al. 2004), but not the proteasome, failed to overcome the lethality of rpn11-1 rpn10Δ, indicating a less important role for its substrate-binding property. These results suggest the possibility of a mechanistic link between proteasome assembly and its interaction with ubiquitinated substrates. In an analogous model in S. pombe, the UIM motif in Pus1 (an Rpn10 homolog) was dispensable in a pus1Δ pad1-1 mutant (which is similar to rpn10Δ rpn11-1) (Wilkinson et al. 2000). Although both UIM and VWA motifs are required for the deubiquitination and degradation of an engineered substate in vitro (Verma et al. 2004), in our genetic system the VWA and UIM motifs exerted differential effects in suppressing rpn11-1 rpn10Δ. We speculate that Rpn10 might promote proteolysis by facilitating proteasome assembly through its interaction with Rpn11, and by steering the multiUb chain to resident deubiquitinating enzymes to initiate substrate degradation.
Materials and methods
Yeast strains and plasmids are described in Tables 1 and 2.
Table 1.
Yeast strains
| Strains | Genotype | Source |
|---|---|---|
| RJD1877 | W303: MATα can1–100, RPN11∷LEU2, his3–11, 15, trp1–1, ura3–1, ade2–1, rho + mpr1–1 | Verma et al. |
| RJD1786 | W303: MATα can1–100, Leu2–3, −112, his3–11, 15, trp1–1, ura3–1, ade2–1, rho +, mpr1–1 | Verma et al. |
| RJD1878 | W303: MATα can1–100, rpn11AXA∷LEU2, his3–11, 15, trp1–1, ura3–1, ade2–1, rho + mpr1–1 | Verma et al. |
| ACY135 | RJD1786 + rpn10D∷HIS3 + pGAL-RPN11∷URA | This study |
| ACY175 | RJD1877 + rad23Δ∷URA3 | This study |
| ACY176 | RJD1786 + rad23Δ∷URA3 | This study |
| ACY177 | RJD1878 + rad23Δ::URA3 | This study |
| MHY961 | MATa his3–200, leu2–3,122, lys2–801, trp1–1, ura3–52, rpn10Δ∷HIS3 | M. Hochstrasser |
Table 2.
Plasmids
| Plasmid | Description | Origin |
|---|---|---|
| pNOY285 | Derivative of pRS316-GAL-Rpn11 | M. Nomura |
| AEP105 | PCUP1–FLAG-Rpn10∷TRP1 | This study |
| AEP80 | PCUP1–FLAG-rpn10-vwa∷TRP1 | This study |
| AEP106 | PCUP1–FLAG-rpn10-uim∷TRP1 | This study |
| AEP107 | PCUP1–FLAG-rpn10-vwa-uim∷TRP1 | This study |
| AEP101 | pGEX-GST–Rpn10 | This study |
| AEP102 | pGEX-GST–rpn10-uim | This study |
| AEP103 | pGEX-GST–rpn10-vwa | This study |
| NEP10 | PCUP1–FLAG-rpn11(1–280)∷TRP1 | This study |
| NEP11 | PCUP1–FLAG-rpn11(1–288)∷TRP1 | This study |
| NEP12 | PCUP1–FLAG-rpn11(1–294)∷TRP1 | This study |
| NEP13 | PCUP1–FLAG-rpn11(1–299)∷TRP1 | This study |
| NEP14 | PCUP1–FLAG-rpn11(1–303)∷TRP1 | This study |
| HEP56 | PCUP1–FLAG-Rpn11(1–306)∷TRP1 | This study |
| LEP385 | pUNI50-Rpn8 | D. Skowyra |
| LEP386 | pUNI50-Rpn9 | D. Skowyra |
| LEP387 | pUNI50-Rpn10 | D. Skowyra |
| LEP389 | pUNI50-Rpn12 | D. Skowyra |
| LEP521 | pET28-his6-Rpn10 | This study |
| LEP545 | pET28-His6-Rpn8 | This study |
| LEP601 | pET28-His6-Rpn11 | This study |
| LEP544 | pGEX-GST-Rpn8 | This study |
| LEP405 | GST-Rpn9 | D. Skowyra |
| LEP406 | GST-Rpn10 | D. Skowyra |
| LEP408 | GST-Rpn12 | D. Skowyra |
Reagents
Suc-Leu-Leu-Val-Tyr-AMC (7-amido-4-methylcoumarin), epoxomicin, multiUb chains (Ub2–7) and anti-His6 antibodies were purchased from Boston Biochem. Anti-Flag and GST-affinity beads were purchased from Sigma Chemical Co., and GE Healthcare, respectively. Polyclonal antibodies against Rpn11 and Rpt1 were generated by Pocono Rabbit Farm and Laboratory, Inc. (Canadensis, PA, USA). Anti-Rpn10 and Rpn12 antibodies were kindly provided by D. Skowyra (St. Louis Univ.). Antibody signals in immunoblots were developed using an enhanced chemiluminenscence reagent (ECL) from Perkin Elmer, and detected using Kodak GelLogic 1500 Imager. Fluorescence released by the hydrolysis of LLVY-AMC was measured using Tecan Infinite F200 plate reader.
In vitro translation reaction and binding assay
In vitro 35S-protein translation reaction was performed as described by the manufacturer (TnT Coupled Reticulocyte Lysate System; Promega). Reactions of 50 µl containing TnT Rabbit Reticulocyte Lysate (25 µl); TnT reaction buffer (2 µl); TnT RNA Polymerase T3 (1 µl); Amino Acid mixture lacking methionine (1 µl); [35S] methionione (1,175 µCi/µMol; Perkin Elmer) (2 µl); Rnase Ribonuclease Inhibitor (40 µ/µl; 1 µl); DNA template (2 µl; 0.5 mg/ ml) and nuclease-free water (16 µl) were incubated at 30°C for 90 min. The reaction was terminated by the addition of 2× SDS-loading buffer (4 µl). The reaction (46 µl) was diluted into 500 µl with protein binding buffer, and mixed with purified GST-proteins at 4°C for 4 h. The GST-protein beads were washed four times with binding buffer to remove unbounded 35S-proteins. Proteins retained on the GST affinity matrix were separated in 10% SDS-PAGE. The gels were stained with Coomassie blue and dried, prior to exposure to X-ray film.
Purified GST-proteins were mixed with bacterial lysate containing recombinant His6-tagged proteins, for 4 h at 4°C. The beads were washed four times with binding buffer, and the bound proteins were separated in 10% SDS-PAGE. Proteins were transferred to nitrocellulose membrane and incubated with antibodies against His6.
DNA oligonucleotides
The following sequences were used for generating carboxyl-terminal truncations in Rpn11. A common 5′-forward primer was used in polymerase chain reactions (PCR) for generating all the Rpn11 truncations. Oligonucleotide sequences are in the conventional 5′→ 3′ orientation.
Forward primer: Oligo #733: CCGAATTCATGGAACGACTACAGAGATT
rpn11 1–280 :Oligo #921: CCGGGTACCTCAGGAAAGGTGCTTCTTTGG
rpn111–288: Oligo #922: CCGGGTACCTCACTCTAGTGTCTCATCTGCTGT
rpn111–294: Oligo #923: CCGGGTACCTCACACAGAAACAATATTGTTCTC
rpn111–299: Oligo #926: CCGGGTACCTCAAACACCCGCCGTCAGCACAGA
rpn111–303 Oligo #925: CCGGGTACCTCATGCCACTGAATTAACACCCGC
To generate the rpn10vwa (D11A) mutation, a forward primer containing an EcoRI DNA restriction site was used (oligo #945: CCGGGAATTCATGGTATTGGAAGC TACAGTGTTAGTGA TTGCTAATTCAGAGTAC). The reverse primer contained a stop codon, followed by a KpnI DNA restriction site (oligo #245: GCGGTACCTATTT GTCTT GGTGTTGTTCAGGCTG).
Construction of the rpn10uim mutation was guided by previous studies, and involved conversion of amino acid residues LAMAL (228 to 332) to NNNNN (5N), using a two-step PCR reaction. In the first amplification cycle, 2 PCR fragments were produced. The following forward and reverse primers (#245) were used for amplifying PCR Fragment 1 (oligo #946: CAATGGACCCAGAAAATAAT AATAATAATCGTCTGTCTATGGAAGAAGAGCAG).
