Abstract
Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2), a direct transcriptional activator of viral and cellular genes, is required for EBV-induced B-cell transformation. The functional role of conserved regions within the amino terminus of the protein preceding the poly-proline region has yet to be fully characterized. Thus, we tested whether the EBNA2 amino-terminal 30 amino acid residues, containing evolutionarily conserved region 1, are required for stimulating viral and cellular gene expression necessary for B-cell transformation in a viral transcomplementation assay. We found that these residues are required for its ability to induce LMP-1 expression in lymphoblastoid cell lines (LCLs), to stimulate LMP-1 promoter reporter plasmids in transient-cotransfection assays, and to rescue LCL growth following inactivation of endogenous wild-type EBNA2 protein. Deletion of amino acid residues 3 to 30 also impaired its ability to self-associate in coimmunoprecipitation assays. These data indicate that EBNA2 residues 3 to 30 comprise an essential domain required for induction of LMP-1 expression and, consequently, for maintenance of the immortalized phenotype of LCLs. The ability to self-associate into dimers or multimers conferred by this domain may be an important mechanism for these effects.
Epstein-Barr virus (EBV) is a ubiquitous human lymphocryptovirus associated with both lymphoid and epithelial cell malignancies as well as lymphoproliferative diseases in immunocompromised patients (27, 41). In vitro infection of primary B cells with EBV drives these cells to become activated proliferating lymphoblasts (27). This process is mediated by the latent proteins EBV nuclear antigen 2 (EBNA2), EBNA3A and -C, EBNA1, and LMP-1 (3, 27). Our laboratory is particularly interested in defining the role of EBNA2 in EBV-mediated B-cell transformation. Understanding the mechanisms by which latent proteins mediate B-cell immortalization will provide insight into general mechanisms of viral persistence and may lead to advances in therapeutics aimed at EBV-associated malignances that would bypass the toxic sequelae of currently available chemotherapeutic agents.
After initial infection of B cells by EBV, the W promoter (Wp) directs transcription of the EBNA-LP and EBNA2 genes that are produced from both bicistronic and alternatively spliced transcripts (43, 60). Following synthesis of EBNA2, transcription switches from Wp to an upstream promoter known as the C promoter (Cp), which is also coincident with expression of the other EBNAs and LMPs (1, 63). EBNA2 is a transcriptional activating protein that stimulates Cp, the LMP2A promoter, and the LMP-1/LMP2B divergent promoter, making it a central regulator of its own expression and of other latency gene products (12, 23, 32, 50, 55, 63, 72). Interestingly, although evolutionarily well-conserved, Cp is not required for EBNA expression during B-cell immortalization in vitro, since EBNA expression can be maintained through usage of the Wp (13, 52, 67, 68). EBNA1 can also be expressed from the latency Q promoter (Qp), while LMP2A is known to be dispensable for B-cell immortalization (29, 35-37, 45). Genetic experiments have demonstrated the importance of LMP-1 for EBV-driven B-cell immortalization, and EBNA2 appears to be a potent regulator of LMP-1 in transient-transfection assays as well as in lymphoblastoid cell lines (LCLs) (11, 12, 23, 24, 28, 61). Thus, of the viral latency proteins required for B-cell immortalization, expression of LMP-1 appears to be the only one highly dependent on EBNA2 for its expression.
EBNA2 has also been shown to induce cellular genes, which include the proto-oncogene c-myc as well as CD21, Hes-1, EBI 1 and 2, and Runx3, but the spectrum of cellular genes directly activated by EBNA2 has not yet been fully elucidated (4, 5, 9, 31, 44, 48, 49). This problem is further complicated by the fact that many cellular genes induced during EBV infection may be the result of cooperative activities between one or more latent proteins (18, 38, 46, 59). It is assumed that activation and maintenance of cellular gene expression by EBNA2 is important for growth proliferation of B cells, since LCLs constitutively expressing LMP-1 from a promoter independent of EBNA2 control still required EBNA2 for continued growth (70). However, it has not yet been demonstrated which cellular gene(s), whose expression is highly dependent on EBNA2, is critical for EBV-driven immortalization.
The EBNA2 protein does not bear any significant sequence homology with cellular proteins. This circumstance has posed a significant obstacle in determination of its functions. Identification of EBNA2 functional domains has been aided by the identification of nine evolutionarily conserved regions (CR1 to -9) among EBNA2 alleles isolated from different primate lymphocryptoviruses (34). Their evolutionary conservation suggests that these regions might mediate protein-protein interactions, which are important for the viral life cycle. The CRs are not evenly distributed throughout the EBNA2 sequence. They form two clusters, one at the amino terminus (CRs 1 to 4) and the other near the carboxy terminus (CRs 5 to 9) (34). Residues between CRs 4 and 5, referred to as the divergent region, vary considerably in length and sequence between different EBNA2s (34). Despite its divergence, this region has been implicated in interactions with cellular ligands potentially involved in mediation of EBNA2 function in immortalization (17, 57, 64, 65).
Biochemical and genetic studies have established that several of the CRs in EBNA2 mediate interactions with cellular proteins required for its function. EBNA2 interacts via CR6 with target promoters through an interaction with a cellular DNA binding protein known as CBF1 (RBPJk) (16, 21, 33, 58, 71). This interaction is facilitated by CR5-mediated binding to SKIP, which likewise interacts with CBF1 (69). EBNA2 proteins recruited to promoters through these interactions activate gene transcription through an acidic activation domain containing CR8 (6-8, 34). The activation domain interacts with several components of the cellular transcription machinery, including TFIIH, TAF40, TFIIB, the coactivator p100 that interacts with TFIIE, p300/CBP, and PCAF (53, 54, 62). The activation domain appears to be somewhat generic, since it can be functionally replaced by another acidic activation domain encoded by the herpes simplex virus VP16 protein (6). EBNA2 also contains two nuclear localization signals encoded within CR9 and a conserved glycine-arginine-rich region (8, 34).
In contrast to the carboxy-terminal half of EBNA2, the roles of CRs 1, 2, 3, and 4 and a long stretch of prolines located near the amino-terminal half of the protein are more poorly understood. The major questions concerning this region are the following. (i) What domains (e.g., CRs) are required for B-cell transformation? (ii) What role does this region play in mediating induction of viral or cellular genes? (iii) Could it mediate important transforming activities independent from its transcriptional functions? (iv) What interactions with cellular proteins occur within this region (if any), and how might they mediate EBNA2 functions?
Genetic studies using a marker rescue approach showed that deletion of amino acids 2 to 280 containing CRs 1 to 4 resulted in viruses unable to transform B cells (20, 66). This suggested that one or more of these regions may mediate interactions required for EBNA2 function and consequently for B-cell immortalization. Recent studies have indicated that EBNA2 may contain two regions mediating homotypic interaction, one located in the first 58 amino acids amino terminal to the poly-proline region (PPR) and the other following the PPR between amino acids 97 and 121 (19, 56). A model has been proposed to explain the genetic and biochemical data (19). The model suggests that the amino-terminal half of EBNA2 comprises two independent homotypic association domains, either of which is sufficient for activity in transient reporter gene activation or B-cell transformation (19). At the present time, however, the ability to self-associate (dimerize or oligomerize) or activate the LMP-1 promoter in reporter gene assays is not always predictive of EBNA2 transforming capability (19, 20, 66). For example, an EBNA2 with amino acids 2 to 88 deleted is severely impaired in its ability to self-associate and activate the LMP-1 promoter, yet it is still functional in B-cell transforming activity as measured in marker rescue experiments (66). In contrast, another mutant with amino acids 2 to 95 deleted is also impaired for its ability to self-associate but can stimulate the LMP-1 promoter to levels similar to wild-type EBNA2, yet it is transformation defective (66). Finally, a mutant lacking the PPR was reported to activate the LMP-1 promoter similar to wild-type EBNA2 and is fully functional for self-association, yet it is transformation defective (18, 65). The inconsistency of these results led us to hypothesize that the first 95 amino acids in EBNA2 might provide important functions required for EBV-mediated transformation in addition to self-association or stimulation of LMP-1 expression.
