Abstract
AD101 and SCH-C are two chemically related small molecules that inhibit the entry of human immunodeficiency virus type 1 (HIV-1) via human CCR5. AD101 also inhibits HIV-1 entry via rhesus macaque CCR5, but SCH-C does not. Among the eight residues that differ between the human and macaque versions of the coreceptor, only one, methionine-198, accounts for the insensitivity of macaque CCR5 to inhibition by SCH-C. Thus, the macaque coreceptor engineered to contain the natural human CCR5 residue (isoleucine) at position 198 is sensitive to HIV-1 entry inhibition by SCH-C, whereas a human CCR5 mutant containing the corresponding macaque residue (methionine) is resistant. Position 198 is in CCR5 transmembrane (TM) helix 5 and is not located within the previously defined binding site for AD101 and SCH-C, which involves residues in TM helices 1, 2, 3, and 7. SCH-C binds to human CCR5 whether residue 198 is isoleucine or methionine, and it also binds to macaque CCR5. However, the binding of a conformation-dependent monoclonal antibody to human CCR5 is inhibited by SCH-C only when residue 198 is isoleucine. These observations, taken together, suggest that the antiviral effects of SCH-C and AD101 involve stabilization, or induction, of a CCR5 conformation that is not compatible with HIV-1 infection. However, SCH-C is unable to exert this effect on CCR5 conformation when residue 198 is methionine. The region of CCR5 near residue 198 has, therefore, an important influence on the conformational state of this receptor.
A new generation of inhibitors of human immunodeficiency virus type 1 (HIV-1) replication is now in clinical trials, based on the blockade of virus entry (28, 43, 54, 59, 62). Among these fusion inhibitors are small molecules targeted at the CCR5 coreceptor, a CC-chemokine receptor that is a member of the 7-transmembrane G-protein-coupled receptor (GPCR) superfamily (27, 53, 62). One such small-molecule CCR5 inhibitor, the RANTES antagonist SCH-C (SCH 351125), has been shown to cause viral load reductions after administration to HIV-1-infected individuals in phase I clinical trials (27, 39, 42, 66). Hence, it is relevant to drug development to know as much as possible about how small molecules interact with CCR5 and thereby prevent HIV-1 from doing so.
Studies with SCH-C, the chemically related compound AD101 (SCH 350581), and the chemically unrelated TAK-779 molecule have shown that all three CCR5 inhibitors block the binding of the HIV-1 envelope glycoprotein gp120 to CCR5 (21, 70). Thus, either there is a direct competition between the small molecule and gp120, or else the small molecule induces a structural change in CCR5 that prevents its recognition by gp120 (21, 63, 70). Moreover, the binding sites for SCH-C, AD101, and TAK-779 have been mapped to a pocket formed between transmembrane (TM) helices 1, 2, 3, and 7 of CCR5; these binding pockets are similar but not identical (21, 63, 70). Members of another set of chemically unrelated small-molecule inhibitors interact with an overlapping binding pocket that involves TM helices 2, 3, 6, and 7 (7). The CCR5 N terminus (NT) and the extracellular loops (ECL) play at most a limited role (more likely, no role) in the binding of the small-molecule inhibitors (7, 21, 63, 70). This contrasts markedly with the important function of the CCR5 external regions in gp120 binding and hence in viral entry (10, 17, 18).
In this study, we show that SCH-C and AD101 have differential effects on CCR5 coreceptor activity in primary human and rhesus macaque peripheral blood mononuclear cells (PBMC), as well as in cell lines transfected with human and rhesus macaque CCR5 (hu-CCR5 and rh-CCR5, respectively). While AD101 was a potent inhibitor of entry mediated by either coreceptor, SCH-C was selective for hu-CCR5, with little or no activity against entry mediated by rh-CCR5. We sought to identify the basis for this difference by first mutating the coding sequences for hu-CCR5 and rh-CCR5 so as to interchange their amino acid differences and then assessing whether AD101 and SCH-C could inhibit the entry of HIV-1 Env-pseudotyped viruses into cells expressing the mutant coreceptors. We found that the differential effects of SCH-C and AD101 on the coreceptor activities of hu-CCR5 and rh-CCR5 are due to a single amino acid difference: the replacement of isoleucine (Ile) at position 198 of hu-CCR5 by methionine (Met) at the same position in rh-CCR5. Thus, when the natural Met at position 198 of rh-CCR5 was altered to Ile to form the rh-CCR5(M198I) mutant, SCH-C could inhibit HIV-1 entry into cells expressing this mutant receptor. Conversely, the hu-CCR5(I198M) mutant was insensitive to SCH-C in this assay. No other amino acid differences between rh-CCR5 and hu-CCR5 had a significant effect on the activities of SCH-C and AD101. Amino acid 198 is in TM helix 5 and is not thought to directly form part of the binding site for either AD101 or SCH-C (63, 70). We further showed that SCH-C is able to bind to rh-CCR5 and to hu-CCR5 containing Met-198. Its inability to act as an entry inhibitor with these coreceptors is due to its inability to induce or stabilize a receptor conformational state that is incompatible with usage by HIV-1. These findings help explain how small-molecule CCR5 inhibitors block HIV-1 entry, and they may have general implications for understanding the actions of other inhibitors of GPCR function.
MATERIALS AND METHODS
Reagents and plasmids.
SCH-C and AD101 were synthesized by Schering-Plough Research Institute. TAK-779 was obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (contributed by the Division of AIDS) (2). The JR-FL, ADA, and YU-2 Env expression plasmids were provided by Tanya Dragic (Albert Einstein College of Medicine, Bronx, N.Y.). The human and macaque CCR5 expression plasmids were pcDNA3-hu-CCR5 (31), provided by David Kabat (Oregon Health Sciences University, Portland, Oreg.), and pc.Rh-CCR5 (AIDS Research and Reference Reagent Program; contributed by Preston Marx) (8), respectively. The fluorescein-conjugated mouse anti-human CCR5 monoclonal antibody (MAb) clone 45523 (33), the fluorescein-conjugated mouse immunoglobulin G2B (IgG2B) isotype control MAb, and recombinant human RANTES were all obtained from R&D Systems (Minneapolis, Minn.). The phycoerythrin (PE)-conjugated mouse anti-human CCR5 MAb clone 2D7 and the PE-conjugated IgG2A isotype control MAb were from BD Pharmingen (San Diego, Calif.).
Inhibition of HIV-1, SIV, and SHIV replication.
The preparation of virus stocks, the replication of HIV-1, simian immunodeficiency virus (SIV), or simian-human immunodeficiency virus (SHIV) in human or macaque PBMC, and the effects of inhibitors were measured as described elsewhere (69, 73). SHIV-162P4 was obtained from Janet Harouse (The Rockefeller University, New York, N.Y.) (24, 25).
Mutagenesis of the CCR5 coding sequence.
Single-residue mutants of hu-CCR5 in pcDNA3.1 were constructed as previously described (20). The rh-CCR5(M198I) expression plasmid was constructed from pc.Rh-CCR5 (8) by the QuikChange site-directed mutagenesis method (Stratagene, La Jolla, Calif.), according to the manufacturer's instructions. Mutations were verified by DNA sequencing of the entire CCR5 coding region (The Rockefeller University Protein/DNA Technology Center, New York, N.Y.).
Env pseudotype assay of HIV-1 infection.
HIV-1 pseudotype particles capable of single-round infection and bearing the firefly luciferase gene were generated by cotransfection of pNL-Luc-E—R+ (12) and an Env expression vector for the JR-FL, ADA, or YU-2 Env protein into 293T cells, as previously described (57). Env pseudotype viruses are represented by the nomenclature HIV-1JR-FL, etc, where the subscript indicates the Env protein used to pseudotype the luciferase reporter construct. Wild-type and mutant CCR5 proteins were expressed in U87MG-CD4 cells (9, 19) by transient transfection, as previously described (21, 63, 70). Entry of Env-pseudotyped HIV-1 reporter viruses into the transfected U87MG-CD4 cells in the presence and absence of CCR5 inhibitors was determined by quantifying luciferase expression (21). The luciferase activity was directly proportional to viral entry, as confirmed by serial dilution of the input virus in the absence of inhibitors (data not shown).