To generate PCR Fragment 2 the forward primer and reverse primer containing an EcoRI DNA restriction site were used (oligo #947: GCTCTTCTTCCATAGACAGA CGGTTATTGTTATTGTTTTCTGGGTCCATTGATGG GTC, and oligo #244: CCGGGAATTCATGGTATTGG AAGCTAC AG). To generate full-length rpn10uim DNA, PCR fragments 1 and 2 were annealed and re-amplified using oligo #244 and oligo #245. To generate the rpn10vwa uim double mutant oligo #945 and oligo #245 were used with rpn10uim template DNA.
PGAL1∷RPN11 shut-off
Initial attempts to generate rpn11-1 rpn10Δ double mutant in Saccharomyces cerevisiae were unsuccessful, and raised the possibility that the double mutant was inviable. We therefore sought to generate a double mutant whose viability was supported by conditional expression of the RPN11 gene. To achieve this we transformed rpn11-1 with a plasmid expressing RPN11 from the GAL1 promoter (PGAL1). This strain was maintained on galactose medium and the RPN10 gene was deleted by homologous recombination. The strain (ACY135) was verified by PCR, to demonstrate loss of the RPN10 gene, as well as by immunoblotting to show loss of Rpn10 protein. As noted in the text, when rpn11-1 rpn10Δ was plated on glucose expression of Rpn11 ceased, and caused cell death. However, viability was rescued by constitutive expression of either Rpn11 or Rpn10 from a second plasmid. To examine the defect caused by the simultaneous loss of Rpn10 and Rpn11 we transferred ACY135 from galactose to glucose medium. A detectable level of Rpn11 was present even after prolonged incubation in liquid, glucose medium, since Rpn11 is a stable protein (Chandra et al. 2010). However, significant inhibition of growth was observed by 48 h on glucose medium, suggesting that growth is not supported following depletion of Rpn11. To improve the termination of Rpn11 expression, following transfer from galactose to glucose medium, we grew the ACY135 for ~ 18 h in 0.6% glucose + 2% galactose. This pre-culture was subsequently transferred to YP-dextrose medium to inhibit PGAL1∷RPN11 transcription. We characterized protein extracts after 48 h incubation in glucose medium, because at this time growth inhibition of rpn11-1 rpn10Δ was clearly evident.
Native PAGE
A native gel was used to examine proteasome assembly and activity as described (Glickman et al. 1998a). Briefly, yeast cells were disrupted with glass beads and the lysate was centrifuged at 12,000×g for 5 min. Fifty micrograms of the lysate was electrophoresed in a native polyacrylamide gel that was prepared with step-gradients of 3, 4 and 5% acrylamide, and electrophoresed (125 V × 3 h). The acrylamide gel was incubated in buffer containing Suc-LLVY-AMC and the fluorescent signal released by AMC was detected with a Kodak Imager. The gel was subsequently incubated with buffer containing Suc-LLVY-AMC and 0.05% sodium dodecylsulfate (SDS) to enhance detection of 20S core peptidase activity.
Immunoprecipitation and immunoblotting
Yeast cells were suspended in buffer A (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA and 1% Triton X-100) containing a protease inhibitor cocktail (Roche), and lysed with a FastPrep disruptor (FP120-Thermo Savant). Cell debris was removed by centrifugation at 12,000×g at 4°C for 2 min. Equal amount of protein extracts (Bradford, Bio-Rad) were incubated with FLAG-M2 affinity agarose beads (Sigma Co.), for 4 h at 4°C. The beads were washed three times with buffer A, and the bound proteins were released by boiling in SDS loading buffer and separated in either 10 or 12% SDS-tricine polyacrylamide gels, transferred to nitrocellulose and examined by immunoblotting.
Measurement of LLVY-AMC hydrolysis
Equal amounts of protein lysate (5 µg in 50 µl) were pre-mixed with 200 ng epoxomicin (prepared in 50% DMSO), or an equivalent volume of DMSO lacking the inhibitor. Proteasome assay buffer (200 µl; 25 mM HEPES pH 7.5, 0.5 mM EDTA) contained 40 µM LLVY-AMC. Reactions were incubated at 30°C for 1 h, and fluorescence was measured using a Tecan Infinite F200 detector. Epoxomicin sensitive activity generated from three independent experiments were quantified and graphed.
Results
Rpn10 and Rpn11 proteins can interact
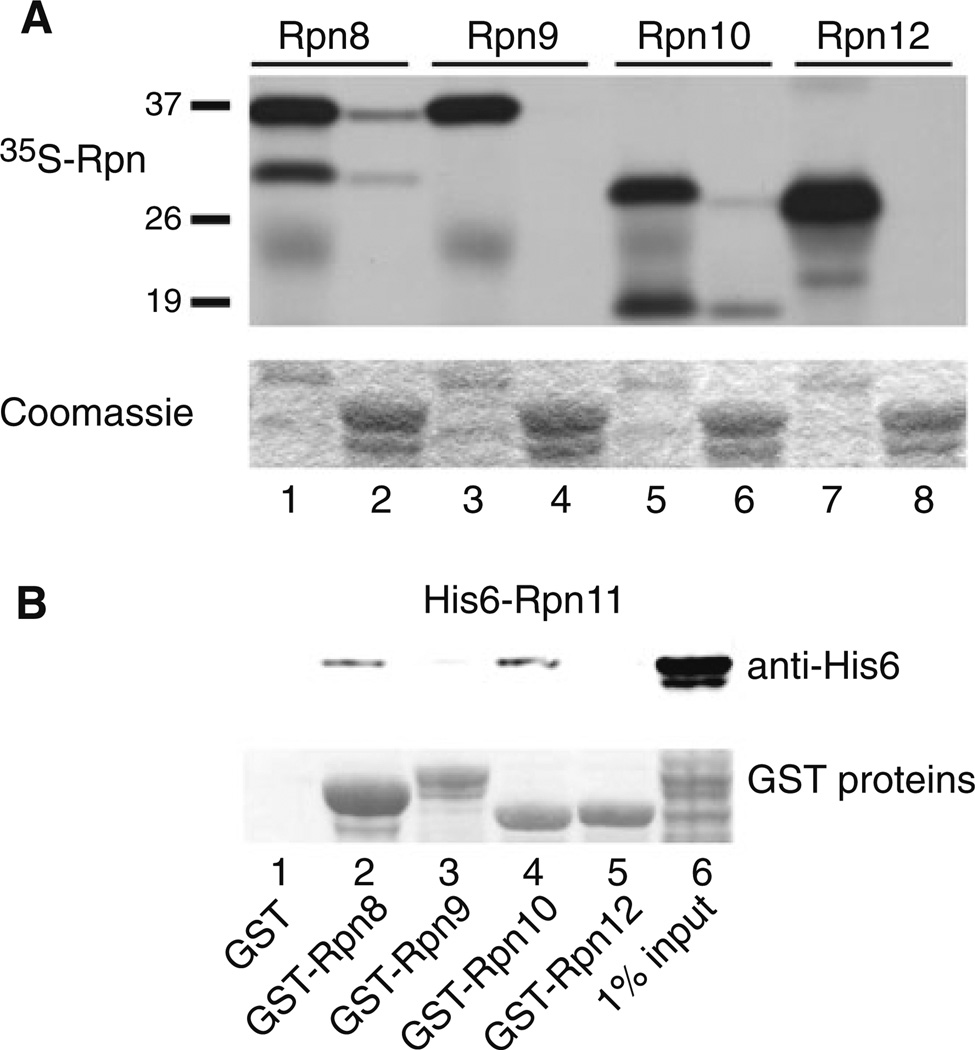
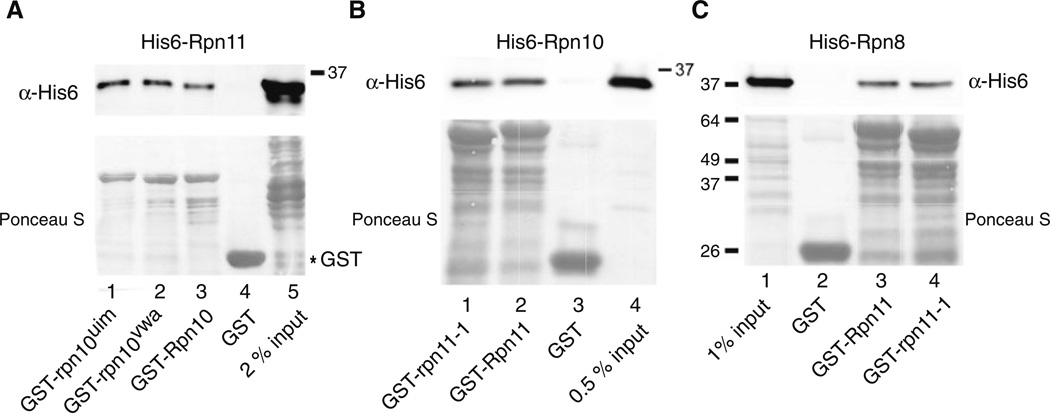
Plasmids expressing subunits of the 19S proteasome regulatory particle (RP), including Rpn8, Rpn9, Rpn10 and Rpn12, were used in an in vitro transcription-coupled translation reaction, and the 35S-labeled proteins were incubated with immobilized (recombinant) GST-Rpn11. Binding was determined by autoradiography, and a weak interaction was detected between Rpn11 and Rpn10 (Fig. 1a; lane 6). A previous report describing an interaction between Rpn11 and Rpn8 (Fu et al. 2001) was confirmed (lane 2). Rpn11 interactions with Rpn8 and Rpn10 were specific, since we did not observe any binding of Rpn11 to either Rpn9 (lane 4) or Rpn12 (lane 8). Truncated fragments of both Rpn8 and Rpn11 that were generated in the translation reactions could bind Rpn11. Intriguingly, the smaller fragment of Rpn10 formed a stronger interaction with Rpn11 than the full-length protein suggesting that key residues that contribute to the