Abstract
Plant growth is regulated by a complex network of signaling events. Points of convergence for the signaling cross-talk between the phytohormones auxin and gibberellin (GA), which partly control overlapping processes during plant development, are largely unknown. At the cellular level, auxin responses are controlled by members of the AUXIN RESPONSE FACTOR (ARF) family of transcription factors as well as AUXIN/INDOLE-3-ACETIC ACID INDUCIBLE (AUX/IAA) proteins that repress the activity of at least a subset of ARFs. Here, we show that the two paralogous GATA transcription factors GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED (GNC) and GNC-LIKE (GNL)/CYTOKININ-RESPONSIVE GATA FACTOR1 (CGA1) are direct and critical transcription targets downstream from ARF2 in the control of greening, flowering time, and senescence. Mutants deficient in the synthesis or signaling of the phytohormone GA are also impaired in greening, flowering, and senescence, and interestingly, GNC and GNL were previously identified as important transcription targets of the GA signaling pathway. In line with a critical regulatory role for GNC and GNL downstream from both auxin and GA signaling, we show here that the constitutive activation of GA signaling is sufficient to suppress arf2 mutant phenotypes through repression of GNC and GNL. In addition, we show that GA promotes ARF2 protein abundance through a translation-dependent mechanism that could serve to override the autoinhibitory negative feedback regulation of ARF2 on its own transcription and thereby further promote GA signaling.
The phytohormone auxin [indole-3-acetic acid (IAA)] regulates virtually all aspects of plant growth and development (1). At the cellular level, auxin responses are mediated by auxin response factors (ARFs) that are identified based on their ability to bind to promoter elements that confer auxin responsive gene expression [so-called auxin response elements (AuxREs)] (2, 3). Auxin responses also require the auxin-induced degradation of Aux/IAAs, which are repressors of a subgroup of ARF family transcription factors that are targeted for auxin-dependent degradation by the auxin receptor and E3 ubiquitin ligase SCFTIR1 and functionally related E3 ligase complexes (4, 5). Aux/IAAs also interact with the corepressor TOPLESS and its relatives, and ARF-targeted gene loci are transcriptionally inactive when ARFs are bound by Aux/IAAs and TOPLESS (6). Several independent studies as well as a larger genome-wide survey of pairwise ARF–Aux/IAA interactions have contributed to a picture where the ARF transcription factor family can be subdivided into ARF+ transcription activator ARFs that are negatively regulated by Aux/IAAs and ARF− repressor ARFs that interact only rarely with selected Aux/IAAs and therefore, would not be expected to be controlled by auxin and Aux/IAAs (3, 4, 7, 8).
Mutants expressing stabilized auxin-insensitive Aux/IAA variants have been described that, at least in some cases, mimic the loss-of-function phenotypes of their ARF interactors (9–13). Based on such phenotypic similarities, the Aux/IAA protein SOLITARY ROOT (SLR/IAA14) was recognized as the repressor of the functionally redundant ARF+ proteins ARF7 and ARF19 (positive regulators of lateral root formation) (7, 10, 11, 14, 15). Loss-of-function mutants of the ARF− protein ARF2 were described as mutants with defects in greening, senescence, flowering time, and floral organ abscission (16–18). Interestingly, arf2 mutant phenotypes are enhanced in mutants of the ARF+ proteins ARF7 and ARF19, suggesting that the Aux/IAA-independent ARF− ARF2 and Aux/IAA-dependent ARF+ ARF7 and ARF19 function together to control the same growth processes, presumably by regulating the same target genes (the identity remains to be determined) (16–19).
Gibberellins (GAs) are another family of plant hormones, and they are well known for their role in the control of germination, greening, and flowering time (20). The GA-labile DELLA proteins are key regulators of GA signaling that function by repressing different types of transcription factors, including the PHYTOCHROME INTERACTING FACTORs (PIFs) (21–23). We have recently identified the two paralogous GATA family transcription factors GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED (GNC) and GNC-LIKE (GNL)/CYTOKININ-RESPONSIVE GATA FACTOR1 (CGA1) as critical transcription targets downstream from GA, DELLAs, and PIFs in Arabidopsis (24). GNC and GNL expression is repressed in response to GA, and this repression is important for proper germination, greening, flowering, and elongation growth.
Here, we show that several phenotypes of GNC and GNL overexpressors can also be observed in mutants of the auxin pathway regulators ARF2 and SLR. This observation has triggered our interest in examining a possible relationship between the two GATA transcription factors and the auxin signaling components. Interestingly, we find that GNC and GNL are critical transcription targets downstream from ARF2 and the Aux/IAA repressor SLR. Importantly, we also find that constitutive activation of GA signaling allows compensation for the loss of ARF2, thus strongly indicating that GNC and GNL integrate auxin and GA signals for the control of plant growth.
Results
GNC and GNL Restrain Growth in the Absence of ARF2.
