Abstract
The plant circadian clock is involved in the regulation of numerous processes. It serves as a timekeeper to ensure that the onset of key developmental events coincides with the appropriate conditions. Although internal oscillating clock mechanisms likely evolved in response to the earth’s predictable day and night cycles, organisms must integrate a range of external and internal cues to adjust development and physiology. Here we introduce three different clock outputs to illustrate the complexity of clock control. Clock-regulated diurnal growth is altered by environmental stimuli. The complexity of the photoperiodic flowering pathway highlights numerous nodes through which plants may integrate information to modulate the timing of phenological outputs. Comparative analyses among ecotypes that differ in flowering response reveal additional environmental cues and molecular processes that have developed to influence flowering. We also explore the process of cold acclimation, where circadian inputs, light quality, and stress responses converge to improve freezing tolerance in anticipation of colder temperatures.
Keywords: circadian clock, photoperiodic flowering, phenology, diurnal growth, cold acclimation, adaptation
1. INTRODUCTION
Numerous physiological and growth processes show daily oscillation patterns, which are often controlled by the circadian clock. In Arabidopsis, about one third of all genes are clock regulated [1]. Likely to have evolved initially in response to the earth’s predictable day and night cycles [2], the circadian clock sets the timing of various transcriptional and posttranscriptional events to specific times of day. This mechanism changes the organisms’ sensitivities and/or responses to various external stimuli throughout the day, thus enabling them to extract specific information that occurred at certain times of the day or year. This clock role is called circadian gating. A well-understood example of this is the external coincidence model of photoperiodic flowering control. To accurately measure differences in day length in the facultative long-day plant, Arabidopsis, regulation of the timing of diurnal CONSTANS (CO) expression is crucial [3]. The circadian clock sets the timing of CO transcription from the late afternoon to night. During the short days of winter, the CO peak occurs at night, and its protein is degraded. In early summer’s long days, the peak coincides with daylight, and CO protein is stabilized to activate transcription of FLOWERING LOCUS T (FT), resulting in earlier flowering [3, 4]. This type of time-keeping mechanism helps guarantee that phenological changes like flowering occur during favorable times of the year.
The circadian clock-regulated biological processes are stable but also adaptable. Dicot leaves maintain a consistent oscillatory diurnal growth pattern despite external temperature changes [5]. Yet, there is growing recognition that diurnal growth oscillations are changeable within the lifetime of a plant, susceptible to external and internal limitations [6, 7]. Among populations and across generations within the same species, clock outputs like flowering, bud break, or onset of dormancy differ [8-10]. The differences persist when growing conditions are the same, indicating genetic adaptations to regional environmental variation. We have a general molecular understanding of how each output is induced as well as how the circadian clock affects them. We still lack information on how endogenous control of clock outputs is adjusted from individual to individual and species to species.
The aim of this review is to address these complexities. We utilize diurnal plant growth to highlight the plasticity of endogenous circadian control and to illustrate that the degree of control is dependent on variation in the immediate environment. We capitalize on the in-depth mechanistic knowledge of seasonal flowering control to explore how numerous external and endogenous cues can modulate a clock-mediated output. We use recent multi-population, genome-wide analyses, which demonstrate underlying genetic differences among phenotypically different populations, to highlight genes and molecular pathways under selective pressure. Finally, we explore the molecular processes of cold-tolerance and highlight areas of uncertainty regarding stress-induced versus clock-mediated cryoprotection.
2. DIURNAL PLANT GROWTH: PLASTICITY OF CIRCADIAN CONTROL
The sensitivity of plants to external and internal stimuli, which regulate diurnal growth, can change depending on growth conditions, plant age, type of organ, and time of day. Dicot leaves and stems display diurnal rhythmic growth that is maintained under constant environmental conditions, indicating the involvement of the circadian clock [5, 11, 12]. However, the timing of peak growth changes from day to night with the developmental age of the leaf, attributable to a shift from metabolic-limited growth in younger leaves to hydraulic-limited growth in mature leaves [13]. Dicot leaves maintain a stable oscillatory growth pattern even under daily temperature cycles [5], perhaps through the ability of the circadian clock to adjust metabolic rates throughout the day. Roots do not show the same degree of circadian growth-rate regulation, as they must quickly alter growth to take advantage of available soil resources and water [14]. The daily rate and pattern of leaf growth is also maintained similarly even under low-light conditions where the carbon source is limited [13]. Arabidopsis can continue growing with carbon limitations by decreasing leaf thickness and maintaining surface area. Conversely, water stress causes growth to slow during the day such that peak growth occurs at night even in very young leaves. Under this condition, decreasing leaf surface area by slowing growth can help reduce water lost through transpiration [13]. The molecular mechanisms by which external and internal factors coordinate to regulate leaf growth and development and how the circadian clock affects this regulation remain largely unknown.
