Abstract
RNA-based biosensors and regulatory devices have received significant attention for their potential in a broad array of synthetic biology applications. One of the primary difficulties in engineering these molecules is the lack of facile methods to link sensory modules, or aptamers, to readout domains. Such efforts typically require extensive screening or selection of sequences that facilitate interdomain communication. Bacteria have evolved a widespread form of gene regulation known as riboswitches that perform this task with sufficient fidelity to control expression of biosynthetic and transport proteins essential for normal cellular homeostasis. In this work, we demonstrate that select riboswitch readout domains, called expression platforms, are modular in that they can host a variety of natural and synthetic aptamers to create novel chimeric RNAs that regulate transcription both in vitro and in vivo. Importantly, this technique does not require selection of device-specific “communication modules” required to transmit ligand binding to the regulatory domain, enabling rapid engineering of novel functional RNAs.
Keywords: riboswitch, aptamer, RNA engineering, biosensors, transcriptional regulation
Devices composed of RNA are emerging as important tools in synthetic biology for applications including in vivo detection of small molecules with genetically encodable biosensors and engineered regulatory elements to control gene expression1. In part, this is because of the ability to select nucleic acids with a desired activity using well-established in vitro evolution methods (SELEX)2. This method subjects a starting pool of random sequences to multiple rounds of a selection pressure and replication of the survivors. The result, when the selection pressure is for binding, is an aptamer capable of binding the target molecule with high affinity and specificity. In the 20 years since its development, SELEX has yielded RNAs capable of binding a chemically diverse assortment of compounds, revealing that aptamers for almost any desired molecule can be obtained3. For most applications, the aptamer must be coupled to a readout module that transduces the binding event into a detectable signal, a significant hurdle in the engineering of practical RNA devices.
Several distinct solutions have emerged that detect binding of a target molecule to an RNA4. One example is the “aptazyme” in which an aptamer is linked to a hammerhead ribozyme such that ligand binding induces the RNA to cleave itself, an event that can be readily detected either in vitro or in vivo5. A striking example of the utility of such devices was a recent use of a theophylline-sensing aptazyme to monitor the directed evolution of a theophylline-degrading protein enzyme6. Engineering of an aptazyme often requires extensive optimization of a “communication module” between the two domains7. However, it has been observed that aptazymes display high background cleavage levels and slow allostery, which can limit their utility8, 9. Other readout modules that have been explored are designed or evolved secondary structural switches10–12, fluorophore binding aptamers13, 14 and trans-acting RNAs that alter translation of target mRNAs in a ligand inducible manner15, 16. As with aptazymes, these devices can be difficult to engineer and suffer from high background or low dynamic ranges. Moreover, these approaches typically require significant reengineering of each readout domain for the particular aptamer in question. Ideally, modular readout domains should be as “plug-and-play” as aptamers domains, which can be easily placed into different contexts with minimal or no re-design.
A potential source of modular readout domains is found in naturally occurring genetic control devices called riboswitches. These RNA elements are generally found in the 5'-leader sequence of mRNAs and are broadly distributed across bacteria regulating a wide assortment of metabolic processes17. Riboswitches are generally composed of two distinct functional domains: the aptamer domain that serves as the receptor for a small molecule and an expression platform containing a secondary structural switch that directs the gene expression machinery (Figure 1a)18. Currently, over twenty riboswitches have been characterized whose aptamers bind a chemically diverse set of molecules including nucleobases and nucleosides, amino acids, protein cofactors, metal ions, second messenger signaling molecules, and aminoglycosides18. These naturally occurring aptamers have been exploited in engineered biosensors, reflecting their modularity and utility19, 20.
Figure 1. Riboswitches regulate transcription through a ligand-dependent secondary structural switch.
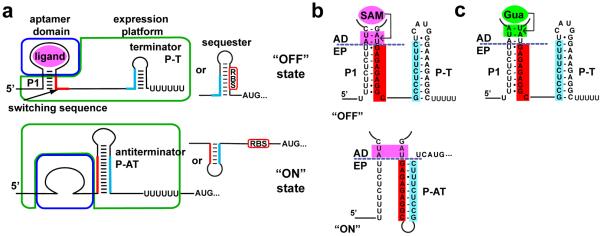
(a) Cartoon of a typical riboswitch that terminates transcription in response to an effector ligand. Effector binding (magenta) to the aptamer domain (blue box) stabilizes incorporation of a switching sequence (red) into the P1 helix, causing the expression platform (green box) to form an intrinsic terminator (P-T) that directs RNA polymerase to abort transcription (“OFF” state). In the absence of ligand binding (bottom), the switching sequence can participate in formation of an alternative structure, the antiterminator (P-AT), disallowing the terminator and transcription continues (“ON” state). Equivalent secondary structural switches can control translation by sequestering or exposing a ribosome binding site (RBS). (b) Sequence of the metE riboswitch P1 helix and the different helix arrangement of the free (bottom) and bound (top) states. The purple dashed line determines the separation between the aptamer domain and expression platform. (c). Sequence of the P1 helix of the xpt/metE chimera, emphasizing the pairs at the 3'-side required for guanine binding (green box).
In contrast to the high degree of structural conservation of each aptamer class, expression platforms are highly variable. These domains regulate by a number of mechanisms, including transcription, translation, mRNA stability and splicing18, typically through their ability to form alternative structures in a ligand-directed fashion17. For example, expression platforms that direct transcription generally contain two alternative hairpin structures: an intrinsic terminator (P-T, Figure 1a) that directs RNA polymerase (RNAP) to abort mRNA synthesis and an antiterminator that enables the RNAP to proceed through the 5'-leader. Analogous hairpin structures that sequester or expose the ribosome binding site (RBS) are used to control expression of the mRNA at the translational level (Figure 1b). However, expression platforms are poorly characterized, both with respect to their structure beyond recognition of the binary switch and how they communicate with the aptamer domain. To date, the potential for the regulatory expression platforms as modular readout elements that can serve as building blocks for novel riboregulatory elements has not been explored.