binding may be more spatially accessible in the fragment. Nonetheless, the weak binding between Rpn11/Rpn10 led us to reexamine this interaction in a reciprocal binding reaction. GST-tagged Rpn proteins (Rpn8, Rpn9, Rpn10 and Rpn12) were immobilized on glutathione-Sepharose and incubated with E. coli extracts containing His6-Rpn11 protein, and binding was measured by immunoblotting against the His6 epitope (Fig. 1b). We note that this reaction differs significantly from the earlier experiment, as the binding was measured in the presence of E. coli whole cell extracts. In agreement with the previous result, we found that both GST-Rpn8 (lane 2) and GST-Rpn10 (lane 4) could bind His6-Rpn11. Despite a weak, non-specific interaction between His6-Rpn11 and GST-Rpn9 (lane 3), convincing interactions were observed between Rpn11, and both Rpn8 and Rpn10. Moreover, these interactions are direct, since no other proteasome subunits were present in the E. coli cell extracts. To further characterize this interaction we generated two mutations in Rpn10, to determine if functionally impaired derivatives retained their ability to bind Rpn11. A site-specific mutation was generated in either the VWA (rpn10vwa; D11 → A11 ) (Fu et al. 2001) or UIM motifs (rpn10uim; 228LAMAL232 → 228NNNNN232) (Elsasser et al. 2004), to disrupt Rpn10 interactions with the proteasome and ubiquitin, respectively (see Supplemental Data; Fig. S1). GST-Rpn10, GST-rpn10vwa and GST-rpn10uim were purified from E. coli (Fig. 2a, lower panel; Ponceau S) and incubated with His6-Rpn11. The bound proteins were resolved by SDS/PAGE and examined by immunoblotting. No significant interaction was detected between GST and His6-Rpn11 (lane 4). In contrast, His6-Rpn11 formed similar interactions with GST-tagged Rpn10, rpn10vwa and rpn10uim (lanes 1–3). In a complementary study His6-Rpn10 was incubated with recombinant GST-Rpn11 and GST-rpn11-1 (Rinaldi et al. 1998), which lacks a critical carboxy-terminal domain that is required for efficient proteasome assembly (Chandra et al. 2010). Immunoblotting showed that His6-Rpn10 could bind both Rpn11 and rpn11-1 (Fig. 2b; lanes 1 and 2), and the non-specific interaction with the GST control beads was minor (lane 3). In a control experiment we verified Rpn11 interaction with Rpn8 (Fig. 2c). As observed with Rpn10, Rpn8 could bind both Rpn11 and rpn11-1 (lanes 3 and 4).
Fig. 1.
Direct interaction between Rpn11 and Rpn10. a A number of proteasome subunits from the 19S regulatory particle were radiolabeled with 35S-methionine, using an in vitro transcription/translation coupled reaction. The 35S proteins were examined to assess the quality of the labeling reaction (2% input: lanes 1, 3, 5 and 7). Following incubation of the 35S proteasome subunits with immobilized GST-Rpn11 (that was purified from E. coli), binding (even numbered lanes) was detected by autoradiography. The lower panel shows Coomassie staining of a polyacrylamide gel to indicate equal levels of the GST-Rpn11 on the glutathione-Sepharose affinity beads. b The weak interaction between Rpn11 and Rpn10 detected in a (lane 6) was verified in a reciprocal binding experiment. Recombinant His6-Rpn11 was purified from E. coli, and incubated with GST-tagged Rpn8, Rpn9, Rpn10 and Rpn12. The bound proteins were released into SDS containing buffer and electrophoresed by SDS/ PAGE. The filter was incubated with anti-His6 antibodies to detect interaction of the immobilized GST proteins with His6-Rpn11 (upper panel). The lower panel is a Ponceau S staining of the nitrocellulose filter to show the levels of the immobilized GST-tagged proteasome subunits
Fig. 2.
Well-characterized mutants do not affect Rpn10/Rpn11 interaction in vitro. a Rpn10 and mutant derivatives rpn10uim and rpn10vwa, were expressed as fusions to GST, purified from E. coli, and immobilized on glutathione-Sepharose beads. His6-Rpn11 was incubated with the beads, and interaction was determined by immuno-blotting. The upper panel shows interaction of His6-Rpn11 with the Rpn10/rpn10 proteins, and the lower panel shows the amount of GST-tagged proteins bound to the affinity matrix. b In a complementary experiment, Rpn11 and rpn11-1 protein mutants were fused to GST and immobilized on glutathione-Sepharose. Interaction with His6-Rpn10 was determined by immunoblotting (upper panel). The lower panel shows the level of GST proteins on the Sepharose beads. c Because both Rpn8 and Rpn10 bind Rpn11, we investigated if the rpn11-1 mutant protein had distinct interaction patterns with another proteasome subunit (Rpn8). Binding reactions were performed as described in b
Lethality caused by loss of both Rpn11 and Rpn10
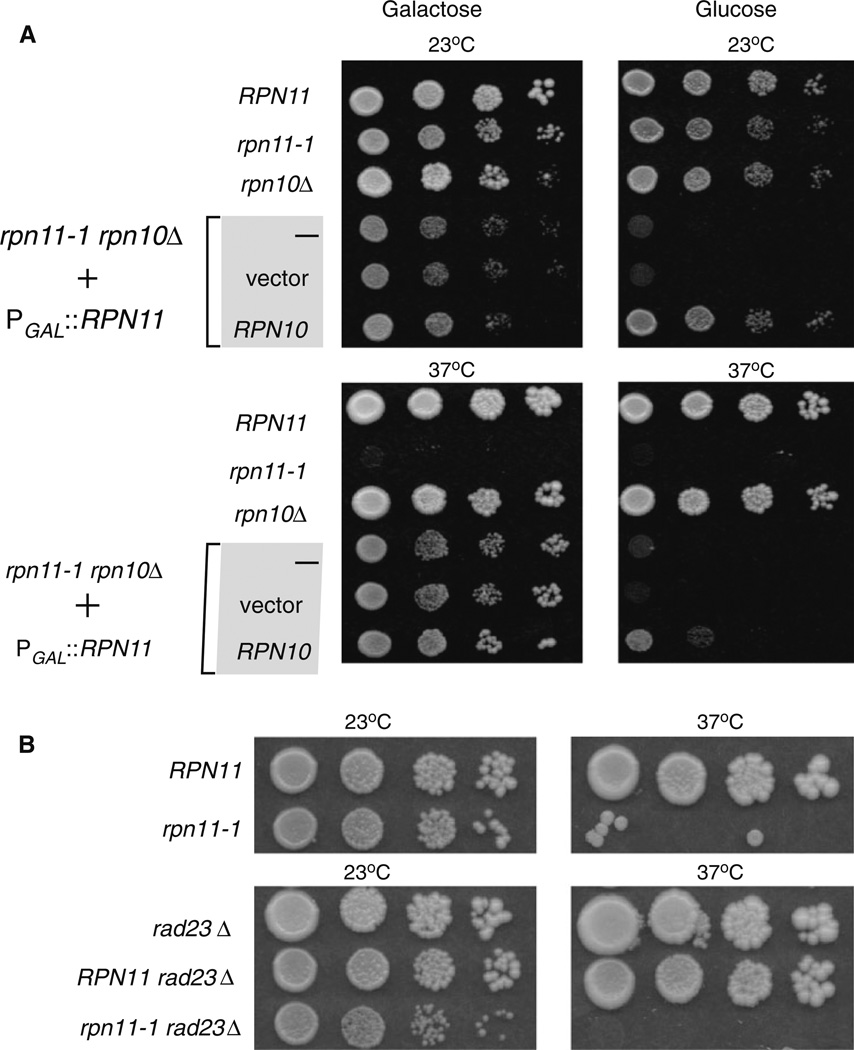