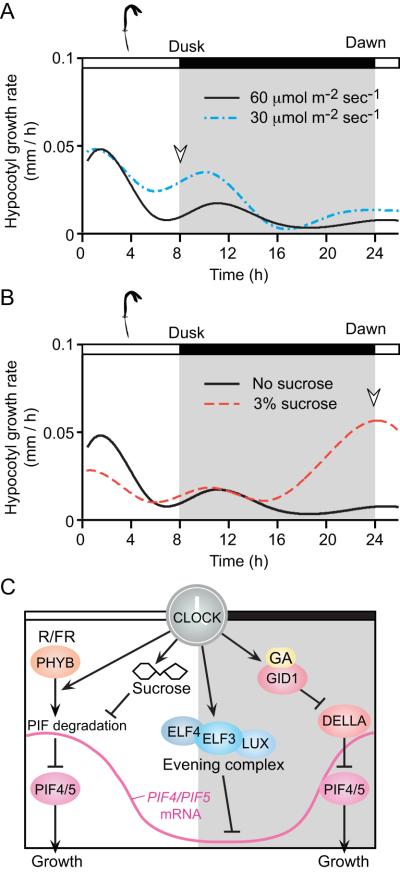
In comparison to the diurnal leaf growth mechanism, we have a better understanding of the mechanisms of diurnal hypocotyl growth. Seedlings respond to changes in light quality and quantity, carbon reserves, and temperature in a time-dependent fashion. During the growth phase, in which Arabidopsis cotyledons fully expand, a change in light intensity most affects hypocotyl growth around dusk (Fig. 1A) [7]. In contrast, the effect of sucrose in the growth media is most pronounced around dawn (Fig. 1B). The mechanisms for diurnal control of growth, best understood in Arabidopsis hypocotyls and reviewed extensively [12, 15], are beginning to shed light on the changes in sensitivity over the course of the day to various parameters regulated by the circadian clock (Fig. 1C). The bHLH transcription factors known as PHYTOCHROME INTERACTING FACTORs (PIFs) are expressed in a clock-dependent manner [16]. The circadian clock components, EARLY FLOWERING 3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX, also known as PHYTOCLOCK1) directly regulate the expression of PIF4 and PIF5 transcription [17]. ELF3, ELF4, and LUX form a protein complex named the evening complex, which directly represses the expression of PIF4 and PIF5 transcripts around dusk, limiting the expression of PIF4 and PIF5 to dawn [17] (Fig. 1C).
Fig. 1. Circadian clock-mediated diurnal hypocotyl growth.
(A, B) Arabidopsis hypocotyl growth for 4 to 5-day-old seedlings, before their cotyledons are fully expanded, is regulated by different inputs at different parts of the day. (A) Hypocotyl growth rate is most affected by light intensity at dusk. (B) Carbon source availability most affects the growth rate at dawn (dusk and dawn are indicated by open arrowheads). The graphs were modified from [7]. (C) The circadian clock coordinates various pathways involved in diurnal growth regulation. These pathways include phytochrome, sucrose and GA signaling, and the components (ELF3, ELF4, and LUX) of the evening complex, ultimately resulting in transcriptional or post-translational regulation of PIF4 and PIF5. Higher sucrose levels reduce the degradation rate of PIF5 resulting in an enlarged morning response to growth. PHYB signals keep PIF levels low under light. The evening complex represses the expression of PIF4 and PIF5 at dusk. Higher amounts of GA-GID1 complex around dawn induces degradation of DELLA proteins, which prevent PIF4 (and possibly PIF5) from binding to target DNA. PIF4/PIF5 mRNA profile is shown by a pink line. Coordinated regulation by the clock contributes to temporally confined growth that occurs at dawn and dusk through convergence of light, hormone, and metabolic signaling.
In addition to clock-dependent transcriptional regulation, time-dependent posttranslational regulation also plays a role. PIF4 and PIF5 proteins are degraded under red light, which is absorbed by PHYTOCHROME B (PHYB) photoreceptor. This light-dependent regulation restricts the activity of these growth-promoting PIF transcription factors into shaded conditions in long days or a period around dawn in short days (reviewed in [12]). The PIF4 proteins are further activated during the night in short days, partly due to the interaction of gibberellin (GA) and its receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1), both of which are modulated by the circadian clock. The clock regulates the timing expression of GA synthetic enzyme gene GA20ox1 and GID1 genes [18, 19]. GID1 binds to GA and this interaction triggers DELLA proteins for proteasomal degradation [20]. The DELLA proteins bind to PIF4 and inhibit its DNA binding ability [21]. DELLA proteins are degraded around dawn in short days [19]. Thus, removing the DELLA proteins enables PIF4 to bind to the G-box to induce the expression of genes involved in hypocotyl growth around dawn [21] (Fig. 1C). Sucrose also stabilizes PIF5 protein throughout the day [7]. This, together with transcriptional regulation, ensures that diurnal hypocotyl growth occurs at a certain time of day (Fig. 1C).
