Abstract
In animals, mtDNA is always transmitted through the female and this is termed “maternal inheritance.” Recently, autophagy was reported to be involved in maternal inheritance by elimination of paternal mitochondria and mtDNA in Caenorhabditis elegans; moreover, by immunofluorescence, P62 and LC3 proteins were also found to colocalize to sperm mitochondria after fertilization in mice. Thus, it has been speculated that autophagy may be an evolutionary conserved mechanism for paternal mitochondrial elimination. However, by using two transgenic mouse strains, one bearing GFP-labeled autophagosomes and the other bearing red fluorescent protein-labeled mitochondria, we demonstrated that autophagy did not participate in the postfertilization elimination of sperm mitochondria in mice. Although P62 and LC3 proteins congregated to sperm mitochondria immediately after fertilization, sperm mitochondria were not engulfed and ultimately degraded in lysosomes until P62 and LC3 proteins disengaged from sperm mitochondria. Instead, sperm mitochondria unevenly distributed in blastomeres during cleavage and persisted in several cells until the morula stages. Furthermore, by using single sperm mtDNA PCR, we observed that most motile sperm that had reached the oviduct for fertilization had eliminated their mtDNA, leaving only vacuolar mitochondria. However, if sperm with remaining mtDNA entered the zygote, mtDNA was not eliminated and could be detected in newborn mice. Based on these results, we conclude that, in mice, maternal inheritance of mtDNA is not an active process of sperm mitochondrial and mtDNA elimination achieved through autophagy in early embryos, but may be a passive process as a result of prefertilization sperm mtDNA elimination and uneven mitochondrial distribution in embryos.
Keywords: assisted reproductive technologies, paternal inheritance, mitophagy
Since Hutchison et al. discovered that mule inherited mitochondria from the female parent (1), it has been found that mtDNA is inherited from one parent in almost all eukaryotic organisms (1, 2). In animals, the offspring’s mtDNA is always inherited from only the female, termed maternal inheritance (3). It is widely believed that maternal inheritance of mtDNA is an advantageously evolutionary event, as sperm mtDNA may be damaged by reactive oxygen species involved in fertilization, thus causing some mitochondrial disorders in individuals (4).
Different mechanisms are involved to ensure uniparental inheritance of mtDNA in different organisms. In Caenorhabditis elegans, several recent reports showed that sperm mitochondria are degraded through autophagy during early embryogenesis, whereas, in autophagy-defective zygotes, paternal mitochondria and their DNA remained even through the first larval stage (5–7). In medaka (Japanese rice fish; Oryzias latipes), through unknown mechanisms, sperm mtDNA was quickly eliminated before degradation of mitochondria after fertilization, leaving vacuolar mitochondria in early embryos (8). It has also been reported that, in Drosophila, sperm mtDNA was eliminated by endonuclease G and removed by a cellular remodeling process during spermatogenesis, thus leaving only vacuolar mitochondria in sperm resulting in maternal inheritance of mtDNA (9).
The mouse is an important model organism in which the patterns and mechanisms of mitochondrial inheritance have not yet been widely studied. Except for a few examples in which biparental mtDNA were observed in the offspring resulting from interspecies mating, maternal inheritance of mtDNA in mice is widely accepted (10–12). A previous study showed that sperm mitochondria were preubiquitination-labeled during spermatogenesis and became degraded in lysosomes during early embryo development (13). However, the mechanism taking place between ubiquitination and lysosome degradation was unclear. Very recently, based on immunofluorescence results showing that P62 protein and microtubule-associated protein 1 light chain 3 (LC3) colocalized to the sperm tail after fertilization, autophagy was speculated to link these two aspects; however, solid evidence is still lacking (5, 14, 15).
To validate the speculation that autophagy may be involved in maternal mitochondrial inheritance, two transgenic mouse strains were used in the present study, one bearing GFP-labeled autophagosomes and the other bearing red fluorescent protein (RFP)-labeled mitochondria. Ubiquitination of the sperm tail and congregation of P62 and LC3 proteins to sperm mitochondria were observed, similar to previous studies (5). However, further investigations showed that sperm mitochondria were not degraded by postfertilization autophagy. Sperm mitochondria were not engulfed by lysosomes for degradation through autophagy until LC3 removal; instead, they even existed in embryos until the morula stage when sperm mitochondria became too dispersed to be detected. Moreover, by using single sperm mtDNA PCR, two more important phenomena were observed: most motile sperm that could reach the oviduct for fertilization had eliminated their mtDNA before fertilization, leaving only vacuolar mitochondria that were unevenly distributed in blastomeres; whereas, if sperm mtDNA was occasionally left and entered the egg, it would be not eliminated, but could be detected in newborn mice. Based on these results, we conclude that maternal inheritance of mtDNA, at least in mice, is a type of passive outcome as a result of the elimination of sperm mtDNA before fertilization and the uneven distribution in early embryo development, not an active process that is achieved through elimination of sperm mitochondria and mtDNA by autophagy after fertilization.
Results and Discussion
Autophagy Did Not Participate in the Degradation of Sperm Mitochondria After Fertilization.