Rpn10 encodes a multi-ubiquitin chain binding receptor (Deveraux et al. 1994; van Nocker et al. 1996), and is present in near stoichiometric levels in proteasomes (Glickman et al. 1998b). However, a major fraction of cellular Rpn10 is not detected in proteasomes (van Nocker et al. 1996), raising the possibility that it might have additional roles. In comparison to other proteasomes subunits, loss of Rpn10 does not cause lethality (Fu et al. 1998; van Nocker et al. 1996), and rpn10Δ cells do not display conspicuous growth defects. However, rpn10Δ shows modest sensitivity to drugs that increase the levels of damaged proteins. Rpn10 also plays a role in stabilizing interactions between sub-complexes in the 19S regulatory particle (Glickman et al. 1998a). In contrast, loss of Rpn11 causes lethality. The rpn11-1 mutant expresses a carboxy-terminal frameshift, in which 31 C-terminal residues are replaced with 13 incorrect residues (Rinaldi et al. 1998). Although this mutant is viable, it displays a strong temperature-sensitive growth defect. Growth at 23 °C is slower than the wildtype strain, and no growth is observed at 37°C. Based on the interaction we observed between Rpn11 and Rpn10 we attempted to construct an rpn11-1 rpn10Δ double mutant. We were unable to generate this strain, which indicated that the double mutant is inviable. We therefore generated an rpn10 deletion in rpn11-1 that expressed wildtype Rpn11 from a plasmid containing a galactose-inducible PGALI1 promoter. The rpn11-1 rpn10Δ + PGAL1∷RPN11 strain (ACY135) was maintained on galactose medium. The expression of Rpn11 from PGAL1∷RPN11 could be turned off by transferring the cells to glucose medium. The conditional expression of Rpn11 in rpn11-1 rpn10Δ allowed us to examine the requirement for various derivatives of Rpn10 and Rpn11. Consistent with previous reports, rpn11-1 was temperature sensitive to growth at 37°C, as shown by tenfold serial dilutions of exponential-phase yeast cultures (Fig. 3a). rpn10Δ showed no obvious growth defect, and its colony-forming ability was indistinguishable from the RPN10 wildtype strain on glucose and galactose medium, at both 23 and 37°C. The synthetic lethality of the double mutant ACY135 (rpn11-1 rpn10Δ) was confirmed on glucose medium at 23 and 37°C, when the expression of Rpn11 was shut off. As expected, PGAL1∷RPN11 could fully overcome the synthetic lethality of rpn11-1 rpn10A on galactose medium, at both 23 and 37°C (Fig. 3a; lower left panel). In contrast, constitutive expression of Rpn10 from a second plasmid permitted growth on glucose medium only at 23 °C (upper right panel), showing that it can overcome the synthetic lethality of rpn11-1 rpn10Δ. However, the temperature sensitive growth defect of rpn11-1 (at 37°C) in rpn11-1 rpn10Δ was not suppressed by Rpn10 (on glucose). Weak growth on glucose medium at 37°C (bottom right panel), suggests that high level expression of Rpn10 might partially alleviate the temperature sensitivity of rpn11-1. But, we note that viability was still ~ 100-fold lower than the wildtype strain. In contrast to Rpn10, expression of Flag-Rpn11 from a second plasmid rescued both the inviability of rpn11-1 rpn10Δ. on glucose, and the temperature sensitivity of rpn11-1 (see below in Fig. 4b).
Fig. 3.
Loss of Rpn10 in rpn11-1 causes synthetic lethality. a A Saccharomyces cerevisiae rpn11-1 rpn10Δ strain was propagated in 2% galactose to maintain expression of Rpn11 from the galactose-inducible GAL1 promoter (left panels). Tenfold dilutions of exponential-phase yeast cells were plated, and growth was examined after 3 days at the indicated temperatures. The bottom three rows show rpn11-1 rpn10Δ + PGAL1∷RPN11 (ACY135), containing either an empty vector, or a plasmid expressing Rpn10. Growth of yeast strains was examined on 2% glucose medium, at both 23 and 37°C. b The specificity of rpn11-1 rpn10Δ associated lethality was determined by examining rpn11-1 rad23Δ
Fig. 4.
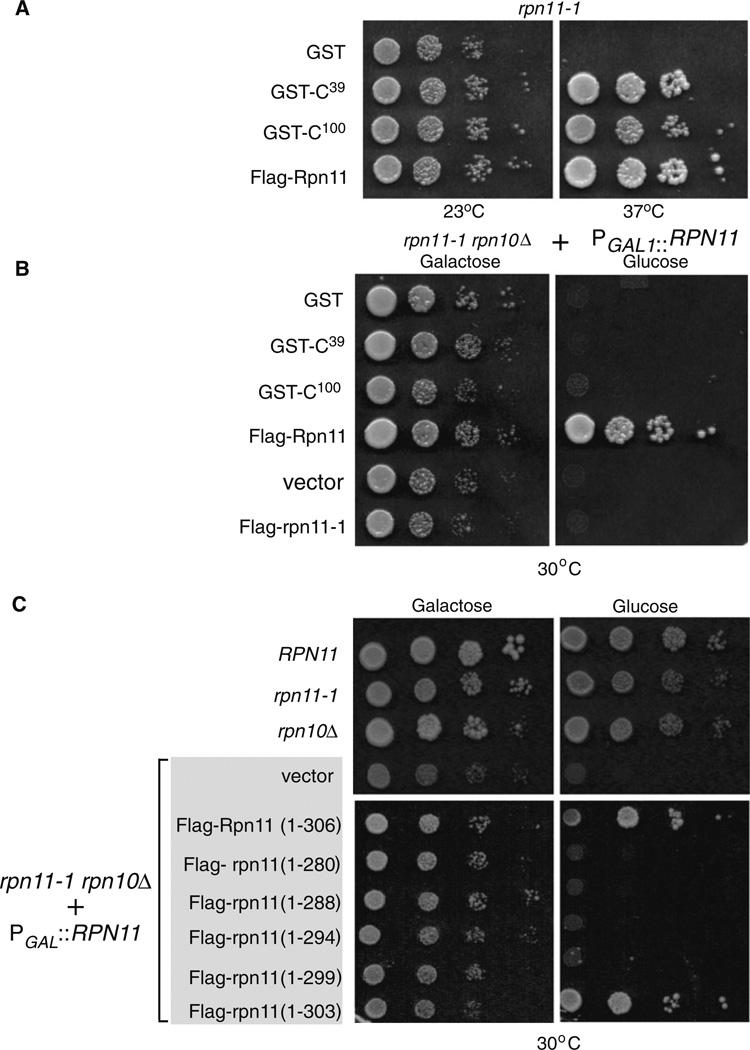
A key role for the carboxy terminus of Rpn11 in proteasome stability. a The carboxyl-terminal domain of Rpn11 can suppress the growth defects of rpn11-1. The C-terminal 39 and 100 residues of Rpn11 were fused to GST and expressed in rpn11-1. A control plasmid expressing full-length Rpn11 (Flag-Rpn11) was also examined. The temperature sensitive (37°C) growth defect of rpn11-1 is evident when only GST is expressed. b The C-terminal GST-fusion proteins were expressed in ACY135. Growth on galactose and glucose medium was investigated at 30°C. For a positive control, we confirmed that Flag-Rpn11 fully suppressed the growth defects. Flag-rpn11-1 was overexpressed to determine if high levels would overcome the defect of rpn11-1 rpn10Δ mutant. c A series of C-terminal truncations in Rpn11 were generated and expressed in ACY135. Yeast cells were plated on galactose and glucose medium, and growth was determined at 30°C
In a previous study we showed that loss of both Rpn10 and Rad23 (rad23Δ rpn10Δ) caused pleiotropic defects (Lambertson et al. 1999). Rad23 encodes a shuttle-factor that can deliver ubiquitinated substrates to proteasome receptors (Chen and Madura 2002), such as Rpn10, and the loss of both proteins caused cold-sensitive growth, and proteolytic deficiencies. Because Rpn10 is functionally linked to both Rpn11 and Rad23, we investigated if Rad23 also contributed to Rpn11 function. Unlike rpn11-1 rpn10Δ, deletion of the gene encoding Rad23 in rpn11-1 (rpn11-1 rad23Δ) had no effect (Fig. 3b). This result supports our view that the genetic and physical association between Rpn10 and Rpn11 is functionally significant and specific.