For the hu-CCR5 mutants with single amino acid substitutions based on the rh-CCR5 sequence, and for the rh-CCR5(M198I) mutant coreceptor, the activities of AD101 and SCH-C were assessed by titrating the inhibitors over the range from 12.8 pM to 1 μM. The extent of HIV-1 entry was calculated as a percentage of luciferase activity in the absence of inhibitor. The 50% inhibitory concentrations (IC50) for SCH-C titration curves were calculated from the sigmoid dose-response curves of entry versus the log value of the inhibitor concentration by nonlinear regression using GraphPad Prism (GraphPad Software, San Diego, Calif.). IC50 for each CCR5 construct are reported relative to the IC50 for hu-CCR5 calculated from the same experiment. In addition to relative IC50, the percent inhibition by SCH-C at a fixed concentration (1 μM) is also reported; this was calculated for a given CCR5 mutant by using the formula 100 × {1 − [(luciferase activity at 1 μM SCH-C)/(luciferase activity without SCH-C)]}.
For the hu-CCR5 constructs with residue 198 mutated, entry supported by each mutant CCR5 was compared to entry supported by hu-CCR5 at a fixed inhibitor concentration, as described previously (21, 63, 70). The following concentrations were used: for TAK-779, 200 nM; for AD101, 100 nM; for SCH-C, 100 nM. The residual entry supported by a mutant CCR5 in the presence of an inhibitor, compared to entry of hu-CCR5, was then calculated as described elsewhere (21, 63, 70).
Modeling of CCR5.
The TM domain model of CCR5 was built by homology modeling using the 2.8 Å crystal structure of bovine rhodopsin (PDB ID 1F88) (50) as a template as described previously (63). Energy-minimized structures of AD101 and SCH-C were generated as described elsewhere (63). Molecular graphics were prepared by using DINO: Visualizing Structural Biology (2002) (http://www.dino3d.org).
Calcium mobilization.
293T cells were transfected with a hu-CCR5 or hu-CCR5(I198M) expression plasmid by using Lipofectamine 2000 according to the manufacturer's instructions. A total of 107 transfected 293T cells were resuspended in EBSS-H buffer, which contains Earles's balanced salt solution (EBSS) (Gibco/Invitrogen Life Technologies, Carlsbad, Calif.) supplemented with 26 mM HEPES, 1 mM MgSO4, 2 mM CaCl2, and 0.1% bovine serum albumin. The cells were then loaded with 2 μM Fluo-3-AM (Molecular Probes, Inc., Eugene, Oreg.) for 1 h at room temperature. After the cells were washed twice with EBSS-H, fluorescence emissions were determined at 505 and 525 nm by using a Hitachi F2500 fluorometer. After 2 min of recording, the cells were treated with SCH-C or buffer for 5 min before addition of RANTES (34 nM). The magnitude of the RANTES-stimulated calcium signal was normalized to the signal obtained from the same cells in response to 1 μM carbachol (Sigma, St. Louis, Mo.).
MAb binding to CCR5.
MAb binding assays were performed essentially as described previously (70), except that 106 CCR5-transfected 293T cells were incubated with the MAb in the presence or absence of AD101 (100 nM) or SCH-C (1 μM). The stained cells were analyzed by flow cytometry. The mean fluorescence intensity derived by using the appropriate isotype control MAb was subtracted from the signal obtained by using each anti-CCR5 MAb. The extent of staining in the presence of an inhibitor was then normalized to that observed in the absence of any inhibitor.
RESULTS
Two related small-molecule CCR5 inhibitors act differently on the human and macaque receptors.
SCH-C and AD101, an earlier-generation compound from the same chemical family of piperidine-based molecules, have been studied as model CCR5 inhibitors (37, 48, 49, 63, 66-70). AD101 is more potent than SCH-C as an HIV-1 entry inhibitor in vitro, but its unfavorable pharmacological properties have prevented it from being pursued as a clinical candidate. While studying AD101 and SCH-C in vitro, we noted that whereas both compounds were active against the hu-CCR5 coreceptor, only AD101 could inhibit viral entry via rh-CCR5.
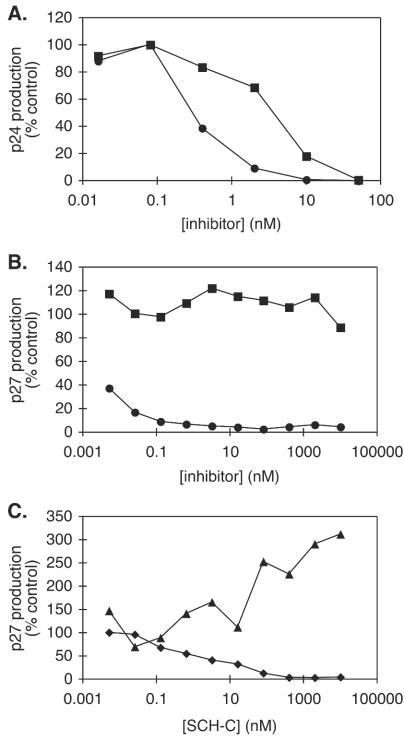
In a standard assay of HIV-1 JR-FL replication in human PBMC from one representative donor (69), the IC50 for AD101 and SCH-C were 0.3 and 5 nM, respectively, a 17-fold difference (Fig. 1A). When PBMC from different donors were used, a difference between the IC50 for AD101 and SCH-C was consistently observed, although the magnitude of the differential varied from 10- to 100-fold among donors (data not shown). Both compounds completely inhibited HIV-1 replication when present at a sufficiently high concentration (Fig. 1A and data not shown). However, when macaque PBMC were used to support SIVmac251 replication (73), the IC50 for AD101 inhibition in a representative experiment was <5 pM but the IC50 for SCH-C was >10 μM, a >106-fold difference (Fig. 1B). Even at an SCH-C concentration as high as 10 μM, the extent of inhibition of SIVmac251 replication in macaque PBMC did not exceed 15% (Fig. 1B).
FIG. 1.
Inhibitory activities of SCH-C and AD101 with human or rhesus macaque PBMC. SCH-C or AD101 was added at the concentrations indicated to mitogen-activated PBMC in the presence of virus. (A) Inhibition of replication of HIV-1 JR-FL in human PBMC by AD101 (circles) and SCH-C (squares). (B) Inhibition of replication of SIVmac251 in macaque PBMC by AD101 (circles) and SCH-C (squares). (C) Inhibition of SHIV-162P4 replication by SCH-C in human PBMC (diamonds) and macaque PBMC (triangles). Results shown are from one representative experiment. All data were normalized to the amount of p27 or p24 produced in the absence of inhibitor (defined as 100%) and are shown as percent virus replication. Similar results were obtained by using PBMC from a minimum of two different macaque and human donors.
To confirm that the differential effect of the two inhibitors was not due to the use of different test viruses (i.e., HIV-1 JR-FL with human cells and SIVmac251 with macaque cells), we tested a virus, SHIV-162P4, that replicates efficiently in both macaque and human PBMC (24, 25). Again, AD101 was a very potent inhibitor with cells from both species. The IC50 were approximately 10 and <5 pM for the macaque and human coreceptors, respectively (data not shown). SCH-C, however, inhibited SHIV-162P4 replication only in human PBMC, with an IC50 of 1.3 nM (Fig. 1C). Indeed, SCH-C consistently caused a modest, dose-dependent enhancement of SHIV-162P4 replication in macaque PBMC that varied in magnitude from two- to fivefold in cells from different macaque donors (Fig. 1C and data not shown). Note, however, that infection enhancement did not occur when SIVmac was the test virus (Fig. 1B), suggesting that the effect on SHIV-162P4 replication has a specific cause. The underlying mechanisms are currently under investigation.
A single-residue difference between human and macaque CCR5 accounts for their differential sensitivity to SCH-C.
