Abstract
The RAD27 gene of Saccharomyces cerevisiae encodes a 5′-3′ flap exo/endonuclease, which plays an important role during DNA replication for Okazaki fragment maturation. Genetic studies have shown that RAD27 is not essential for growth, although rad27Δ mutants are temperature sensitive. Moreover, they exhibit increased sensitivity to alkylating agents, enhanced spontaneous recombination, and repetitive DNA instability. The conditional lethality conferred by the rad27Δ mutation indicates that other nuclease(s) can compensate for the absence of Rad27. Indeed, biochemical and genetical analyses indicate that Okazaki fragment processing can be assured by other enzymatic activities or by alternative pathways such as homologous recombination. Here we present the results of a screen that makes use of a synthetic lethality assay to identify functions required for the survival of rad27Δ strains. Altogether, we confirm that all genes of the Rad52 recombinational repair pathway are required for the survival of rad27Δ strains at both permissive (23°C) and semipermissive (30°C) temperatures for growth. We also find that several point mutations that confer weaker phenotypes in mitotic than in meiotic cells (rad50S, mre11s) and additional gene deletions (com1/sae2, srs2) exhibit synthetic lethality with rad27Δ and that rad59Δ exhibits synergistic effects with rad27Δ. This and previous studies indicate that homologous recombination is the primary, but not only, pathway that functions to bypass the replication defects that arise in the absence of the Rad27 protein.
The RAD27/RTH1 gene of Saccharomyces cerevisiae (1, 2) encodes a functional homolog of mammalian FEN1/DNaseIV, a structure-specific 5′-3′ exo/endonuclease (flap endonuclease) that functions in multiple DNA metabolic processes including replication and repair (reviewed in ref. 3). Eukaryotic chromosomal replication is asymmetric, with continuous synthesis of the leading strand and discontinuous synthesis of the lagging strand. The latter involves the formation of numerous Okazaki fragments that consist of a short initiator RNA (7–14 nt) that primes DNA synthesis carried out by polymerase α. After a polymerase switch, this primer is further extended by polymerase δ to the next downstream fragment (reviewed in refs. 4 and 5). To achieve the continuity of the lagging strand, the primer RNA must be removed, as has been demonstrated by in vitro reconstitution experiments showing that simultaneous action of a DNA polymerase, RNase H1, FEN1/Rad27, and DNA ligase I result in correct Okazaki fragment processing (6–8). In vitro studies also suggest that the endonuclease RNase H1 cleaves the initiator RNA 1 nt upstream of the RNA–DNA junction and that the FEN1 nuclease completes the removal of the initiating RNA primer by cleavage of the unannealed 5′ “flap” structure (Fig. 1). The generation of this 5′ tail, which requires strand separation, is likely performed by a DNA helicase such as Dna2, which associates with Rad27 (9, 10), or by a DNA polymerase (4, 5). The role of Rad27 in DNA repair is likely relevant to the resolution of similar flap structures that may form during the processing of certain DNA lesions (reviewed in ref. 4).
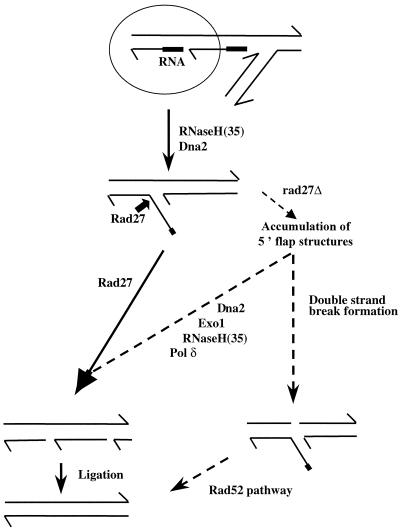
Figure 1.
Simplified steps of Okazaki fragment maturation during DNA lagging strand synthesis (for further details see refs. 3–5). In rad27Δ cells, unprocessed replication intermediates could be resolved either by using an alternative process involving Dna2, Exo1, RNase H(35), and Polδ activities or by the Rad52 recombinational repair pathway.