We have recently developed a genetic system that can be used for analysis of EBNA2 regulation of viral and cellular gene expression during immortalization (14). This assay uses the EBV-immortalized EREB cell line expressing a conditional estrogen receptor-EBNA2 fusion protein (EREBNA2) (23, 25, 26). In the absence of estrogen, EREBNA2 function is inactivated and EREB cells undergo growth arrest and die (26). We have previously shown that transduction of EREB cells with a lentivirus expressing wild-type EBNA2 was able to rescue cellular growth when the endogenous EREBNA2 protein was inactivated by removing estrogen from the growth medium (14). In contrast, expression of an immortalization-incompetent mutant EBNA2 gene encoding a protein unable to interact with CBF1 (WW326SR) failed to rescue the EREB cells after estrogen withdrawal (14). Thus, the transcomplementation approach can be used to study the effect of mutations in EBNA2 on cellular growth and/or phenotype. We have previously used this approach to examine the effects of activated Notch, the functional cellular homolog of EBNA2, and a mutant EBNA2 lacking the PPR to support B-cell immortalization (14, 15). The transcomplementation approach offers an advantage over the previously used marker rescue approach, as it allows analysis of EBNA2 target gene expression in the context of EBV-immortalized cell lines. By correlating effects of EBNA2 mutations on immortalization phenotype with changes in cellular or viral EBNA2 target gene expression, it should be possible to determine the functional significance of the amino-terminal CRs.
The goal of this study was to express a mutant EBNA2 lacking CRs 1 and 2 and the PPR (amino acid residues 1 to 103) in our transcomplementation assay. This region was of particular interest, since it apparently fails to affect EBNA2's ability to stimulate the LMP-1 promoter but is absolutely required for the ability of EBNA2 to support immortalization independently of LMP-1 induction (66). Based on these results, we hypothesized that this region of EBNA2 was necessary for induction of cellular genes or pathways critical for immortalization. Unexpectedly, in transcomplementation assays, the results indicated that in addition to residues 3 to 103, amino acids 3 to 30 of EBNA2 are critical for its ability to transactivate LMP-1 and immortalize B cells.
MATERIALS AND METHODS
Cell culture.
EREB2.5, DG75, and Eli-BL cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone) and 15 mM HEPES, pH 7.4, and grown in 5% CO2 at 37°C. 293T cells were maintained in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% fetal bovine serum (HyClone) and grown in 5% CO2 at 37°C. For EREB2.5 cells, the culture medium contained β-estradiol (Sigma) unless otherwise specified.
Expression plasmids.
EBNA2 and EBNA-LP expression plasmids pPDL176A and pSG5EBNA-LP have been described previously (18, 39). pPDL176A was further modified by PCR so that the EBNA2 protein contained an epitope tag on its carboxy-terminal end (pAG155). pPDL176A was used as template to introduce the hemagglutinin (HA) affinity tag (YPYDVPDYAS) at the carboxy terminus of EBNA2 using the following oligonucleotide primers: 5′-CGAGATCTTTAGCTAGCGTAATCTGGAACATCGTATGGGTACTGGATGGAGGG GCAGGTCTT-3′ and 5′-CCAGGGGTCATCAACGACCAAC-3′. The PCR-derived fragment was digested with AvaI and BglII restriction enzymes and cloned into pPDL176A replacing the wild-type AvaI/BglII fragment, resulting in plasmid pAG155. Nested amino-terminal deletions were generated by PCR using pPDL176A as a template. Initially, all PCR-generated fragments were subcloned into the T-easy vector (Promega), and sequences were verified. The PCR products in T-easy plasmids were digested with EcoRI and BamHI and cloned into the same sites in pAG155, replacing the wild-type sequences. The plasmids containing the amino-terminal deletions were as follows: pAG190 (Δ3-30), pAG191 (Δ3-52), pAG177 (Δ3-97), pAG167 (Δ3-103), pAG163 (Δ3-115), and pAG164 (Δ3-156). Sequences upstream of initiation methionine codons in the mutant EBNA2 expression plasmids were identical to that in pPDL176A. The 5′ primer used for PCR with the Δ3-30 and Δ3-52 deletions was 5′-CATCCAAAGCATTCGCATAGCAGATGC-3′, while the 3′ primers were 5′-CGGAATTCCATCATGCCTATTCCCTCGAATCCCTACCAGGAAC-3′ (Δ3-30) and 5′-CGGAATTCCATCATGCCTGGGGAAAACACGGGGGTGCC-3′ (Δ3-52). The 5′ primer used for PCR with the Δ3-115, Δ3-156, Δ3-103, and 3-97 deletions was 5′-GTCGCAGATGGGTGGCCACCAT-3′, while the 3′ primers were 5′-CGGAATTCCATCATGCCTCAGCGCAGGGATGCCTGGAC (Δ3-103), 5′-CGGAATTCCATCATGCCTCTTGATAGGGATCCGCTAG (Δ3-115), 5′-CGGAATTCCATCATGCCACAAGGCCCACAAACAGCC (Δ3-156), and 5′-CGGAATTCCATCATGCCTCCACCACCCCCACCACCTCAGCGCAGGGATGC (Δ3-97).
The expression plasmid F199 expressing EBNA2 amino acid residues 1 to 199 with a carboxy-terminal Flag epitope tag was made by PCR amplification and cloning of the appropriate sequence into the vector pSG5. This plasmid is similar to one previously described (19), except that it was derived from EBV strain B95-8, which contains several extra proline residues in the PPR relative to strain W91.
Transient transfections, electroporations, and reporter gene assays.
For reporter gene assays, 0.5 μg of a reporter plasmid with indicated amounts of effector plasmids encoding the wild-type or mutant EBNA2 proteins were transiently transfected into DG75 cells by a DEAE-dextran method as described previously (39). pJT123A and LMPLUC0 firefly luciferase reporter plasmids have been described previously (32, 39). Luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's recommendations. For LMP-1 protein induction in EBV-positive type I Burkitt's lymphoma cells, 107 Eli-BL cells were electroporated with 10 μg of each effector plasmid encoding EBNA-LP and wild-type or mutant EBNA2 proteins at 200 V and 975 μF using the Genepulser II (Bio-Rad) and allowed to recover in 10 ml of culture medium. Cell lysates were made 48 h after electroporation and processed for LMP-1 expression analysis by Western blotting.
Lentiviral vector construction and production.
The lentivirus vector pLIG has been described previously (14). To generate a lentiviral vector expressing wild-type or mutant EBNA2 proteins, pAG155 or the mutant derivatives (pAG163, pAG164, pAG167, pAG177, pAG190, and pAG191) were digested with EcoRI and BglII to release fragments containing their coding sequences. The fragments were gel purified, blunt ended with Klenow fragment (Gibco BRL), and ligated into XbaI-cleaved and blunt-ended pLIG proviral vector. Lentiviruses were produced by transient transfection of 293T cells as described previously (14, 51). Briefly, 50% confluent 293T cells were transfected with a mixture of plasmid DNAs, including proviral plasmid, a human immunodeficiency virus Tat-encoding plasmid, pBC12-Tat, and a vesicular stomatitis virus G protein-encoding plasmid, pME-VSVG, by the Ca-phosphate coprecipitation method. The cells were incubated with the DNA precipitate for at least 8 h, shocked with 10% dimethyl sulfoxide, and further incubated in Dulbecco's modified Eagle's medium. Forty-eight hours later, lentivirus-containing supernatants were collected, cleared of cellular debris by centrifugation at 3,000 × g for 10 min, and stored at −70°C until use.