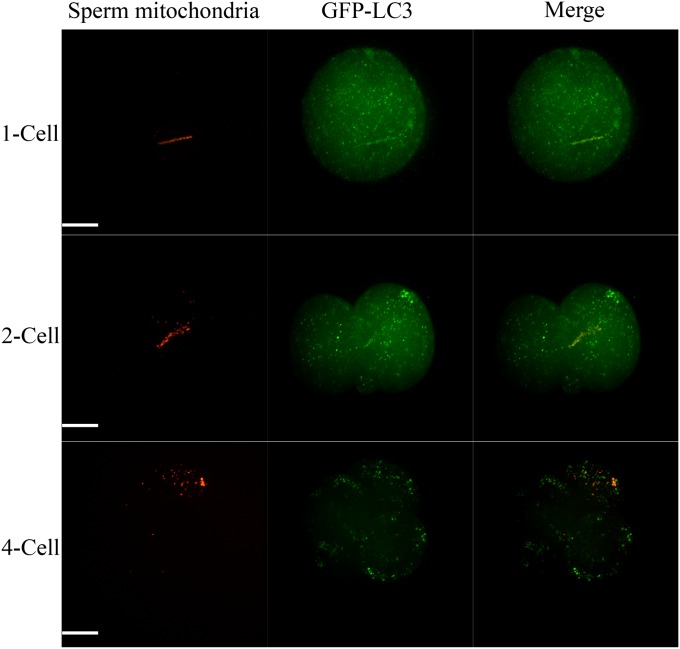
Autophagy is a lysosome-dependent pathway to eliminate unwanted constitutes in eukaryotic cells. Briefly, phosphatidylethanolamine lapidated LC3 proteins, termed LC3-II, congregate to target substrates and form double membrane autophagosomes, then fuse with lysosomes and degrade the engulfed substrates (16). Thus, congregated LC3-II, always existing in punctuate form, is an important indicator for the existence of autophagosomes (17). Here, by using transgenic mice, which expressed GFP-labeled LC3 protein, we found that LC3 proteins congregated to sperm mitochondria at the one-cell stage but dissociated at the four-cell stage (Fig. 1). There are several explanations for this phenomenon. The first was that LC3 proteins were degraded earlier than sperm mitochondria in lysosomes, and the second was that the fluorescence of GFP but not RFP was quenched because of the acid environment in lysosomes (18). Therefore, we used LysoTracker Blue DND-22 to label lysosomes to observe the localization in relation to sperm mitochondria and lysosomes. Strikingly, as shown in Fig. S1, no colocalization between lysosomes and sperm mitochondria was observed until the morula stages. This indicated that sperm mitochondria were never ingested into lysosomes through autophagy. It is likely that LC3 labeled other elements localized nearby mitochondria. This was verified partly by the fact that not only sperm mitochondria specifically but the entire sperm tail was labeled with ubiquitin and P62 proteins as shown in Figs. S2 and S3, respectively. Also, results from C. elegans showing that nematode-specific sperm membranous organelles but not the mitochondria are ubiquitinated before autophagosome formation support our speculation indirectly (5, 6).
Fig. 1.
Live cell fluorescence imaging of GFP-LC3 and sperm mitochondria in early embryos. GFP-LC3 congregated to sperm mitochondria immediately after fertilization, but dissociated at the four-cell stage. (Scale bars: 20 µm.)
Although the aforementioned fluorescence imaging results of LC3 and lysosomes indicated that sperm mitochondria were never wrapped into autophagosomes, it was unclear whether autophagy was involved in the disaggregation of the sperm tail, thereby impacting the fate of sperm mitochondria. To investigate this, autophagy-defective embryos were obtained from oocyte-specific Atg5-deficient mice. As shown in Fig. S4, despite autophagy being inhibited by KO Atg5, the full dynamic process including the detachment and distribution of sperm mitochondria was still similar to the results seen in WT mice (Figs. 1 and 2). Therefore, no effect of autophagy on the fate of sperm mitochondria after fertilization in mice was confirmed.