In this study, we demonstrate the modularity of expression platforms of select riboswitches that control gene expression at the transcriptional level. These chimeric riboswitches comprising novel combinations of riboswitch aptamer domains and expression platforms retain the regulatory properties of biological sequences. Importantly for synthetic biology, we find that the secondary structural switch of these expression platforms can be directed by synthetic aptamers. The engineering strategy that we employ to couple aptamers with these modular expression platforms is conceptually simple and requires minimal screening of variants to find an effective riboswitch. These switches are functional in both a basic in vitro transcription assay and as a regulatory element controlling expression of a reporter gene in E. coli. Our data reveal a robust strategy for generating a wide variety of practical RNA devices.
RESULTS AND DISCUSSION
Considerations in design of modular expression platforms
The two primary criteria considered in selecting modular expression platforms from biological sequences were: i) the two domains of the riboswitch must not overlap in their sequence requirements and ii) easy and rapid assays for their in vitro and in vivo function of readout must exist. To determine whether the domains of a riboswitch are separable, the alternative secondary structures must be examined. For the archetypal “OFF” switch two distinct forms exist: a ligand-bound form comprising the intact ligand-bound aptamer and a rho-independent transcriptional terminator/ribosome binding site (RBS) sequester hairpin or an unbound aptamer that is partially disrupted by formation of the antiterminator/RBS antisequester hairpin (Figure 1a, b). In this model, the two domains intersect at a secondary structural element called the P1 helix. Within this helix are sequence elements crucial for productive ligand binding to the aptamer domain or part of the secondary structural switch of the expression platform.
For a large subset of riboswitches, the sequence requirements of the two domains do not overlap within the P1 helix, allowing for assignment of a functional boundary. For example, in one class of S-adenosylmethionine (SAM) binding riboswitches (known as SAM-I) only two A-U base pairs near the 3'-end of the P1 helix are required for ligand binding as revealed by the crystal structure of an aptamer-SAM complex (Figure 1c; Supplementary Figure 1a, b)21. Of the eleven SAM-I riboswitches in the Bacillus subtilis genome22, 23, seven present slight or no overlap between these A-U pairs and the switching sequence. Since crystal structures of most known riboswitch aptamers have been determined, information about the ligand binding requirements for almost all riboswitches is available for this analysis21. It is important to note, however, that a number of riboswitches cannot meet the above requirement for several reasons. For example, riboswitches in which the penultimate helix is part of a pseudoknot generally require a sequence that is essential for both ligand recognition and for secondary structural switching24, 25. In the most extreme cases of the SAM-II and SAM-III classes, the two domains completely overlap24. Well-characterized classes of riboswitches that are populated by suitable candidates include the purine, SAM-I, lysine, thiamine pyrophosphate and flavin mononucleotide (FMN)18.
The second consideration is that the regulatory domains function by a well-understood mechanism and whose activity could be rapidly assessed. While structural switches that control translation have been more frequently employed in synthetically designed riboswitches10, 26, we have concluded that the RNA sequence requirements of intrinsic (rho-independent) transcriptional termination are mechanistically better understood, and thus more amenable to rational design. In addition, well-established and simple in vitro single turnover transcriptional termination assays using commercially available E. coli RNAP can be performed to rapidly assess activity of novel riboswitches prior to their introduction into a cell27. In our implementation of this assay, a riboswitch is placed downstream of a strong T7A1 promoter in a DNA template and RNAP is allowed to transcribe only a single round to ensure that the ligand and NTP concentrations remain constant. We have further strived to match physiological ionic conditions as closely as possible, with the exception of low NTP concentrations (50 μM) compared to intracellular concentrations measured during log-phase growth (>1 mM)28. Although this simplifies the implementation of this assay, these conditions significantly slow the rate of transcription, a key parameter determining the concentration of ligand required to elicit a regulatory response29–32. Finally, E. coli RNAP holoenzyme was used rather than native RNAP (B. subtilis for the riboswitches that are used in this study). Previous work has shown that the difference in activity between riboswitches transcribed with B. subtilis and E. coli polymerases is moderate22, 32, 33. Further, since E. coli is an organism of choice for many synthetic biology applications, it is important to assess the activity of artificial riboswitches with this RNAP.
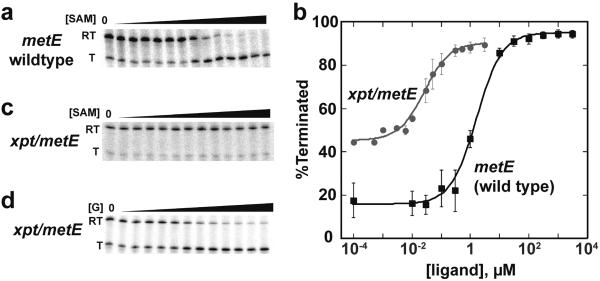
Using the above considerations, we focused upon three candidate riboswitches for further investigation: those that regulate the B. subtilis metE, yitJ and lysC transcriptional units (Supplementary Figure 1). All three riboswitches are “OFF” switches that terminate transcription in the presence of the effector ligand (SAM for metE and yitJ, lysine for lysC) and efficiently terminate transcription in a ligand-dependent fashion in a single turnover transcription assay27 (Figure 2a) consistent with previous reports22, 23, 34, 35. In this assay, the 32P-labeled transcriptional products are visualized by separating the RNAs on a denaturing polyacrylamide gel. The amount of RNA corresponding to the terminated (T) and fully transcribed read-through products (RT) are quantified (see Materials and Methods) and the data fit to a two-state binding equation (Figure 2b). This analysis yields three important parameters: T50 (the amount of ligand required to elicit the half-maximal regulatory response), the maximal transcriptional termination at high ligand concentration (%Tmax), and the dynamic range or DR (the amplitude of the response as the difference between percent terminated at low and high ligand concentrations) (Table 1). To directly compare T50 and affinity of each aptamer for its ligand, the apparent equilibrium dissociation constant (KD) was measured under transcription conditions (buffer matched and at 37 °C) by isothermal titration calorimetry (Table 1). Four candidate guanine-sensing riboswitches from B. subtilis were also tested--those controlling the yxjA, pbuG, purE, and xpt-pbuX genes--but showed no riboswitching activity in the transcription assay (data not shown). Since three of these riboswitches have been shown to function in vivo36, differences between cellular and minimal in vitro transcription conditions likely explain this observed loss of function.