Overall, the pattern of the results outlined above suggests that the reason for the differential effect of AD101 and SCH-C on virus replication in cells from humans and rhesus macaques lies at the level of the CCR5 receptor, not the test virus. Inspection of the sequences of hu-CCR5 and rh-CCR5 revealed a total of eight single amino acid differences between the two proteins (Table 1). Three of these are within the ECL or NT of CCR5, three are located in TM helices 1 and 2 near the putative binding sites for AD101 and SCH-C, one is intracellular, and one is in TM5, a helix which is not thought to directly form a part of the AD101 or SCH-C binding site (63, 70).
TABLE 1.
Amino acid differences between hu-CCR5 and rh-CCR5
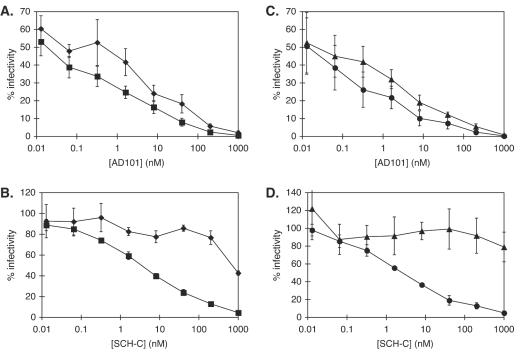
To determine which of the sequence variations between hu-CCR5 and rh-CCR5 were responsible for the difference in phenotype, we performed a mutagenesis study and used an Env pseudotype assay of HIV-1 entry (21, 63, 70). We first expressed the human and macaque CCR5 proteins in transfected U87MG-CD4 cells (9, 19) and then measured the entry of Env-pseudotyped HIV-1JR-FL, HIV-1ADA, and HIV-1YU-2 in the presence and absence of CCR5 inhibitors (21, 63, 70). In this assay, AD101 was again a potent inhibitor of HIV-1JR-FL entry via both coreceptors (Fig. 2A) and also of HIV-1ADA and HIV-1YU-2 entry (data not shown). In contrast, SCH-C was again a poor inhibitor of the entry of all three Env-pseudotyped viruses mediated by rh-CCR5, with IC50 values 240- to >490-fold higher than those for entry via hu-CCR5 (Table 2; Fig. 2B). However, SCH-C efficiently inhibited the entry of the same viruses via hu-CCR5, with average IC50 of 3.7 ± 0.9, 3.1 ± 2.2, and 1.2 ± 0.2 nM for HIV-1JR-FL, HIV-1ADA, and HIV-1YU-2, respectively (Fig. 2B and data not shown).
FIG. 2.
Inhibitory activities of SCH-C and AD101 in an Env pseudotype assay of HIV-1 entry. SCH-C or AD101 was added at the concentrations indicated to U87MG-CD4 cells that transiently expressed CCR5 or a CCR5 mutant. The entry of HIVJR-FL Env pseudotypes was then measured in the transfected cells. Shown is inhibition by AD101 (A and C) and SCH-C (B and D) of entry into cells expressing hu-CCR5 (squares), rh-CCR5 (diamonds), hu-CCR5(I198M) (triangles), or rh-CCR5(M198I) (circles). Data were normalized to the amount of luciferase expressed in the absence of inhibitor and are shown as percent luciferase activity. Error bars, standard errors of the means for values derived from three to five independent experiments.
TABLE 2.
Effects of amino acid substitutions in hu-CCR5 and rh-CCR5 on HIV-1 Env pseudotype infectivity in a single-round assay
| CCR5 construct | Result for the following Env-pseudotyped virus:
|
|||||
|---|---|---|---|---|---|---|
| HIVJR-FLa
|
HIVADAb
|
HIVYU2c
|
||||
| Relative IC50d | % Inhibitione | Relative IC50 | % Inhibition | Relative IC50 | % Inhibition | |
| hu-CCR5 | 1 | 95.3 ± 0.6 | 1 | 97.9 ± 0.7 | 1 | 98.7 ± 0.6 |
| rh-CCR5f | 240 ± 15 | 57.3 ± 2.1 | >490 | 51.5 ± 7.4 | >390 | 61.8 ± 8.2 |
| hu-CCR5(I9T) | 0.6 ± 0.1 | 97.4 ± 1.4 | 0.2 | 99 | 2.3 | 98 |
| hu-CCR5(N13D) | 2.1 ± 0.1 | 95.9 ± 1.2 | 2.0 | 98 | 0.7 | 99 |
| hu-CCR5(M49I) | 1.0 ± 0.6 | 95.2 ± 0.3 | 0.9 | 99 | 1.5 ± 1.0 | 99.1 ± 0.7 |
| hu-CCR5(I52V) | 0.3 ± 0.1 | 97.1 ± 1.9 | ND | ND | 1.5 | 100 |
| hu-CCR5(F78L) | 0.6 ± 0.1 | 97.5 ± 1.2 | 0.1 | 100 | 0.6 ± 0.3 | 100 |
| hu-CCR5(V130I) | 3.7 ± 3.5 | 96.0 ± 2.5 | 0.5 | 98 | 1.8 | 97 |
| hu-CCR5(K171R) | 0.6 ± 0.4 | 96.4 ± 2.0 | 1.8 | 98 | 0.2 | 98 |
| hu-CCR5(I198M) | >250 | 21.0 ± 16.6 | >3,700 | 49 | 450 | 54 |
| rh-CCR5(M198I) | 0.8 ± 0.2 | 95.3 ± 3.1 | ND | ND | 2.1 | 93 |
n = 5 for hu-CCR5; n = 4 for rh-CCR5(M198I); n = 3 for rh-CCR5 and hu-CCR5(I198M); n = 2 for hu-CCR5(I9T), hu-CCR5(N13D), hu-CCR5(M49I), hu-CCR5(I52V), hu-CCR5(F78L), hu-CCR5(V130I), and hu-CCR5(K171R). Values are means ± standard errors of the means where n is >2 and means ± ranges where n is 2.
n = 5 for hu-CCR5 and n = 4 for rh-CCR5; values are means ± standard errors of the means. n = 1 for all other CCR5 constructs. ND, not done.
n = 5 for hu-CCR5 and n = 4 for rh-CCR5; values are means ± standard errors of the means. n = 2 for hu-CCR5(M49I) and hu-CCR5(F78L); values are means ± ranges. n = 1 for all other CCR5 constructs.
IC50 were obtained from individual data sets fit to a sigmoidal dose-response curve by nonlinear regression using the Prism program (GraphPad Software) and were normalized to the IC50 for hu-CCR5 in the same experiment.
Percent inhibition is the average percent reduction in luciferase activity at 1 μM SCH-C.
Boldfaced values indicate those CCR5 constructs for which inhibition by SCH-C was significantly reduced relative to that for hu-CCR5.
We next made eight single-residue mutants of hu-CCR5 by introducing individually each of the eight residues that were different in the macaque version. Each of these hu-CCR5 mutants was tested for its ability to mediate Env-pseudotyped HIV-1 entry. Most of the mutant receptors could efficiently support virus entry at levels similar to that with hu-CCR5; with six of the eight mutants, HIV-1JR-FL entry levels were on average between 80 and 130% that with hu-CCR5. The exceptions were hu-CCR5(I52V) and hu-CCR5(K171R), which, respectively, supported HIV-1JR-FL entry on average at 50 and 230% the levels seen with hu-CCR5. However, the hu-CCR5(I52V) and hu-CCR5(K171R) mutants supported the entry of HIV-1YU-2 at 160 and 60% of the level with hu-CCR5, respectively, yet the results of SCH-C inhibition experiments were broadly similar with both HIV-1YU-2 and HIV-1JR-FL (Table 2). Hence, the modest differences in entry efficiency mediated by these two mutant receptors are not likely to affect our analyses. Furthermore, all the hu-CCR5 mutants had similar sensitivities to AD101 (Fig. 2A and C; also data not shown), suggesting that each contained an intact, unimpaired binding pocket for this small-molecule inhibitor.