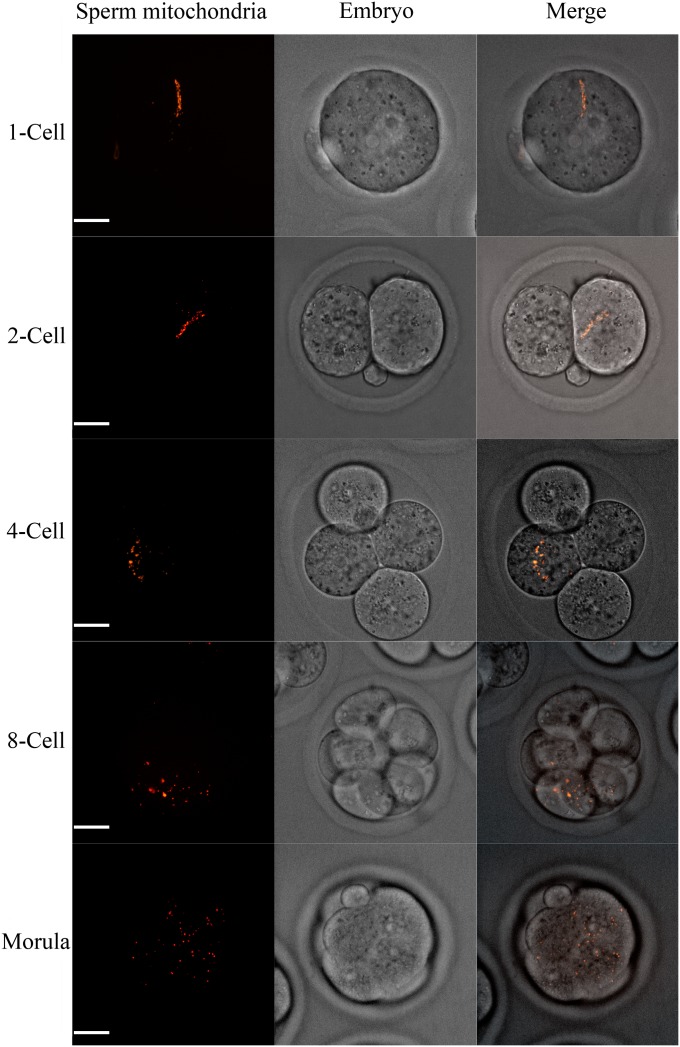
Fig. 2.
Live cell fluorescence imaging of sperm mitochondria (red) in early embryos. Sperm mitochondria disaggregated and became restricted to only one blastomere during one-cell to four-cell stages. Sperm mitochondria distributed in several cells after the eight-cell stage and could be detected until morula stages. (Scale bars: 20 µm.)
Sperm Mitochondria Persisted and Dispersed Unevenly in Several Cells Until the Morula Stages.
Previous work with the use of mitochondria-tracking dyes to investigate the fate of sperm mitochondria after fertilization had demonstrated that fluorescence dyes labeling sperm mitochondria disappeared primarily before the two-cell stages (19). However, when fluorescence dyes were used in in vitro studies, some problems such as photobleaching or molecule dispersion could influence the results, and eventually the conclusions. In the present study, transgenic mice that had RFP-labeled mitochondria were used to monitor the fate of sperm mitochondria after fertilization in real time. Because sperm mitochondria are tightly packed helical structures (20), as shown in Fig. 2 upon fertilization, sperm mitochondria first disaggregated during the one- to four-cell stages, and, at the same time, they were always restricted to only one blastomere. After the four-cell stages, sperm mitochondria existed disproportionally in several cells that originated from the former blastomere, which inherited the sperm mitochondria. However, perhaps because of high dilution and disaggregation, fluorescence of the sperm mitochondria was no longer detected after the morula stage. The persistence of sperm mitochondria until at least the morula stage confirmed the aforementioned conclusion that sperm mitochondria were not eliminated through autophagy.
Our results were obtained on BALB/c and C57BL/6j mice crosses that were intraspecific, but Kaneda et al. reported that paternal mitochondria in embryos was a leakage that occurred only in interspecific crosses (12), so these results were contradictory. To exclude the possibility that our observations may also be an interspecific crossing result because of the multiple rounds of interbreeding that render these two mice genetically distant, sisters and brothers mating experiments were carried out. Our results shown in Fig. S5 indicated that sperm mitochondria could also be observed until morula stages in sister–brother crosses; there was no difference observed between this and BALB/c–C57BL/6j crosses (Fig. 2). Thus, the possibility that our results may come from interspecific effects was excluded. We further propose that the results regarding paternal mitochondria elimination at the early pronuclear stages in the intraspecific crosses in the studies of Kaneda et al. may be a combined effect of the nuclear genome of C57BL/6 and mitochondrial genome of Mus spretus.
Moreover, because only two blastomeres of a four-cell embryo contribute to the cells of an animal body in early development (21, 22), the uneven distribution of only one of four cells obtaining sperm mitochondria in early embryo development prompted us to argue that, even though sperm mitochondria and their DNA are not degraded during development, there were only 50% of offspring and a few tissues that had the opportunity to inherit paternal mtDNA from the paternal parent, which was supported by examining paternal mtDNA in placentas and newborn mice, as described later, and by the results obtained by Shitara et al. (10) and Kaneda et al. (12), in which paternal mtDNA was detected in offspring from interspecies mating 17 of 38 and 25 of 45, respectively. Thus, uneven distribution may be one of the mechanisms of maternal mitochondrial inheritance. Additionally, the uneven distribution of mitochondria would likely result in concentrated effects. If sperm mtDNA is transmitted to the next generation, it has a greater opportunity compared with simple dilution to influence the tissues that inherit sperm mtDNA, and this may induce some mitochondrial inherited diseases in a few tissues rather than affecting the whole body (23).
Sperm mtDNA Was Detected in only a Small Proportion of Hybrids.