Lentiviral transduction, flow cytometry, and enrichment for GFP-positive cells.
Lentiviral transduction of EREB cells was performed as described previously (14). Briefly, 5 × 105 log-phase EREB2.5 cells were transduced with 2 ml of lentivirus-containing supernatant and further incubated in estrogen-containing medium. Five days posttransduction, cells were analyzed for green fluorescent protein (GFP) expression by fluorescence-activated cell sorting (FACS). Enriched GFP-positive populations were established by positively sorting for the brightest 17.5% of all GFP-positive cells in the transduced cell pools. GFP-expressing cells were sorted or analyzed using a Becton Dickinson or Cytomation flow cytometer. Generally, 3.5 × 105 to 5 × 105 output cells were collected by FACS followed by expansion under standard culture conditions in the presence of antibiotic-antimycotic (Gibco BRL). Once sufficient numbers of cells were obtained, they were then maintained under standard cell culture conditions. The mean fluorescence intensity of GFP and EBNA2 levels expressed in the transduced cells was stable for at least 6 weeks in all cell lines, and this was the time period during which analysis of the cell pools was performed (14).
Immunoblot assays.
Transiently transfected, transduced, or nontransduced cells were lysed in Laemmli sample buffer, and proteins were resolved by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 7.5% gel as described previously (30). The protein load per lane was equalized by quantification using Bradford's method (Bio-Rad DC protein assay) according to the manufacturer's recommendations. After gel separation, the proteins were transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dried milk in phosphate-buffered saline. Wild-type and mutant EBNA2 proteins were detected by incubation of the blots with either EBNA2-specific rat monoclonal antibody R3 or anti-HA mouse monoclonal antibody (Covance) followed by horseradish peroxidase-conjugated anti-rat (Jackson Laboratories) or anti-mouse (Amersham) antibodies, respectively. Detection of LMP-1 using monoclonal antibody S12 has been described previously (40). c-myc was detected by a monoclonal anti-c-myc antibody (Santa Cruz) followed by the horseradish peroxidase-conjugated anti-mouse secondary antibody. After secondary antibody incubation, the proteins were visualized by using an enhanced chemiluminescence kit (Pierce).
Proliferation assays.
To measure proliferation of transduced and nontransduced EREB2.5 cells in the absence of estrogen, 5 × 106 log-phase cells were washed twice with 10 ml of estrogen-free medium and resuspended in the estrogen-free culture medium at 5 × 105/ml concentration. After 24 h, the cells were recounted, washed once again, and seeded in 96-well plates in 200 μl of estrogen-free culture medium/well at the indicated concentrations. Proliferation was monitored as described before by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) conversion assay using the MTT cell proliferation kit I (Molecular Roche) as described by the manufacturer.
Immunoprecipitation assays.
Cell lysis and immunoprecipitations were carried out as described by Harada et al. (19).
RESULTS
EBNA2 lacking amino acids 3 to 103 (Δ3-103) fails to rescue EREB cell proliferation or maintain expression of LMP-1 and c-myc.
It has been previously reported that an EBNA2 protein with amino acids 2 to 95 deleted, containing CRs 1 and 2 and all but 2 of the 37 prolines in the PPR from a W91 strain of EBV, results in an EBNA2 mutant capable of stimulating the LMP-1 promoter with wild-type efficiency yet incapable of supporting EBV-mediated immortalization of primary B cells in vitro (66). LMP-1 is the only viral gene essential for immortalization whose expression in immortalized cells requires EBNA2 (25-27, 69). Thus, we hypothesized that perhaps the portion of the protein encompassing CRs 1 and 2 and the PPR may be required to stimulate cellular genes or to modulate cellular pathways critical for B-cell proliferation and/or survival in B cells. To test this hypothesis, we introduced a similar mutation into EBNA2 from the prototype B95.8 strain of EBV by deleting amino acids 3 to 103. The larger size of the deletion was due to the fact that the B95.8 EBNA2 PPR described by Dambaugh et al. (10) possesses eight additional proline residues compared to the W91 EBNA2 and three more than the B95.8 EBNA2 sequence published by Baer et al. (2, 8, 66). Sequence analysis of our EBNA2 cDNA revealed that it was identical to the one described by Dambaugh et al. (10).
Following transduction of EREB cells with lentiviruses expressing either wild-type or mutant EBNA2 proteins and enrichment for GFP-positive cells by FACS, we showed that 90% of the cells expressed either wild-type or mutant EBNA2 proteins (Fig. 1A and data not shown). The transduced cell pools were all sorted using a similar gating strategy for GFP fluorescence that resulted in similar mean fluorescence intensities of GFP expression (reference 13 and data not shown). In these transduced cell pools, several minor polypeptides migrating at 64,000 to 70,000 were invariably detected by immunoblot analysis with either the anti-HA or EBNA2-specific monoclonal antibodies (Fig. 1A). These bands have been observed previously and do not appear to interfere with EBNA2 function in immortalization maintenance (15). The Δ3-103EBNA2 protein, which migrated closely with some of these polypeptides, was expressed at wild-type levels in the transduced cells (Fig. 1A).
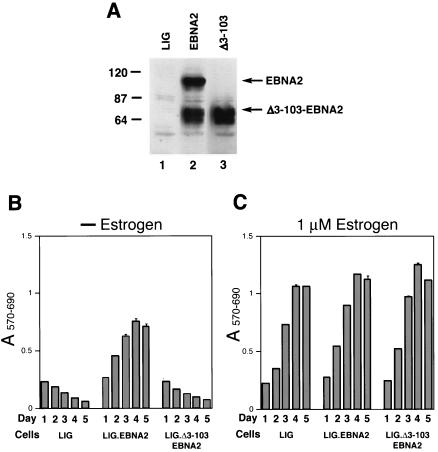
FIG. 1.
Δ3-103-EBNA2 fails to support survival and proliferation of EREB cells upon EREBNA2 inactivation. (A) Western blot analysis of pLIG (lane 1), pLIG.EBNA2 (lane 2), and pLIG.Δ3-103-EBNA2 (lane 3) transduced cell pools after enrichment by FACS were grown in the presence of 1 μM estrogen for 2 weeks prior to Western blotting. EBNA2 expression was analyzed using an anti-HA monoclonal antibody. Equal amounts of cellular protein for each cell pool were analyzed. Molecular mass markers are shown in kilodaltons on the left. (B) EREB cell pools transduced with the pLIG vector alone (LIG), pLIG.EBNA2 (LIG.EBNA2), or pLIG.Δ3-103-EBNA2 (LIG.Δ3-103EBNA2) lentiviruses after enrichment by FACS were grown in the presence of 1 μM estrogen for 2 weeks prior to the experiment. At day 0, the cells were washed of estrogen and placed in the estrogen-free medium. Twenty-four hours later, the cell pools were plated in 200-μl aliquots of estrogen-free medium in 96-well plates at 2 × 104 cells/well and monitored for survival and proliferation by MTT assay daily over the course of 5 days, starting with day 1 (day of plating) after estrogen withdrawal. Optical density at 570 nm (A570) and 690 nm (A690) wavelengths of completed MTT reactions were determined to calculate the A570-690 values by subtracting A690 from A570. (C) The same experiment as that shown in panel B, except that cells were grown in medium containing estrogen.