Among the eight hu-CCR5 mutants, hu-CCR5(I198M) was uniquely insensitive to SCH-C (Table 2; Fig. 2D). This observation suggests that a single amino acid difference between the macaque and human forms of CCR5 accounts for their differential susceptibility to inhibition by SCH-C. To confirm this supposition, we made the converse (M198I) change in rh-CCR5 and tested whether it was sufficient to confer SCH-C sensitivity on the macaque coreceptor. Clearly, it was. Infection by HIV-1 pseudotypes was efficiently supported by the rh-CCR5(M198I) mutant and was sensitive to both AD101 and SCH-C, with a concentration dependence similar to that seen with hu-CCR5 (Table 2, Fig. 2C and D, and data not shown).
Impact of other changes at residue I198 in hu-CCR5 on the efficacy of SCH-C and AD101.
To increase our understanding of the role that residue I198 might play in the interactions of AD101 and SCH-C with CCR5, we made additional amino acid substitutions and then analyzed whether the mutant coreceptors were still sensitive to the inhibitors (Table 3). We also included TAK-779 in this analysis because, although it is chemically unrelated to AD101 and SCH-C, the TAK-779 binding site on CCR5 is very similar to those of AD101 and SCH-C (21, 63, 70). Previous studies have shown that an alanine substitution at amino acid I198 of hu-CCR5 substantially impairs the inhibitory activities of AD101 and SCH-C but has only a modest effect on TAK-779 action (21, 70).
TABLE 3.
Effects of conservative and polar substitutions of residue I198 in hu-CCR5 on HIV- 1 Env pseudotype infectivity in a single-round assay
| CCR5 construct | % HIV-1JR-FL entrya in the presence of the following inhibitor:
|
||
|---|---|---|---|
| TAK-779 | AD101 | SCH-C | |
| hu-CCR5 | 0 | 0 | 0 |
| hu-CCR5(I198A) | 19b | 76.6 ± 6.7c | 72.2 ± 3.3c |
| hu-CCR5(I198V) | 3.2 ± 2.1 | 15.4 ± 4.5 | 11.0 ± 2.3 |
| hu-CCR5(I198L) | 1.6 ± 1.6 | 10.2 ± 2.6 | 50.2 ± 8.5 |
| hu-CCR5(I198M) | 0d | 2.4 ± 0.6 | 67.6 ± 2.3 |
| hu-CCR5(I198T) | 0.5 ± 0.5 | 33.0 ± 2.3 | 28.8 ± 1.8 |
The entry of HIV-1JR-FL in the presence of the indicated inhibitors is normalized to entry in the absence of inhibitors. All values are means ± standard errors of the means from at least three independent experiments. A value of 100% means that a mutant is insensitive to the inhibitor; a value of 0% means that a mutant is as sensitive to the inhibitor as wild-type CCR5. As described elsewhere, mutants with entry levels above 14% are considered to have significantly reduced sensitivity for the respective inhibitor (21). Values for these mutants are boldfaced. Inhibitors were used at the following concentrations: TAK-779, 200 nM; AD101, 100 nM; SCH-C, 100 nM.
Data from reference 21.
Data from reference 70.
Occasionally, values calculated by this method may be less than zero, which would indicate that a mutation enhances the action of an inhibitor. In this case, however, the value is not significantly different from that for hu-CCR5, and it has been rounded to zero for clarity, as described elsewhere (21).
We first evaluated the consequences of conservative amino acid replacements. Replacement of I198 with valine had no effect on the inhibition of viral entry by TAK-779 or SCH-C and only a very minor impact on AD101 inhibition (Table 3). However, replacement of isoleucine with leucine resulted in a mutant coreceptor [hu-CCR5(I198L)] with properties similar to those of the hu-CCR5(I198M) mutant; the sensitivity of hu-CCR5(I198L) to SCH-C was severely reduced, but its responses to AD101 and TAK-779 were not significantly affected. Hence, altering the position of only the single methyl group that differs between Ile and Leu is sufficient to interfere with the activity of SCH-C but not with that of AD101 or TAK-779. Conversely, the introduction of a polar side chain, to make the hu-CCR5(I198T) mutant, modestly affected the inhibitory activities of both AD101 and SCH-C but not that of TAK-779 (Table 3).
SCH-C inhibits RANTES signaling via CCR5.
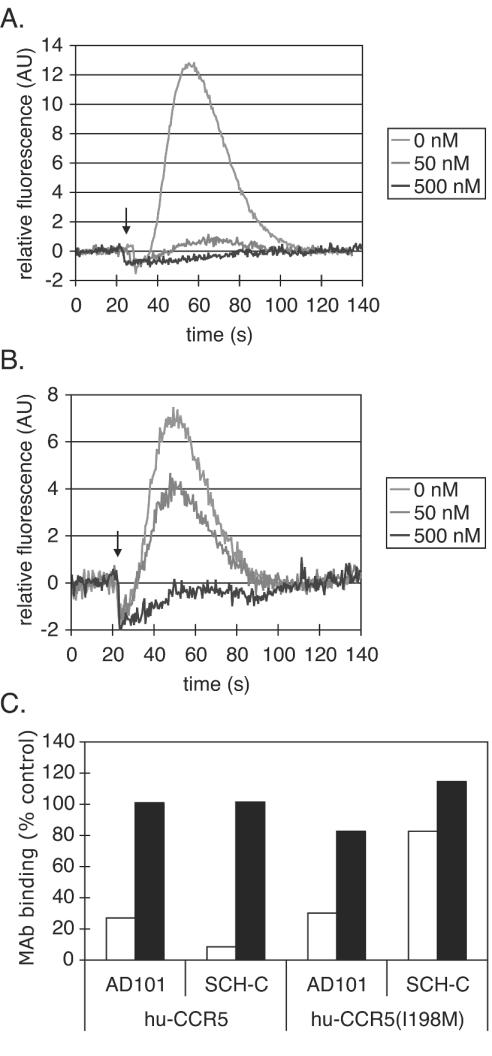
The results described above show that viral entry via rh-CCR5 or the hu-CCR5(I198M) mutant is insensitive to inhibition by SCH-C. The simplest explanation of these observations would be that the presence of a Met residue at position 198 prevents SCH-C from binding to CCR5. To confirm or refute this hypothesis, we assessed whether SCH-C could interfere with RANTES-induced calcium mobilization by the hu-CCR5(I198M) receptor. The calcium response of the wild-type hu-CCR5 receptor to RANTES was completely inhibited by SCH-C at 500 nM and partially inhibited at 50 nM (Fig. 3A). RANTES efficiently induced calcium mobilization in 293T cells transfected with the hu-CCR5(I198M) mutant receptor, showing that the I198M substitution did not impair the ability of CCR5 to act as a chemokine receptor. RANTES signaling via the hu-CCR5(I198M) receptor was also inhibited in a dose-dependent manner by SCH-C; complete inhibition occurred at 500 nM (Fig. 3B). Similar results were obtained with rh-CCR5 (data not shown).
FIG. 3.
SCH-C inhibition of RANTES signaling via CCR5 and MAb binding to CCR5. (A and B) Calcium mobilization in response to RANTES in 293T cells transfected with hu-CCR5 (A) or hu-CCR5(I198M) (B). RANTES (34 nM) was added 5 min after the indicated concentration of SCH-C. The recordings begin ∼25 s before RANTES addition (indicated by the arrow). Signals are expressed as a percentage of the response induced by carbachol (1 μM) in the same cells and have been corrected for baseline signals. Results of one representative experiment are shown. (C) Binding of MAb 45523 (open bars) or MAb 2D7 (solid bars) to 293T cells transfected with the CCR5 expression construct indicated in the presence or absence of AD101 (100 nM) or SCH-C (1 μM). MAb binding is expressed as a percentage of that occurring in the absence of an inhibitor (defined as 100% binding). Data shown were derived from a single representative experiment.
There may be quantitative, and fairly subtle, differences among hu-CCR5, hu-CCR5(I198M), and rh-CCR5 in their abilities to mediate RANTES signaling and their responses to SCH-C inhibition. However, it is clear that, at 500 nM, SCH-C can completely inhibit RANTES signaling through hu-CCR5(I198M) and rh-CCR5 (Fig. 3B and data not shown). To be able to inhibit RANTES signaling, it is obvious that SCH-C must actually bind to the receptor. Yet at the same 500 nM concentration, SCH-C is incapable of inhibiting virus entry via hu-CCR5(I198M) or rh-CCR5 (Fig. 2). The defect in the inhibitory process must, therefore, be exerted postbinding.