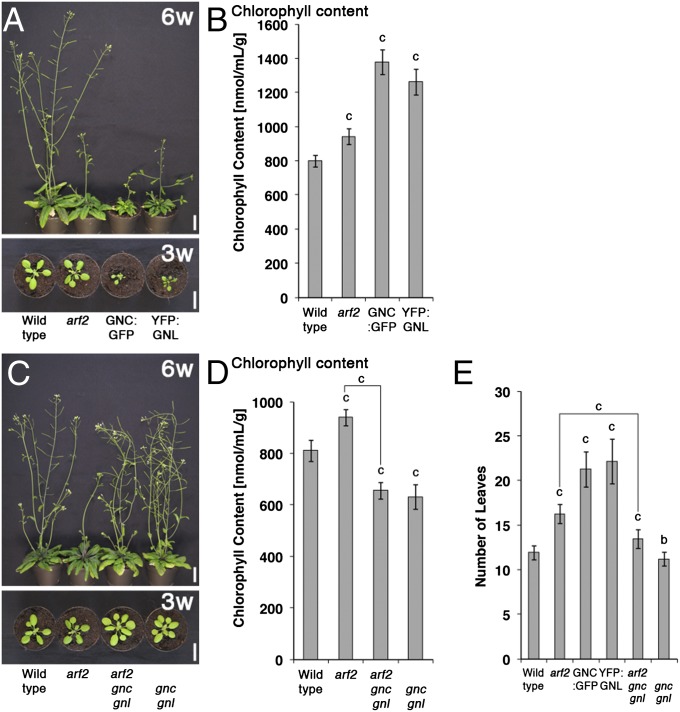
We have previously generated and characterized loss-of-function mutants and overexpression lines of the two GATA factors GNC (GNC:GFP) and GNL (YFP:GNL) (24). During the characterization of the GNC:GFP and YFP:GNL overexpression lines, we noticed that the GATA-overexpressing plants share a number of phenotypes with previously described arf2 loss-of-function mutants (16, 18). Similarly to arf2 mutants, GNC and GNL overexpressors accumulate chlorophyll and are delayed in senescence as judged by the degradation of chlorophyll after the transfer of leaves from light-grown plants to the dark (Fig. 1A and B and Fig. S1A) (25). Importantly, these arf2 phenotypes are strongly suppressed when we introduce gnc and gnl loss-of-function alleles into arf2, indicating that GNC and GNL are important regulators downstream from ARF2 (Fig. 1 C and D and Figs. S1B and S2). Additional phenotypic analyses revealed additional shared phenotypes between the GNC and GNL overexpressors on the one side and arf2 on the other side in the control of flowering time, stamen elongation, and floral organ abscission as well as seed size; in each case, the defects of the arf2 mutant are suppressed in arf2 gnc gnl triple mutants (Fig. 1E and Fig. S1 C–E). The suppression of the arf2 phenotype in arf2 gnc gnl is also apparent at the gene expression level, because an expression analysis of the PROTOCHLOROPHYLLIDE OXIDOREDUCTASE genes (PORA, PORB, and PORC) that encode important enzymes in chlorophyll biosynthesis as well as a global gene expression analysis showed a partial or full suppression of the gene expression defects of arf2 in arf2 gnc gnl (Fig. S1 F and G) [e.g., 45% (376 genes) of 840 genes that we found to be differentially regulated in arf2 compared with the WT are antagonistically regulated in arf2 gnc gnl (Fig. S1G and Dataset S1)].
Fig. 1.
Genetic interaction of ARF2 with GNC and GNL. (A and C) Representative photographs of 6- (6w) and 3-wk-old (3w) plants. (Scale bars: 2 cm.) (B and D) Absolute chlorophyll content of 10-d-old light-grown seedlings. (E) Number of leaves until bolting as a measure of flowering time. The average and SE of three replicate measurements are shown in B, D, and E. Student t test: b = P ≤ 0.01; c = P ≤ 0.001.
GNC and GNL Promoters Are Bound by ARF2 and ARF7.
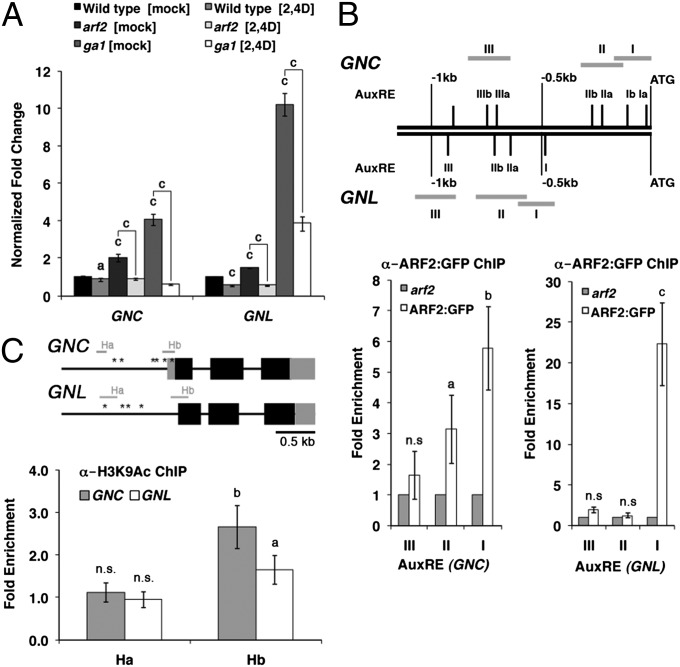
The phenotypic similarities between GNC and GNL overexpressors and arf2 mutants could potentially be explained by increased expression of the GATA genes in arf2, and indeed, we detected increased transcript levels of both GATAs in arf2 mutants (Fig. 2A). To test whether ARF2 can bind to the GNC and GNL promoters, we performed ChIP with ARF2:GFP using a transgenic line that expresses the ARF2:GFP fusion protein under control of the ARF2 promoter (26). In this experiment, we detected binding of ARF2:GFP to two promoter regions that span a total of four predicted AuxREs (TGTCTN) in the GNC promoter and one predicted AuxRE in the GNL promoter (Fig. 2B and Table S1) (2). We also performed ChIP analysis using an antibody directed against K9-acetylated HISTONE3 (H3K9Ac), which marks open chromatin, to find evidence on whether increased transcript abundance is a result of increased transcription (27). In this experiment, we detected increased H3K9Ac binding in arf2 mutants at promoter sites (Hb) located in proximity of the GNC and GNL transcription start sites (Fig. 2C). At the same time, H3K9Ac binding was unaltered at distal promoter sites (Ha), and therefore, we concluded that the increased GNC and GNL abundance in arf2 is the result of increased GNC and GNL transcription (Fig. 2C). In summary, these findings invite the conclusion that ARF2 binds directly to the GNC and GNL promoters and represses the expression of the two GATA genes.