In S. cerevisiae, genetic studies have shown that RAD27 is not essential for cell survival but that it plays an important role in numerous aspects of DNA metabolism. Several distinct phenotypes that have been identified in rad27Δ mutants include temperature sensitivity for growth (11) (the mutant does not grow at 37°C and cells exhibit an elongated nucleus stretching between mother and daughter cells due to nuclear division defects), sensitivity to UV radiation (2), and increased sensitivity to methyl methanesulfonate (MMS) but not to γ-ray or x-ray irradiation (2, 11, 12). Furthermore, rad27Δ mutants have an elevated rate of spontaneous mutagenesis that is characterized by a unique mutational signature of duplications between separated short direct repeats (12). They also exhibit elevated spontaneous recombination rates (100-fold over wild-type rates) (13), increased microsatellite (14–19), and minisatellite [three tandem repeats of a 20-nt motif (16)], instability, destabilization of telomeric repeats (20), a 4-fold reduction in nonhomologous end joining (21), and a deficiency in base excision repair (22, 23). This constellation of phenotypes, and the physical evidence that single-stranded DNA fragments accumulate during replication in rad27Δ mutants (20, 24), strongly suggests that the Rad27 protein is involved in the correct processing of Okazaki fragments during lagging-strand DNA synthesis, as well as in base excision repair, and emphasizes the biological consequences of disturbances of these processes. However, the viability of the rad27Δ mutant suggests that cells have ways to bypass the rad27Δ defect. The current interpretation of biochemical in vitro and in vivo results supports two classes of bypass mechanisms for processing Okazaki fragments (Fig. 1): partially redundant biochemical activities may act at the replication fork to remove RNA–DNA flaps (class I), and accumulated DNA intermediates may be redirected into alternative repair pathways, in particular the recombinational repair pathway (class II). These possibilities are not mutually exclusive.
Class I bypass process(es) includes several mechanisms for the removal of the initiator RNA; these have been explored by a combination of genetical and biochemical approaches in S. cerevisiae. The current view is that the combined action of RNase H(35) and Rad27 activities in large part accounts for the processing of Okazaki fragments. This process is underscored by the observation that simultaneous deletion of the RNH35 and RAD27 genes has a strong synergistic effect, resulting in extremely slow growth of double mutant cells (25). In the absence of Rad27p alone, it is envisaged that RNase H(35), which normally eliminates the RNA primer up to the last ribonucleotide, could remove the remaining monoribonucleotide at low efficiency, which therefore would facilitate subsequent DNA ligation (25). In addition, a recent study showed that in vitro the human Exo1 protein, a functional homolog of the Schizosaccharomyces pombe and S. cerevisiae Exo1 5′-3′ exonuclease (26, 27), was as efficient in removal of ribonucleotides as deoxynucleotides (28). This result and the observation that the exo1Δ rad27Δ double mutant exhibits synthetic lethality or sublethality at 30°C (29) indicates that Exo1p (also involved in mismatch repair) is a good candidate to allow the survival of the rad27Δ single mutant and rad27Δ rnh35Δ double mutant (28). Other structure-specific nucleases that may be functionally redundant also have been investigated. The Fen1/Rad27 protein belongs to a family of nucleases and is related to the S. cerevisiae exonucleases Rad2 (30) and Exo1 (27, 31), which are implicated in repair and recombination, as well as to two other proteins, Yen1 of unknown function, and Din7 with a mitochondrial function (32). Single or multiple deletions of the RAD27, RAD2, YEN1, and DIN7 genes were not found to confer synthetic lethality or synergistic effects on cell viability, indicating that none of these genes substitutes for the rad27Δ deficiency (22, 33). Additional studies have shown that overexpression of Exo1 (29) or Dna2, which has helicase (34) and flap endonuclease activity (10, 35), can compensate for the rad27Δ growth defect at 37°C. This result suggests that these proteins, alone or with other partners, are able to overcome the replication defect, either by their direct action on replication intermediates (reviewed in ref. 4) or as essential players in the alternative pathway(s). In addition, the synthetic lethality of rad27Δ with the pol3–01 mutation affecting the 5′-3′ exonuclease proofreading domain of DNA polymerase δ also suggests an additional mean to process the intermediate DNA structures accumulating at the border between two Okazaki fragments (16, 36).
Class II processes include those that compensate for the defect of rad27Δ by channeling blocked replication intermediates into the homologous recombinational repair pathway. This proposal is consistent with the hyperrecombinogenic phenotype of rad27Δ cells and the significant result that rad27Δ exhibits synthetic lethality in combination with deletions of several genes of the Rad52 epistasis group (12, 37). The colethality of the rad27Δ mutation with deletions of several genes of DNA damage checkpoint pathways, including RAD9, RAD17, RAD24, and MEC3 (13, 38), is also consistent with the accumulation of single-stranded DNA in rad27Δ cells (21), and possibly of double-strand breaks (DSBs) (12), which must be processed by the DNA repair machinery to avoid the lethal segregation of damaged or broken chromosomes. In this paper, we review and present additional evidence, based on extensive synthetic lethality approaches, for the essential role of the homologous recombinational-repair pathway (Rad52 pathway) in the survival of rad27Δ mutants. This analysis provides an extended example of the links between replication, repair, and recombination processes.