We next tested the transduced cells for their ability to proliferate after inactivation of the endogenous EREBNA2 protein by withdrawal of estrogen from the culture medium. Briefly, cell pools expressing either wild-type EBNA2 or Δ3-103EBNA2 were expanded for 2 weeks after initial sorting for GFP expression and then subjected to estrogen withdrawal. While the wild-type EBNA2-expressing EREB cells continued to proliferate, the Δ3-103EBNA2-expressing cells ceased to proliferate and died out at a rate similar to control cell pools transduced with an empty lentiviral vector (Fig. 1B). Therefore, the Δ3-103EBNA2 mutant protein failed to support maintenance of EBV-mediated immortalization. In contrast, all three transduced cell pools proliferated with similar efficiency when cultured in the presence of estrogen (Fig. 1C). In an attempt to explain the failure of Δ3-103EBNA2-expressing cells to proliferate in the absence of estrogen, we analyzed expression of the viral LMP-1 and the cellular proto-oncoprotein c-myc in these cells following estrogen withdrawal for 3 days. The results showed that, whereas cells expressing wild-type EBNA2 maintained LMP-1 and c-myc expression, cells expressing Δ3-103EBNA2 lacked detectable LMP-1 and c-myc (Fig. 2). Thus, Δ3-103EBNA2 was significantly impaired at maintaining LMP-1 and c-myc expression in transformed B cells.
FIG. 2.
Δ3-103-EBNA2 fails to support expression of key viral and cellular EBNA2 target genes in EREB cells following EREBNA2 inactivation. EREB cell pools transduced with the pLIG vector alone (LIG), pLIG.EBNA2 (WT), or pLIG.Δ3-103-EBNA2 (Δ3-103.1 and Δ3-103.2) lentiviruses after enrichment by FACS were grown in the presence of estrogen for 2 weeks prior to the experiment. Δ3-103.1 and Δ3-103.2 represent two independently generated cell lines. At day 0, the cells were washed of estrogen and incubated in either estrogen-free (lanes 1 to 4) or estrogen-containing (lanes 5 to 8) medium for 4 days. Cell lysates were prepared, and equal amounts of cellular proteins were separated by SDS electrophoresis. The separated proteins were transferred to nitrocellulose membranes and probed with monoclonal antibodies against LMP-1 (A) and c-myc (B). Migration of LMP-1 and c-myc is indicated by arrows on the right.
Δ3-103EBNA2 fails to stimulate LMP-1 expression in transient-cotransfection assays.
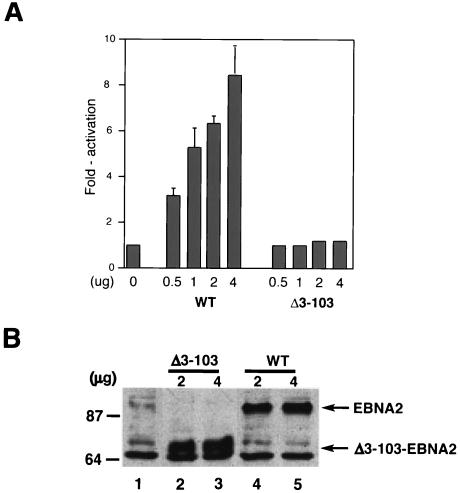
Because a similar EBNA2 mutant was previously shown to transactivate the LMP-1 promoter similar to the wild-type protein in transient-cotransfection assays (66), we also evaluated the ability of Δ3-103EBNA2 to transactivate an LMP-1 promoter luciferase reporter plasmid in transient-cotransfection assays. Consistent with our results presented in Fig. 2, Δ3-103EBNA2 also failed to appreciably stimulate LMP-1 promoter activity, even at the highest concentration of EBNA2 expression plasmid (Fig. 3A). Western blot analysis of wild-type and mutant EBNA2 expression in this assay showed that both proteins were expressed at comparable levels (Fig. 3B). Therefore, Δ3-103EBNA2 is also severely defective in stimulating the LMP-1 promoter in transient-expression assays.
FIG. 3.
Δ3-103-EBNA2 cannot transactivate an LMP-1 promoter reporter plasmid. (A) A 0.5-μg aliquot of an LMP-1 promoter-luciferase reporter plasmid was cotransfected into DG75 cells with the indicated amounts of wild-type EBNA2 (WT) or Δ3-103-EBNA2 (Δ3-103) expression plasmids. The amount of expression plasmid in each transfection reaction was equilibrated with the empty SG5 expression plasmid. The results are presented as fold activation of the promoter in the presence of an effector. Average transactivation efficiencies representing three independent experiments are presented. Standard error bars are shown. (B) Representative Western blot analysis of wild-type and Δ3-103-EBNA2 expression in samples from the transient-transfection experiments described in the legend for panel A. At 48 h after transfection, cells transfected with the SG5 plasmid alone (lane 1), the indicated amounts of Δ3-103-EBNA2 (Δ3-103; lanes 2 and 3), or wild-type EBNA2 (WT; lanes 4 and 5) expression plasmids were subjected to SDS-PAGE and Western blot analysis using an anti-HA monoclonal antibody. Cell lysate aliquots containing equal amounts of total cellular protein were analyzed in each lane. Molecular mass markers are shown in kilodaltons on the left. Arrows on the right indicate migrations of WT and Δ3-103 proteins.
Amino acids 3 to 30 of EBNA2 are required for transactivating the LMP-1 promoter but not a promoter with multimerized CBF1-binding sites.
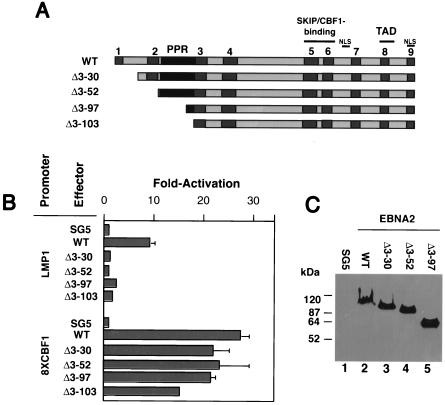
We have previously demonstrated that the PPR is a modulatory domain dispensable for both EBNA2-mediated activation of the LMP-1 promoter and for immortalization maintenance. The Δ3-103 deletion removes CRs 1 and 2, and we hypothesized that perhaps at least one of these regions might be required for these EBNA2 activities. In a previous study, a mutant EBNA2 lacking CRs 1 and 2 (Δ2-88 [W91] EBNA2) could still support immortalization, albeit with a significantly lower-than-wild-type efficiency, possibly due to its impaired ability to transactivate LMP-1 expression (66). Therefore, we examined a similar mutant EBNA2 in our experiments as well as three additional deletion mutants missing CR1 (Δ3-30), CR1 and most of CR2 (Δ3-52), and CRs 1 and 2 and all but 7 of the 45 prolines (Δ3-97; equivalent to Δ2-88 W91 EBNA2) (Fig. 4A). In transient-cotransfection assays the wild type, but none of the mutants, was able to stimulate an LMP-1 reporter plasmid (Fig. 4B). With the exception of Δ3-103EBNA2, however, the mutants were capable of stimulating a reporter plasmid containing multiple (eight) CBF1-binding sites similar to wild-type levels, indicating that the mutant proteins were not globally defective for transactivating function (Fig. 4B). Δ3-103EBNA2 was consistently able to stimulate the CBF1-responsive promoter to within 60% of the wild-type protein level (Fig. 4B). The expression levels of the wild-type and mutant EBNA2s were similar (Fig. 4C). These results demonstrated that amino acids 3 to 30 of EBNA2 specifically drive expression of LMP-1 in B cells.