Although sperm mitochondria were detected up to the morula stages, whether the sperm mitochondrial genome remained was uncertain. To investigate the existence of sperm mtDNA in early embryos, allele-specific PCR and restriction enzyme analysis were used in this study. As shown in Fig. 3, this method could detect as few as one copy of C57/6j (paternal) mouse mtDNA, even when mixed with one BALB/c (female) mouse oocyte mtDNA. Sperm mtDNA in different-stage embryos and newborn mice was investigated. Unexpectedly, as shown in Table 1, sperm mtDNA could be detected in only a few embryos or newborn mice, even in the early pronuclear stage, at which the positive ratio was only 13 in 115. However, the similar ratios from early pronuclear stages to newborn mice indicated that sperm mtDNA were not subjected to degradation until birth, which also confirmed the results described earlier that sperm mitochondria were not degraded during the embryo stage.
Fig. 3.
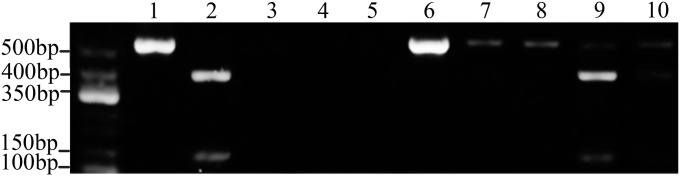
Single copy of paternal (C57/6j) mtDNA could be selectively amplified and distinguished from female (BALB/c) mtDNA of an oocyte. Lane 1, PCR product of BALB/c mtDNA could not be digested with Tth111I restriction enzyme and was 520 bp; lane 2, PCR product of C57/6j mtDNA was digested to 383- and 157-bp segments by Tth111I restriction enzyme; lanes 3–6, PCR products amplified from single copy of C57/6j mtDNA (as a result of Poisson distribution effect, C57/6j mtDNA was detected in lane 6 but not in lanes 3–5); lanes 7–10, PCR products amplified from single copy of C57/6j mtDNA and a BALB/c oocyte mtDNA were digested with Tth111I restriction enzyme (as a result of Poisson distribution effect, C57/6j mtDNA was detected only in lanes 9 and10).
Table 1.
Detection of sperm mtDNA from oviductal sperm, different stages of embryos, and newborn mice
| Group | Sperm from oviduct | One-cell embryos | Eight-cell embryos | Hatched blastocysts | 10.5 d embryo | Newborn mice |
| Total no. of samples | 124 | 115 | 116 | 120 | 102 | 93 |
| Positive | 14 | 13 | 8 | 9 | 11 | 7 |
Paternal mtDNA genome could be detected in only a few embryos, newborn mice, and sperm from oviduct. The similar positive rate indicated that, when sperm with mtDNA had fertilized an oocyte, paternal mtDNA was not eliminated until birth of the mouse.
Furthermore, as discussed earlier, sperm mitochondria were always limited to one blastomere before the four-cell stage embryo and distributed unevenly in several cells until morula stage. To test whether this uneven distribution was still kept after implantation and birth, paternal mtDNA in the fetus and placenta of 10.5-d embryos and several tissues of newborn mice was examined, and the results are shown in Table 2 and Table S1, respectively. In 10.5-d samples (n = 102), paternal mtDNA was detected in three fetuses and eight placentas; meanwhile, in the 93 newborn mice, paternal mtDNA was detected in several different tissues in seven newborn individuals. Thus, we have verified that the uneven distribution of sperm mitochondria may be one of the mechanisms of maternal mitochondria inheritance and may cause paternal mtDNA being transmitted only in a few tissues rather than the whole body. Additionally, in mouse 83 (Table S1), paternal mtDNA was detected even in ovary tissue, which implied that paternal mtDNA transmission to the progenies through germ cells may be possible (11). However, considering the preelimination of mtDNA in sperm (as discussed later) and mtDNA bottleneck effect during oocyte maturation, this possibility may be low (10, 24).
Table 2.
Sperm mtDNA detected in 10.5-d placenta and fetus
| Individual no. of embryos | Fetus | Placenta |
| 17 | + | |
| 26 | + | |
| 34 | + | |
| 53 | + | |
| 56 | + | |
| 60 | + | |
| 66 | + | |
| 81 | + | |
| 85 | + | |
| 89 | + | |
| 90 | + | |
| Total | 3 | 8 |
A total of 102 embryos (10.5 d) were subjected to examination of paternal mtDNA, and the plus signs indicate that paternal mtDNA was detected. Among the embryos examined, three fetuses and eight placentas were detected to contain paternal mtDNA. Paternal mtDNA detected in placentas and fetuses indicated that uneven distribution of sperm mitochondria before the four-cell stage was one of the mechanisms used to ensure maternal mitochondrial inheritance.
Most Motile Sperm Eliminated Their mtDNA Before Fertilization.
The reason for the result that little sperm mtDNA was detected even in early pronuclear embryos may be a result of the possibility that sperm had eliminated its mtDNA before fertilization. Because of the exponential increase effect of PCR, even a single copy mtDNA may induce a false positive result; to test this possibility, we investigated mtDNA from single sperm that were obtained from the oviduct. Results shown in Table 1 indicated that the majority of sperm obtained from the oviduct did not contain any mtDNA, validating the speculation that sperm mtDNA was eliminated before fertilization.