Materials and Methods
Media and Sporulation Conditions.
Growth and sporulation of yeast cells were performed by standard methods (39). Standard medium (yeast extract/peptone/dextrose) was used for vegetative growth. For sporulation, cells were grown in presporulation media at 23°C or 30°C, washed in water, resuspended, and incubated in sporulation medium (1% potassium acetate supplemented with required amino acids) at 23°C or 30°C.
Yeast Strains.
The names of diploids used in this study are shown in Tables 1 and 2. The origin and complete genotype of each can be supplied on request. In most cases, strains are of the S288C background, including the collection of single-deletion strains derived from FY1679 [MATa/MATα, ura3–52/ura3–52, trp1Δ63/TRP1, leu2Δ1/LEU2, his3Δ200/HIS3 (40)] and used in the EUROFAN deletion project, and the collection derived from BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) strains (41) and used for the international systematic S. cerevisiae gene disruption project (42). In FY and BY strains (obtained from EUROSCARF, Frankfurt), deleted genes are replaced with the KanMX marker (providing resistance to G418). These strains were found to be highly compatible with our laboratory MGD strains, which also share the S288C background (43), based on the high viability of meiotic products from hybrid crosses. All mre11 strains (44, 45) are of the SK1 background. The sgs1Δ strain, provided by S. Gangloff (Commissariat à l'Energie Atomique, Paris), is a W303 derivative. The rad27Δ strains used for synthetic lethality assays are FW2612 [MATα rad27∷HIS3 (12)], FW2617 [MATa rad27∷HIS3 (12)], and LSY702–2A (MATa rad27∷TRP1) and LSY702–6B (MATα rad27∷TRP1), which are W303 derivatives (37). We constructed the SK1 strains ORD5902–1A (MATa rad27∷URA3, ura3, trp1, arg4ΔHpa1) and ORD5902–1C (MATα rad27∷URA3, ura3) by one-step gene replacement of the RAD27 locus with the rad27∷URA3 EcoRI–SphI restriction fragment of plasmid pMRΔrad27∷URA3 (2). All RAD27 disruptions in this study were verified by Southern blot analysis, and the expected relevant phenotypic traits, such as cell division morphology, temperature sensitivity, and colethality with a rad51 deletion, also were confirmed.
Table 1.
Genetic interactions of rad27Δ
| Mutation assayed | Diploid strain | Growth temperature, °C | Results of tetrad analysis
|
Nature of genetic interaction with rad27Δ | |||
|---|---|---|---|---|---|---|---|
| Number of spores
| |||||||
| Wild type | rad27Δ single mutant | xΔ single mutant | rad27ΔxΔ double-mutant* | ||||
| rad51Δ | ORD 5324 | 30 | 15 | 12 | 14 | 0 | Colethal |
| ORD 5324 | 23 | 22 | 22 | 23 | 0 | Colethal | |
| rad52Δ | ORD 5377 | 30 | 13 | 7 | 11 | 1* | Colethal |
| ORD 7108 | 23 | 15 | 26 | 24 | 0 | Colethal | |
| rad54Δ | ORD 4887 | 30 | 10 | 8 | 26 | 5* | Colethal |
| ORD 5694 | 30 | 18 | 21 | 23 | 0 | Colethal | |
| ORD 5694 | 23 | 19 | 28 | 29 | 0 | Colethal | |
| rad55Δ | ORD 4886 | 30 | 9 | 16 | 31 | 0 | Colethal |