Fig. 2.
Transcriptional regulation of GNC and GNL by ARF2. (A) Quantitative RT-PCR (RT-qPCR) analyses of GNC and GNL expression in WT and arf2 as well as ga1 mutant seedlings after a 30-min treatment with 5 µM 2,4D (2,4D) or mock (mock) solution. The fold change relative to transcript levels of mock-treated WT seedlings is shown. (B) Schemes of the GNC and GNL promoter regions (Upper). Roman numbers indicate the predicted AuxREs; gray bars indicate promoter regions selected for amplification after ChIP. The fold enrichment (ARF2:GFP/arf2) of AuxRE amplification after ChIP-PCR (Lower). (C) Schemes of the GNC and GNL genomic loci (Upper). Black boxes indicate exons; gray boxes indicate UTRs. Ha and Hb correspond to two regions amplified after H3K9Ac ChIP. Asterisks indicate the positions of the AuxRE binding sites examined in B. Please note that there is no 5′ UTR known for GNL. The fold enrichment (arf2/wild type) of Ha and Hb amplification after ChIP-qPCR (Lower); 10-d-old seedlings were used for all experiments, and the average and SE of three replicate measurements are shown. Student t test: a = P ≤ 0.1; b = P ≤ 0.05; c = P ≤ 0.01; n.s., not significant.
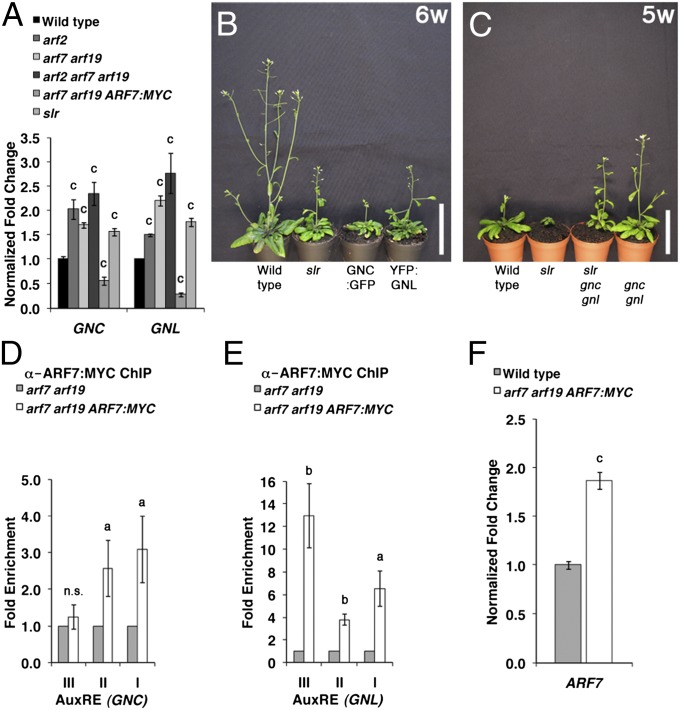
To understand the regulation of GNC and GNL transcription by ARF2 and auxin, we examined the effect of auxin on the expression of GNC and GNL in the WT and arf2 mutants. We found that GNC and GNL transcript abundance is strongly decreased after auxin treatment, suggesting that the expression of the two GATAs is under control of an auxin-regulated transcription repressor (Fig. 2 and Fig. S3). Interestingly, the effect of auxin on the repression of the GATAs was much more pronounced in the arf2 and ga1 mutants, where GNC and GNL expression is derepressed (Fig. 2A). Together, these observations gave rise to the hypothesis that other auxin-dependent regulators must control GNC and GNL transcription in the absence of ARF2. When examining the phenotypes of other relevant mutants, we noticed that slr gain-of-function mutants also share phenotypes with the GNC and GNL overexpressors as well as arf2 mutants (11, 16); slr mutants were similar to GNC and GNL overexpressors at least with regard to their increased chlorophyll content, delayed flowering, and increased seed size (Fig. S4) (11). In line with the idea that SLR/IAA14 as well as its established interacting ARFs ARF7 and ARF19 are involved in GATA regulation, we detected increased GNC and GNL transcript levels in the slr gain-of-function mutant and the arf7 arf19 loss-of-function mutant, and compared with arf2 or arf7 arf19, even greater increased transcript levels were detected in the arf2 arf7 arf19 triple mutant (Fig. 3A). We then tested genetically whether the slr phenotypes are caused by increases in GNC and GNL abundance and found that slr mutant phenotypes are partially or fully suppressed in slr gnc gnl (Fig. 3C and Fig. S4). Taken together, these data suggested that GNC and GNL repress growth downstream from SLR/IAA14. At the same time, the transcript abundance of the GATAs was decreased to levels even below those levels detected in the WT in a transgenic line expressing ARF7:MYC in an arf7 arf19 background (Fig. 3A). Because these data indicated that ARF7 and ARF19 may act as repressors of GNC and GNL expression, we examined next whether ARF7 can bind to the GNC and GNL promoters. Indeed, ChIP studies provided proof for the direct binding of ARF7:MYC to five of six AuxRE-containing sites of the GNC and GNL promoters (Fig. 3 D and E). Because we used a 35S cauliflower mosaic virus (CaMV) promoter-driven ARF7:MYC line for this experiment and because transcription factor overexpression may lead to the binding and regulation of off-target gene promoters, we also examined the degree of ARF7 overexpression in ARF7:MYC. However, because only a twofold increase in the expression of ARF7 was detectable in the ARF7:MYC transgenic line compared with WT, we consider it very unlikely that the observed repression of GNC and GNL is the result of an off-target binding event (Fig. 3F).