Figure 2. A chimeric xpt/metE riboswitch regulates transcriptional termination.
(a) Single round in vitro transcription assay of the wild type B. subtilis metE riboswitch as a function of S-adenosylmethionine (SAM) concentration. The two observed bands correspond to the read through (RT) product in which the riboswitch allows transcription of the full mRNA and the terminated product (T) where transcription is stopped at the terminator. (b) In vitro transcription data of the wild type B. subtilismetE SAM-responsive riboswitch and the xpt/metE guanine-responsive chimera fit to a two-state transition. The raw data for these two titrations are shown in panels (a) and (d). (c) Titration of the xpt/metE chimera with SAM. (d) Titration of the xpt/metE chimera with guanine.
Table 1.
Binding and regulatory activity measurements of aptamers and riboswitches.
| expression platform | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B. sub metE | B.sub yitJ | B. sub lysC | |||||||||
| Ligand | aptamer1 | KD2 (μM) | T50 (μM) | DR3 | %Tmax | T50 (μM) | DR | %Tmax | T50 (μM) | DR | %Tmax |
| SAM | metE | 2.0 ± 0.5 | 2± 0.2 | 83 | 95±1 | 12±4 | 45 | 85±5 | 3.5±0.7 | 47 | 95±3 |
| SAM | yitJ | 1.5 ± 0.3 | 2.3±0.5 | 38 | 97±2 | 2.5±0.2 | 83 | 95±2 | 0.5±0.1 | 62 | 97±1 |
| lysine | lysC | 18 ± 2 | 160±10 | 23 | 54±1 | 180±10 | 24 | 69±1 | 61±6 | 52 | 78±5 |
| guanine | xpt | 0.024 ± 0.003 | 0.030±0.003 | 47 | 92±3 | 0.071±0.004 | 55 | 87±3 | 0.073±0.006 | 72 | 83±4 |
| 2-AP | xpt(C74U) | 8.8 ± 0.8 | 30±5 | 34 | 96±1 | 7.3±1.1 | 57 | 72±2 | 32±4 | 52 | 87±2 |
| FMN | ribD | 0.91 ± 0.1 | 8.8±1.9 | 46 | 72±3 | 19±1 | 42 | 51±3 | 1.3±0.1 | 43 | 53±1 |
| theophylline | SELEX | 20±6 | 130±10 | 59 | 91±4 | 13±3 | 58 | 93±2 | 280±20 | 77 | 82±2 |
gene regulated by the riboswitch in B. subtilis, with the exception of the theophylline aptamer. All measurements were performed in triplicate with the error defined as the standard deviation obtained from three experiments.
KD obtained through ITC matching conditions of in vitro transcription assay.
DR is the dynamic range (%Tmax − %Tmin), where %Tmax is the maximum fraction terminated at high ligand concentration and %Tmin is the minimum fraction terminated at low ligand concentration.
Several characteristics can be noted about these three riboswitches. First, the SAM riboswitches are efficiently regulated by S-adenosylmethionine in this assay--both riboswitches show a high dynamic range and are almost completely “OFF” at high effector concentrations (95% terminated, Table 1). The lysC riboswitch displays a lesser degree of efficiency both with respect to the dynamic range (52%) and the degree of termination at saturating ligand concentration (78%). The lower efficiency of transcriptional termination may indicate that this RNA may be less suitable for in vivo applications due to the potential for leaky expression. Second, both SAM riboswitches under our experimental conditions are regulated by ligand concentrations equivalent to the aptamer's affinity for SAM. This “thermodynamic control” reflects the ability of the aptamer to fully equilibrate with respect to ligand binding prior to a regulatory decision being made31, 32. This probably reflects our choice to perform the assay at low NTP concentration, which decreases the rate of transcription and increases the time the aptamer has to equilibrate with respect to ligand concentration. In contrast, the lysine riboswitch appears to be under weak “kinetic control,” which is defined by T50>KD31. This is the result of the aptamer having insufficient time to reach equilibrium prior to the RNAP reaching the intrinsic terminator and a regulatory decision being made.
Chimeric riboswitches regulate transcription in vitro
The strategy for creating chimeric riboswitches is to splice a minimal aptamer domain into the metE, yit, and lysC expression platforms at the boundaries shown in Supplementary Figure 1. For each aptamer, structural and biochemical analysis yielded sufficient information to determine the minimal sequence element required for ligand recognition. Each minimal aptamer includes all direct ligand-RNA contacts observed in crystal structures and all nucleotides involved in ligand-induced conformational changes as judged by in-line, RNase or chemical probing. This sequence was substituted for the wild type aptamer in each expression platform such that the full switching sequence was preserved as well as the number of Watson-Crick base pairs of the P1 helix. It has been observed that the length of the P1 helix is important for efficient functioning of an adenine and a lysine riboswitches33, 37.
To determine whether expression platforms are capable of being modular, we created a fusion between the well-characterized B. subtilis xpt-pbuX guanine riboswitch aptamer domain38 and the metE expression platform (full secondary structure is shown in Supplementary Figure 2). The resulting chimera, called xpt/metE (aptamer domain/expression platform), showed no response to SAM (Figure 2c), but strongly regulates in the presence of guanine (Figure 2d) (the sequences of all riboswitches used in this work are listed in Supplementary Table 1). The T50 of the guanine-responsive chimera correlates strongly with the KD of the natural xpt aptamer for guanine under transcription conditions (0.030 and 0.024 nM, respectively; Table 1). Furthermore, a single point mutation (U51C) that is highly deleterious to guanine binding to the xpt aptamer39 abrogates ligand-dependent regulation (data not shown), demonstrating that the chimera's regulatory activity is due to the same specific interaction with guanine as the wild type riboswitch. Chimeras between the xpt aptamer domain and yitJ or lysC expression platform exhibit similar behavior (Table 1). These data, along with a fusion of the Streptococcus mutans tetrahydrofolate (THF) aptamer to the metE expression platform presented in a prior study40, demonstrate the modularity of some riboswitch expression platforms.