FIG. 4.
LMP-1 promoter transactivation by EBNA2 mutants. (A) Schematic of EBNA2 deletion mutants generated for this study. CRs are shown as gray boxes. Nuclear localization signals (NLS), the transcriptional activation domain (TAD), and promoter targeting domain (Skip/CBF1) are indicated. The PPR is shown in black. (B) A 0.5-μg aliquot of an LMP-1 promoter-luciferase reporter plasmid (LMP-1) or JT123A reporter plasmid (8XCBF1) was cotransfected into DG75 cells with 4-μg amounts of expression plasmids encoding wild-type EBNA2 (WT), Δ3-30-EBNA2 (Δ3-30), Δ3-52-EBNA2 (Δ3-52), Δ3-97-EBNA2 (Δ3-97), and Δ3-103-EBNA2 (Δ3-103). The amount of expression plasmid in each transfection reaction was equilibrated with the empty SG5 expression plasmid. The results are presented as fold activation of the promoter in the presence of the effector. Average transactivation efficiencies representing three independent experiments are presented. Standard errors are also shown. (C) A representative Western blot analysis of wild-type and mutant EBNA2 expression in samples from the transient-transfection experiments described in the legend for panel B. At 48 h after transfection, cell lysates were prepared and equivalent amounts of proteins for each sample were subjected to SDS-PAGE and Western blot analysis using the anti-EBNA2 monoclonal antibody R3. Molecular mass markers are shown in kilodaltons on the left.
EBNA2 amino acids 3 to 30 are critical for induction of LMP-1 in type I Burkitt's lymphoma cells.
To exclude the possibility that the EBNA2 mutants were unable to transactivate the LMP-1 promoter-reporter plasmid because it lacks essential EBNA2-responsive cis-acting elements, we also tested the EBNA2 mutants for their ability to induce LMP-1 expression in type I Burkitt's lymphoma cells. The type I cell line Eli-BL does not express LMPs or EBNAs except EBNA-1. Ectopic expression of EBNA2 has been shown to result in a modest induction of LMP-1 that is dramatically enhanced by coexpression with the EBNA-LP protein (38, 39). We showed that wild-type EBNA2, but not the mutant EBNA2s, induced LMP-1 expression in Eli-BL cells (Fig. 5A), despite the fact that all EBNA2 proteins were expressed at similar levels in these cells (Fig. 5B).
FIG. 5.
Induction of LMP-1 gene expression in type I Burkitt's lymphoma cells. (A) A 10-μg aliquot of SG5 plasmid, SG5-EBNA-LP expression plasmid, SG5-wild-type EBNA2 (WT), or expression plasmids encoding Δ3-30-EBNA2 (Δ3-30), Δ3-52-EBNA2 (Δ3-52), Δ3-97-EBNA2 (Δ3-97), or Δ3-103-EBNA2 (Δ3-103) were introduced into Eli-BL cells by electroporation. The amount of expression plasmid in each transfection reaction mixture was equilibrated with SG5 plasmid (empty expression vector). At 48 h after electroporation, cell lysates were prepared and equivalent amounts of proteins for each sample were subjected to SDS-PAGE and Western blot analysis using anti-LMP-1 monoclonal antibody. Results from a representative experiment are shown. Molecular mass markers are shown in kilodaltons on the left. (B) A representative Western blot analysis of wild-type and mutant EBNA2 expression in samples from the transient-transfection experiments described in the legend for panel A. At 48 h after electroporation, cell lysates were prepared and equivalent amounts of proteins for each sample were subjected to SDS-PAGE and Western blot analysis using an anti-EBNA2 R3 monoclonal antibody. Molecular mass markers are shown in kilodaltons on the left.
EBNA2 amino acids 3 to 30 are required for immortalization maintenance.
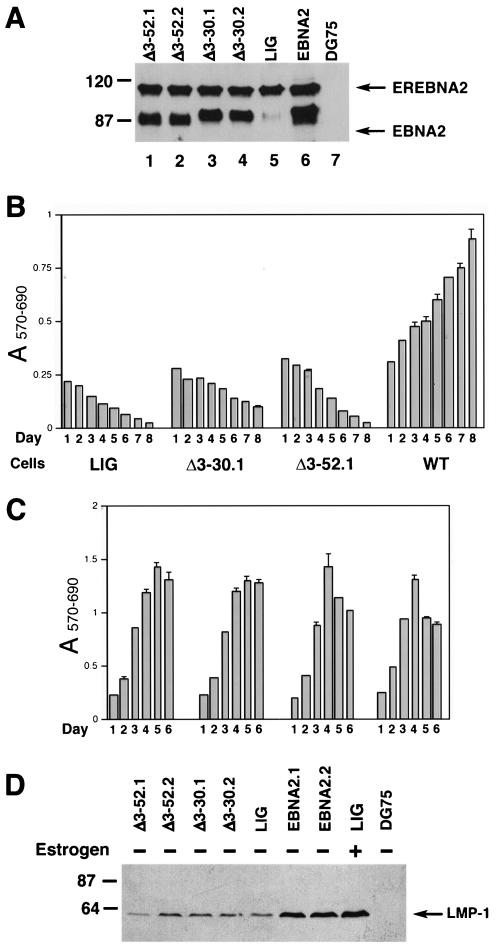
We next performed transcomplementation assays to determine whether the deletion mutants were defective in activating LMP-1 in EBV-infected cells and also to address the ability of the mutants to support cellular proliferation. Two independently generated EREB cell pools expressing each mutant protein were generated, and each expressed an EBNA2 protein that exhibited the expected electrophoretic mobility (Fig. 6A and data not shown). The cell pools were then grown in medium without estrogen and subsequently monitored for their proliferative capacity. Whereas wild-type EBNA2 efficiently supported cell proliferation in the absence of estrogen, deletion mutants Δ3-30 and Δ3-52 failed to do so (Fig. 6B). In addition, neither the Δ3-30 nor Δ3-52 EBNA2 mutant could appreciably maintain LMP-1 expression at day 4 after estrogen withdrawal compared to WTEBNA2 in EREB cells (Fig. 6D). Δ3-30EBNA2 appeared to promote limited survival of EREB cells, as judged by the MTT assay (Fig. 6B), and prolonged persistence of GFP-positive cells under estrogen-free conditions (data not shown). However, even after 2 weeks of culture in the estrogen-free medium, these cell pools failed to produce a detectable outgrowth (data not shown). As a control, we showed that, in the presence of estrogen, all of the cell pools proliferated with compatible efficiency (Fig. 6C). Mutant EBNA2s containing the larger deletion Δ3-97 or deletions from amino acids 3 to 115 and 3 to 156 likewise were unable to complement EBNA2 function upon inactivation of endogenous EREBNA2 by estrogen withdrawal (data not shown). The data from the transcomplementation and LMP-1 promoter transactivation assays using the mutant EBNA2s are summarized in Table 1.
FIG. 6.