Fig. 3.
Interaction between SLR1/IAA14 and ARF7 with GNC and GNL. (A) Detection of GNC and GNL by RT-qPCR from 10-d-old light-grown WT, slr, and arf mutant seedlings. Shown is the fold change relative to WT levels. (B and C) Representative photographs of 6- (6w) and 5-wk-old (5w) plants grown in long-day conditions. (Scale bars: 5 cm.) (D and E) qPCR result after ChIP with an anti-MYC antibody (ARF7:MYC). Shown is the fold enrichment (ARF7:MYC/arf7 arf19) by qPCR after ChIP of individual AuxRE-containing promoter regions present in the (D) GNC and (E) GNL promoters (Fig. 2B). (F) RT-qPCR analysis of ARF7 expression in WT and arf7 arf19 ARF7:MYC; 10-d-old seedlings were used for all experiments, and the average and SE of three replicate measurements are shown. Student t test: a = P ≤ 0.1; b = P ≤ 0.05; c = P ≤ 0.01; n.s., not significant.
Suppression of Auxin Mutant Phenotypes by Promoting GA Signaling.
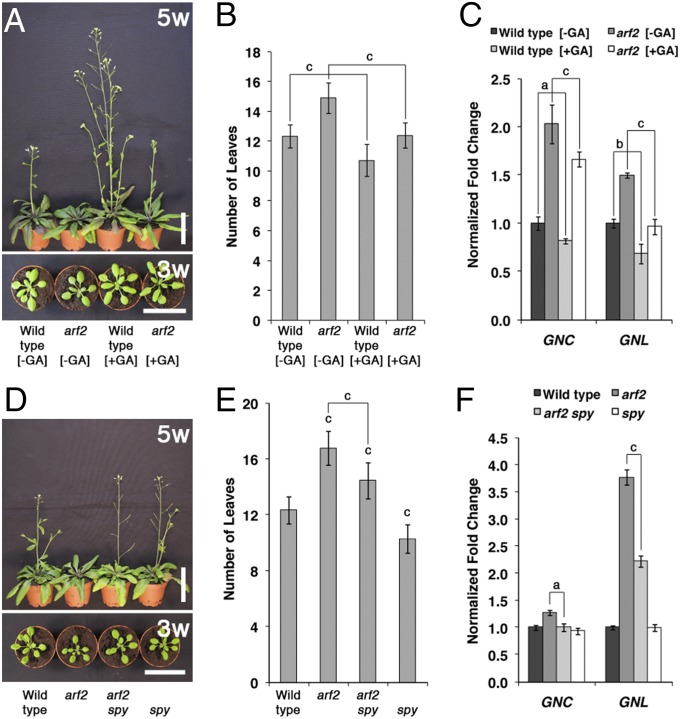
We had previously shown that GNC and GNL are repressors of GA signaling downstream from DELLAs and PIFs (20, 24). Because our analysis of GID1 GA receptor gene expression and GA-induced DELLA protein degradation provided no evidence for an impairment of GA signaling in arf2 mutants (Fig. S5), we hypothesized that it may be possible to suppress arf2 growth defects by promoting GA and PIF signaling. Indeed, when we examined the arf2 flowering phenotype in mock and GA-treated arf2 mutant plants, we observed a suppression of the arf2 flowering time delay, which correlated with a decrease in GNC and GNL transcript abundance in the GA-treated arf2 mutants (Fig. 4 A–C). To obtain additional support for a suppression of auxin pathway defects through activation of the GA signaling cascade, we introduced the constitutive GA response mutant spindly (spy) into arf2 (28); spy mutants are deficient in the function of the O-linked N-acetylglucosamine transferase SPY, they resemble plants that have been grown in the presence of high doses of GA, and they accumulate DELLA proteins in a seemingly inactive state (28–30). Interestingly, we found that the loss of SPY function is sufficient to at least partially suppress the flowering time phenotype of arf2 mutants and that the suppression of the arf2 phenotype correlates with decreases in GNC and GNL transcript abundance (Fig. 4 D–F). In summary, these findings suggested that the repression of GNC and GNL gene expression through constitutive activation of the GA pathway can, indeed, suppress defects of mutants with a defect in auxin signaling.