Assaying the regulatory activity of a diverse set of aptamer domain/expression platform chimeras reveals that the ability to reprogram regulation is both general and robust (Figure 3 and Table 1). In all, eighteen chimeras were created that used expression platforms of the metE, yitJ and lysC riboswitches in combination with aptamers from the guanine-sensing B. subtilis xpt riboswitch38, an xpt(C74U) aptamer that imparts adenine and 2-aminopurine (2AP) responsiveness41, and the B. subtilis flavin mononucleotide (FMN) responsive ribD aptamer domain42. For every chimera we were able to measure an effector-dependent regulatory response, further supporting the generality of the mix-and-match strategy (examples of titration data for metE expression platform containing different aptamers are shown in Figure 3a).
Figure 3. Diverse chimeric riboswitches are functional.
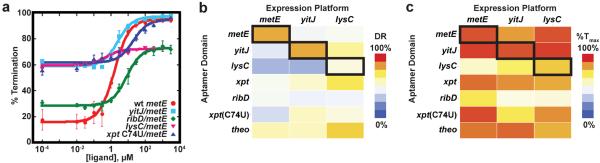
(a) Comparison of the in vitro transcription data quantified, plotted and fit to a two-state transition as a function of ligand concentration. The B. subtilis metE riboswitch wild type (wt) in red circles, compared to chimeras containing a different SAM-binding aptamer (yitJ/metE, cyan), a flavin mononucleotide (FMN) aptamer (ribD/metE, green), a lysine binding aptamer (lysC/metE, pink), and the xpt(C74U) aptamer (blue). The values for T50, dynamic range (DR) and % termination at saturating ligand concentrations are given in Table 1. In all experiments the riboswitch was titrated with the cognate effector of the aptamer. Error bars represent the standard deviation of at least three independent experiments. (b) Heat map of the dynamic range for eighteen chimeric riboswitches. The heavy box denotes the wild type metE, yitJ and lysC riboswitches. (c) Heat map for the maximal termination at high effector concentration for the riboswitches of this study.
Comparing the regulatory response of these chimeric RNAs to their wild type counterparts, we observed two general trends. First, there is there is a strong correlation between the KD of the isolated aptamer domain and the observed T50 in the majority of riboswitches under in vitro transcription conditions for riboswitches containing the metE and lysC expression platforms (Supplementary Figure 3). Conversely, chimeras with the yitJ expression platform show a weak correlation between KD and T50, likely the result of slow binding equilibration to the aptamer domain relative to the rate of transcription through the expression platform31, 32. Secondly, many of the chimeras are able to achieve dynamic ranges and/or maximal transcriptional termination levels that rival or exceed that of wild type riboswitches (Figure 3b, c). Several aptamers appear to perform well, such as yitJ SAM-I aptamer that reaches ≥95% transcriptional termination when coupled to the metE and lysC expression platforms. Conversely, several aptamers consistently underperform, such as the ribD FMN responsive aptamer, which only achieves 50–70% transcriptional termination at saturating FMN concentrations, consistent with observations of the wild type B. subtilis ribD riboswitch32. These observations indicate that the aptamer domain strongly influences both T50 and maximal extent of transcriptional termination. The response of the chimera can potentially be improved by “tuning” the effector binding properties of the aptamer domain43.
Synthetic aptamers can control the regulatory switch
The above data reveals the modular nature of certain expression platforms whose regulatory switch can be directed by structurally diverse natural aptamers. However, these biological aptamers have presumably evolved in the context of a riboswitch and therefore may contain inconspicuous features that enable them to interface with the secondary structural switch of the expression platform. In fact, it has been proposed that the greater structural complexity of biological receptors as compared to their in vitro selected aptamer counterparts is important for their role in regulation44. Synthetic aptamers are selected in the absence of any pressure for regulatory function and thus may not be predisposed to interface with the expression platform. To determine whether biological RNA receptors have unique properties that enable them to direct secondary structural switching, we tested the theophylline (theo)45 and tetracycline (tet)46 aptamers as chimeras with expression platforms.
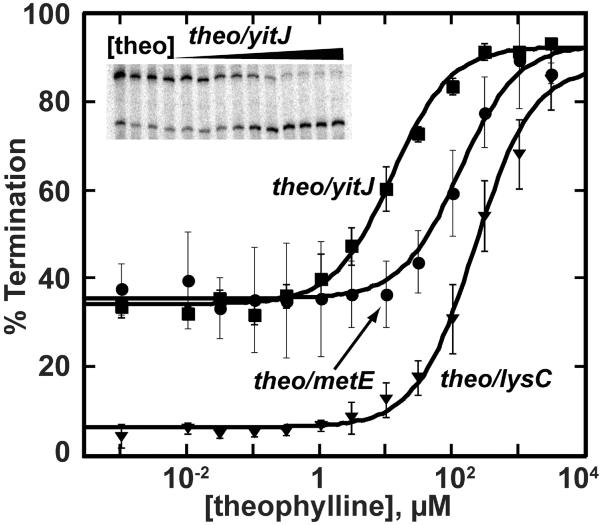
As with biological aptamers, we observed that in vitro selected aptamers are capable of effector-dependent transcriptional regulation. Each of the three theophylline chimeras is functional to varying degrees in a single turnover transcription assay (Figure 4 and Table 1). While the theo/yitJ chimera showed a T50 close to KD, the same aptamer in the context of metE and lysC showed considerably higher T50's. Furthermore, the theo/metE and theo/yitJ chimeras showed a >90% repression at high theophylline concentrations, indicating that the in vitro selected aptamer can achieve a level of transcriptional termination, comparable to wild type riboswitches. A similar chimera created with the tet aptamer and the metE regulatory domain resulted in a functional chimera responsive to tetracycline (vida infra). Importantly, these chimeras were created without the need for a second selection step to create an aptamer-specific expression platform as in other approaches5, 10, 26.
Figure 4. An in vitro selected aptamer can be incorporated into a functional chimera.
Titrations of chimeric riboswitches with the theophylline aptamer fused to the metE, yitJ and lysC expression platform. In the left corner is an image of a single turnover transcription assay of theophylline dependent termination of the theo/yitJ chimera. While the chimera displays a small level of termination at low ligand concentration, more importantly, nearly complete termination is observed at high ligand concentration.