SCH-C inhibits MAb binding to CCR5.
We next examined the ability of a conformation-dependent anti-CCR5 MAb (45523) with a complex epitope spanning the NT and ECL2 domains of CCR5 (33) to bind to hu-CCR5 and hu-CCR5(I198M) in the presence and absence of SCH-C. It has previously been shown that the binding of MAb 45523 to hu-CCR5 expressed on the surfaces of mouse L1.2-CCR5 cells is inhibited by both AD101 and SCH-C (70). The binding of MAb 45523 to hu-CCR5 on transfected 293T cells is also inhibited by both SCH-C and AD101 (Fig. 3C). However, the binding of the same MAb to cells expressing hu-CCR5(I198M) was efficiently inhibited only by AD101; SCH-C had much less of an effect even when added at 1 μM (Fig. 3C). In contrast, the binding of the conformation-independent MAb, 2D7, directed against ECL2 (33, 47, 71), to either hu-CCR5 or hu-CCR5(I198M) was not affected by either AD101 or SCH-C (Fig. 3C).
Clearly, then, the hu-CCR5(I198M) and hu-CCR5 receptors can be distinguished by how they respond to the binding of SCH-C: respectively, they continue to display, or no longer efficiently display, the CCR5 conformation-dependent epitope for MAb 45523. The binding of SCH-C to the two versions of the receptor must, therefore, have different consequences for the conformation of CCR5.
Location of I198 in a model of the hu-CCR5 TM domains.
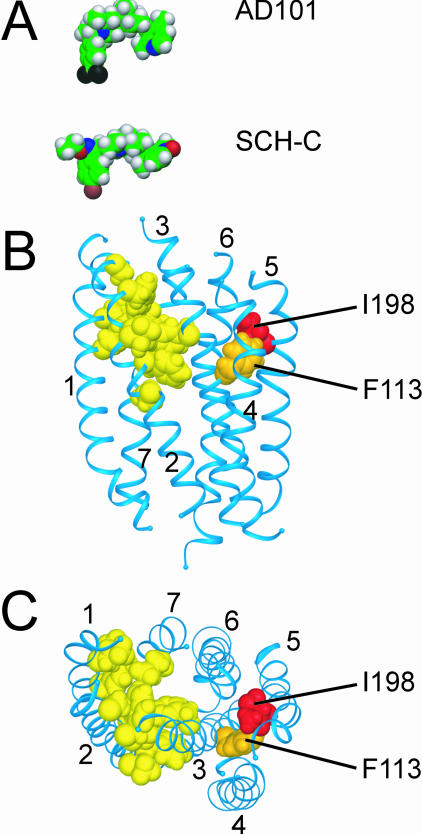
The results for the hu-CCR(I198T) and hu-CCR5(I198A) mutants clearly show that I198 is a key residue for the CCR5 interactions of both SCH-C and AD101. However, SCH-C differs from AD101 in being sensitive to small changes in the size and structure of the amino acid 198 side chain, as shown by the differential results obtained with the hu-CCR5(I198M) and hu-CCR5(I198L) mutants. A simple explanation for these observations would be that residue I198 interacted with AD101 and SCH-C directly. A model of the CCR5 TM domain suggests that this is an unlikely scenario, however (Fig. 4) (63). The model predicts that the small-molecule binding site is located within a cavity formed by TM helices 1, 2, 3, and 7 (Fig. 4B and C). Residues L33, Y37, V83, W86, A90, and E283, which are directly associated with this cavity (Fig. 4B and C), have been shown to be crucial for the inhibitory activities of both AD101 and SCH-C (63, 70). Hence, it is very likely that these residues interact directly with common elements in AD101 and SCH-C (63, 70). Residues D76, F79, Y108, and G286 are also associated with the putative small-molecule binding pocket, forming a second layer of residues which could interact with peripheral groups in the inhibitor molecules (Fig. 4B and C). Alternatively, these residues could indirectly affect the conformation of the binding site (63, 70). While replacement of D76 or G286 reduces the inhibitory activities of both AD101 and SCH-C, changing F79 or Y108 affects only AD101 (63, 70). In contrast to all the above residues, I198, which is located in TM5, is predicted to be outside the small-molecule binding cavity formed by TM helices 1, 2, 3, and 7 (Fig. 4B and C). Hence, we believe that I198 is not likely to be directly involved in the binding of AD101 or SCH-C and that a Met substitution at this position has an indirect effect (see below).
FIG. 4.
Structural model of the TM domain of CCR5 with energy-minimized structures of AD101 and SCH-C. (A) Energy-minimized structures of AD101 and SCH-C were calculated by using the PM3 semiempirical method of the HyperChem software (Hypercube, Inc.) (63) and are depicted in space-filling representation. Atoms are color-coded: carbon, green; oxygen, red; nitrogen, blue; hydrogen, grey; bromine, brown; fluorine, black. (B) Structural model of the TM domain of CCR5 viewed from within the plane of the membrane. The extracellular surface is oriented toward the top of the figure; the cytoplasmic surface is oriented toward the bottom. The seven α-helical TM segments are depicted as blue ribbons. Amino acid side chains of residues involved in the interaction of CCR5 with AD101 and/or SCH-C are shown in space-filling representation. Red, residue I198; orange, F113; yellow, L33, Y37, D76, F79, W86, V83, A90, Y108, E283, and G286. (C) View of the CCR5 model from the extracellular side of the membrane after rotation of the model by approximately 90° out of the paper plane from the orientation shown in panel B. Labeling and color-coding are the same as for panel B. The CCR5 model is based on homology with rhodopsin by using the crystal structure of bovine rhodopsin as a template (63). Models in all panels are shown at the same scale.
A similar situation might apply to residue F113, where replacement by alanine (F113A) substantially impairs the activity of AD101 but has only a marginal effect on inhibition by SCH-C (70). The side chain of F113 is located close to that of I198, and it too is predicted to be outside the putative small-molecule binding site (Fig. 3B and C). Hence, the F113A and I198M substitutions have similar consequences, in that they have differential effects on the inhibitory potencies of AD101 and SCH-C. But the two changes also have dissimilar outcomes: the F113A substitution impairs only the activity of AD101, while I198M impairs only that of SCH-C.
DISCUSSION
In broad terms, we now know why rh-CCR5 is insensitive to the antiviral activity of SCH-C while remaining vulnerable to the structurally related compound AD101. Thus, a single amino acid change between the two receptors determines whether SCH-C does or does not act as an entry inhibitor. This finding may have implications for the clinical use of SCH-C and other small-molecule CCR5 inhibitors as antiviral therapies. More significantly, our studies further our understanding of how small molecules interact with CCR5 to inhibit HIV-1 entry.
Possible mechanisms of the I198M substitution on the action of SCH-C.
How does the Ile-Met difference between hu-CCR5 and rh-CCR5 at position 198 affect the actions of SCH-C? Residue 198 is not located within the binding pocket between TM helices 1, 2, 3, and 7 that has been defined for SCH-C, AD101, and TAK-779 (21, 63, 70). Nor is it located in proximity to the overlapping binding site for the chemically unrelated 2-aryl-4-(piperidin-1-yl)butanamines and 1,3,4-trisubstituted pyrrolidines, which involves TM helices 2, 3, 6, and 7 (7). Based on the molecular model of the TM domains of CCR5, it is unlikely that this residue directly contacts SCH-C (Fig. 4) (63). One obvious explanation was that the I198M substitution acted indirectly to alter the conformation of the SCH-C binding site and thereby prevent the inhibitor from binding to rh-CCR5 or the hu-CCR5(I198M) mutant. Any such effect on the SCH-C binding site would be extremely subtle, because the activity of the closely related AD101 molecule is unaffected, but the possibility could not be excluded a priori. Thus, the position of even a single methyl group on CCR5 [the hu-CCR5(I198L) mutant] can affect SCH-C activity.