Fig. 4.
Suppression of arf2 by activation of the GA signaling pathway. (A and D) Representative photographs of 5- (5w) and 3-wk-old (3w) plants. Plants in A were watered every other day with a mock (−GA) or 100 µM GA3-containing solution (+GA). (B and E) Flowering time analysis. Number of leaves until bolting. (C and F) RT-qPCR analyses to examine the levels of GNC and GNL transcript after a mock (−GA) or 30-min treatment with 10 µM GA3 (+GA). Fold change relative to (mock-treated) WT levels; 10-d-old seedlings were used for all experiments, and the average and SE of three replicate measurements are shown. Student t test: a = P ≤ 0.05; b = P ≤ 0.01; c = P ≤ 0.001.
To verify that auxin and GA signaling act independently on the control of GNC and GNL expression, we additionally examined the effects of ARF2 and PIF1 (as a representative PIF protein) on GNC and GNL expression. To this end, we introduced a PIF1 overexpression line (PIF1-TAP) into the arf2 background and examined the effects of the presence and absence of the ARF2 and PIF1 repressors on the transcript abundance of the GATAs. In line with ARF2 and PIF1-TAP being independent repressors of GNC and GNL, we observed intermediate GNC and GNL expression levels in the PIF1-TAP arf2 background compared with the arf2 mutant and the PIF1-TAP line, where GNC and GNL expression is derepressed and strongly repressed, respectively (Fig. S6). Additionally, we found, as presented earlier in this study, that the auxin-induced repression of GNC and GNL does not require GA, because the transcript abundance of the GATAs could be efficiently down-regulated by auxin in the GA-deficient ga1 mutant (Fig. 2A). In summary, our findings strongly support the notion that GA and auxin signaling act independently to control GNC and GNL expression and that these two GATAs are critical for the control of GA- and auxin-controlled growth responses.
GA Promotes ARF2 Abundance.
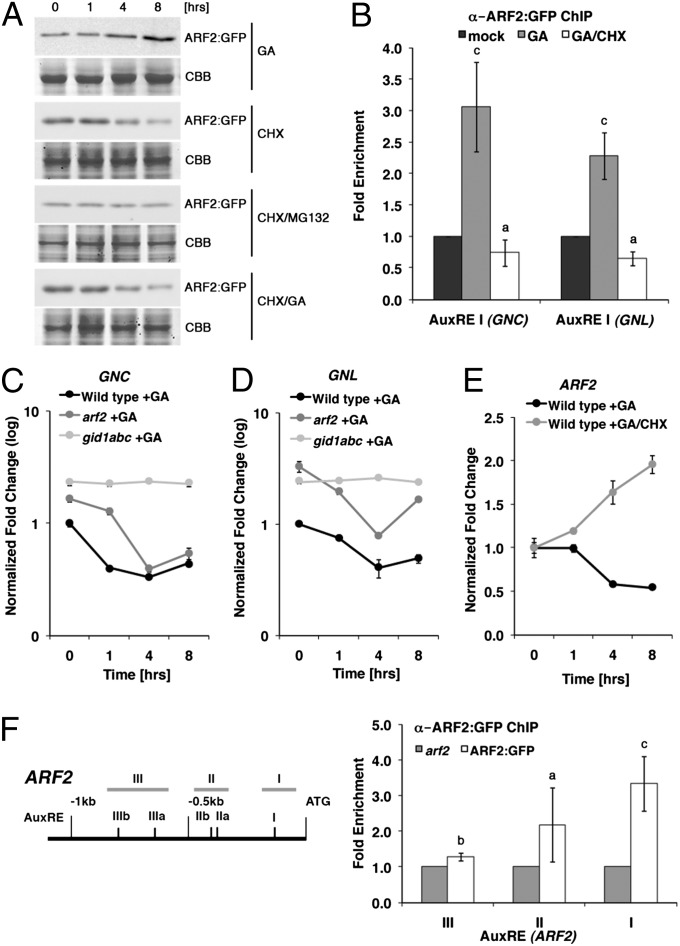
We also examined putative effects of auxin and GA on ARF protein abundance. Interestingly, although we found no evidence for a regulation of ARF2:GFP or ARF7:MYC protein by auxin (Fig. S7), we noted that the abundance of ARF2:GFP but not ARF7:MYC increases in response to GA treatment (Fig. 5A and Fig. S8). Importantly, ARF2:GFP abundance decreased after treatment with the protein biosynthesis inhibitor cycloheximide (CHX) (Fig. 5A). Because concomitant treatment with the proteasome inhibitor MG132 could prevent this degradation, we concluded that ARF2 is degraded by the 26S proteasome (Fig. 5A). Additionally, because combined treatments with CHX and GA were not sufficient to inhibit the ARF2:GFP turnover, we concluded that ARF2:GFP translation may be under GA control or alternatively, that a de novo synthesized and unknown GA-responsive protein may regulate ARF2:GFP abundance (Fig. 5A). In agreement with the role of ARF2 as a repressor of GNC and GNL, we found that the GA-promoted increase in ARF2:GFP abundance correlated with an increased binding of ARF2:GFP to AuxREs of the GNC and GNL promoters on the one side and increased GNC and GNL repression on the other side (Fig. 5 B–D). Importantly, this GA-induced transcriptional repression of ARF2 could not be detected in the GA-insensitive gid1abc triple mutant, where ARF2 abundance cannot be promoted by GA because of the absence of all three GID1 GA receptor genes that confer complete GA insensitivity (Fig. 5 C and D) (31). We, therefore, concluded that ARF2:GFP levels are positively regulated by the GA signaling pathway. Interestingly, we also found that the increase in ARF2:GFP abundance after GA treatment and the decrease in ARF2:GFP abundance after GA/CHX treatment correlated negatively and positively, respectively, with a decrease in ARF2 transcript abundance (Fig. 5E). Because our subsequent ChIP analyses then revealed that ARF2:GFP can bind to AuxREs in the ARF2 promoter, we concluded that ARF2 protein may repress its own transcription as part of a negative feedback regulatory loop (Fig. 5F). Thus, the GA-dependent increase in ARF2 abundance may serve as a translation-dependent mechanism that promotes ARF2 abundance and overrides the autoinhibitory negative feedback of ARF2 on its own transcription in the presence of GA.