Hypocotyl growth is also susceptible to changes in ambient temperature. High ambient temperatures cause pre-dawn expression of PIF4 in long days similar to that observed for short days, helping to explain why hypocotyl growth occurs in such conditions [16]. The presence of PIF4 and PIF5 proteins are required both in short days and in 28 °C long days for pre-dawn expression of a suite of genes associated with gibberellin, auxin, brassinosteroid, ethylene, and cytokinin, and which are also important for the shade avoidance response [16, 22]. In sum, we are beginning to get a picture of a flexible, clock-mediated system that coordinates growth-regulating genes, turgor, and resource availability with external and internal stimuli.
3. SEASONAL FLOWERING
3.1 THE PHOTOPERIODIC FLOWERING PATHWAY IN ARABIDOPSIS
In addition to diurnal growth regulation, the circadian clock plays an important role in the regulation of photoperiodic flowering. The photoperiodic flowering pathway controls the amount of florigen, which determines flowering time. In Arabidopsis, FT protein is a major part of florigen synthesized in the leaf vasculature [23]. FT protein is translocated to the shoot apical meristem and, together with FD and 14-3-3 [24], initiates transcription of floral identity genes that regulate floral development [23, 25]. To selectively induce FT transcription in long days, restricting CO expression by the circadian clock to the long-day afternoon is essential. In the morning, CYCLING DOF FACTORs (CDFs) are highly expressed and directly repress CO transcription [26, 27]. At the same time, the core clock components, CCA1 and LHY, repress the transcription of FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and GIGANTEA (GI), both of which negatively regulate CDFs [28]. FKF1 and GI proteins peak in the afternoon and form a complex in a blue-light dependent manner [28]. The FKF1-GI complex removes CDF repressors by proteasomal degradation in the long-day afternoon [28]. Simultaneously, the levels of CDF transcripts also decrease due to repression by other clock components, PSEUDO RESPONSE REGULATOR 9 (PRR9), PRR7, and PRR5 [29, 30]. These mechanisms determine the timing of daytime CO gene expression in long days. Recently, four transcriptional activators of CO, named FLOWERING BHLH (FBH), were identified [31]. Interestingly, FBH transcriptional activators control the amplitude of CO expression, which also affects the expression levels of FT [31].
Another important mechanism for day-length sensing is specific stabilization of CO protein [4]. Light signals perceived by phytochromes (PHY) and cryptochromes (CRY) stabilize CO protein only in long-day afternoons [4]. This time-dependent stabilization of CO protein is regulated by the combinational regulation of light and the clock. The PHYB signal and the E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) are involved in the degradation of CO during the morning [4, 32], although it is not known whether the PHYB signal regulates CO through HOS1. CRY2 binds to SUPPRESSOR OF PHYA-105 1 (SPA1) and the CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) E3 ubiquitin ligase complex under blue light to suppress the activity of the complex [33]. In the dark, even though CO mRNA is highly accumulated, CO protein is actively degraded by the COP1/SPA1 complex [4, 34, 35]. The clock-regulated blue-light photoreceptor, FKF1, stabilizes CO in the long-day afternoon. When FKF1 is expressed under light, FKF1 binds to and stabilizes CO protein in a blue light-dependent manner [36]. The complex interplay between light and clock ensures that CO protein exists only in long day afternoons when FT is induced. Thus, the circadian clock controls CO transcriptional and posttranscriptional mechanisms to achieve proper day-length sensing for flowering.
The photoperiodic pathway also serves to integrate numerous signals, and many of these signals act through independent or partially independent pathways to regulate flowering. Members of the PHY and CRY photoreceptor families not only regulate flowering under different light conditions, but also differentially control flowering depending on ambient temperature (reviewed in [37]). One of the components that integrates photoreceptor signaling with flowering time regulation is PHYTOCHROME AND FLOWERING TIME 1 (PFT1). Acting downstream of PHYB, PHYD, and PHYE, PFT1, a component (MED25) of the Mediator complex, integrates light quality and possibly temperature signals for a subset of flowering genes [38]. In response to changes in ambient temperature, HOS1 regulates FT and TWIN SISTER OF FT (TSF) independently of CO through the autonomous pathway genes, FVE and FLK [39]. The expression of some of the genes listed above is regulated by abiotic and biotic stimuli. For instance, the simultaneous application of drought and heat stresses induces the expression of FBH3 [40]. A carbon supplement in the growth media reduces PRR5 and CDF2 expression [41]. Metabolic sucrose seems to adjust the circadian clock through GI activity [42]. As the addition of 5% sucrose to the growth media represses FT expression and delays flowering [43], GI may also regulate flowering via sucrose signaling (see details in Haydon et al. in this issue). These findings indicate that precise environmental information can be integrated into the photoperiodic flowering pathway to fine tune flowering time in nature.