Chimeric riboswitches function in vivo
While the above experiments reveal that artificial chimeras regulate transcriptional termination in vitro, these conditions are different from those encountered by natural riboswitches in the cell. Transcription conditions were chosen to match physiological ionic conditions as closely as possible including lowering the [Mg2+], but the NTP concentrations are low (50 μM) in comparison to their intracellular concentration in rapidly growing bacteria (>1 mM)28. Further, accessory transcription factors such as NusA influence the elongation rate in vivo47, 48. Previous work has shown that the absence of NusA in the in vitro transcription assay can reduce riboswitch activity; for the B. subtilis FMN riboswitch this was approximately 2-fold32. Thus, in vitro activity of a chimera may not correlate with its performance in the cell.
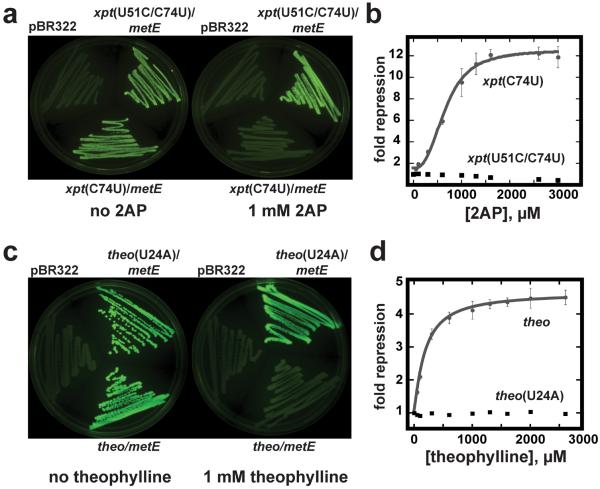
To determine whether these artificial riboswitches function in a cellular environment, the ability of several chimeras to regulate expression of a gfpuv reporter gene was examined. We focused on two metE regulatory domain chimeras containing either the C74U variant of the xpt guanine riboswitch aptamer or the theophylline aptamer. These chimeras were chosen because E. coli does not rapidly metabolize their effectors. Further, we used a strain of E. coli harboring a deletion of the purine efflux pump (nep) to prevent potential export of these molecules. As negative controls, a single point mutation was introduced into the ligand binding pocket of each small molecule binding site (U51C for the xpt aptamer39 and U24A for the theophylline aptamer49) that abrogates binding. Both chimeric riboswitches display strong regulatory activity in E. coli (Figure 5) as indicated by reduction in fluorescence to a background level upon addition of 1 mM effector to a defined medium (Figure 5a, c). Despite the use of the strong constitutive promoter (tac) both chimeras achieve complete repression of reporter in comparison to background cellular fluorescence, underscoring the effectiveness of these regulatory RNA elements. Furthermore, aptamers containing a single point mutation preventing ligand binding fail to diminish gfpuv expression.
Figure 5. Chimeric riboswitches regulates gfpuv expression in E. coli.
(a) 2-aminopurine (2AP) dependent regulation of gfpuv expression by the xpt(C74U)/metE chimera in E. coli. Cells transformed with either the control parental vector (pBR322) that does not contain gfpuv, the xpt(C74U)/metE riboswitch in the 5'-leader of an mRNA encoding gfpuv, and a mutant of this riboswitch (U51C) that specifically abrogates 2AP binding to the aptamer were plated on defined medium-agar in the absence (left) or presence (right) of 1 mM 2AP. (b) Quantification of the fluorescence of E. coli transformed with the xpt(C74U)/metE chimera (grey circles) or the mutant (U51C, black squares) grown in defined medium with increasing concentrations of 2AP. (c) Theophylline-dependent control of gfpuv expression in E. coli. Cells were grown under the same media conditions as in (a), but supplemented with 1 mM theophylline in the right plate. A single point mutation in the theophylline aptamer (U24A) serves a control for potential theophylline-dependent effects on gfpuv expression not directly related to its binding to the aptamer. (d) Quantification of the fluorescence of E. coli transformed with the theo/metE chimera (grey circles) or the point mutation thereof (black squares).
Titration of the effector into a defined liquid medium enabled measurement of an EC50, defined as the concentration of ligand in the media that elicits half-maximal repression of reporter gene expression. For the xpt(C74U)/metE riboswitch an EC50 of 730±80 μM was measured, with a 12-fold repression of expression (Figure 5b); this EC50 is ~24-fold higher than the T50 measured in vitro (Table 1). The theo/metE riboswitch displayed a response of 190±10 μM, similar to the observed T50 (130±10 μM) and ~10-fold above the KD for the aptamer (20±6 μM) (Table 1 and Figure 5d). The observed EC50 for this riboswitch is consistent with the performance of other theophylline-dependent synthetic riboswitches8, 50–54. It must be noted that although we used a strain of E. coli deficient in purine efflux, the cells are still capable of purine degradation and thus the intracellular 2AP concentration does not necessarily tightly correlate with that in the medium, confounding making conclusions regarding the lack of tight correlation between EC50 and T50. We also did not account for the small concentration of adenine in the cell (~1 μM55), which further complicates this comparison. Nonetheless, these observations reinforce the validity of using the transcription assay to screen and optimize variants prior to introduction into a cell.
Optimizing riboswitch performance by altering P1 helix stability
It is not obvious why these expression platforms can be directed by a number of distinct aptamers. One hypothesis regarding folding of alternative RNA secondary structures called “encoded co-transcriptional folding”56 may provide an explanation. This model proposes that a combination of the 5'-to-3' sequential order and relative stability of secondary structural elements directs the folding outcome. A recent study of synthetic RNA switches demonstrated that accounting for the thermodynamics and kinetics of RNA folding proved to be sufficient for design of RNAs that fold almost exclusively into one of two mutually exclusive structures despite each being isoenergetic56. In application of this concept to transcriptional riboswitches, the competing P1 and antiterminator (P-AT) structures constitute the regulatory switch (Figure 1a), while the terminator (P-T) is merely a readout of the P1/P-AT outcome. This hypothesis is consistent with our observations that the regulatory switches of B. subtilis metE, yitJ and lysC expression platforms are not hard-wired to a specific receptor. Further, the information required to direct RNA folding along one of two folding trajectories should be primarily encoded within the P1/P-AT switch of the expression platform. Any mechanism that changes the stability of the P1 helix relative to P-AT in response to a specific signal should therefore be able to act as an efficient effector for the riboswitch.