Fig. 5.
GA promotes ARF2 abundance. (A) ARF2:GFP immunoblot analyses of 10-d-old light-grown seedlings treated with 10 µM GA3, 50 µM CHX, and 100 µM MG132 as indicated. CBB, Coomassie Brilliant Blue (loading control). (B) ARF2:GFP ChIP on selected AuxRE-containing promoter elements of the GNC and GNL promoters (Fig. 2B). The fold enrichment (ARF2:GFP/arf2) of AuxRE amplification is shown. (C–E) RT-qPCR analyses for ARF2, GNC, and GNL after 10 µM GA3 treatment; gid1abc is a GA receptor loss-of-function triple mutant (31). (F) Scheme of the ARF2 promoter with predicted AuxREs and promoter regions used for ChIP. The fold enrichment (ARF2:GFP/arf2) of regions containing AuxREs after qPCR amplification is shown. Student t test: a = P ≤ 0.05; b = P ≤ 0.01; c = P ≤ 0.001.
Discussion
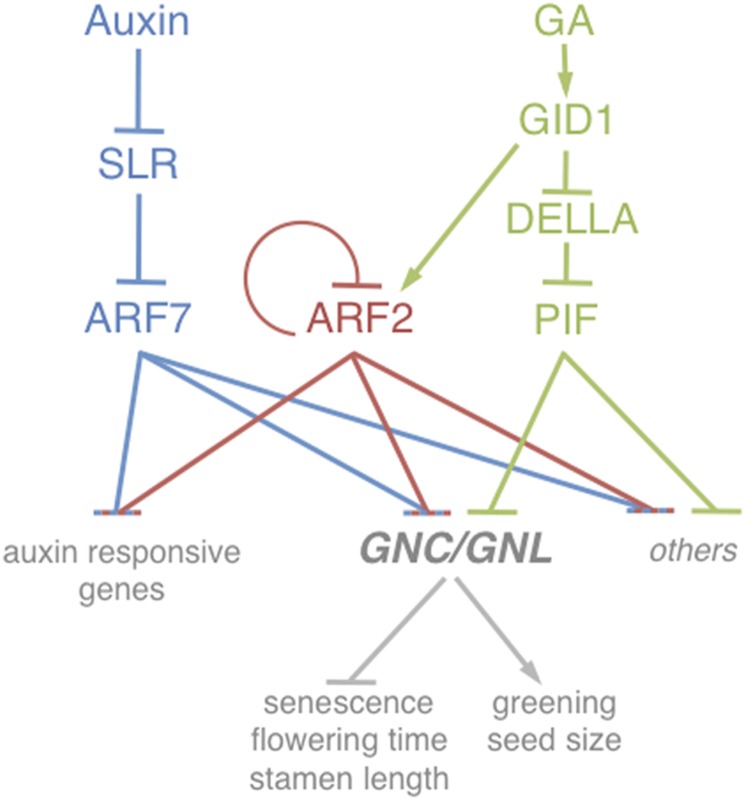
In our study, we identify the Arabidopsis GATA transcription factors GNC and GNL as critical transcription targets downstream from auxin signaling in the control of different physiological growth processes, including greening, flowering time control, and senescence (Fig. 6). We have previously established GNC and GNL as transcription targets of GA signaling and show here that the two GATAs are also critical and direct transcription targets of the auxin pathway (24). We reveal that both GA and auxin signaling function independently in the repression of GNC and GNL and that the activation of the GA pathway allows suppressing phenotypes of auxin response mutants (Fig. 6). Based on our observations, we conclude that GNC and GNL are threshold-dependent regulators that are targeted by both pathways to control plant growth.
Fig. 6.
Model of the control mechanisms acting on the transcriptional control of GNC and GNL. GNC and GNL expression is repressed by ARF2 (red) and ARF7 (blue). In addition, the abundance of their transcripts is under the direct repression control of the GA signaling cascade and the PIF repressors as well as indirect GA control by the stabilization of ARF2 (green). Other genes that are under ARF and PIF control may also be regulated in a manner similar to GNC and GNL. In addition, gene expression events that are under control of the auxin pathway may also be regulated by GA signaling because of the control of ARF2 abundance by GA. The individual pathways are shown in distinct colors.