To confirm these single sperm results, real-time PCR was used to examine mtDNA copy number of sperm in different activities from a colony angle. Methods as described by Furimsky et al. (25) and May-Panloup et al. (26) were used in this study. Briefly, 45% and 90% Percoll gradient centrifugation was used to separate less-motile and motile sperm, and mtDNA/β-globin ratio was used to determine the sperm mtDNA copy number. As shown in Table 3, the mtDNA/β-globin ratio assay revealed that mouse sperm contained 29.12 copies mtDNA per sperm, which was in agreement with previous results that mouse sperm contained 10∼100 copies mtDNA per sperm (27, 28). However, when sperm were separated into motile and less-motile categories, we found that motile sperm located in the 90% Percoll gradient layer contained only 1.29 copies of mtDNA per sperm whereas less motile sperm located in the 45% Percoll gradient layer contained 45.93 copies mtDNA per sperm. This was similar to results obtained in human sperm studies that demonstrated that motile sperm had less than one copy of mtDNA (26). mtDNA copies of sperm that were not used for gradient separation, and those found in the 45% Percoll layer varied largely among different mice, which might be a result of the different qualities of sperm in different individuals. The fact that motile sperm contained very few copies of mtDNA per sperm implied that the majority of effective sperm that could get to the oviduct to undergo fertilization may not have any mtDNA, which was verified by examination of mtDNA in single sperm that had different motile activities as shown in Table S2. A very recent report in Drosophila also showed that sperm eliminated their mtDNA before fertilization (9). Thus, another important pathway to ensure maternal mitochondrial inheritance in mice might be the elimination of sperm mtDNA before fertilization. This mechanism may imply that, when assisted reproductive technology is used, because of the deletion of natural selection of mtDNA-free sperm in the process of sperm swimming to the oviduct, the rate of embryos inheriting paternal mtDNA may be increased, which has been observed in human abnormal embryos (29).
Table 3.
Comparison of sperm mtDNA/β-globin ratio among 45% and 90% density gradient layers and samples that were not subjected to gradient centrifugation
| Mouse | 45% layer | 90% layer | Sperm not subjected to gradient centrifugation |
| 1 | 70.26 | 0.55 | 44.27 |
| 2 | 31.49 | 0.27 | 1.50 |
| 3 | 5.83 | 0.12 | 40.12 |
| 4 | 16.01 | 0.74 | 32.91 |
| 5 | 106.04 | 4.79 | 26.81 |
| Average | 45.93 | 1.29 | 29.12 |
Sperm mtDNA/β-globin ratio in sperm samples from 90% Percoll gradient layer was much lower than that from sperm in 45% Percoll gradient layer or that from sperm not subjected to gradient centrifugation. Moreover, the most absolute values were <1 in the 90% Percoll gradient layer, implying that many of the sperm located in this layer might not have any mtDNA.
Conclusions
Keeping homogeneous mtDNA is very important for organisms because, even when two normal mtDNAs are mixed, they may produce adverse physiological effects (30). As one of the most important mechanisms to maintain mtDNA homogeneity, maternal inheritance working at the beginning of life is very conservative and has great significance for evolution. To ensure maternal inheritance of mtDNA, different mechanisms are involved in different organisms; however, all the mechanisms are attributed to an active elimination of sperm mitochondria or mtDNA after fertilization (8, 13, 15). In this study, by using mitochondria-labeled transgenic mice and analysis of sperm mtDNA from single sperm or embryo, we described that maternal inheritance is a passive result that relies on two aspects, the preelimination of mtDNA in sperm and the uneven distribution of sperm mitochondria during early embryo development. The unique mechanism of maternal mitochondrial inheritance demonstrated here is hopefully useful in the interpretation and prevention of some inherited mitochondrial diseases. In particular, when assisted reproductive technologies are used, we suggest that paternal mtDNA inheritance effects should be considered because of the concentrated effects from their uneven distribution. In addition, the use of centrifugation to select mtDNA-free sperm would likely be a effective method to prevent some inherited mitochondrial diseases as a result of biparental mtDNA inheritance.
Materials and Methods
If not stated specifically, all chemicals and reagents used in this study were purchased from Sigma. All mice were maintained in accordance with the Animal Experiment Standard of the Institute of Zoology, Chinese Academy of Sciences. Briefly, mice were bred in a 12/12 h light/dark period and killed by cervical vertebra dislocation.
Mouse Strains.
To observe the role of autophagy in maternal mitochondrial inheritance after fertilization, two transgenic mouse strains from RIKEN BioResource Center were used. The first, named GFP-LC3#53 (no. 00806) mice, which expressed GFP-labeled LC3 proteins as female mice, were used to provide autophagosome GFP-labeled oocytes (31). The second, B6D2-Tg(CAG/Su9-DsRed2,Acr3-EGFP)RBGS002Osb (no. 03743) mice, which expressed RFP mitochondria and GFP-labeled acrosomes, were used to provide RFP mitochondria-labeled sperm (32). In addition, Atg5flox/flox mice and Zp3-cre were used to produce oocyte-specific Atg5-deficient mice (Atg5flox/−; Zp3-Cre mice) (33). To analyze the fate of paternal mtDNA in sperm and embryos, BALB/c female mice (Institute for Laboratory Animal Resources, National Institutes for Food and Drug Control, China) and C57BL/6j male mice (Beijing HFK Bioscience) were used in this study, as these two strains have three different bases in their mtDNA genomes that could be used for allele-specific PCR and restriction enzyme analysis as described later.