We can, however, reject the above explanation with some certainty. SCH-C must be able to bind to macaque CCR5 and the hu-CCR5(I198M) mutant, because it inhibits RANTES signaling via these receptors (Fig. 3B and data not shown). We argue, then, that SCH-C does bind to rh-CCR5 and to hu-CCR5(I198M), but in a way that does not block HIV-1 entry. This could happen if the geometry of SCH-C within its binding site were altered by the presence of Met at position 198, such that the inhibitor did not now block gp120 binding to CCR5. If the action of SCH-C on gp120 binding were competitive, the geometry of the inhibitor-binding site would have to be altered such that SCH-C no longer sufficiently overlapped an as yet undefined gp120-binding site in the core of CCR5.
However, our preferred explanation is that in the presence of SCH-C, or a related CCR5 inhibitor, the inhibitor binds within the TM domains of hu-CCR5 and prevents the coreceptor from adopting a conformation that is required for productive gp120 binding (Fig. 5) (70). If this model is correct, we could extend the argument to suggest that when a Met residue is present at position 198 (as in rh-CCR5), SCH-C becomes incapable of blocking the conformational change in CCR5 (Fig. 5). The latter scenario is supported by the finding that SCH-C does not inhibit the binding of a conformation-dependent anti-CCR5 MAb to hu-CCR5(I198M) but does block the binding of the same antibody to hu-CCR5 (Fig. 3C). The implication is that, when residue 198 is methionine, SCH-C cannot prevent the transition of CCR5 to a configuration that is required for productive gp120 binding. The latter, gp120-reactive configuration of CCR5 is efficiently recognized by MAb 45523. Exactly how the I198M change could alter the ability of CCR5 to undergo a conformational change is not known, but again the effect must be both subtle and indirect. By analogy, the F113A substitution would have a similar, adverse effect on the activity of AD101 only.
FIG. 5.
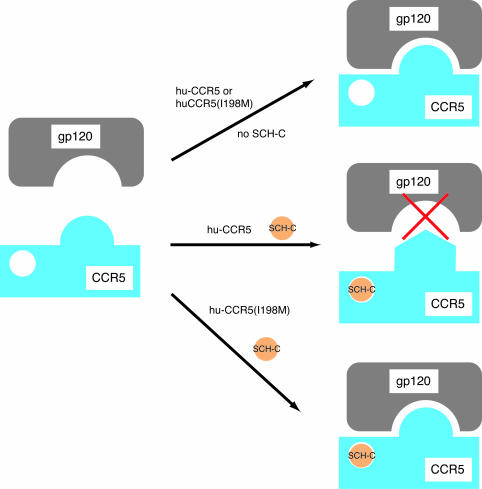
Model for the mechanism of action of the I198M substitution on SCH-C inhibition of HIV-1 infection. Orange circle, SCH-C; grey, gp120; blue, CCR5. The interaction between gp120 and CCR5 is shown in a simple, one-site form, with no intent to identify any individual gp120 domain(s) that might be involved in the overall interaction. (Top) hu-CCR5 or hu-CCR5(I198M) in the absence of SCH-C adopts a conformation permissive for gp120 binding, as indicated by the fit between gp120 and the extracellular regions of CCR5. (Center) Binding of SCH-C to hu-CCR5 stabilizes a conformation of CCR5 that does not productively interact with gp120, as represented by the change in the shape of the extracellular regions of CCR5 and the red “X” indicating that productive interaction does not occur. (Bottom) SCH-C binds to the mutant CCR5 as in the center scenario, but it does not alter the conformation of the extracellular regions so as to block infection.
We could extend the above model if we consider that the binding of gp120 to CCR5 might be a multistage process. Suppose, then, that CCR5 normally exists in a conformational state that is recognized by only one domain of gp120, but the initial contact between that region of gp120 and CCR5 converts the latter into a configuration that is now recognized by a second gp120 domain. In this argument, the secondary interaction must take place before further conformational changes are induced in the CCR5-Env complex to activate the final stages of the gp41-mediated fusion process. If this model is correct, the small-molecule CCR5 inhibitors might act to prevent the transition of CCR5 into the configuration that can be recognized by the secondary binding site on gp120. Mutagenesis studies on gp120 do suggest that its CCR5 binding site has two components: (i) a relatively conserved surface involving the bridging sheet and (ii) elements of the V3 loop (13, 14, 60).
If chemokines interact with their receptors in a broadly similar manner, by using two distinct binding domains sequentially (3, 40, 65), then HIV-1 gp120 may have hijacked a natural process for its own purposes. However, the suggestion that the gp120 V3 loop is itself a structural and functional mimic of the 40s loop region of chemokines (64) is not supported by most of the available evidence on the structure of chemokines and the function of the 40s loop, which is important for chemokine binding to glycosoaminoglycans but not GPCRs (29, 30, 32, 35, 45, 55).
Our studies on CCR5 may have a wider implication for the understanding of chemokine receptor structure-function relationships. Residue I198 is highly variable among members of the chemokine receptor family (51). The highly variable nature of this residue might contribute to the receptor selectivity of chemokine receptor inhibitors in general. Attention might, therefore, usefully be paid to this particular amino acid in efforts to understand how and why small molecules inhibit the natural or subverted functions of chemokine receptors.
Implications of CCR5 sequence variation for the clinical use of CCR5 inhibitors.
Natural variation in the responses of different individuals to CCR5 inhibitors should be anticipated, due, at least in part, to factors that influence virus-cell fusion (42, 58). Increasing the rate of fusion by, for example, increasing the density of CCR5 on target cells inversely affects the potency of coreceptor inhibitors (42, 58). CCR5 expression varies over at least a 20-fold range among individuals homozygous for CCR5 alleles that encode the wild-type CCR5 protein (41, 72). While a direct link has yet to be shown, some of this variation is expected to be related to promoter polymorphisms (5, 15, 22, 23, 36, 38, 44, 46). Another factor that may influence the efficacy of CCR5 inhibitors is the local concentration of the natural CC-chemokine ligands for CCR5 (11, 42, 52). More relevant to the present study is the fact that the coding region of the CCR5 gene does vary among humans (1, 4-6, 15, 22, 23). One very well characterized coding polymorphism, CCR5-Δ32, truncates the CCR5 protein and prevents its cell surface expression (4, 16, 34, 61). The frequency of the CCR5-Δ32 allele in Caucasian populations is typically around 15%, although it varies according to precisely where in Europe different individuals originated; the frequency of CCR5-Δ32 homozygosity is ∼1% (1, 6, 15, 46). Individuals homozygous for CCR5-Δ32 are strongly protected from HIV-1 infection, and disease progression is delayed in infected heterozygous individuals (5, 16, 22, 23, 44, 46). Another, much rarer polymorphism, T303A (m303), also prevents CCR5 expression via the premature introduction of a stop codon; in CCR5-Δ32/T303A heterozygotes, it is protective against HIV-1 infection (4, 6, 56).
By analogy to what we observed with SCH-C and rh-CCR5, polymorphisms that result in the substitution of single amino acids in hu-CCR5 could have a direct effect on inhibitor activity. Several single-residue polymorphisms have been described, some located within the CCR5 NT and ECL, others within the TM domain (1, 6, 15). The allele frequency is usually low (<1%) and variable, but for some single-residue polymorphisms it can be as high as 4% in certain human populations (1, 6, 15). Most of the variant CCR5 proteins that have been tested for biological activity are functional as chemokine receptors and/or HIV-1 entry coreceptors (4, 26). Few functional differences from wild-type CCR5 have been reported, but some mutant receptors do differ in how they bind chemokines or mediate HIV-1 infection. Among these, the I12T, C20S, and C178R variants differ only in the NT and ECL2 (4, 26), extracellular regions distinct from the binding sites for the small-molecule inhibitors (7, 21, 63, 70). However, there are three reported CCR5 variants with single amino acid changes in TM1 (A29S, I42F, L55Q) and one with a change in TM2 (A73V), and both these TM helices form part of the binding site for all the small-molecule CCR5 inhibitors characterized to date (7, 21, 63, 70). The biological effect of the I42F, L55Q, and A73V variants is to increase the affinity of CCR5 for RANTES but not for macrophage inflammatory protein 1β, whereas the A29S variant is defective in chemokine binding (4, 26). While the A29S, I42F, and A73V variants are relatively rare (allele frequency, <1%), the L55Q allele occurs with a frequency of approximately 4% in U.S. Caucasians (4, 15). On the CCR5 model, L55 lies at the end of TM1 proximal to the cytoplasm; it is therefore unlikely to be directly involved in inhibitor binding. Among the naturally occurring polymorphic residues, alanine substitutions have been made experimentally at positions C20, L55, and C178. None of these alanine mutant coreceptors differed significantly from wild-type CCR5 in their sensitivities to AD101 and SCH-C (70). Leucine and serine substitutions have also been made at A29; the A29L substitution affects the action of TAK-779 but not that of AD101 or SCH-C (63; C. Seibert and T. P. Sakmar, unpublished data).