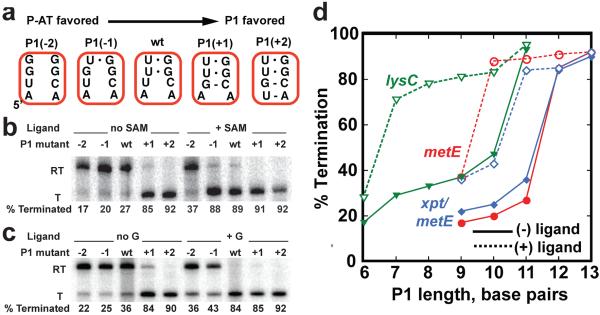
If the encoded co-transcriptional folding hypothesis accounts for how the regulatory switch is directed by the aptamer domain, then the intrinsic stability of the P1 helix is a primary site for tuning the ligand-responsiveness of the secondary structural switch. A prior study demonstrated that weakening the P1 helix by removing base pairs forces the lysC riboswitch to become constitutively “ON” while strengthening the helix by adding base pairs results in a constitutively “OFF” switch37. To more precisely define how helix length/stability directs the switch to fold into one of the two regulatory states, we introduced a series of point mutations in the wild-type metE and lysC riboswitches and the xpt/metE chimera at the 5'-terminus of their P1 helix (metE mutations shown in Figure 6a). These mutations alter the stability of the P1 helix while sequence elements required for receptor binding and conformational switching are unaltered. Both the metE and xpt/metE switches exhibit a sharp boundary between predominantly folding in the “OFF” (terminated) and “ON” (read through) states in the absence of ligand (Figure 6b). In the case of the xpt/metE riboswitch, this transition occurs over a single base pair (Figure 6c). In contrast, the wild type lysC riboswitch is more tolerant to mutations; lysine-dependent switching occurs at four different P1 lengths (Supplementary Figure 4) in general agreement with a previous report37.
Figure 6. The length of the P1 helix can dictate the switch.
(a) Variation of the length of the P1 helix was achieved by introducing mutations on the 5'-side of the helix that either remove (P1(−1), P1(−2)) or add (P1(+1), P1(+2)) Watson-Crick base pairs. Variants are ordered from left to right as those expected to favor the antiterminator P-AT to those favoring formation of P1/P-T. (b) Separation of read through and terminated transcription products of wild type and P1 mutants of the metE riboswitch by PAGE. Reactions were performed in the presence and absence of SAM and the percentage of termination shown below each lane. (c) Identical representation as in (b), for the xpt/metE chimeric riboswitch. (d) Plot of the length of the P1 helix (number of contiguous Watson-Crick and G•U pairs) for the metE (red), lysC (green) and xpt/metE (blue) riboswitches as a function of percentage termination in the absence (solid lines) or presence (dashed lines) of ligand. Where the two values differ significantly (>20%) is the hallmark of ligand-dependent riboswitching.
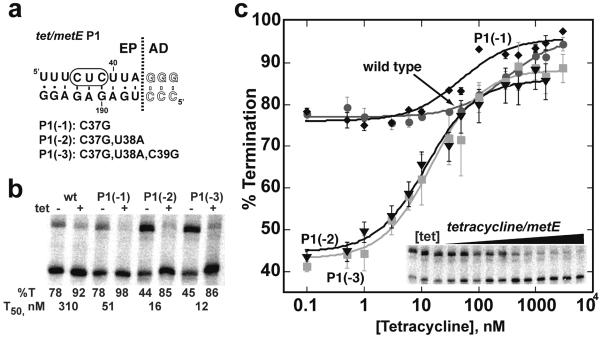
The importance of P1 helix length variation in the design of new chimeras is illustrated in the tet/metE chimera (Figure 7). In the first design of this chimera, we took a conservative approach with the aptamer and retained three strong G-C base pairs in the aptamer portion of the P1 helix. This created a chimera whose P1 helix was twelve base pairs (Figure 7a) and showed very low tetracycline responsiveness (“wt”, Figure 7b, c). The strong transcriptional termination in the absence of ligand was evidence for an overly stable P1 helix. To design a better performing riboswitch, we introduced a set of mutations on the 5'-side of the P1 helix that do not disrupt ligand binding or P-AT formation. While the tet/metE(P1(−1)) chimera still displayed a low dynamic range, the P1(−2) and P1(−3) switches showed substantial switching ability and lower T50s (Figure 7b, c). Thus, modulation of the stability of the P1 helix is a simple and highly effective design strategy to optimize the activity of a new riboswitch. This approach is conceptually similar to the process for computationally designing riboswitches that terminate transcription by modulating the thermodynamic stability of P-T12. However, in our case, we cannot calculate the strength of P1 because of the influence of tertiary interactions with the aptamer domain, and thus rely on an empirical approach.
Figure 7. P1 helix length can be used to optimize chimera performance.
(a) The sequence of the P1 regulatory helix for a tetracycline aptamer (tet) fused to the metE expression platform. Mutations introduced into the 5'-side of the P1 helix to destabilize formation of the helix to favor formation of the competing antiterminator helix are shown below. (b) Transcription reactions at low and high tetracycline concentrations as a function of P1 helix length. For each RNA, the T50 and percent termination at low and high effector concentrations are given as the average of three independent experiments. (c) Quantification of the regulatory response of the four chimeras shown in (b). The gel of a complete titration of tet/metE(P1(−3)) is shown in the inset of the graph.