Sperm, Oocyte, and Embryo Collection.
Spermatozoa used for fluorescence imaging or real-time PCR were prepared by extruding and cutting spermiduct and cauda epididymidis in human tubal fluid (HTF) medium as described elsewhere (34). Oviduct sperm was released by cutting oviducts from mated mouse at 14 h of injection with human chorionic gonadotropin (hCG), and placed into lysis buffer with a capillary sucking device under a phase-contrast microscope. Motile and less-motile sperm were separated by Percoll gradient centrifugation as described by Furimsky et al. (25). Primarily, fresh sperm prepared from spermiduct and cauda epididymis were kept in Krebs Ringer bicarbonate medium buffered with Hepes for 30 min, allowing sperm dispersion, and then loaded onto discontinuous two-layer Percoll (Pharmacia) density gradients (45% and 90%) for 30 min centrifugation (650 × g, 25 °C). The 45% and 90% Percoll solutions were then separated into individual centrifugation tubes, and equal volumes of 0.9% physiological saline solution was added. The tube was then centrifuged at 430 × g for 10 min to allow sperm to sediment as pellet. After carefully removing Percoll solutions, sperm sediment was resuspended in 0.9% physiological saline solution and centrifuged at 430 × g for 10 min once, and separated sperm were finally incubated at 37 °C and 5% CO2 for 30 min, allowing active sperm to swim up in HTF medium. As normal cells contained significantly more mtDNA than sperm, even when contaminated with a few cells, the results concerning sperm mtDNA may be reliably estimated. In this study, besides centrifugation, we further eliminated contaminated cells by treating collected samples with 200 µL of 50 mM Tris⋅HCl buffer (pH 6.8) at 8 °C for 20 min before lysis, as sperm is resistant to this treatment but contaminated cells are lysed in this process (35).
Superovulation was conducted to prepare oocytes and embryos. Briefly, 8- to 12 wk-old GFP-LC3 or 12- to 15-wk-old BALB/c female mice were intraperitoneally injected with 10 IU pregnant mare serum gonadotropin (Ningbo Animal Hormone Factory), followed by 10 IU hCG (Ningbo Animal Hormone Factory) 48 h later. Oocytes and embryos were collected from oviducts after injection with hCG at 14 h, and the cumulus mass was removed with 0.15% (wt/vol) hyaluronidase in M2 medium for 5 min. If needed, embryos were cultured in potassium simplex optimization medium drops covered with mineral oil at 37 °C and 5% CO2. If embryos were collected for PCR, incubation was performed in acidic tyrode solution to remove sperm that was localized at the zonae and in the perivitelline space, and the embryos were examined under a phase-contrast microscope before lysis.
DNA and mRNA Preparation and Microinjection.
Lysis buffer containing 50 mM Tris⋅HCl, pH 8.5, 1 mM EDTA, 0.5% Tween-20, and 100 mg⋅mL−1 proteinase K, which efficiently released mtDNA, was used in this study, and 10 mM DTT was added to break the disulphide bonds in sperm. mtDNA except that used for real-time PCR was prepared by incubating samples in 3 µL lysis buffer for 2 h at 55 °C and 15 min at 95 °C. To be certain that paternal mitochondria would be not lost, lysis and the first-cycle PCR were completed in the same PCR tube. For real-time PCR, samples were prepared by using the same method, except that lysis buffer was used at 7 µL per tube.
For analysis of the paternal mtDNA in different tissues and placentae and fetuses, newborn mice younger than 5 d after birth were killed by ethyl ether anesthesia, or 10.5 d pregnant mice were killed by cervical vertebra dislocation, respectively. Twelve tissues (Table S1) and placentas and fetuses were obtained by using a dissecting microscope. All samples were subjected to lysis in 150 µL tissue lysis buffer (50 mM KCl, 10 M Tris⋅HCl, pH 8,1.5 mM MgCl2, 0.1% gelatin, 0.45% Nonidet P-40, 0.45% Tween-20,and 150 mg/mL Proteinase K) for 24 h at 55 °C. Total DNA then was purified with a genome purification kit (General AllGen Kit; CoWin Biotech) and kept at −20 °C until used for PCR.
To investigate the role of P62 protein in maternal mitochondrial inheritance, CFP-P62 was expressed in embryos by microinjection of zygotes with CFP-P62 mRNA. The P62 cDNA was amplified from total cDNA, which was obtained from the liver of C57BL/6j female mice with a PrimeScript First-Strand cDNA Synthesis Kit (Takara Biotechnology). The DNA was then inserted into PCS2-MT vector forming plasmids, which were transfected and replicated in TOP10 Escherichia coli (TianGen Biotech). Purified plasmids then were linearized by NotI restriction enzyme (Takara Biotechnology) and used as template to express mRNA with an mMESSAGE mMACHINE SP6 Kit (Ambion). Finally, CFP-P62 mRNA was purified with a RNeasy Micro Kit (Qiagen) and kept at −80 °C until use. All procedures used were performed strictly in accordance with the product instructions.