Unless new CCR5 variants with unusual properties are discovered at significant frequencies in as yet poorly sampled population groups, it seems unlikely that single amino acid polymorphisms will play a major role in influencing host-dependent variable responses to CCR5 inhibitors in vivo. However, as seen with the hu-CCR5(I198M) mutant and SCH-C, substitutions distant from the drug binding site can affect the action of a CCR5 inhibitor in a way that is completely unpredictable from mere sequence inspection. It may therefore be prudent to sequence the coding regions of CCR5 genes from any individuals who respond poorly to these drugs during clinical trials. Any unusual polymorphisms could be rapidly evaluated with the Env pseudotype-based entry assays used in this study. Sequencing of noncoding regions of the CCR5 gene to identify promoter polymorphisms that could affect CCR5 expression, and hence perhaps the response to therapy, should also be considered (42, 58).
Acknowledgments
We thank Tanya Dragic for providing reagents and advice. We thank Preston Marx, Janet Harouse, Anandan Palani, Jayaram Tagat, Dennis Nazareno, John Clader, and Stuart McCombie for reagents. We are grateful to Tom Morgan, Amy Snyder, Anthony Sanfiz, Jia Liu, Cathy Buontempo, and Yan Hou for technical support.
This work was supported by NIH grant R01 AI41420 (to J.P.M.), NIH Immunology training grant T32 AI07621 (to S.E.K.), NIH MSTP grant GM07739 (to E.B.), and Schering Plough Research Institute. C.S. has been an Associate and T.P.S. is an Associate Investigator of the Howard Hughes Medical Institute; J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Ansari-Lari, M. A., X. M. Liu, M. L. Metzker, A. R. Rut, and R. A. Gibbs. 1997. The extent of genetic variation in the CCR5 gene. Nat. Genet. 16:221-222. [DOI] [PubMed] [Google Scholar]
- 2.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanpain, C., B. J. Doranz, A. Bondue, C. Govaerts, A. De Leener, G. Vassart, R. W. Doms, A. Proudfoot, and M. Parmentier. 2003. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278:5179-5187. [DOI] [PubMed] [Google Scholar]
- 4.Blanpain, C., B. Lee, M. Tackoen, B. Puffer, A. Boom, F. Libert, M. Sharron, V. Wittamer, G. Vassart, R. W. Doms, and M. Parmentier. 2000. Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood 96:1638-1645. [PubMed] [Google Scholar]
- 5.Carrington, M., M. Dean, M. P. Martin, and S. J. O'Brien. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. 8:1939-1945. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, M., T. Kissner, B. Gerrard, S. Ivanov, S. J. O'Brien, and M. Dean. 1997. Novel alleles of the chemokine-receptor gene CCR5. Am. J. Hum. Genet. 61:1261-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castonguay, L. A., Y. Weng, W. Adolfsen, J. Di Salvo, R. Kilburn, C. G. Caldwell, B. L. Daugherty, P. E. Finke, J. J. Hale, C. L. Lynch, S. G. Mills, M. MacCoss, M. S. Springer, and J. A. DeMartino. 2003. Binding of 2-aryl-4-(piperidin-1-yl)butanamines and 1,3,4-trisubstituted pyrrolidines to human CCR5: a molecular modeling-guided mutagenesis study of the binding pocket. Biochemistry 42:1544-1550. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., P. Zhou, D. D. Ho, N. R. Landau, and P. A. Marx. 1997. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J. Virol. 71:2705-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesebro, B., R. Buller, J. Portis, and K. Wehrly. 1990. Failure of human immunodeficiency virus entry and infection in CD4-positive human brain and skin cells. J. Virol. 64:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi, F., A. L. DeVico, R. Yarchoan, R. Redfield, F. Cleghorn, W. A. Blattner, A. Garzino-Demo, S. Colombini-Hatch, D. Margolis, and R. C. Gallo. 2000. Higher macrophage inflammatory protein (MIP)-1α and MIP-1β levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proc. Natl. Acad. Sci. USA 97:13812-13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 13.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, M., M. Carrington, and S. J. O'Brien. 2002. Balanced polymorphism selected by genetic versus infectious human disease. Annu. Rev. Genomics Hum. Genet. 3:263-292. [DOI] [PubMed] [Google Scholar]
- 16.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 17.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 18.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 19.Dragic, T., and M. Alizon. 1993. Different requirements for membrane fusion mediated by the envelopes of human immunodeficiency virus types 1 and 2. J. Virol. 67:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez, E., M. Bamshad, N. Sato, S. Mummidi, R. Dhanda, G. Catano, S. Cabrera, M. McBride, X. H. Cao, G. Merrill, P. O'Connell, D. W. Bowden, B. I. Freedman, S. A. Anderson, E. A. Walter, J. S. Evans, K. T. Stephan, R. A. Clark, S. Tyagi, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 1999. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. USA 96:12004-12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harouse, J. M., A. Gettie, T. Eshetu, R. C. Tan, R. Bohm, J. Blanchard, G. Baskin, and C. Cheng-Mayer. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J. Virol. 75:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harouse, J. M., A. Gettie, R. C. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816-819. [DOI] [PubMed] [Google Scholar]
- 26.Howard, O. M., A. K. Shirakawa, J. A. Turpin, A. Maynard, G. J. Tobin, M. Carrington, J. J. Oppenheim, and M. Dean. 1999. Naturally occurring CCR5 extracellular and transmembrane domain variants affect HIV-1 co-receptor and ligand binding function. J. Biol. Chem. 274:16228-16234. [DOI] [PubMed] [Google Scholar]
- 27.Kazmierski, W., N. Bifulco, H. Yang, L. Boone, F. DeAnda, C. Watson, and T. Kenakin. 2003. Recent progress in discovery of small-molecule CCR5 chemokine receptor ligands as HIV-1 inhibitors. Bioorg. Med. Chem. 11:2663-2676. [DOI] [PubMed] [Google Scholar]
- 28.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 29.Koopmann, W., C. Ediriwickrema, and M. S. Krangel. 1999. Structure and function of the glycosaminoglycan binding site of chemokine macrophage-inflammatory protein-1β. J. Immunol. 163:2120-2127. [PubMed] [Google Scholar]
- 30.Koopmann, W., and M. S. Krangel. 1997. Identification of a glycosaminoglycan-binding site in chemokine macrophage inflammatory protein-1α. J. Biol. Chem. 272:10103-10109. [DOI] [PubMed] [Google Scholar]
- 31.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J. Virol. 71:8642-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurence, J. S., C. Blanpain, A. De Leener, M. Parmentier, and P. J. LiWang. 2001. Importance of basic residues and quaternary structure in the function of MIP-1β: CCR5 binding and cell surface sugar interactions. Biochemistry 40:4990-4999. [DOI] [PubMed] [Google Scholar]
- 33.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 34.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 35.Martin, L., C. Blanpain, P. Garnier, V. Wittamer, M. Parmentier, and C. Vita. 2001. Structural and functional analysis of the RANTES-glycosaminoglycans interactions. Biochemistry 40:6303-6318. [DOI] [PubMed] [Google Scholar]