3.2 DIFFERENCES IN FLOWERING TIME WITHIN AND BETWEEN POPULATIONS
Numerous field studies have reported variations among individuals and populations in the timing of key phenological events. Early studies noted significant site and regional differences in the timing of bud break [8, 9], which in Populus shares components of the Arabidopsis photoperiodic flowering pathway [44]. Some of these differences could be explained by environmental differences like photoperiod or temperature [8, 9]. Others persisted when plants of the same species were collected from different sites and grown together [8, 9]. These observations indicate that adaptation to regional climates contributes to genetic variation in internal regulatory mechanisms among populations. The capacity of a population to adapt depends on the genetic material available, and some studies have noted within population variation as well. Mertensia fusiformis correlates its flowering to snowmelt, displaying a skewed population distribution [10]. End-of-season drought selects for early-flowering plants, while the chance of late spring frost favors maintenance of a few that flower later.
Much remains to be learned about the mechanisms underlying flowering time modulation by parameters other than photoperiod and vernalization. However, it is likely the circadian clock plays a role in regulating responses to other predictable environmental cues. For instance, daytime temperatures better correlate with flowering times than nighttime temperatures for Arabidopsis strains planted in the field, except for populations sown in fall [45]. This implies that the clock may influence the timing of temperature sensitivity within a day, and that the circadian gating function differs by seasons. Plant size, which itself is affected by the environment, also impacts timing of flowering in biennial species (see [46] for further discussion). For biennial species, plant size likely correlates to a threshold level of resources needed to induce flowering. Consistent with this idea, flowering in Arabidopsis and Sinapis alba is coupled with an increase in sucrose and the carbon:nitrogen ratio at the shoot apical meristem [47, 48]. Taken together, these examples indicate there are adaptations among individuals and populations that modulate clock outputs. Yet few studies have demonstrated concrete mechanistic differences among them [44].
With the available data of single nucleotide polymorphisms (SNPs) from a large number of wild-type Arabidopsis accessions, comparative analyses of natural populations provide clues by which to understand the environmental drivers of adaptation [49-51]. Several environmental parameters correlate with an enrichment of amino acid-changing SNPs [50, 51]. After removing SNPs related to specific geographic locations, representative SNPs in early-flowering plant populations correlated to summer precipitation, whereas growing-season temperatures drove most of the explained SNP variation in late-flowering plants [50]. These findings are supported by a reciprocal transplant between Swedish and Italian Arabidopsis populations [52]. The strains had higher fitness in their home sites than representatives from the other population. The authors postulate that the Swedish plants were better able to tolerate cold temperatures while Italian plants could flower early to avoid late-season droughts. Similar early- and late-flowering differentiation can occur within a population over time. Seeds of Brassica Rapa plants collected from the same site after multi-year periods of above-then below-average rainfall were grown together in a common garden experiment [53, 54]. The drought years selected for plants that flowered earlier when they were smaller in stature. These results demonstrate that selection can rapidly occur, and that flowering times are affected when growing seasons are curtailed by drought. These data also provide clues by which to understand the molecular events that plants must simultaneously integrate when they flower in natural settings.
3.3 FLOWERING AND THE CLOCK: MOLECULAR TARGETS OF SELECTION
Comparative analyses of natural populations that vary in their phenotypic responses to environmental cues are tools by which to identify molecular pathways under selective pressure in nature [49-51]. Hormone signaling pathways may be targets for natural selection and have some links to the circadian clock. A recent comprehensive microarray study compared the expression levels in response to photoperiod among three Arabidopsis populations from locales with different photoperiods, light quality, winter severity, and precipitation in Norway [55]. They differed in their photoperiodic sensitivity when grown together, indicating underlying genetic differences in phenological regulation. Genes involved in biosynthesis of and response to abscisic acid (ABA), a drought-signaling molecule, were differentially regulated among populations. There are also surprising correlations among the expression patterns of genes involved in ethylene and auxin response and flowering [55]. The transcription of EIN3-BINDING F-BOX PROTEIN 1 (EBF1) and EBF2, both involved in circadian control of ethylene response [56], was the highest in the population showing the greatest sensitivity to photoperiod [55].