Most research on auxin signal transduction is concerned with the role of auxin in the control plant development, differentiation, and morphology. In the case of ARF7, ARF19, and SLR/IAA14, the control of lateral root formation by activation of the genes LATERAL ORGAN BOUNDARIES DOMAIN16 and LATERAL ORGAN BOUNDARIES DOMAIN18 has been given particular attention (32). Our study now emphasizes the role of auxin in the regulation of physiological growth responses, such as greening, flowering time, and senescence. Although such phenotypes were already described for arf2 mutant combinations several years ago, the underlying molecular mechanisms remained elusive (16–18). Through the analysis of slr and arf7 arf19 mutants, we emphasize here the fact that other auxin mutants have—other than their already well-understood defects in controlling lateral root development—defects in physiological growth responses (11, 16–18). At the same time, we identify GNC and GNL as critical target genes downstream from these auxin pathway regulators. Unlike the ARF+ proteins ARF7 and ARF19, several pieces of evidence support the notion that the ARF− protein ARF2 is not controlled by Aux/IAA repressors (3, 7). These findings suggest that ARF− proteins, such as ARF2, may negatively control GNC and GNL expression in an auxin-independent manner and that auxin, through derepression of ARF+-type ARFs, can only partially modulate the expression of the two GATAs (7). Obviously, it can be expected that other ARF target genes are also regulated by auxin-independent and -dependent ARFs that compete for the same AuxRE binding sites in the promoters of the respective genes (Fig. 6).
Interestingly, we show that GNC and GNL are repressed by the Aux/IAA SLR as well as ARFs. Auxin-induced transcriptional repression has been reported already in several studies (33–36) but cannot be explained by the current model of Aux/IAA repressor function, according to which the auxin-dependent degradation of the Aux/IAAs leads to the derepression of ARFs acting as transcriptional activators (6). Our data on the regulation of GNC and GNL and data in the published literature, thus, suggest that other mechanisms must exist to control the repressor activity of Aux/IAA-controlled ARFs. It can be envisioned that Aux/IAAs inhibit the DNA binding activity of repressor ARFs, Aux/IAA degradation allows for the regulation of ARFs by posttranslational modifications (37), or Aux/IAA degradation allows for the formation of repressive heterodimers of ARFs with other ARFs or other transcription regulators (7, 38). Which mechanism operates in the context of the transcriptional repression of GNC and GNL is unclear at present.
Our study also suggests a molecular mechanism for the GA–auxin cross-talk, where GA promotes the abundance of ARF2 and thereby enhances the repression of GNC and GNL transcription in response to GA (Fig. 6). At the same time, we also establish that ARF2 represses its own transcription through negative feedback regulation (Fig. 6). Because the effect of GA on ARF2 protein abundance must be dominant over the repressive effects of ARF2 on its own transcription, this feedback regulatory mechanism should help to ensure the dominant role of GA pathway activation over ARF2-dependent signaling (Fig. 6). Interestingly, GA does not increase the abundance of the ARF+ protein ARF7, and therefore, the GA-dependent increase of ARF2 could represent a molecular mechanism that serves to enhance GA signaling through the recruitment of auxin and Aux/IAA-independent ARFs. This GA-dependent regulatory mechanism may, thus, allow for the uncoupling of ARF-controlled gene expression from its regulation by auxin, Aux/IAAs, and ARF+ regulators.
In summary, our analysis identifies GNC and GNL as critical repressors of plant growth downstream from auxin and GA signaling. GNC and GNL had originally been identified based on their transcriptional regulation by nitrate and by cytokinin and light, respectively (39, 40). Although the light control of GNL expression could be mediated by PIFs, which are degraded in response to light-dependent interactions with the phytochrome red light receptors (41), the signaling pathways that control responses to the growth-promoting nitrate and cytokinin hormone signals remain to be identified. In fact, the spy mutant had also been described as a cytokinin-insensitive mutant (42, 43). It may, thus, be envisioned that the regulation of GNC and GNL by cytokinin is mediated by SPY and that the role of SPY in cytokinin signaling may interfere with the interpretation of our genetic interaction analysis between ARF2 and SPY. However, we found that GNC and GNL transcript levels were unaltered in spy compared with the WT (Fig. 4F), and thus, altered cytokinin signaling in spy should not interfere with the suppression of arf2 in arf2 spy. Regardless of the underlying molecular mechanisms, the control of GNC and GNL expression by three hormone signaling pathways (auxin, GA, and cytokinin) as well as their regulation by nitrate suggest that the transcriptional regulation of these GATA factors is critical for the integration of multiple growth-controlling signals during plant growth and development. Identification of their transcription targets will further add to the understanding of this intriguing signaling network.
Materials and Methods
Biological Material.
The following mutants and transgenic lines were used in this study: arf2-5, arf2-8, and arf2 arf7 arf19 (16); ARF2:GFP (ARF2:ARF2:GFP) (26); arf7 arf19 (10); ARF7:MYC (35S:ARF7:MYC; gift from Hidehiro Fukaki, Kobe University, Kobe, Japan); gnc, GNC:GFP (35S:GNC:GFP), gnl, YFP:GNL (35S:YFP:GNL), and ga1 (24); slr (11); gid1abc (31); and PIF1-TAP (44). All mutants and transgenic lines are in the A. thaliana ecotype Columbia.