| ORD 5322 | 23 | 29 | 25 | 31 | 0 | Colethal | |
| rad57Δ | ORD 4883 | 30 | 17 | 14 | 16 | 0 | Colethal |
| ORD 5309 | 23 | 16 | 20 | 18 | 0 | Colethal | |
| rad50Δ | ORD 5314 | 30 | 9 | 12 | 11 | 1* | Colethal |
| ORD 5314 | 23 | 15 | 17 | 20 | 0 | Colethal | |
| rad50S-KE6 | ORD 5397 | 30 | 15 | 17 | 18 | 0 | Colethal |
| rad50S-KI81 | ORD 5312 | 30 | 10 | 10 | 13 | 1* | Colethal |
| rad50S-QK99 | ORD 5904 | 30 | 11 | 7 | 7 | 0 | Colethal |
| mre11Δ | ORD 5311 | 30 | 15 | 18 | 16 | 0 | Colethal |
| ORD 5311 | 23 | 19 | 23 | 24 | 0 | Colethal | |
| xrs2Δ | ORD 5313 | 30 | 9 | 15 | 11 | 3* | Colethal |
| ORD 5313 | 23 | 7 | 28 | 24 | 2** | Colethal | |
| com1/sae2Δ | ORD 5347 | 30 | 12 | 13 | 10 | 0 | Colethal |
| ORD 5688 | 30 | 11 | 13 | 12 | 0 | Colethal | |
| srs2Δ | ORD 5308 | 30 | 26 | 28 | 37 | 0 | Colethal |
| ORD 5663 | 30 | 23 | 23 | 22 | 1** | Colethal | |
| ORD 5663 | 23 | 22 | 22 | 26 | 3** | Colethal | |
| sgs1Δ | ORD 5380 | 30 | 6 | 8 | 14 | 3* | Viable |
| rad59Δ | ORD 5672 | 30 | 22 | 21 | 21 | 0 | Colethal |
| ORD 5672 | 23 | 19 | 24 | 25 | 20 | Synergistic | |
In the case of tetrad analysis giving rise to a low number of double-mutant (*), the conclusion (colethal or viable) is based on the results of an additional random spore analysis in which we determined the number of viable spores with the double-mutant marker phenotype among spores selected for either the rad27Δ or the xΔ marker (see Materials and Methods). The results, expressed as the ratio of rad27ΔxΔ/rad27Δ or rad27ΔxΔ/xΔ, were as follows: ORD 5377, 0/113; ORD 4887, 0/100; ORD 5314, 1/72; ORD 5312, 3/63; ORD 5313, 9/137; ORD 5380, 56/145. In the cases of ORD 5313 (23°C) and ORD 5663 (30°C and 23°C), additional random spore analysis was not performed (marked by the double asterisk).
Table 2.
Genetic interactions of rad27Δ
| Mutation assayed | Diploid strain | Growth temperature, °C | Results of tetrad analysis
|
Nature of genetic interaction with rad27Δ | |||
|---|---|---|---|---|---|---|---|
| Number of viable spores per tetrad (number of tetrads)
| |||||||
| 4 | 3 | 2 | 1+0 | ||||
| rad52-Δ251 | ORD 5946 | 30 | 3 | 9 | 3 | 2 | Colethal |
| rad52-Δ327 | ORD 5947 | 30 | 6 | 17 | 13 | 0 | Colethal |
| mre11-D16A | ORD 5936 | 30 | 5 | 25 | 9 | 0 | Colethal |
| mre11-6 | ORD 5916 | 30 | 4 | 32 | 11 | 2 | Colethal |
| mre11-58S | ORD 5918 | 30 | 3 | 11 | 6 | 0 | Colethal |
| mre11-5 | ORD 5917 | 30 | 16 | 0 | 4 | 0 | Viable |
| mre11ΔC49 | ORD 5938 | 30 | 25 | 13 | 2 | 0 | Viable |
| rfa1-t11 | ORD 5310 | 30 | 4 | 21 | 10 | 0 | Colethal |
| ORD 5310 | 23 | 3 | 16 | 5 | 0 | Colethal | |
| rfa1-t48 | ORD 5319 | 30 | 0/32 rad27Δ colonies were double-mutant | Colethal | |||
| ORD 5319 | 23 | 0/23 rad27Δ colonies were double-mutant | Colethal | ||||
Results of sporulation of diploid in which the rad27 deletion and the x mutation could not be individually distinguished (see Materials and Methods). The data indicate the number of viable spores per tetrad.
Synthetic Lethality Assay.