Δ3-30EBNA2 and Δ3-52EBNA2 are defective in supporting proliferation of EREB cells following EREBNA2 inactivation. EREB cell pools transduced with the pLIG vector alone (LIG) or pLIG.EBNA2 (WT), as well as two independently generated EREB cell pools transduced with the pLIG.Δ3-30EBNA2 (Δ3-30.1 and Δ3-30.2) or pLIG.Δ3-52EBNA2 (Δ3-52.1 and Δ3-52.2) lentiviruses after enrichment by FACS were grown in the presence of estrogen for 2 weeks prior to the experiment. (A) Western blot analysis of pLIG (LIG), pLIG.EBNA2 (WT), and pLIG.Δ3-30EBNA2 (Δ3-30.1 and Δ3-30.2) and pLIG.Δ3-30EBNA2 (Δ3-52.1 and Δ3-52.2) transduced EREB cell pools using an EBNA2 R3 monoclonal antibody. Equal amounts of cellular protein for each cell pool were analyzed. Molecular mass markers are shown in kilodaltons on the left. (B) At day 0, logarithmically growing cell pools expressing pLIG, pLIG.Δ3-30EBNA2 (Δ3-30.1), or pLIG.Δ3-52EBNA2 (Δ3-52.1) were washed of estrogen and placed in the estrogen-free medium. Twenty-four hours later, the cell pools were plated in 200-μl aliquots of estrogen-free medium in 96-well plates at 2 × 104 cells/well and monitored for survival and proliferation by MTT assay daily over the course of 8 days starting with day 1 (day of plating) after estrogen withdrawal. Optical densities at 570 nm (A570) and 690 nm (A690) of completed MTT reaction mixtures were determined to calculate the A570-690 values by subtracting A690 from A570. (C) Same experiment as for panel B except that cells were grown in medium containing estrogen and proliferation was monitored over the course of 6 days. (D) Δ3-30- and Δ3-52-EBNA2s fail to support expression of the LMP1 gene in EREB cells following EREBNA2 inactivation. EREB cell pools transduced with the pLIG vector alone (LIG), pLIG.EBNA2 (EBNA2.1 and EBNA2.2), pLIG.Δ3-30-EBNA2 (Δ3-30.1 and Δ3-30.2), or pLIG.Δ3-30-EBNA2 (Δ3-52.1 and Δ3-52.2) lentiviruses after enrichment by FACS were grown in the presence of estrogen for 2 weeks prior to the experiment. At day 0, the cells were washed of estrogen and incubated in either estrogen-free (lanes 1 to 7 and 9) or estrogen-containing (lane 8) medium for 4 days. DG75 is an EBV-negative Burkitt's lymphoma cell line. Cell lysates were prepared, and equal amounts of cellular proteins were separated by SDS-PAGE. The separated proteins were transferred to nitrocellulose membranes and probed with monoclonal antibodies against LMP-1. Molecular mass markers are shown in kilodaltons on the left.
TABLE 1.
Stimulation of LMP1 expression and LCL proliferation by mutant EBNA2 proteins
| Mutation | Proliferationa | LMP1 transactivationb |
|---|---|---|
| Δ3-30 | − | − |
| Δ3-52 | − | − |
| Δ3-97 | − | − |
| Δ3-103 | − | − |
| Δ3-115 | NSc | NSc |
| Δ3-156 | NSc | NSc |
| Wild type | ++ | ++ |
| Δ59-103 (ΔPPR)d | ++ | ++++++ |
As determined by the transcomplementation assay.
As determined by transient-cotransfection assays in DG75 and Eli-BL cells.
NS, data not shown.
From reference 14.
EBNA2 amino acids 3 to 30 are impaired for self-association.
Recent studies have suggested that EBNA2 possesses two redundant self-association domains located in the amino-terminal 210 amino acid residues (19). To determine whether amino acid residues 3 to 30 contribute to self-association, we performed coimmunoprecipitation assays from transfected cell extracts expressing a smaller EBNA2 polypeptide truncated for the carboxy-terminal 291 amino acid residues (F199) and either wild-type EBNA2 or EBNA2 deletion proteins. The results indicated that the Δ3-30 mutant EBNA2 was much less efficient at associating with the truncated F199 polypeptide than wild-type EBNA2 (Fig. 7). As expected, a larger deletion in EBNA2 removing amino acid residues 1 to 156 resulted in a polypeptide that was completely unable to coprecipitate with F199. Further studies will be required to determine whether residues 3 to 30 mediate direct EBNA2-EBNA2 interactions or if a cellular intermediate is required.
FIG. 7.
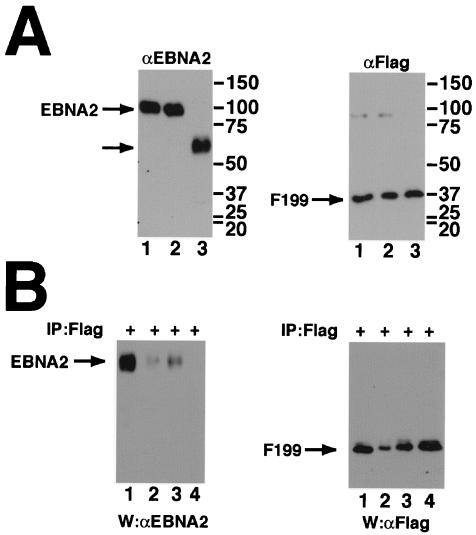
Δ3-30EBNA2 is impaired in its ability to self-associate. EBV-negative DG75 cells were cotransfected with 5 μg each of a plasmid expressing a truncated EBNA2 (F199; amino acids 1 to 199 with a carboxy-terminal Flag epitope tag) and a plasmid expressing wild-type or amino-terminal deletion variants of full-length EBNA2 proteins. (A) Western blots indicating equivalent expression of EBNA2 and F199 in cotransfected cells. The panel on the left was probed with a monoclonal antibody to EBNA2, and the one on the right was probed with a monoclonal antibody to the Flag epitope tag. Equal amounts of transfected cell lysates containing wild-type (lane 1) and amino-terminal deletions Δ3-30EBNA2 and Δ3-156EBNA2 (lanes 2 and 3, respectively) were compared. (B) Cell lysates were precipitated using the M2 anti-Flag antibody (+), divided into two equal parts, resolved by SDS-PAGE, and transferred to nitrocellulose. One blot was probed using the R3 anti-EBNA2 antibody, while the other was probed using an anti-Flag antibody. Migration of EBNA2 and F199 proteins is indicated. Migration of molecular mass standards is shown on the left. Lysates expressing wild-type EBNA2 and F199 (lane 1), expressing Δ3-30EBNA2 and F199 (lanes 2 and 3), and Δ3-156EBNA2 and F199 were tested. Lane 2 was precipitated using 50% of the lysate used in lanes 1, 3, and 4.
DISCUSSION
Our results define a novel domain between amino acid residues 3 and 30 of EBNA2 that is required for stimulation of LMP-1 expression and proliferation of EBV-immortalized cells. This region, which includes CR1, is distinct from other domains required for B-cell immortalization that include the Skip/CBF1 interaction domains located in CRs 5 and 6 and the transactivation domain.
The requirement of an EBNA2 region within amino acids 3 to 30 for transactivation of LMP-1 has been suggested previously in other studies (8, 47). Cohen et al. introduced a linker insertion at EBNA2 amino acid 19 and also deleted amino acids 19 to 34, which lie just outside of CR1 (8). Both mutations resulted in a decreased ability to transactivate LMP-1 and reduced the overall efficiency of B-cell immortalization by recombinant viruses expressing the mutant proteins (8). Sjoblom et al. introduced a deletion in amino acids 2 to 19 and tested the mutant protein in transient-cotransfection assays (47). Interestingly, they observed a marked reduction in the protein's ability to stimulate the LMP-1 promoter that had little overall effect on stimulation of the viral latency C promoter. Our results are the first to fully characterize the role of the first 30 amino acid residues containing CR1 in transient-reporter gene assays for LMP-1 promoter induction, to correlate this activity to LMP-1 expression in LCLs, and to determine that this domain is required for LCL proliferation.