Suppression of the rpn11-1 rpn10Δ growth defect requires the carboxy-terminus of Rpn11
The carboxy-terminal region that is deleted in rpn11-1 protein is caused by a frame-shift mutation that results in the replacement of 31 C-terminal residues by 13 non-native residues (Rinaldi et al. 1998). This domain plays an important role in promoting proteasome assembly and stability. Intriguingly, expression of high levels of 39 carboxy-terminal residues of Rpn11 (GST-C39) significantly improved proteasome assembly, and alleviated the growth defects of rpn11-1. Similarly, a previous study showed that the C-terminal 100 amino acid residues of Rpn11 (GST-C100) could suppress the defects of rpn11-1 (Rinaldi et al. 2008). We reproduced these findings to show that both GST-C39 and GST-C100 could significantly overcome the growth defect of rpn11-1 at 37°C (Fig. 4a). We therefore determined if the carboxy-terminal domain of Rpn11 could also overcome the inviability of rpn11-1 rpn10Δ. We transformed ACY135 with plasmids expressing glutathione S-transferase (GST), GST-C39 and GST-C100. Yeast cells were plated on galactose and glucose media, and growth was examined after 3 days of incubation at 30°C (Fig. 4b). The carboxy-terminal domains failed to overcome the synthetic lethality of rpn11-1 rpn10Δ at both 23°C (data not shown) and 30°C in glucose (Fig. 4b), although they fully suppressed the temperature sensitive growth defect of rpn11-1 (Fig. 4a). This is an intriguing result, because rpn10Δ by itself lacks a conspicuous growth defect and yet its loss in an rpn11-1 genetic background contributes to a significant defect.
Because the carboxy-terminus of Rpn11 promotes proteasome stability, we transformed ACY135 with plasmids encoding rpn11 proteins containing small, carboxy-terminal truncations. Only wildtype Flag-Rpn11 (1–306) and Flag-rpn111–303 complemented the growth defect of rpn11-1 rpn10Δ. on glucose medium at 30°C (Fig. 4c). In contrast, Flag-rpn111–299 and other truncations failed to suppress the synthetic lethality of rpn11-1 rpn10Δ., indicating a key requirement for these carboxy-terminal residues. All the truncated proteins showed comparable expression in yeast cells (see Supplemental Data; Fig. S2), so their inability to suppress the growth defect of rpn11-1 rpn10Δ is not due to protein instability, or reduced expression. Flag-Rpn11 was constitutively expressed in ACY135, and full growth was observed on glucose medium at 30°C. This finding showed that Flag-Rpn11 can replace Rpn11 that was expressed from the Pgal1 promoter. Because rpn11-1 is an unstable protein, we investigated if higher expression would suffice to restore growth of the double mutant. rpn11-1 rpn10Δ containing either an empty vector or expressing Flag-rpn11-1 grew on galactose but not on glucose medium, when the expression of Rpn11 was turned off (Fig. 4b). This finding shows that high-level expression of rpn11-1 protein cannot overcome the defects caused by the rpn11-1 mutation.
VWA, but not UIM, is required for suppressing rpn11-1 rpn10Δ
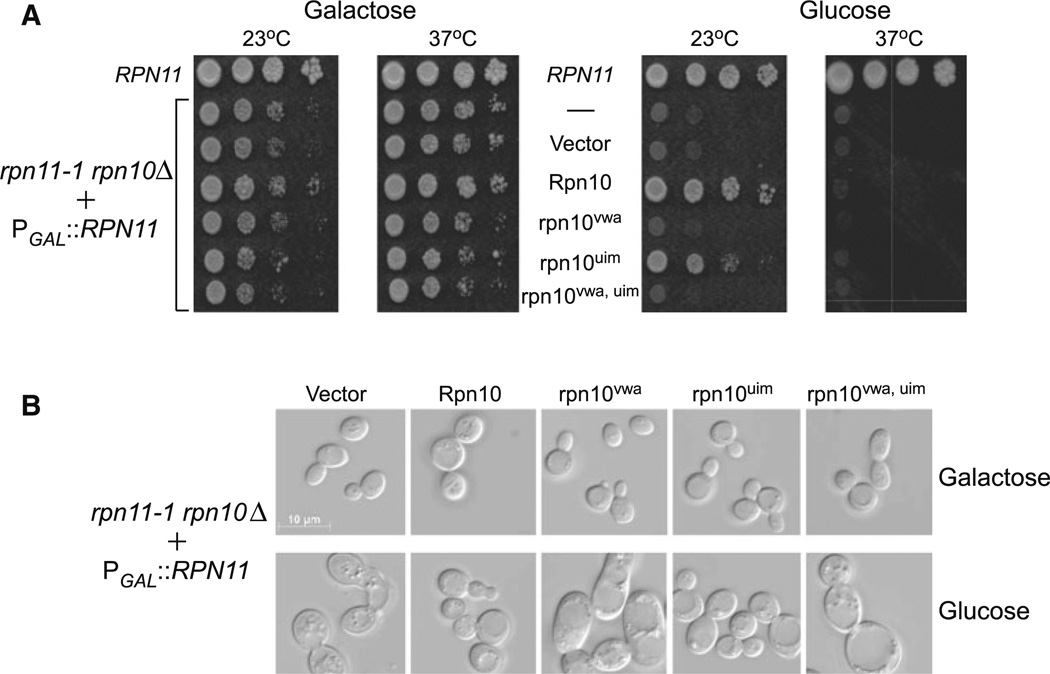
The Rpn10 protein contains two well-characterized motifs. A ubiquitin-interacting-motif (UIM) forms preferential interactions with multiubiquitin chains (Hofmann and Falquet 2001), and is required for the degradation of ubiquitinated proteins (Verma et al. 2004). A von Willebrand A (VWA) domain in the amino half of Rpn10 was reported to promote proteasome stability and/or assembly. We generated mutations in both domains, confirmed their binding properties (see Supplemental Data; Fig. S1), and expressed plasmids encoding these Flag-tagged derivatives in ACY135. Yeast cells were grown on galactose and glucose medium, at both 23 and 37°C (Fig. 5a). The yeast strains were adjusted to equal density and tenfold serial dilutions were spotted on agar medium, and growth was examined after 3 days of incubation. As expected, efficient growth of all strains was observed on galactose medium at both 23 and 37°C, since Rpn11 was expressed from the PGALI1 promoter. However, on glucose medium, Rpn11 expression ceased and Rpn10 rescued growth at 23°C, but not 37°C (also see Fig. 3a). Similarly, rpn10uim supported growth on glucose medium only at 23°C. However, neither rpn10vwa nor a double mutant (rpn10vwa uim) supported growth at 23°C. These findings suggest that the ubiquitin-binding properties of Rpn10 (mediated by UIM) are not required for overcoming the synthetic lethality of rpn11-1 rpn10Δ, while the residues contributing to Rpn10/proteasome interaction (VWA domain) are required.
Fig. 5.