CONCLUSION
Robust methods for connecting sensory and readout domains with minimal experimental or computational design remains a significant hurdle in the development novel RNA-based devices for synthetic biology. Through a careful consideration of the sequence features of biological riboswitches, we have found three expression platforms capable of being directed by a diverse set of aptamers--both natural and synthetic. Since properties of co-transcriptional folding guide alternative RNA secondary structure formation, altering the stability of one of the helices of the regulatory switch in the expression platform is an easy route to optimizing the performance of a designed chimera of aptamer and regulatory modules. This approach provides a simple route to “plug-and-play” secondary structural switch modules that can be directly coupled to aptamers to yield practical RNA devices.
Although the three expression platforms used in this study effectively couple ligand binding to regulation for a variety of aptamers, there are a few notable limitations related to the use of expression platforms that terminate transcription. First, the effector concentration required to elicit a detectable change in gene expression in the transcription assay and in vivo cannot be readily correlated. Differences such as the influence of transcriptional pause sites in the expression platform, accessory pausing factors such as NusA, and NTP concentrations can significantly alter transcription kinetics and thus the response to the effector. This is in contrast to riboswitches that control at the level of translation, whose response is generally believed to be independent of transcription kinetics. Second, some of the chimeric RNAs have lower dynamic range and termination efficiencies than their native counterparts, another property difficult to predict. One interpretation of this observation is that riboswitch chimeras have not co-evolved for optimal folding and the resultant misfolding leads to a population of unproductive riboswitches trapped in either the “ON” or “OFF” state. Because of these limitations, creating a useful chimeric riboswitch using this modular approach requires that the aptamer be empirically tested with several expression platforms to find the optimal combination.
Nonetheless, the simplicity with which naturally evolved expression platforms can be employed to build transcriptional regulatory devices offers a number of advantages for applications in synthetic biology. For example, transcriptional regulators could be easily arranged in tandem to form more complicated logic gates such as those that occur naturally in bacterial mRNAs57. Furthermore, we envision that the large repository of natural riboswitches can be prospected for functional readouts in bacteria or eukarya to generate device libraries that regulate across a broad range of ligand concentrations, or to implement control at different levels of the information transfer process. Such devices could be readily combined in various ways to establish complex regulatory circuits that respond to a wide variety of orthogonal, user defined chemicals for which in vitro aptamers exist or can be selected. Artificial riboregulatory devices have already been implemented in a broad assortment of applications including driving bioremediation of an environmental pollutant10, guiding directed protein evolution6 and elucidating metabolic pathways58, and the ability to rapidly actualize novel riboswitches should be a powerful technique in the synthetic biologist's toolbox.
METHODS
Synthesis of DNA templates for transcription
The DNA templates used for single turnover transcription assays were synthesized by PCR amplification from B. subtilis genomic DNA for wild type sequences (listed in Supplementary Figure 1 and Supplementary Table 1). These sequences were placed immediately downstream of the T7A1 promoter using recombinant PCR. Chimeric riboswitches were constructed in two different pieces. The first piece consisted of the T7A1 promoter, the 5'-side of the expression platform of choice (metE, yitJ or lysC), and the chosen aptamer domain. The second piece was composed of the 3'-side of the expression platform. Recombinant PCR was used to fuse the two segments. Mutant templates were obtained by altering the desired position/s in the DNA by standard site directed mutagenesis. The sequences of all DNAs were verified prior to use in transcription assays.
In vitro transcription assays
DNA templates containing a chimeric riboswitch were transcribed as previously described40. Briefly, 100 ng of DNA were incubated at 37 °C for 15 minutes in 25 μl of 2× transcription buffer (140 mM Tris-HCl, pH 8.0, 140 mM NaCl, 0.2 mM EDTA, 28 μM β-mercaptoethanol and 70 mg/mL BSA), 5 μl of 25 mM MgCl2, 1 mCi of α-32P-ATP and 0.5 units of E. coli RNA polymerase 70 holoenzyme (Epicentre Biotechnologies) to a final volume of 35 μl. The reactions were initiated with the addition of 15 μl of NTP mix (NTPs in equimolar ratios of 165 μM), 0.2 mg/ml heparin and the desired concentration of ligand. After incubation at 37 °C for 15 minutes the reactions were quenched with equal volume of 8 M urea and 2 minutes incubation at 65 °C. The species were separated by denaturing PAGE, dried and exposed to a phosphorimager screen. Quantitation of radioactive counts in each band was performed with ImageQuant software (Molecular Biosystems) and data fit to a two state model with nonlinear least squares analysis.
In vivo assay
A reporter plasmid for testing riboswitch function in E. coli with a gfpuv reporter gene was constructed using pBR322 as the parental vector using standard molecular biological techniques. The sequence of the 5'-leader sequence of the gfpuv reporter and the sequence and map of the entire plasmid is shown in Supplementary Figure 5 and Supplementary Table 2. The sequence of the chimeric riboswitch insert for the xpt(C74U)/metE and theo/metE reporters are identical to those used for in vitro transcription assays. The resultant vectors were transformed into E. coli strain BW25113(Δnep). Cells were transformed with pRR1 plasmid containing the different riboswitch chimeric constructs. Single colonies were picked and grown overnight in 3 ml of CSB media (Supplementary Table 3) + 100 μg/ml ampicillin. This saturated culture was used to inoculate 100 ml of fresh media, and allowed to grow to early exponential phase (OD600 = 0.1–0.5). Samples of these growths were taken in triplicate (3 ml each sample) and ligand was added to the media at the concentrations indicated in the titration. The cells were allowed to grow for 6 hours at 37 °C. At this point 300 μl of the cultures were used to measure the OD600 and their fluorescence intensity in a plate reader (Infinite M200 Pro, Tecan). Fluorescent measurements were taken at an excitation wavelength of 395 nm and the average fluorescence was taken from 513–515 nm, where the maximum emission for GFP was observed. Optical density normalized fluorescent values were plotted as a function of ligand concentration and fitted to a two state binding equation to extract the EC50 values. The background fluorescence was obtained by performing a ligand titration into cells carrying the parental plasmid pBR322 and calculating the OD600 normalized fluorescence (raw data is shown in Supplementary Figure 6). An average of the background was subtracted to the values of the cultures containing the plasmids with the chimeric riboswitch constructs.