Not surprisingly, known components of the photoperiod pathway are represented in population studies that examine flowering [49, 55]. CRY2, GI, LHY, FKF1, FT and PHYA, PHYB, and PHYC, as well as SPA2 and SPA4 are highlighted. PHYA, GI, and FT are mentioned in more than one study. As we described above, many of these play roles in temperature and light quality perception along with photoperiod. DWARF IN LIGHT 2 (DFL2) has no known role in flowering; however, it was implicated as a target candidate in two studies [49, 55]. Both studies also highlight SHORT VEGETATIVE PHASE (SVP). SVP, a MADS-domain transcription factor, is a known mediator of ambient temperature response and an upstream regulator of FT [57]. An area of interest for molecular studies is analyzing the molecular natures of representative synonymous and non-synonymous (=amino-acid-changing) SNPs associated with climate [50, 51]. Synonymous mutations represent changes in DNA sequence that could affect cis-acting elements, whereas non-synonymous mutations may alter protein function. Many downstream flowering integrators are differentially expressed among populations of Arabidopsis [55], thus this is an area warranting further research.
4. DIURNAL AND SEASONAL COLD RESPONSE
4.1 CIRCADIAN REGULATION OF COLD RESPONSE
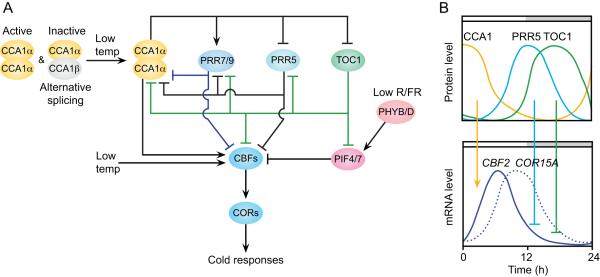
Cold temperatures have a drastic impact on survival for plants in temperate or boreal environments. While the adaptive response that allows plants to survive freezing conditions is well understood, only recently has a comprehensive mechanism for cold acclimation come into focus. Circadian regulation, light quality perception, and stress responses all converge to activate cold response. Adaptation to freezing temperatures is accomplished through cold acclimation, the process by which exposure to cold, non-freezing temperatures instills subsequent freezing tolerance. The canonical cold-response pathway has been studied extensively [58] and involves the C-REPEAT BINDING FACTOR (CBF also known as DREB) genes, which are transcriptional activators of their COLD REGULATED (COR) downstream targets [59] (Fig. 2A). Overexpression of the three CBF genes in Arabidopsis resulted in enhanced freezing tolerance upon induction of cold treatment [58]. A global transcriptome analysis revealed that nearly a quarter of cold inducible genes have CBF/DREB cis-regulatory elements in their promoters, implying that CBFs have a broad class of targets in initiating cold tolerance [60]. Cold-inducible transcriptome contains genes involved in processes including biosynthesis of secondary metabolites (raffinose, proline, antioxidants etc.), lipid desaturation and biosynthesis, cryoprotective compounds as well as drought stress [58, 60].
Fig. 2. Circadian clock regulation of the cold acclimation pathway in Arabidopsis.
(A) Inputs from the circadian clock, light signaling, and temperature activate COR genes through the CBF transcriptional activators, which induce the process of cold acclimation in Arabidopsis. In the diagram, colored lines are given to help distinguish interactions between different components. Cold temperatures induce CBF expression through CCA1 alternative splicing. The accumulation of the functional CCA1α splice variant in low temperatures increases CBF expression. Competition for the non-functional CCA1β variant at higher temperatures attenuates the cold response. TOC1 and PRR5 directly repress CBF expression in the evening and PRR7 and PRR9 likely act in a similar manner. Changes in the red/far-red light ratio are sensed through PHYB and PHYD, which in low red/far-red conditions drive transcriptional activation of CBF genes through PIF4 and PIF7. TOC1 protein binds to the PIF4 promoter and may physically interact with PIF7 protein [70, 74]. Through these mechanisms, the circadian clock, temperature, and light inputs mediate and amplify environmental signals to induce cold acclimation. (B) Relative abundance of clock component proteins at different times of the day dictates temporal CBF expression. In the morning, CCA1 activates CBFs, which in turn activate COR downstream genes. In the evening, TOC1 and PRR5 repress CBF expression. Schematic protein and mRNA levels are drawn from data in which plants were grown in continuous light conditions [64, 76, 77].