Differential effect of rpn10 mutant proteins. a Rpn10, rpn10vwa and rpn10uim were expressed in rpn11-1 rpn10Δ mutant (+PGAL1 RPN11), and growth was determined on galactose and glucose medium, at both 23 and 37°C. ACY135, with or without an empty vector (second and third rows), showed a similar loss of viability on glucose. The effect of expressing an Rpn10 double mutation (rpn10vwa uim) in rpn11-1 rpn10Δ was also determined. b The ability of the mutant rpn10 proteins to suppress the morphological defects of rpn11-1 rpn10Δ was examined by microscopy. Yeast cells were plated on glucose medium for 48 h and then examined with a Zeiss Axioplan microscope. A representative view of yeast cells, from both galactose and glucose medium (23 °C cultures) is shown
The synthetic lethality associated with loss of both Rpn11 and Rpn10 is preceded by significant morphological changes. rpn11-1 rpn10Δ cells expressing Rpn10 and mutant derivatives were plated on synthetic complete glucose medium for 2 days, and then suspended in YPD liquid medium for 2 h and visualized by microscopy. We found that cells expressing empty vector (Fig. 5b) showed loss of cell integrity, and the accumulation of small vesicles (see Glucose). In contrast, cells expressing either Rpn10 or rpn10um showed normal morphology. Consistent with the results described in Fig. 5a, cells expressing rpn10vwa or rpn10vwa uim showed significant morphological changes including large size, multiple buds, and the presence of one or more large vacuoles.
The proteasome interaction domain of Rpn10 is required for suppression of rpn11-1 rpn10Δ
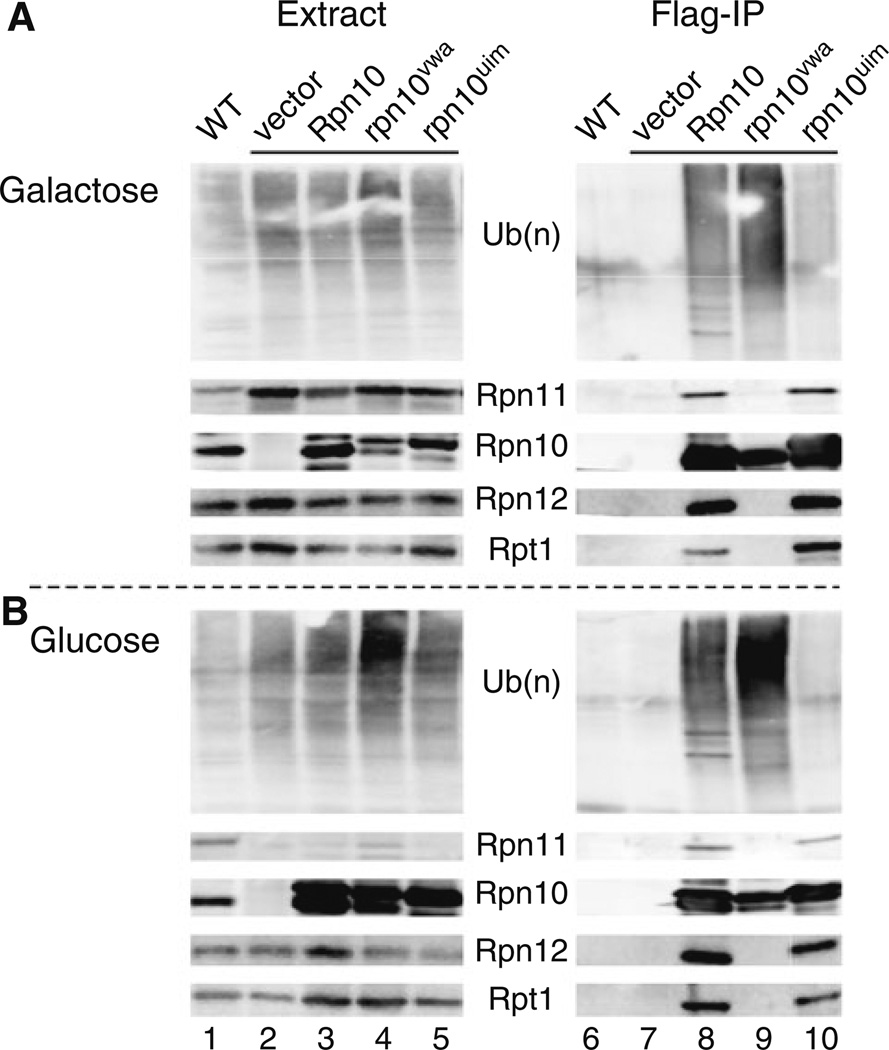
To more clearly delineate the requirement for the VWA and UIM domains we expressed Flag-rpn10uim and Flag-rpn10vwa mutant proteins in ACY135, and measured their interactions with multiubiquitin chains and the proteasome. Yeast cells were grown in either galactose (expressing Rpn11), or in glucose medium (lacking Rpn11) at 23°C, and protein extracts were prepared. An equal amount of protein was resolved in an SDS-polyacrylamide gel and characterized by immunoblotting (Fig. 6a). The absence of Rpn10 is seen in Fig. 6a (Extract; lane 2). Endogenous Rpn11 was detected in a wildtype strain (WT; lane 1), and higher levels were present in rpn11-1 rpn10Δ over-expressing Rpn11 (PGAL1∷RPN11: lanes 2–5). The levels of other proteasome subunits (Rpn12 and Rpt1) were essentially unaffected, indicating that overall proteasome abundance is not significantly altered by the increased levels of Rpn11 (compare lanes 1 and 2). Under conditions leading to cell death (galactose → glucose), we detected increased abundance of multiubiquitinated species in cell extracts expressing Flag-rpn10vwa (Fig. 6b; lane 4). As expected, the abundance of Rpn11 was reduced in glucose medium. We note that a protein band detected in glucose grown cells is expected to represent a mixture of endogenous rpn11-1 protein, and residual Rpn11 that persists after transfer to glucose medium.
Fig. 6.
Suppression of rpn11-1 rpn10Δ by Rpn10 requires the VWA domain. Flag-epitope tagged derivatives of Rpn10 and rpn10 mutants were expressed from plasmids, and transformed into ACY135. Protein extracts were prepared from galactose a, and glucose b grown cultures. Equal amount of protein was resolved by SDS/PAGE and characterized by immunoblotting with antibodies against the proteins indicated (Extracts; left panels). An equal amount of protein extract was also incubated with Flag-agarose to immunoprecipitate the Flag-tagged proteins. The co-purified proteins were detected by immunoblotting, using antibodies against the proteasome subunits shown
Protein extracts were incubated with Flag-agarose to immunoprecipitate the Flag-tagged Rpn10/rpn10 proteins (Fig. 6a, b; lanes 6–10). Both Flag-Rpn10 and Flag-rpn10vwa formed strong interactions with multiubiquitinated proteins, while much lower binding was detected with Flag-rpn10uim, in both galactose and glucose grown cells. The higher amounts of ubiquitinated proteins that were purified with Flag-rpn10vwa could be due to the increased abundance of ubiquitinated proteins (Fig. 6b; Extract), or to an inability to transfer them to the proteasome. Flag-rpn10vwa interaction with the proteasome was defective, as revealed by its failure to co-purify Rpn11, Rpn12 and Rpt1. This finding is consistent with previous studies showing a requirement of the VWA motif for binding the proteasome (Verma et al. 2004). In contrast, Flag-rpn10uim and Flag-Rpn10 formed similar interactions with the proteasome (compare lanes 8 and 10). These findings also verify that the VWA and UIM domains in Rpn10 have independent and separable functions. The control lanes (6 and 7) showed no non-specific precipitation of proteasome subunits, or ubiquitin. An intriguing finding is that the abundance of Rpn10 increased significantly following transfer to glucose medium (Fig. 6b; lanes 3–5). We speculate that this effect might reflect a compensatory response to the depletion of Rpn11 following transfer from galactose → glucose medium. A higher molecular weight derivative of Rpn10 could represent a mono-ubiquitinated form that was recently described (Isasa et al. 2010), and shown to affect Rpn10 function and stability.