Supplementary Material
ACKNOWLEDGEMENTS
This work has been funded by grants to R.T.B. from the National Institutes of Health (R01 GM073850 and R01 GM083953) and an Innovative Seed Grant from the University of Colorado, Boulder. J.G.M.-V. and A.D.G. were supported in part through a grant from the NIH/CU Molecular Biophysics Training Program (T32 GM065103).
ABBREVIATIONS
- FMN
flavin mononucleotide
- G
guanine
- GFP
green fluorescent protein
- NTP
nucleotide triphosphate
- RBS
ribosome binding site
- RNAP
RNA polymerase
- SAM
S-adenosylmethionine
Footnotes
Supporting Information Supplementary figures show the complete secondary structure of the wild type metE, yitJ and lysC and the chimeric xpt/metE riboswitches, graphs of the correlation between KD and T50, transcriptional analysis of P1 length variants of the wild type lysC riboswitch, plasmid map of the parental reporter vector pRR1, and raw fluorescence data for in vivo experiments. Supplementary tables list all of the sequences of the wild type and chimeric riboswitches used in this study, the complete sequence of the parental reporter vector for assaying the in vivo activity of riboswitches, and defined medium used for cell growth. This material is available free of charge via Internet at http://pubs.acs.org.
Author contributions P.C. and A.D.G. helped to develop the idea of chimeric riboswitches and implement the in vitro transcription assay. P.C. performed all in vitro transcription assays except for those relating to the lysC aptamer, which were performed by A.D.G. J.G.M.-V. performed the in vivo activity assays. P.C., A.D.G. and R.T.B. wrote the manuscript and all authors participated in its revision.
The authors declare no competing financial interests.
REFERENCES
- 1.Isaacs FJ, Collins JJ. Plug-and-play with RNA. Nat Biotechnol. 2005;23:306–307. doi: 10.1038/nbt0305-306. [DOI] [PubMed] [Google Scholar]
- 2.Joyce GF. Directed evolution of nucleic acid enzymes. Annual review of biochemistry. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 3.Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Aptamer database. Nucleic acids research. 2004;32:D95–100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Famulok M, Mayer G. Aptamer modules as sensors and detectors. Accounts of chemical research. 2011;44:1349–1358. doi: 10.1021/ar2000293. [DOI] [PubMed] [Google Scholar]
- 5.Soukup GA, Breaker RR. Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Structure. 1999;7:783–791. doi: 10.1016/s0969-2126(99)80102-6. [DOI] [PubMed] [Google Scholar]
- 6.Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab Eng. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic acids research. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Silva C, Walter NG. Leakage and slow allostery limit performance of single drug-sensing aptazyme molecules based on the hammerhead ribozyme. RNA. 2009;15:76–84. doi: 10.1261/rna.1346609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chemistry & biology. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachsmuth M, Findeiss S, Weissheimer N, Stadler PF, Morl M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic acids research. 2012 doi: 10.1093/nar/gks1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. Journal of the American Chemical Society. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 15.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nature biotechnology. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 16.Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP. Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic acids research. 2012;40:5775–5786. doi: 10.1093/nar/gks168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite binding riboswitches. Genome biology. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. Journal of the American Chemical Society. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- 20.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angewandte Chemie. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 21.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 22.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci U S A. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 24.Batey RT. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdiscip Rev RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nature chemical biology. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Artsimovitch I, Henkin TM. In vitro approaches to analysis of transcription termination. Methods. 2009;47:37–43. doi: 10.1016/j.ymeth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. Journal of bacteriology. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garst AD, Porter EB, Batey RT. Insights into the regulatory landscape of the lysine riboswitch. Journal of molecular biology. 2012;423:17–33. doi: 10.1016/j.jmb.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 32.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Molecular cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 33.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS genetics. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 35.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic acids research. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blouin S, Chinnappan R, Lafontaine DA. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic acids research. 2011;39:3373–3387. doi: 10.1093/nar/gkq1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert SD, Montange RK, Stoddard CD, Batey RT. Structural studies of the purine and SAM binding riboswitches. Cold Spring Harb Symp Quant Biol. 2006;71:259–268. doi: 10.1101/sqb.2006.71.015. [DOI] [PubMed] [Google Scholar]
- 42.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoddard CD, Widmann J, Trausch JJ, Marcano-Velazquez JG, Knight R, Batey RT. Nucleotides adjacent to the ligand-binding pocket are linked to activity tuning in the purine riboswitch. Journal of molecular biology. 2013 doi: 10.1016/j.jmb.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha J, Topp S, Gallivan JP. From SELEX to Cell Dual Selections for Synthetic Riboswitches. Methods in enzymology. 2011;497:207–220. doi: 10.1016/B978-0-12-385075-1.00009-3. [DOI] [PubMed] [Google Scholar]
- 45.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 46.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorganic & medicinal chemistry. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 47.Grundy FJ, Henkin TM. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. Journal of bacteriology. 2004;186:5392–5399. doi: 10.1128/JB.186.16.5392-5399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yakhnin AV, Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism invitro. Proc Natl Acad Sci U S A. 2002;99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 50.Feng X, Liu L, Duan X, Wang S. An engineered riboswitch as a potential gene-regulatory platform for reducing antibacterial drug resistance. Chemical communications. 2011;47:173–175. doi: 10.1039/c0cc00980f. [DOI] [PubMed] [Google Scholar]
- 51.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. Journal of the American Chemical Society. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 52.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa A. Rational construction of eukaryotic OFF-riboswitches that downregulate internal ribosome entry site-mediated translation in response to their ligands. Bioorganic & medicinal chemistry letters. 2012;22:1639–1642. doi: 10.1016/j.bmcl.2011.12.118. [DOI] [PubMed] [Google Scholar]
- 54.Auslander S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst. 2010;6:807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 55.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature chemical biology. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xayaphoummine A, Viasnoff V, Harlepp S, Isambert H. Encoding folding paths of RNA switches. Nucleic acids research. 2007;35:614–622. doi: 10.1093/nar/gkl1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 58.Fowler CC, Brown ED, Li Y. Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli. Chemistry & biology. 2010;17:756–765. doi: 10.1016/j.chembiol.2010.05.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.