Recent studies have highlighted the importance of the circadian clock in gating cold acclimation in Arabidopsis. Clock control in genome-wide cold response was first inferred when transcriptome surveys differed significantly based on time of sampling [61]. Expanded analysis of the promoter regions of cold-inducible genes showed the prevalence of Evening Elements (EE), cis-elements recognized by CCA1 and LHY. The EE elements in CBF promoters are often coupled to ABA responsive cis-elements (ABRE) and both elements are necessary for cold-inducibility of COR genes [62], indicating the clock components are indispensible pieces in the cold response mechanism (Fig. 2A). CBF expression levels oscillate throughout the day with a peak at midday [63]. In cca1 lhy double mutant backgrounds, CBF genes have significantly reduced expression, which results in diminished freezing tolerance [64]. Temperature-dependent alternative splicing of CCA1 also plays a role in the establishment of cold tolerance [65]. CCA1 and LHY form homodimers or CCA1/LHY heterodimers; the formation of the paired transcriptional complex increases their DNA binding affinity and ability to activate or repress transcription through the EE. Functional CCA1 alternative splice variants are preferentially produced at lower temperatures, which allows for increased induction of CBF genes under cold acclimation conditions [65]. LHY is also affected by temperature dependent alternative splicing, but the active variant is preferentially transcribed at higher temperatures [66]. How the alternative splicing of these genes in concert functionally affects the cold acclimation process is not known (for further discussion, see Henriquez and Mas in this issue).
While CCA1/LHY is the best-characterized link between the clock and cold-acclimation, other clock genes also affect the response. PRR5 binds to the CBF promoter regions and represses their transcription (Fig 2A and B) [30]. In addition, prr5 prr7 prr9 triple mutants, which have an arrhythmic clock phenotype, constitutively express CBFs and have increased freezing tolerance [67]. The gi-3 mutants have freezing susceptibility, which may be linked to a decrease in endogenous sugar concentrations [68]. The circadian clock-regulated CONSTANS-LIKE genes, COL1 and COL2, are implicated in cold-tolerance in genome-wide transcriptome studies [60]. Furthermore, ABA responses, which influence cold acclimation, are also regulated by the core clock protein TOC1 [69]. TOC1 also directly binds to the CBF1 and CBF2 promoters [70], so TOC1 may regulate CBFs in both ABA-dependent and independent manners (Fig 2A and B). Additional connections between known clock components and cold acclimation are likely to be established.
4.2 PHOTOPERIOD, LIGHT QUALITY, AND COLD RESPONSE
In addition to the circadian clock, light perceived by PHYs affects cold response. Sensing changes in the red/far-red ratio may be important for coordinating cold responses in anticipation of cold nighttime temperatures or seasonally cold temperatures. Interestingly, a 16 °C growth condition coupled with a low red/far-red ratio is sufficient to induce CBF expression [63]. PHYB and PHYD mediate this light specific CBF induction at ambient temperatures [63]. PHYB regulation of the transcription of CBF genes occurs through PIF4 and PIF7 [71] (Fig. 2A). Additionally, exposure to shorter photoperiods also induces the cold acclimation response in temperate zone perennial species [72, 73]. Even in Arabidopsis, the amplitude of CBF gene oscillations in short days is higher than in long days [71]. Either the phyB or the pif4 pif7 double mutations abolish the higher amplitude in CBF expression in short days, indicating the same components were recruited to sense changes in red/far-red ratio, temperature and day length, all of which occur when winter is approaching [71]. Furthermore, PIF7 binds to G-box elements in CBF promoters [71, 74]. Thus, integration between light, temperature and clock output pathways at the CBF promoter through EE and G-box elements constitutes a novel mechanism by which these pathways assimilate various environmental information to prepare for winter by initiating cold acclimation.
5. CONCLUSION
Each clock output that we have described provides insight into the complexity and adaptability of circadian control tempered by external and internal stimuli. To help us understand how external and internal limitations interact in modulating circadian outputs, an interesting avenue of research would be to pair ecophysiological estimates of respiration and stomatal conductance with diurnal growth and molecular assays. As different climate variables affect early-versus late-flowering accessions of Arabidopsis, genome-wide association studies that separate these groups could be useful [49, 55]. Several downstream flowering integrators have been implicated in population-based studies, and both synonymous and non-synonymous SNPs are enriched with climate [50, 51]. It is possible this is due to linkage disequilibrium among synonymous and non-synonymous mutations. However, QTL analysis of two parental lines mapped ambient temperature and photoperiod sensitivity to the FT promoter [75]. A feasible next step is to sequence the promoters of these genes as well as measure the expression levels of these genes to determine whether these SNPs alter gene expression by mutating cis-acting elements. We do not understand how the circadian clock works in the ABA pathway to directly or indirectly affect cold acclimation, nor do we understand how ABA, the circadian clock, and light-quality sensing through PHYB interact to regulate cold response. It is likely that each factor of the cold-response pathway plays a greater or lesser role in activation of CBFs depending on the length of cold exposure. Mechanistic studies that assess the effects of both short- and long-term cold treatments on each of these components could help clarify their roles. In nature, plants must integrate information about external variables like climate, light intensity and surrounding vegetation, as well as endogenous carbon, nutrients, hydraulic status, and developmental age. To understand the complex regulation of physiology and development that occurs in nature, further experiments should be carried out under a wide variety of conditions, including those that more closely reflect the natural environment.
Highlights.
The circadian clock regulates various physiological and developmental processes.