- 36.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. R. O'Brien, M. W. Hilgartner, D. Vlahov, S. J. O'Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907-1911. [DOI] [PubMed] [Google Scholar]
- 37.McCombie, S. W., J. R. Tagat, S. F. Vice, S. I. Lin, R. Steensma, A. Palani, B. R. Neustadt, B. M. Baroudy, J. M. Strizki, M. Endres, K. Cox, N. Dan, and C. C. Chou. 2003. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. III. Synthesis, antiviral and pharmacokinetic profiles of symmetrical heteroaryl carboxamides. Bioorg. Med. Chem. Lett. 13:567-571. [DOI] [PubMed] [Google Scholar]
- 38.McDermott, D. H., P. A. Zimmerman, F. Guignard, C. A. Kleeberger, S. F. Leitman, P. M. Murphy, et al. 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Lancet 352:866-870. [DOI] [PubMed] [Google Scholar]
- 39.Meanwell, N. A., and J. F. Kadow. 2003. Inhibitors of the entry of HIV into host cells. Curr. Opin. Drug Discov. Dev. 6:451-461. [PubMed] [Google Scholar]
- 40.Monteclaro, F. S., and I. F. Charo. 1996. The amino-terminal extracellular domain of the MCP-1 receptor, but not the RANTES/MIP-1α receptor, confers chemokine selectivity. Evidence for a two-step mechanism for MCP-1 receptor activation. J. Biol. Chem. 271:19084-19092. [DOI] [PubMed] [Google Scholar]
- 41.Moore, J. P. 1997. Coreceptors: implications for HIV pathogenesis and therapy. Science 276:51-52. [DOI] [PubMed] [Google Scholar]
- 42.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 100:10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell. Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 44.Mummidi, S., S. S. Ahuja, E. Gonzalez, S. A. Anderson, E. N. Santiago, K. T. Stephan, F. E. Craig, P. O'Connell, V. Tryon, R. A. Clark, M. J. Dolan, and S. K. Ahuja. 1998. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat. Med. 4:786-793. [DOI] [PubMed] [Google Scholar]
- 45.Nardese, V., R. Longhi, S. Polo, F. Sironi, C. Arcelloni, R. Paroni, C. DeSantis, P. Sarmientos, M. Rizzi, M. Bolognesi, V. Pavone, and P. Lusso. 2001. Structural determinants of CCR5 recognition and HIV-1 blockade in RANTES. Nat. Struct. Biol. 8:611-615. [DOI] [PubMed] [Google Scholar]
- 46.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 47.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palani, A., S. Shapiro, J. W. Clader, W. J. Greenlee, K. Cox, J. Strizki, M. Endres, and B. M. Baroudy. 2001. Discovery of 4-[(Z)-(4-bromophenyl)-(ethoxyimino)methyl]-1′-[(2,4-dimethyl-3-pyridinyl)carbonyl]-4′-methyl-1,4′- bipiperidine N-oxide (SCH 351125): an orally bioavailable human CCR5 antagonist for the treatment of HIV infection. J. Med. Chem. 44:3339-3342. [DOI] [PubMed] [Google Scholar]
- 49.Palani, A., S. Shapiro, H. Josien, T. Bara, J. W. Clader, W. J. Greenlee, K. Cox, J. M. Strizki, and B. M. Baroudy. 2002. Synthesis, SAR, and biological evaluation of oximino-piperidino-piperidine amides. 1. Orally bioavailable CCR5 receptor antagonists with potent anti-HIV activity. J. Med. Chem. 45:3143-3160. [DOI] [PubMed] [Google Scholar]
- 50.Palczewski, K., T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima, B. A. Fox, I. Le Trong, D. C. Teller, T. Okada, R. E. Stenkamp, M. Yamamoto, and M. Miyano. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739-745. [DOI] [PubMed] [Google Scholar]
- 51.Paterlini, M. G. 2002. Structure modeling of the chemokine receptor CCR5: implications for ligand binding and selectivity. Biophys. J. 83:3012-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paxton, W. A., A. U. Neumann, S. Kang, L. Deutch, R. C. Brown, R. A. Koup, and S. M. Wolinsky. 2001. RANTES production from CD4+ lymphocytes correlates with host genotype and rates of human immunodeficiency virus type 1 disease progression. J. Infect. Dis. 183:1678-1681. [DOI] [PubMed] [Google Scholar]
- 53.Pierce, K. L., R. T. Premont, and R. J. Lefkowitz. 2002. Seven-transmembrane receptors. Nat. Rev. Mol. Cell. Biol. 3:639-650. [DOI] [PubMed] [Google Scholar]
- 54.Pöhlmann, S., and R. W. Doms. 2002. Evaluation of current approaches to inhibit HIV entry. Curr. Drug Targ. Infect. Dis. 2:9-16. [DOI] [PubMed] [Google Scholar]
- 55.Proudfoot, A. E., S. Fritchley, F. Borlat, J. P. Shaw, F. Vilbois, C. Zwahlen, A. Trkola, D. Marchant, P. R. Clapham, and T. N. Wells. 2001. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 276:10620-10626. [DOI] [PubMed] [Google Scholar]
- 56.Quillent, C., E. Oberlin, J. Braun, D. Rousset, G. Gonzalez-Canali, P. Metais, L. Montagnier, J. L. Virelizier, F. Arenzana-Seisdedos, and A. Beretta. 1998. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet 351:14-18. [DOI] [PubMed] [Google Scholar]
- 57.Rabut, G. E., J. A. Konner, F. Kajumo, J. P. Moore, and T. Dragic. 1998. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J. Virol. 72:3464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 60.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 61.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 62.Schwarz, M. K., and T. N. Wells. 2002. New therapeutics that modulate chemokine networks. Nat. Rev. Drug Discov. 1:347-358. [DOI] [PubMed] [Google Scholar]
- 63.Seibert, C., W. Ying, S. Gavrilov, F. Tsamis, S. E. Kuhmann, A. Palani, J. R. Tagat, J. W. Clader, S. W. McCombie, B. M. Baroudy, S. O. Smith, T. Dragic, J. P. Moore, and T. P. Sakmar. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. J. Biol. Chem., in press. [DOI] [PubMed]
- 64.Sharon, M., N. Kessler, R. Levy, S. Zolla-Pazner, M. Gorlach, and J. Anglister. 2003. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure 11:225-236. [DOI] [PubMed] [Google Scholar]
- 65.Siciliano, S. J., T. E. Rollins, J. DeMartino, Z. Konteatis, L. Malkowitz, G. Van Riper, S. Bondy, H. Rosen, and M. S. Springer. 1994. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 91:1214-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tagat, J. R., S. W. McCombie, R. W. Steensma, S. Lin, D. V. Nazareno, B. Baroudy, N. Vantuno, S. Xu, and J. Liu. 2001. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. I. 2(S)-Methyl piperazine as a key pharmacophore element. Bioorg. Med. Chem. Lett. 11:2143-2146. [DOI] [PubMed] [Google Scholar]
- 68.Tagat, J. R., R. W. Steensma, S. W. McCombie, D. V. Nazareno, S. I. Lin, B. R. Neustadt, K. Cox, S. Xu, L. Wojcik, M. G. Murray, N. Vantuno, B. M. Baroudy, and J. M. Strizki. 2001. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. II. Discovery of 1-[(2,4-dimethyl-3-pyridinyl)carbonyl]-4-methyl-4-[3(S)-methyl-4-[1(S)-[4-(trifluoromethyl)phenyl]ethyl]-1-piperazinyl]-piperidine N1-oxide (SCH-350634), an orally bioavailable, potent CCR5 antagonist. J. Med. Chem. 44:3343-3346. [DOI] [PubMed] [Google Scholar]
- 69.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsamis, F., S. Gavrilov, F. Kajumo, C. Seibert, S. Kuhmann, T. Ketas, A. Trkola, A. Palani, J. W. Clader, J. R. Tagat, S. McCombie, B. Baroudy, J. P. Moore, T. P. Sakmar, and T. Dragic. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]