Loss of Rpn10 in rpn11-1 alters proteasomes assembly
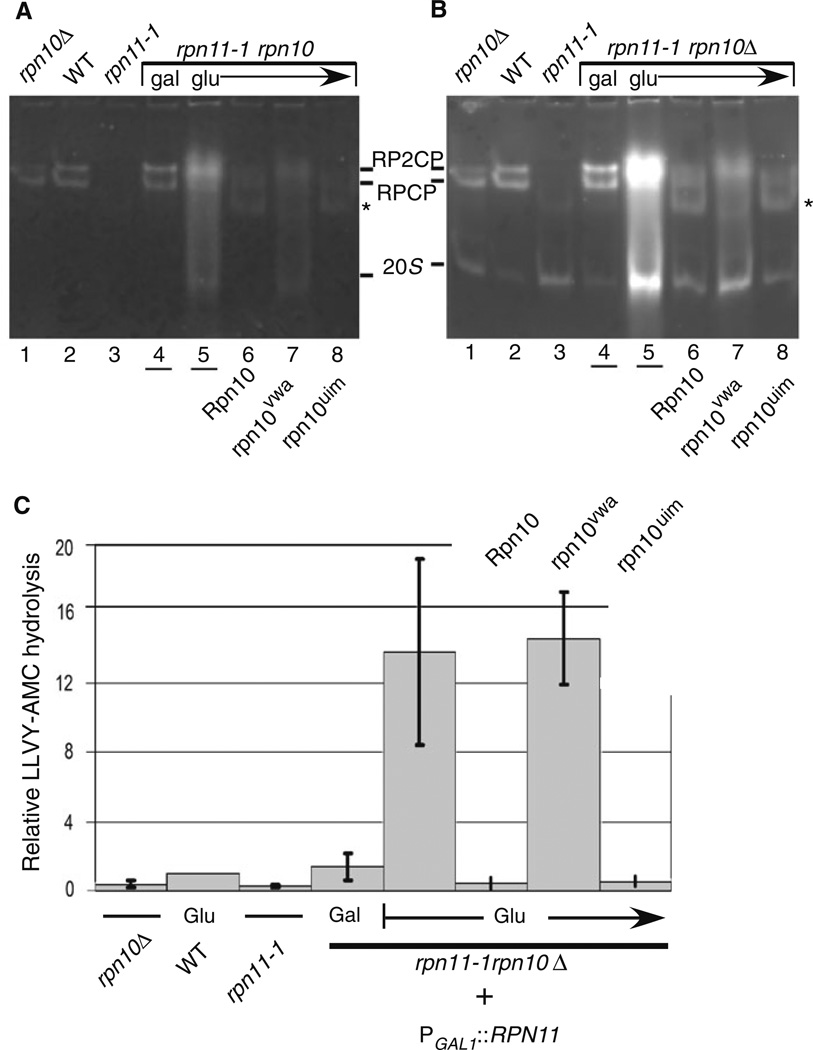
We and others have shown that proteasomes are unstable, or poorly assembled in rpn11-1 (Chandra et al. 2010; Rinaldi et al. 2008; Verma et al. 2002). Specifically, we reported that components of the 19S regulatory particle are vulnerable to dissociation, and reduced levels of 19S components were co-purified with 20S catalytic particles in rpn11-1 (Chandra et al. 2010). Moreover, lower amounts of ubiquitinated proteins were co-purified with proteasomes from rpn11-1, using an epitope-tagged subunit in the 20S core particle. Based on these results, we investigated if the synthetic lethality of rpn11-1 rpn10Δ was caused by further destabilization of proteasomes. To determine if proteasome instability was exacerbated in rpn11-1 rpn10Δ we performed an in-gel analysis (Chandra et al. 2010). Yeast cells were collected from glucose plates after 48 h, and equal amounts of protein (30 µg) were resolved in a native polyacrylamide gel, and then incubated in a buffer containing the fluorogenic proteasome substrate Suc-Leu-Leu-Val-Tyr-AMC (LLVY-AMC). A control sample representing cells that were maintained in galactose medium was also examined (Fig. 7a; lane 4). In the absence of detergent (Fig. 7a) the activity of intact proteasomes (RP2CP and RPCP) was detected. As reported previously, the abundance of fully assembled 26S proteasomes (RP2CP and RPCP) was reduced in rpn11-1 (lane 3) (Chandra et al. 2010). An apparent reduction in RP2CP level in rpn10Δ was not consistent, and no significant assembly defect was reliably observed in native conditions (lane 1). Proteasome assembly and activity in galactose-grown ACY135 (lane 4) was similar to the wildtype strain (lane 2), demonstrating complete rescue of the double mutant defect. However, after transfer to glucose medium we detected a slightly higher molecular weight form of the proteasome in rpn11-1 rpn10Δ containing empty vector (lane 5). This unexpected pattern is markedly different from that observed in rpn11-1 (lane 3), and rpn10Δ (lane 1). Moreover, this high molecular weight form of the proteasome had detectable peptidase activity (lane 5), in contrast to rpn11-1 which showed not significant activity at the positions of RPCP/ RP2CP.
Fig. 7.
Assembly defects associated with loss of both Rpn10 and Rpn11. a A representative native in-gel assay for proteasome activity is shown. Equal amounts of whole cell extracts were separated, and the positions of proteasome complexes containing the 20S core particle (CP) were detected by chymotryptic peptidase activity, using the fluorogenic substrate LLVY-AMC. RP2CP represents a single CP bound to two regulatory particles (RP), while RPCP represents CP bound to one RP. b To detect activity associated with the free 20S core particle, the gel shown in a was incubated in 0.05% SDS, and hydrolysis of LLVY-AMC was reexamined. The images were captured with Kodak GelLogic imager. c Data were generated from three independent measurements of LLVY-AMC hydrolysis, and epoxomycin-sensitive values were quantified. The values were normalized to the level measured in the wildtype strain (lane 2). The order of the samples is as shown in a
The gel described in Fig. 7a was subsequently treated with buffer containing 0.05% SDS, to detect the peptidase activity of the activated 20S core particle (CP). Overall chymotryptic activity was strongly increased following Rpn11 depletion in ACY135 (Fig. 7b; lane 5). High activity was associated with both the 20S core particle, and the high molecular weight form. The apparent size of the high molecular weight species differs slightly from the pattern seen in the wildtype strain (lane 2; RP2CP). Moreover, the significantly increased peptidase activity generated by this complex is quite unlike the activity observed with the wildtype 26S proteasome, which showed only modest activation by detergent (compare lanes 2 and 5). Because the abundance of proteasome subunits was not altered in ACY135, following transfer to glucose medium (Fig. 6b), we speculate that the increased LLVY-AMC hydrolysis in rpn11-1 rpn10Δ is due to higher specific activity of proteasomes, conceivably through persistent and futile stimulation. Similarly, the peptidase activity in rpn11-1 rpn10Δ expressing Flag-rpn10vwa was also elevated (Fig. 7b, lane 7). Furthermore, the migration pattern of proteasome sub-complexes differed from rpn11-1, and more closely resembled rpn11-1 rpn10Δ that only expressed vector. The expression of Flag-rpn10uim in rpn11-1 rpn10Δ (lane 8), showed an assembly profile that resembled rpn11-1 (lane 3), and the chymotryptic activity was similarly reduced. Expression of Flag-Rpn10 also restored the peptidase activity, and electrophoresis pattern to that observed in rpn11-1 (Fig. 7b; compare lanes 3 to 6).
Based on results indicating a failure of rpn10vwa to bind proteasomes (Fig. 6), we measured peptidase activity in ACY135 expressing specific rpn10 mutant proteins. We measured chymotryptic activity by monitoring the hydrolysis of the fluorogenic substrate LLVY-AMC (Fig. 7). Yeast strains were grown in galactose medium, transferred to medium containing 2% galactose + 0.6% glucose, and finally incubated in 2% glucose. Protein lysates (5 µg) were combined with 200 ng of proteasome inhibitor (epoxomicin), or an equivalent volume of dimethyl sulfoxide (DMSO) lacking the inhibitor. Reactions were incubated at 30°C for 1 h, and the fluorescence signal was measured. All measurements were collected in duplicate from three experiments, and the data representing epoxomicin sensitive signal were plotted (Fig. 7c). The y-axis shows relative level of chymotryptic activity, with the data standardized to the wildtype strain (arbitrary value of 1). Both rpn10Δ and rpn11-1 showed reduced total proteasome activity. Strikingly, rpn11-1 rpn10Δ expressing PGAL1∷RPN11 showed near normal levels of peptidase activity, consistent with full suppression of the functional defects caused by proteasome disassembly. Following transfer to glucose medium, rpn11-1 rpn10Δ expressing either Flag-Rpn10 or Flag-rpn10uim showed reduced peptidase activity, comparable to that detected in rpn11-1. In contrast, expression of an empty vector or Flag-rpn10vwa resulted in dramatic increase in LLVY-AMC hydrolysis, consistent with the in-gel analysis (Fig. 7b). The experimental values in these two strains were variably, possibly a result of rapid cellular responses to the depletion of functional proteasomes. Nonetheless, the high levels of chymotryptic activity were consistently >fivefold over the control strains.