Circadian clock-controlled diurnal growth is adaptive and responsive to various stimuli.
The photoperiodic flowering pathway can integrate other environmental signals.
Circadian clock, light, and abiotic stress converge to regulate freezing tolerance.
Acknowledgements
This work was supported by funding from the National Institutes of Health (GM079712) and the University of Washington Royalty Research Fund to TI, and the National Science Foundation Graduate Research Fellowship Program to HKS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–64. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–12. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- [4].Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–6. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- [5].Poire R, Wiese-Klinkenberg A, Parent B, Mielewczik M, Schurr U, Tardieu F, et al. Diel time-courses of leaf growth in monocot and dicot species: endogenous rhythms and temperature effects. J Exp Bot. 2010;61:1751–9. doi: 10.1093/jxb/erq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pantin F, Simonneau T, Muller B. Coming of leaf age: control of growth by hydraulics and metabolics during leaf ontogeny. New Phytol. 2012;196:349–66. doi: 10.1111/j.1469-8137.2012.04273.x. [DOI] [PubMed] [Google Scholar]
- [7].Stewart JL, Maloof JN, Nemhauser JL. PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS ONE. 2011;6:e19894. doi: 10.1371/journal.pone.0019894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kozlowski TT. Shoot growth in woody plants. Bot Rev. 1964;30:335–92. [Google Scholar]
- [9].Lechowicz M. Why do temperate deciduous trees leaf out at different times? adaptation and ecology of forest communities. Amer Nat. 1984;124:821–42. [Google Scholar]
- [10].Forrest J, Thomson JD. Consequences of variation in flowering time within and among individuals of Mertensia fusiformis (Boraginaceae), an early spring wildflower. Am J Bot. 2009;97:38–48. doi: 10.3732/ajb.0900083. [DOI] [PubMed] [Google Scholar]
- [11].Walter A. Dynamics of leaf and root growth: endogenous control versus environmental impact. Ann Botany. 2005;95:891–900. doi: 10.1093/aob/mci103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Farré EM. The regulation of plant growth by the circadian clock. Plant Biology. 2012;14:401–10. doi: 10.1111/j.1438-8677.2011.00548.x. [DOI] [PubMed] [Google Scholar]
- [13].Pantin F, Simonneau T, Rolland G, Dauzat M, Muller B. Control of leaf expansion: a developmental switch from metabolics to hydraulics. Plant Physiol. 2011;156:803–15. doi: 10.1104/pp.111.176289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Walter A, Silk WK, Schurr U. Environmental effects on spatial and temporal patterns of leaf and root growth. Annu Rev Plant Biol. 2009;60:279–304. doi: 10.1146/annurev.arplant.59.032607.092819. [DOI] [PubMed] [Google Scholar]
- [15].Ruts T, Matsubara S, Wiese-Klinkenberg A, Walter A. Diel patterns of leaf and root growth: endogenous rhythmicity or environmental response? J Exp Bot. 2012;63:3339–51. doi: 10.1093/jxb/err334. [DOI] [PubMed] [Google Scholar]
- [16].Nomoto Y, Kubozono S, Yamashino T, Nakamichi N, Mizuno T. Circadian clock and PIF4-controlled plant growth: A coincidence mechanism directly integrates a hormones-signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1950–64. doi: 10.1093/pcp/pcs137. [DOI] [PubMed] [Google Scholar]
- [17].Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hisamatsu T, King RW, Helliwell CA, Koshioka M. The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 2005;138:1106–16. doi: 10.1104/pp.104.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Arana MV, Marin-de la Rosa N, Maloof JN, Blazquez MA, Alabadi D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:9292–7. doi: 10.1073/pnas.1101050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–98. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- [21].de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–4. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- [22].Nomoto Y, Kubozono S, Miyachi M, Yamashino T, Nakamichi N, Mizuno T. A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1965–73. doi: 10.1093/pcp/pcs141. [DOI] [PubMed] [Google Scholar]
- [23].Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–3. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- [24].Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–5. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- [25].Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–6. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- [26].Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–7. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- [27].Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- [28].Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–5. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nakamichi N, Kita M, Niimura K, Ito S, Yamashino T, Mizoguchi T, et al. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–32. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- [30].Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci U S A. 2012;109:17123–8. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:3582–7. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lazaro A, Valverde F, Pineiro M, Jarillo JA. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012;24:982–99. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21:841–7. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, et al. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–22. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- [35].Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. Embo J. 2008;27:1277–88. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–9. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Franklin KA. Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol. 2009;12:63–8. doi: 10.1016/j.pbi.2008.09.007. [DOI] [PubMed] [Google Scholar]