The failure of Δ3-103EBNA2 to upregulate LMP-1 was unexpected, as a similar mutant was reported to transactivate the LMP-1 gene comparable to wild-type EBNA2. The discrepancy between our results and those previously published could be due to differences in the EBNA2 allele used (B95.8 versus strain W91). The W91 EBNA2 PPR is eight proline residues shorter than the B95.8 EBNA2 PPR (hence, Δ2-95 versus Δ3-103) (8, 10). Although our deletion closely matches the Δ2-95EBNA2 mutant described before, it is unclear whether the other single 16 amino acid differences between W91 and B95.8 EBNA2 also contributed to our disparate results. In addition, Yalamanchili et al. used an LMP-1 reporter containing a significantly shorter promoter sequence extending from +40 to −234 (as opposed to +40 to −327, used in our experiments) that may lack an element required for transactivation by wild-type EBNA2 (66). However, this does not seem likely, as various LMP-1 promoter-reporter gene constructs extending from +40 to between −234 and −512 in the LMP-1 promoter sequence exhibit similar EBNA2 responsiveness (32, 55). Our finding that Δ3-103EBNA2 was severely defective in transactivation of the LMP-1 promoter was further strengthened by Δ3-103EBNA2's inability to maintain LMP-1 expression in LCLs (Fig. 2) or to stimulate LMP-1 expression in type I Burkitt's lymphoma cells (Fig. 5), where the entire LMP-1 promoter is present. In addition, all other deletion mutant EBNA2s tested in the present study were unable to stimulate LMP-1 expression (Fig. 4, 5, and 6). While the reporter gene assay results differed between our study and that of Yalamanchili et al., it is quite possible that within the context of primary B-cell infection that the Δ2-95 (W91) EBNA2, like our Δ3-103 (B95-8) EBNA2, may be unable to stimulate LMP-1 expression, giving rise to the null immortalization phenotype observed in the marker rescue assays (66).
Another unexpected finding of our study was that the smaller deletion, mutant Δ3-97EBNA2, which retains seven prolines of the PPR, was unable to complement loss of EBNA2 function in the EREB cells. In marker rescue assays, a similar W91 EBNA2 mutant (Δ2-88) has previously been shown to be immortalization competent, even though it was unable to efficiently stimulate the LMP-1 promoter in reporter gene assays (66). One possible reason for the different outcomes may be due to inherent differences in the transcomplementation and marker rescue assays. Induction of various growth-promoting activities, such as autocrine loops due to epigenetic changes in cellular gene expression, appears to contribute to overall efficiency of immortalization and growth of EBV-infected cells (22, 42). EREB cells may be already epigenetically programmed and restricted in their potential to develop compensatory changes in gene expression, which would not allow survival and outgrowth of mutant EBNA2-expressing cell pools after estrogen withdrawal. It is possible that primary B cells possess an inherently higher potential for epigenetically driven compensatory events, which could make up for the deficiency in the function of the EBNA2 deletion mutant. Since EREBNA2 possesses an intact amino terminus, the established EREB cells may not have been under selective pressure during initial stages of immortalization to develop compensatory epigenetic changes similar to those which may have been required for cells infected with EBVs expressing Δ2-88EBNA2 (W91 strain) in marker rescue assays. Thus, the differences in outcomes between those results reported here versus earlier marker rescue assay results may be due to different physiological and developmental states of the B cells at the initial starting points of the assays.
Based on a coimmunoprecipitation assay, amino acid residues 3 to 30 appear to function as a self-association domain, suggesting that EBNA2 dimerization or multimerization is critical for LMP-1 induction and B-cell immortalization. However, further studies will be required to determine whether this domain facilitates direct EBNA2-EBNA2 interactions or if a cellular cofactor is involved. Given that distinct domains conferring dimerization and multimerization often exist within a polypeptide, additional amino-terminal mutants need to be tested to determine how the domain of amino acids 3 to 30 contributes to these activities.
Based on data presented here, we have refined identification of a domain within the amino terminus of the EBNA2 molecule that is important for induction of LMP-1 expression and LCL proliferation (Fig. 8). The model provides a foundation from which more refined mutagenesis of the amino acid 3 to 30 region can be pursued to determine the precise mechanism by which it contributes to transactivation of viral or cellular genes essential for EBV-mediated transformation.
FIG. 8.
Functional roles of EBNA2 regions critical for immortalization. The carboxy-terminal CRs 5, 6, and 8 function to mimic an activated Notch protein (Notch-IC). The amino-terminal CRs of EBNA2 contribute to EBV-induced immortalization in several ways. The PPR modulates the EBNA2's ability to stimulate LMP-1 expression (14). Residues 3 to 30 containing CR1 comprise a genetic determinant critical for EBNA2 transactivation of the LMP1 gene and for proliferation of EBV-immortalized cells. It remains unknown whether this region is required for transactivation of EBNA2 cellular target genes. The two self-association domains (SAD1 and 2) have previously been mapped to residues 1 to 58 and 97 to 121 in EBNA2.
Acknowledgments
We thank Ron Javier for critical reading of the manuscript.
This work was supported by a National Space Biomedical Research Institute grant NCC (9-58) (cooperative agreement) and ACS grant RPG-00-099-01.
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. (Erratum, 185:946, 1991.) [DOI] [PubMed] [Google Scholar]
- 2.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 3.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. London B 356:437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgstahler, R., B. Kempkes, K. Steube, and M. Lipp. 1995. Expression of the chemokine receptor BLR2/EBI1 is specifically transactivated by Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 215:737-743. [DOI] [PubMed] [Google Scholar]
- 5.Callahan, J., J. Aster, J. Sklar, E. Kieff, and E. S. Robertson. 2000. Intracellular forms of human NOTCH1 interact at distinctly different levels with RBP-jκ in human B and T cells. Leukemia 14:84-92. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. I. 1992. A region of herpes simplex virus VP16 can substitute for a transforming domain of Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 89:8030-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I., and E. Kieff. 1991. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J. Virol. 65:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, J. I., F. Wang, and E. Kieff. 1991. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J. Virol. 65:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordier-Bussat, M., M. Billaud, A. Calender, and G. M. Lenoir. 1993. Epstein-Barr virus (EBV) nuclear-antigen-2-induced up-regulation of CD21 and CD23 molecules is dependent on a permissive cellular context. Int. J. Cancer 53:153-160. [DOI] [PubMed] [Google Scholar]
- 10.Dambaugh, T., K. Hennessy, L. Chamnankit, and E. Kieff. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. USA 81:7632-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirmeier, U., B. Neuhierl, E. Kilger, G. Reisbach, M. L. Sandberg, and W. Hammerschmidt. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982-2989. [PubMed] [Google Scholar]
- 12.Fahraeus, R., A. Jansson, A. Ricksten, A. Sjoblom, and L. Rymo. 1990. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc. Natl. Acad. Sci. USA 87:7390-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes-Panana, E. M., S. Swaminathan, and P. D. Ling. 1999. Transcriptional activation signals found in the Epstein-Barr virus (EBV) latency C promoter are conserved in the latency C promoter sequences from baboon and rhesus monkey EBV-like lymphocryptoviruses (cercopithicine herpesviruses 12 and 15). J. Virol. 73:826-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordadze, A. V., R. Peng, J. Tan, G. Liu, R. Sutton, B. Kempkes, G. W. Bornkamm, and P. D. Ling. 2001. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J. Virol. 75:5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordadze, A. V., D. Poston, and P. D. Ling. 2002. The EBNA2 polyproline region is dispensable for Epstein-Barr virus-mediated immortalization maintenance. J. Virol. 76:7349-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 18.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada, S., R. Yalamanchili, and E. Kieff. 2001. Epstein-Barr virus nuclear protein 2 has at least two N-terminal domains that mediate self-association. J. Virol. 75:2482-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada, S., R. Yalamanchili, and E. Kieff. 1998. Residues 231 to 280 of the Epstein-Barr virus nuclear protein 2 are not essential for primary B-lymphocyte growth transformation. J. Virol. 72:9948-9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 22.Jochems, G. J., M. R. Klein, R. Jordens, D. Pascual-Salcedo, F. van Boxtel-Oosterhof, R. A. van Lier, and W. P. Zeijlemaker. 1991. Heterogeneity in both cytokine production and responsiveness of a panel of monoclonal human Epstein-Barr virus-transformed B-cell lines. Hum. Antibodies Hybridomas 2:57-64. [PubMed] [Google Scholar]