Physiological Experiments.
For chlorophyll measurements, chlorophyll was extracted and quantified as previously described; three independent replicates and measurements were performed (45). Basal chlorophyll levels were quantified from 10-d-old seedlings. The senescence assay was performed using a previously established method (25). Chlorophyll was extracted from leaf numbers 3 and 5 of 21-d-old plants (set to 100%) and plants that were subsequently kept for 4 d in liquid medium in the dark. For flowering time analyses, plants were randomly arranged and grown in 150 μmol m−2 s−1 white light in MobyLux GroBanks (CLF Plant Climatics) in long-day conditions (16/8 h at 21 °C/18 °C). The time of bolting was scored from at least 18 plants by counting the number of rosette leaves (46). Seed size was determined from at least 200 seeds per genotype. Floral organ abscission was determined as previously described by counting the last floral bud that still retained sepals and/or petals starting from the top of the inflorescence (16).
Quantitative Real-Time PCR.
Total RNA was isolated with a NucleoSpin RNA Plant Kit (Machery-Nagel). DNA was removed by an on-column treatment with rDNase (Machery-Nagel), and 2 µg total RNA were reverse transcribed with an oligo(dT) primer and M-MuLV Reverse Transcriptase (Fermentas). The cDNA equivalent of 60–80 ng total RNA was used in a 10 μL PCR in a CFX96 Real-Time System Cycler with iQ SYBR Green Supermix (BioRad). A 40-cycle two-step amplification protocol (10 s at 95 °C and 25 s at 60 °C) was used for all measurements. Relevant primers are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Inês Barbosa, Pascal Braun, and Erika Isono for critical comments on the manuscript and Jutta Elgner for excellent technical support. We thank Hidehiro Fukaki and Dolf Weijers for the ARF7:MYC and ARF2:GFP lines, respectively. This work was supported by Deutsche Forschungsgemeinschaft Grants Schw751/9-1, Schw751/10-1, and SFB924.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35730).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304250110/-/DCSupplemental.
References
- 1.Teale WD, Paponov IA, Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7(11):847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 2.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276(5320):1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 3.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA. 1999;96(10):5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 6.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319(5868):1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 7.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19(3):309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 9.Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16(13):1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29(2):153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 12.Weijers D, et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10(2):265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Ubeda-Tomás S, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10(5):625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Tiwari SB, Hagen G, Guilfoyle TJ. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell. 2005;17(7):1979–1993. doi: 10.1105/tpc.105.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilmoth JC, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43(1):118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- 16.Ellis CM, et al. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132(20):4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 17.Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant J. 2005;43(1):29–46. doi: 10.1111/j.1365-313X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- 18.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133(2):251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 19.Lim PO, et al. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot. 2010;61(5):1419–1430. doi: 10.1093/jxb/erq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- 21.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451(7177):475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 23.Schwechheimer C. Gibberellin signaling in plants—the extended version. Front Plant Sci. 2011;2:107. doi: 10.3389/fpls.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter R, Behringer C, Müller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010;24(18):2093–2104. doi: 10.1101/gad.594910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver LM, Amasino RM. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 2001;127(3):876–886. [PMC free article] [PubMed] [Google Scholar]
- 26.Rademacher EH, et al. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011;68(4):597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 27.Ha M, Ng DW, Li WH, Chen ZJ. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 2011;21(4):590–598. doi: 10.1101/gr.116467.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5(8):887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverstone AL, et al. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143(2):987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson RN, Somerville CR. Phenotypic Suppression of the Gibberellin-Insensitive Mutant (gai) of Arabidopsis. Plant Physiol. 1995;108(2):495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willige BC, et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;19(4):1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009;151(3):1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, et al. Transcriptome profiling of cytokinin and auxin regulation in tomato root. J Exp Bot. 2013;64(2):695–704. doi: 10.1093/jxb/ers365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman EJ, et al. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE. 2012;7(5):e36210. doi: 10.1371/journal.pone.0036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DJ, Park JW, Lee HW, Kim J. Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J Exp Bot. 2009;60(13):3935–3957. doi: 10.1093/jxb/erp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paponov IA, et al. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant. 2008;1(2):321–337. doi: 10.1093/mp/ssm021. [DOI] [PubMed] [Google Scholar]
- 37.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105(28):9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin R, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19(8):2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T. Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2007;71(6):1557–1560. doi: 10.1271/bbb.60692. [DOI] [PubMed] [Google Scholar]
- 40.Bi YM, et al. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005;44(4):680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 41.Castillon A, Shen H, Huq E. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12(11):514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Greenboim-Wainberg Y, et al. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17(1):92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steiner E, et al. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24(1):96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon J, Zhu L, Shen H, Huq E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA. 2008;105(27):9433–9438. doi: 10.1073/pnas.0803611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inskeep WP, Bloom PR. Extinction coefficients of chlorophyll a and B in n,n-dimethylformamide and 80% acetone. Plant Physiol. 1985;77(2):483–485. doi: 10.1104/pp.77.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development. 2011;138(4):677–685. doi: 10.1242/dev.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.