- [38].Inigo S, Alvarez MJ, Strasser B, Califano A, Cerdan PD. PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 2012;69:601–12. doi: 10.1111/j.1365-313X.2011.04815.x. [DOI] [PubMed] [Google Scholar]
- [39].Lee JH, Kim JJ, Kim SH, Cho HJ, Kim J, Ahn JH. The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 2012:1802–14. doi: 10.1093/pcp/pcs123. [DOI] [PubMed] [Google Scholar]
- [40].Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004;134:1683–96. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Usadel B, Blasing OE, Gibon Y, Retzlaff K, Hohne M, Gunther M, et al. Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 2008;146:1834–61. doi: 10.1104/pp.107.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci U S A. 2011;108:5104–9. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001;127:252–61. doi: 10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–3. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- [45].Chew YH, Wilczek AM, Williams M, Welch SM, Schmitt J, Halliday KJ. An augmented Arabidopsis phenology model reveals seasonal temperature control of flowering time. New Phytol. 2012;194:654–65. doi: 10.1111/j.1469-8137.2012.04069.x. [DOI] [PubMed] [Google Scholar]
- [46].Bernier G, Périlleux C. A physiological overview of the genetics of flowering time control. Plant Biotechnol J. 2005;3:3–16. doi: 10.1111/j.1467-7652.2004.00114.x. [DOI] [PubMed] [Google Scholar]
- [47].Corbesier L, Bernier G, Perilleux C. C : N ratio increases in the phloem sap during floral transition of the long-day plants Sinapis alba and Arabidopsis thaliana. Plant Cell Physiol. 2002;43:684–8. doi: 10.1093/pcp/pcf071. [DOI] [PubMed] [Google Scholar]
- [48].Bernier G, Havelange A, Houssa C, Petitjean A, Lejeune P. Physiological signals that induce flowering. Plant Cell. 1993;5:1147–55. doi: 10.1105/tpc.5.10.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–31. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lasky JR, Des Marais DL, McKay JK, Richards JH, Juenger TE, Keitt TH. Characterizing genomic variation of Arabidopsis thaliana : the roles of geography and climate. Mol Ecol. 2012;21:5512–29. doi: 10.1111/j.1365-294X.2012.05709.x. [DOI] [PubMed] [Google Scholar]
- [51].Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–6. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- [52].Ågren J, Schemske DW. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 2012;194:1112–22. doi: 10.1111/j.1469-8137.2012.04112.x. [DOI] [PubMed] [Google Scholar]
- [53].Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci U S A. 2007;104:1278–82. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Franks SJ, Weis AE. A change in climate causes rapid evolution of multiple life-history traits and their interactions in an annual plant. J Evol Biol. 2008;21:1321–34. doi: 10.1111/j.1420-9101.2008.01566.x. [DOI] [PubMed] [Google Scholar]
- [55].Lewandowska-Sabat AM, Winge P, Fjellheim S, Dørum G, Bones AM, Rognli OA. Genome wide transcriptional profiling of acclimation to photoperiod in high-latitude accessions of Arabidopsis thaliana. Plant Sci. 2012;185-186:143–55. doi: 10.1016/j.plantsci.2011.10.009. [DOI] [PubMed] [Google Scholar]
- [56].Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, et al. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell. 2007;19:509–23. doi: 10.1105/tpc.106.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Gene Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Van Buskirk HA, Thomashow MF. Arabidopsis transcription factors regulating cold acclimation. Physiol Plant. 2006;126:72–80. [Google Scholar]
- [59].Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–6. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- [60].Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1:e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–79. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–39. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- [63].Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410–3. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- [64].Dong MA, Farré EM, Thomashow MF. CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:7241–6. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Seo PJ, Park M-J, Lim M-H, Kim S-G, Lee M, Baldwin IT, et al. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–42. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–81. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–62. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- [68].Cao SQ, Song YQ, Su L. Freezing sensitivity in the gigantea mutant of Arabidopsis is associated with sugar deficiency. Biol Plant. 2007;51:359–62. [Google Scholar]
- [69].Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. Embo J. 2009;28:3745–57. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–9. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- [71].Lee CM, Thomashow MF. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2012;109:15054–9. doi: 10.1073/pnas.1211295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Welling A, Moritz T, Palva ET, Junttila O. Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol. 2002;129:1633–41. doi: 10.1104/pp.003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Puhakainen T, Li C, Boije-Malm M, Kangasjärvi J, Heino P, Palva ET. Short-day potentiation of low temperature-induced gene expression of a C-repeat-binding factor-controlled gene during cold acclimation in silver birch. Plant Physiol. 2004;136:4299–307. doi: 10.1104/pp.104.047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009;151:2046–57. doi: 10.1104/pp.109.147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Schwartz C, Balasubramanian S, Warthmann N, Michael TP, Lempe J, Sureshkumar S, et al. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics. 2009:723–32. doi: 10.1534/genetics.109.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–17. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- [77].Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–22. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]