Fluorescence Imaging.
In this study, immunofluorescence and live cell fluorescence imaging were primarily used to study the role of ubiquitin, P62 protein, and autophagy in maternal mitochondrial maternal inheritance. The first antibodies used in this study included Lys63-specific anti-ubiquitin antibody (also termed K63 antibody; 05–1308; Merck Millipore) and Lys48-specific anti-ubiquitin antibody (also termed K48 antibody; 05–1307; Merck Millipore), and the second antibody was Cy5-conjugated and FITC-conjugated goat anti rabbit IgG (Zhongshan Goldenbridge Biotechnology). Samples were first washed in PBS solution three times, and then subjected to fixation in 4% paraformaldehyde in PBS solution for 30 min, permeabilization in 0.2% Triton X-100 in PBS solution for 20 min, and blocking in solution containing 2% BSA, 0.05% PBS solution/Tween-20 for 1 h step by step. Subsequently, samples were incubated with the first antibodies overnight at 4 °C and the second antibodies for 1 h at room temperature. After washing three times with 0.05% PBS solution/Tween-20 for 10 min after the second antibody incubations, samples were mounted in 90% glycerol, 0.1 M Tris⋅HCl, pH 8.0, and 2.3% 1,4-diazobicyclo-2,2,2-octane for fluorescence imaging. For live cell fluorescence imaging, samples incubated with 5 µM LysoTracker Blue DND-22 (L-7525; Invitrogen) for 20 min or from transgenic mice or expressing exogenous CFP-P62 after microinjection of CFP-P62 mRNA were used. All fluorescence imaging was performed by using a 60× 1.4 NA oil immersion objective on a spinning-disk confocal microscope (UltraVIEW-VoX; PerkinElmer).
PCR and Restriction Enzyme Identification.
By blasting the mtDNA of C57BL/6j and BALB/c mice, three different bases between them were found (Fig. S6). The different base localized on position 9347 allowed mtDNA from C57BL/6j mice but not from BALB/c mice to be cut by Tth111I enzyme. So, when C57BL/6j male mice were mated to BALB/c female mice, paternal mtDNA could be distinguished from maternal mtDNA with Tth111I enzyme. Considering that the oocyte possesses significantly more mtDNA than sperm (∼105 times as much), the other two different bases, which localized on positions 9460 and 9820, were used to specifically amplify C57BL/6j mtDNA from BALB/c by two-cycle allele-specific PCR reactions. Briefly, two primers, antisense primers 1 and 2 (Table S3), were selected in this study for the allele-specific PCR reactions, which would selectively amplify one copy of paternal mtDNA even when mixed with one of the oocyte’s mtDNAs. Therefore, to detect single sperm mtDNA from single oviduct sperm or embryo, samples were first specifically amplified to produce 520 bp DNA by two allele-specific PCR reactions with the 2× Taq MasterMix PCR kit (CoWin Biotech); subsequently, the PCR products were cut to 383 and 157 bp DNA by Tth111I restriction enzyme (Takara Biotechnology) reaction and examined by agarose gel electrophoresis.
Real-time PCR by examining fluorescence of SYBR Green I was used to quantify the number of mtDNAs and β-globin. Sperm mitochondrial number was determined by comparing the number of mtDNAs to β-globin, which was a single copy in the genome, and oocyte mitochondrial number was determined directly by real-time PCR. For quantification of the mtDNA number and β-globin, two standard amplification curves were established. To decrease the error, besides keeping the PCR condition completely the same, a group of standard samples was operated at the same time for each experiment to correct amplification efficiency. Templates used for standard samples were amplified from C57BL/6j mice by PCR with primers as shown in Table S3. Sperm sample (1 µL) or 1/100 oocyte sample was used as template for real-time PCR, and when standard samples or oocyte samples were used as template, 1 µL lysis buffer was added to the reaction system to keep the reaction system the same. The real-time PCR system contained 10 µL 2× reaction mix (UltraSYBR Mixture with ROX; CoWin Biotech), 1 µL template or lysis buffer, 0.25 mM and 0.5 mM primers for mitochondrial PCR and β-globin PCR separately, adjusted to 20 µL by complementation with pure water. Both reactions were 40 cycles containing 15 s denaturation at 95 °C and 60 s extension at 60 °C and completed on a Rotor-Gene Q (Qiagen). All data were analyzed with Rotor-Gene Q Series software.
Supplementary Material
Acknowledgments
We thank Prof. Noboru Mizushima (Tokyo Medical and Dental University) and Prof. Masaru Okabe (Osaka University) for providing the GFP-LC3#53 and B6D2-Tg(CAG/Su9-DsRed2,Acr3-EGFP)RBGS002Osb transgenic mice, respectively; and Profs. Shaorong Gao (National Institute of Biological Sciences) and Wei Li (Institute of Zoology, Chinese Academy of Sciences) for providing the Zp3-Cre and Atg5flox/flox mice, respectively. This study was supported by Major Basic Research Program Grants 2012CB944404 and 2011CB944501 (to Q.-Y.S.) and National Natural Science Foundation of China Grant 30930065 (to Q.-Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303231110/-/DCSupplemental.