Discussion
Substrate-interaction with the proteasome might represent the first committed step in protein turnover, since the degradation of unbound multiubiquitinated proteins can be avoided by deubiquitination (Chen et al. 2002). A general understanding of the mechanism by which proteolytic substrates bind the proteasome, become deubiquitinated, and unfolded is now available (Glickman and Ciechanover 2002; Pickart and Cohen 2004). A number of subunits have been reported to bind multiubiquitin chains (Deveraux et al. 1994; Husnjak et al. 2008; Lam et al. 2002; Schreiner et al. 2008; van Nocker et al. 1996; Verma et al. 2004), although their relative contributions to substrate selectivity, and preference for specific ubiquitin chain linkages are poorly defined. That substrate deubiquitination is closely coupled to degradation underscores a potentially cooperative and directed channeling of proteolytic substrates into the catalytic particle (Koulich et al. 2008; Verma et al. 2002; Yao and Cohen 2002). Moreover, substrate unfolding is required for translocating the deubiquitinated polypeptide chain into the 20S catalytic particle (Elsasser and Finley 2005; Zhang et al. 2009). However, the detailed sequence of events that lead from substrate interaction with the proteasome to degradation has not been described.
Previous studies showed that sub-complexes within the 19S regulatory particle were more readily detected in an rpn10Δ mutant (Glickman et al. 1998b). These studies indicated that Rpn10 could stabilize the lid and base complexes in the 19S particle. However, since an rpn10Δ mutant is viable, other proteasome subunits appear to augment this function. A number of studies have either directly, or through proteomic approaches, defined both the composition and activity of base subunits (Elsasser et al. 2002; Rubin et al. 1998), and their general arrangement within the 19S base complex. In contrast, the arrangement of subunits in the 19S lid sub-complex is still uncertain.
Rpn10 is a major multiubiquitin chain binding protein in the proteasome, and could play a key role in presenting ubiquitinated substrates to deubiquitinating enzymes in the proteasome, such as Rpn11. An intriguing possibility is that Rpn10 might interact with a multiubiquitin chain in an orientation that specifically promotes deubiquitination. A similar mechanism might underlie the recognition and processing of multiubiquitinated substrates by Rpn13 and Uch37 (Koulich et al. 2008; Schreiner et al. 2008).
In studies attempting to further characterize the mechanism of substrate recognition and deubiquitination we detected a weak interaction between Rpn11 and Rpn10. This interaction was verified using multiple methods, and validated genetically by showing synthetic lethality of the double mutant. The interaction between Rpn10 and Rpn11 described here has also been observed previously (Ferrell et al. 2000; Hendil et al. 2009). The interactions observed with purified proteins were not verified in vivo, although genetic studies implicate a strong functional link between these two multi-ubiquitin chain interacting proteins.
The synthetic lethality of rpn11-1 rpn10Δ was fully suppressed by either Rpn10 or Rpn11. Surprisingly, rpn10uim, which can bind the proteasome but not multi-ubiquitin chains, rescued the synthetic lethality of rpn11-1 rpn10Δ, while rpn10vwa, which binds multiubiquitin chains but not the proteasome, failed to suppress the growth defect. Moreover, expression of rpn10vwa was associated with altered proteasome assembly. Based on these findings we propose that inefficient proteasome assembly, rather than reduced interaction with ubiquitinated proteins, underlies the defect of rpn11-1 rpn10Δ. One interpretation of these results is that following successful assembly of the proteasome by rpn10uim (facilitated by the VWA domain), other multiUb chain binding proteins will facilitate substrate binding and degradation. This conclusion is in agreement with previous studies which showed that yeast expressing an rpn10 mutant that is unable to bind multiUb chains can restore degradation of substrates that are stabilized in rpn10Δ (Fu et al. 1998).
Rpn10 and Rpn11 may together improve the stability of the 19S regulatory particle by promoting the interaction between the base and lid sub-complexes. Although the 19S regulatory particle is more readily dissociated from the 20S particle in rpn11-1 we detected an altered form of the proteasome in rpn11-1 rpn10Δ. Specifically, a high molecular weight form, whose peptidase activity was strongly induced by detergent, was present at elevated levels. This property is strikingly different from wildtype 26S proteasomes, whose peptidase activity is not significantly increased in the presence of SDS. The S. pombe counterparts (Pus1; Pad1) also play a closely coupled role, as evidenced by the defects of pus1Δ pad1-1 (Wilkinson et al. 2000), and it is likely that these two factors also contribute to proteasome stability in the fission yeast. Other studies have also detected Rpn11 in various proteasome assembly intermediates (Hendil et al. 2009; Isono et al. 2007), further implicating an important role for this protein in proteasome assembly.
Rpn10/Rpn11 interactions differed in vitro and in vivo, since strong binding was observed between Rpn11 and rpn10vwa in vitro but not in vivo. One interpretation of this result is that the in vivo interaction between rpn10vwa and Rpn11 is inhibited by the high levels of multiubiquitin chains that are bound to rpn10vwa (Fig. 6). This would not occur in vitro, as the assay was performed in the absence of exogenous multiubiquitin chains. Another possibility is that Rpn10 might normally form only a transient interaction with Rpn11, to transfer substrate-linked multiubiquitin chains to this resident DUB. Finally, it is conceivable that Rpn10 has other partners in the cell, and the rpn10vwa mutant might form a persistent interaction with another factor in vivo. Taken together our findings suggest that a productive interaction between Rpn10 and the proteasome contributes to efficient assembly, and/or stability. The synthetic lethality caused by loss of both Rpn11 and Rpn10 (rpn11-1 rpn10Δ) suggests that the interaction between Rpn11 and Rpn10 is also functionally important. Intriguingly, the synthetic lethality of the double mutant was suppressed by an rpn10 mutant that could bind proteasomes, but not multiubiquitin chains, further supporting our assertion that a critical need for Rpn10 is to support proteasome assembly.
Supplementary Material
Acknowledgments
We thank D. Skowyra (St. Louis University) and M. Nomura (University of California, Irvine) for providing strains, plasmids and antibodies. We thank N. Torres for generating carboxy-terminal truncations in Rpn11. V. Tournier is thanked for review of the manuscript. These studies were supported by a grant from the National Institutes of Health (CA083875) to KM.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00294-010-0321-3) contains supplementary material, which is available to authorized users.
References
- Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- Chandra A, Chen L, Liang H, Madura K. Proteasome assembly influences interaction with ubiquitinated proteins and shuttle factors. J Biol Chem. 2010;285:8330–8339. doi: 10.1074/jbc.M109.076786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang B, Fischer JA. A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets. Genes Dev. 2002;16:289–294. doi: 10.1101/gad.961502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Mueller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004 doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Ferrell K, Wilkinson CR, Dubiel W, Gordon C. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem Sci. 2000;25:83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- Fu H, Sadis S, Rubin DM, Glickman M, van Nocker S, Finley D, Vierstra RD. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998a;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998b;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hendil KB, Kriegenburg F, Tanaka K, Murata S, Lauridsen AM, Johnsen AH, Hartmann-Petersen R. The 20S proteasome as an assembly platform for the 19S regulatory complex. J Mol Biol. 2009;394:320–328. doi: 10.1016/j.jmb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isasa M, Katz EJ, Kim W, Yugo V, Gonzalez S, Kirkpatrick DS, Thomson TM, Finley D, Gygi SP, Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, Toh-e A. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae . Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K, Prakash S. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD23 increases in response to UV light and in meiosis but remain constant in the mitotic cell cycle. Nucleic Acids Res. 1990;18:4737–4742. doi: 10.1093/nar/18.16.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi T, Ricordy R, Bolotin-Fukuhara M, Frontali L. Mitochondrial effects of the pleiotropic proteasomal mutation mpr1/rpn11: uncoupling from cell cycle defects in extragenic revertants. Gene. 2002;286:43–51. doi: 10.1016/s0378-1119(01)00799-5. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Pick E, Gambadoro A, Zilli S, Maytal-Kivity V, Frontali L, Glickman MH. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi T, Hofmann L, Gambadoro A, Cossard R, Livnat-Levanon N, Glickman MH, Frontali L, Delahodde A. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol Biol Cell. 2008;19:1022–1031. doi: 10.1091/mbc.E07-07-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Ferrell K, Penney M, Wallace M, Dubiel W, Gordon C. Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J Biol Chem. 2000;275:15182–15192. doi: 10.1074/jbc.275.20.15182. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.