- 23.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kempkes, B., M. Pawlita, U. Zimber-Strobl, G. Eissner, G. Laux, and G. W. Bornkamm. 1995. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-Jκ in a conditional fashion. Virology 214:675-679. [DOI] [PubMed] [Google Scholar]
- 26.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2574. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed., vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 28.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, O. J., and J. L. Yates. 1993. Mutants of Epstein-Barr virus with a selective marker disrupting the TP gene transform B cells and replicate normally in culture. J. Virol. 67:7634-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Larcher, C., B. Kempkes, E. Kremmer, W. M. Prodinger, M. Pawlita, G. W. Bornkamm, and M. P. Dierich. 1995. Expression of Epstein-Barr virus nuclear antigen-2 (EBNA2) induces CD21/CR2 on B and T cell lines and shedding of soluble CD21. Eur. J. Immunol. 25:1713-1719. [DOI] [PubMed] [Google Scholar]
- 32.Laux, G., F. Dugrillon, C. Eckert, B. Adam, S. U. Zimber, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling, P. D., J. J. Ryon, and S. D. Hayward. 1993. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J. Virol. 67:2990-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longnecker, R., C. L. Miller, X. Q. Miao, A. Marchini, and E. Kieff. 1992. The only domain which distinguishes Epstein-Barr virus latent membrane protein 2A (LMP2A) from LMP2B is dispensable for lymphocyte infection and growth transformation in vitro; LMP2A is therefore nonessential. J. Virol. 66:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longnecker, R., C. L. Miller, X. Q. Miao, B. Tomkinson, and E. Kieff. 1993. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J. Virol. 67:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longnecker, R., C. L. Miller, B. Tomkinson, X. Q. Miao, and E. Kieff. 1993. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J. Virol. 67:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, R., A. V. Gordadze, E. M. Fuentes Panana, F. Wang, J. Zong, G. S. Hayward, J. Tan, and P. D. Ling. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J. Virol. 74:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, R., J. Tan, and P. D. Ling. 2000. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed., vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 42.Rochford, R., M. J. Cannon, R. E. Sabbe, K. Adusumilli, G. Picchio, J. M. Glynn, D. J. Noonan, D. E. Mosier, and M. V. Hobbs. 1997. Common and idiosyncratic patterns of cytokine gene expression by Epstein-Barr virus transformed human B cell lines. Viral Immunol. 10:183-195. [DOI] [PubMed] [Google Scholar]
- 43.Rogers, R. P., M. Woisetschlaeger, and S. H. Speck. 1990. Alternative splicing dictates translational start in Epstein-Barr virus transcripts. EMBO J. 9:2273-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakai, T., Y. Taniguchi, K. Tamura, S. Minoguchi, T. Fukuhara, L. J. Strobl, U. Zimber-Strobl, G. W. Bornkamm, and T. Honjo. 1998. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J. Virol. 72:6034-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1995. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc. Natl. Acad. Sci. USA 92:10565-10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sjoblom, A., A. Nerstedt, A. Jansson, and L. Rymo. 1995. Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters. J. Gen. Virol. 76:2669-2678. [DOI] [PubMed] [Google Scholar]
- 48.Spender, L. C., G. H. Cornish, A. Sullivan, and P. J. Farrell. 2002. Expression of transcription factor AML-2 (RUNX3, CBFα-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J. Virol. 76:4919-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strobl, L. J., H. Hofelmayr, G. Marschall, M. Brielmeier, G. W. Bornkamm, and U. Zimber-Strobl. 2000. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J. Virol. 74:1727-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung, N. S., S. Kenney, D. Gutsch, and J. S. Pagano. 1991. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J. Virol. 65:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutton, R. E., H. T. M. Wu, R. Rigg, E. Bohnlein, and P. O. Brown. 1998. Human immunodeficiency virus type I vectors efficiently transduce human hematopoietic stem cells. J. Virol. 72:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swaminathan, S. 1996. Characterization of Epstein-Barr virus recombinants with deletions of the BamHI C promoter. Virology 217:532-541. [DOI] [PubMed] [Google Scholar]
- 53.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong, X., R. Drapkin, R. Yalamanchili, G. Mosialos, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15:4735-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsang, S. F., F. Wang, K. M. Izumi, and E. Kieff. 1991. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein expression. J. Virol. 65:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsui, S., and W. H. Schubach. 1994. Epstein-Barr virus nuclear protein 2A forms oligomers in vitro and in vivo through a region required for B-cell transformation. J. Virol. 68:4287-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voss, M. D., A. Hille, S. Barth, A. Spurk, F. Hennrich, D. Holzer, N. Mueller-Lantzsch, E. Kremmer, and F. A. Grasser. 2001. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J. Virol. 75:11781-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, F., L. Petti, D. Braun, S. Seung, and E. Kieff. 1987. A bicistronic Epstein-Barr virus mRNA encodes two nuclear proteins in latently infected, growth-transformed lymphocytes. J. Virol. 61:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, F., S. F. Tsang, M. G. Kurilla, J. I. Cohen, and E. Kieff. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woisetschlaeger, M., X. W. Jin, C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. 1991. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc. Natl. Acad. Sci. USA 88:3942-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, D. Y., A. Krumm, and W. H. Schubach. 2000. Promoter-specific targeting of human SWI-SNF complex by Epstein-Barr virus nuclear protein 2. J. Virol. 74:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yalamanchili, R., S. Harada, and E. Kieff. 1996. The N-terminal half of EBNA2, except for seven prolines, is not essential for primary B-lymphocyte growth transformation. J. Virol. 70:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yandava, C. N., and S. H. Speck. 1992. Characterization of the deletion and rearrangement in the BamHI C region of the X50-7 Epstein-Barr virus genome, a mutant viral strain which exhibits constitutive BamHI W promoter activity. J. Virol. 66:5646-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoo, L. I., M. Mooney, M. T. Puglielli, and S. H. Speck. 1997. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J. Virol. 71:9134-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, S., M. Fujimuro, J. J. Hsieh, L. Chen, and S. D. Hayward. 2000. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol. 74:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimber-Strobl, U., B. Kempkes, G. Marschall, R. Zeidler, C. Van Kooten, J. Banchereau, G. W. Bornkamm, and W. Hammerschmidt. 1996. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 15:7070-7078. [PMC free article] [PubMed] [Google Scholar]
- 71.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimber-Strobl, U., K. O. Suentzenich, G. Laux, D. Eick, M. Cordier, A. Calender, M. Billaud, G. M. Lenoir, and G. W. Bornkamm. 1991. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J. Virol. 65:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]