References
- 1.Hutchison CA, 3rd, Newbold JE, Potter SS, Edgell MH. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974;251(5475):536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- 2.Basse CW. Mitochondrial inheritance in fungi. Curr Opin Microbiol. 2010;13(6):712–719. doi: 10.1016/j.mib.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Ankel-Simons F, Cummins JM. Misconceptions about mitochondria and mammalian fertilization: Implications for theories on human evolution. Proc Natl Acad Sci USA. 1996;93(24):13859–13863. doi: 10.1073/pnas.93.24.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford WC. Regulation of sperm function by reactive oxygen species. Hum Reprod Update. 2004;10(5):387–399. doi: 10.1093/humupd/dmh034. [DOI] [PubMed] [Google Scholar]
- 5.Al Rawi S, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334(6059):1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 6.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334(6059):1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Li H, Xue D. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 2011;21(12):1662–1669. doi: 10.1038/cr.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura Y, et al. Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc Natl Acad Sci USA. 2006;103(5):1382–1387. doi: 10.1073/pnas.0506911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLuca SZ, O’Farrell PH. Barriers to male transmission of mitochondrial DNA in sperm development. Dev Cell. 2012;22(3):660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shitara H, Hayashi JI, Takahama S, Kaneda H, Yonekawa H. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics. 1998;148(2):851–857. doi: 10.1093/genetics/148.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gyllensten U, Wharton D, Josefsson A, Wilson AC. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991;352(6332):255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 12.Kaneda H, et al. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92(10):4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutovsky P, et al. Ubiquitin tag for sperm mitochondria. Nature. 1999;402(6760):371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 14.Sato M, Sato K. Maternal inheritance of mitochondrial DNA: Degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy. 2012;8(3):424–425. doi: 10.4161/auto.19243. [DOI] [PubMed] [Google Scholar]
- 15.Levine B, Elazar Z. Development. Inheriting maternal mtDNA. Science. 2011;334(6059):1069–1070. doi: 10.1126/science.1215480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 19.Cummins JM, Wakayama T, Yanagimachi R. Fate of microinjected spermatid mitochondria in the mouse oocyte and embryo. Zygote. 1998;6(3):213–222. doi: 10.1017/s0967199498000148. [DOI] [PubMed] [Google Scholar]
- 20.Sutovsky P, Tengowski MW, Navara CS, Zoran SS, Schatten G. Mitochondrial sheath movement and detachment in mammalian, but not nonmammalian, sperm induced by disulfide bond reduction. Mol Reprod Dev. 1997;47(1):79–86. doi: 10.1002/(SICI)1098-2795(199705)47:1<79::AID-MRD11>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 21.Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132(3):479–490. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- 22.Tarkowski AK, Ozdzeński W, Czołowska R. How many blastomeres of the 4-cell embryo contribute cells to the mouse body? Int J Dev Biol. 2001;45(7):811–816. [PubMed] [Google Scholar]
- 23.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med. 2002;347(8):576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 24.Khrapko K. Two ways to make an mtDNA bottleneck. Nat Genet. 2008;40(2):134–135. doi: 10.1038/ng0208-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furimsky A, et al. Percoll gradient-centrifuged capacitated mouse sperm have increased fertilizing ability and higher contents of sulfogalactosylglycerolipid and docosahexaenoic acid-containing phosphatidylcholine compared to washed capacitated mouse sperm. Biol Reprod. 2005;72(3):574–583. doi: 10.1095/biolreprod.104.036095. [DOI] [PubMed] [Google Scholar]
- 26.May-Panloup P, et al. Increased sperm mitochondrial DNA content in male infertility. Hum Reprod. 2003;18(3):550–556. doi: 10.1093/humrep/deg096. [DOI] [PubMed] [Google Scholar]
- 27.Wai T, et al. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod. 2010;83(1):52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shitara H, et al. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics. 2000;156(3):1277–1284. doi: 10.1093/genetics/156.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St John J, et al. Failure of elimination of paternal mitochondrial DNA in abnormal embryos. Lancet. 2000;355(9199):200. doi: 10.1016/s0140-6736(99)03842-8. [DOI] [PubMed] [Google Scholar]
- 30.Sharpley MS, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151(2):333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasuwa H, et al. Transgenic mouse sperm that have green acrosome and red mitochondria allow visualization of sperm and their acrosome reaction in vivo. Exp Anim. 2010;59(1):105–107. doi: 10.1538/expanim.59.105. [DOI] [PubMed] [Google Scholar]
- 33.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321(5885):117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 34.Chao SB, et al. Heated spermatozoa: Effects on embryonic development and epigenetics. Hum Reprod. 2012;27(4):1016–1024. doi: 10.1093/humrep/des005. [DOI] [PubMed] [Google Scholar]
- 35.Kao SH, Chao HT, Liu HW, Liao TL, Wei YH. Sperm mitochondrial DNA depletion in men with asthenospermia. Fertil Steril. 2004;82(1):66–73. doi: 10.1016/j.fertnstert.2003.11.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.