Haploid strains containing a null allele of RAD27 (rad27∷HIS3, rad27∷URA3, or rad27∷TRP1) were crossed to appropriate strains containing a point mutation or a deletion of the gene to be assayed (gene X). The resulting heterozygotes (RAD27/rad27Δ X/xΔ) then were sporulated at 23°C or 30°C, and the meiotic products were analyzed. A minimal sample of at least 10 tetrads was dissected, and the haploid colonies were genotyped with respect to the biosynthetic and G418R markers. A high frequency of spores that form viable haploid colonies, as well as the presence of double mutant colonies (≈25% of the total), indicates that the function of gene X is not required for the survival of a rad27Δ strain. In contrast, the failure of a significant number of colonies (≈25%) to develop, and the presence of two or three viable haploids per tetrad (tetrads ratio could be made available on request), none of which has the double mutant genotype (rad27Δ xΔ), is indicative of synthetic lethality. In these cases, or when the general spore viability was low due to the poor germination of the single mutants, or when an exceptional colony with a double mutant phenotype was obtained (marked by an asterisk in Table 1), more tetrads were dissected and random spore analyses were performed to increase the sample size. In random spore analysis, we determined the ratio of viable spores with the double-mutant marker phenotype (rad27Δ xΔ) among spores first selected for one single mutant marker, either rad27Δ or xΔ. If the double mutant is viable, the expected ratio of doubly mutant to singly mutant haploids is 1/2, and much lower if the two mutations confer synthetic lethality (“colethal”). The results for progeny of diploids in which both single mutations were differently marked, so that all four haploid products (wild type, rad27Δ, xΔ, and rad27Δ xΔ) could be individually identified, are grouped in Table 1. When both mutations were similarly marked or one of them was not genetically marked, we could only analyze spore viability per tetrad, and these data are grouped in Table 2. Colethality is deduced by the presence of numerous tetrads with only two or three viable spores. To test the rfa1-t48 mutation, the parental strain with a replacement of the RFA1 gene by the TRP1 marker and the complementing rfa1-t48 mutation (46) on the replicative plasmid pKU1 (with the LEU2 marker), was crossed to a rad27Δ∷HIS3 haploid. After sporulation, the haploid colonies were genotyped, and the absence of His+ (rad27Δ), Trp+ (rfa1Δ), and Leu+ (rfa1-t48) colonies was indicative of the synthetic lethality of the rad27Δ rfa1-t48 double mutant.
Results and Discussion
Synthetic Lethality Assay.
To identify functions that are required for the viability of cells defective for RAD27, we have performed analyses of synthetic lethality by testing whether haploid cells containing a rad27Δ null allele in combination with a deletion or point mutation of the tested gene X are viable. As described in Materials and Methods, for each gene to be assayed, we constructed diploids heterozygous for both mutations and carried out tetrad analysis at both 23°C (a permissive temperature for growth of the rad27Δ single mutant) and 30°C (a semipermissive temperature). The absence of the double mutant genotype (rad27Δ xΔ) among the haploid products indicates a case of synthetic lethality. For viable double mutant haploids, an examination of the growth phenotype of the double mutant in comparison to each single mutant allows synergistic effects to be detected.
Absolute Requirement of the Rad52 Recombinational Repair Pathway for the Viability of rad27Δ Cells.
Numerous studies in S. cerevisiae have described the central role of the Rad52 recombinational repair pathway for the repair of DNA damage caused by ionizing radiation as well as for homologous recombination (reviewed in refs. 47 and 48). Strains with deletions of any gene of the Rad52 pathway (RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, MRE11, or XRS2) are viable but highly sensitive to ionizing radiation and to the radiomimetic compound MMS, and are deficient in DNA DSB recombinational repair (reviewed in ref. 49). Previous studies (12, 37) demonstrated that the rad27Δ mutation confers synthetic lethality in combination with null mutations in Rad52 group genes. This result indicates that homologous recombination is crucial in compensating for the rad27Δ replication defect. The results of our parallel synthetic lethal study confirmed all these observations. Sporulated double mutant heterozygotes yielded a significant fraction of tetrads with only two or three viable colonies after growth at 23°C and 30°C, and spore inviability systematically correlated with the absence of the double mutant phenotype, as expected for lethality of two unlinked mutations (Table 1). Altogether, this set of results obtained in two different genetic backgrounds (S288C and W303) demonstrates that the process(es) that allows the rad27Δ growth defect to be bypassed at permissive temperature absolutely depends on homologous recombination, as defined by the activity of the Rad52 pathway.
Synthetic Lethality of rad27Δ with Core Alleles of Rad52p.
Biochemical studies have shown that yeast Rad52p binds DNA and promotes annealing of complementary single-stranded DNA (50), and that it works with the single-strand DNA binding activity of replication protein A (RPA, see below) to promote Rad51p-mediated strand exchange (51). Numerous rad52 mutations have been characterized for their effects on mitotic and meiotic DSB repair in S. cerevisiae (50). To further explore the requirement of Rad52p for survival of rad27Δ cells, we focused on the rad52-Δ251 and rad52-Δ327 truncation alleles, which encode proteins that consist of the N-terminal 251 and 327 aa of the Rad52p polypeptide, respectively, and that have residual Rad52 activities (52). These mutants are ≈80- and 200-fold more resistant to MMS exposure than are rad52Δ null mutants but they are not fully wild type. As reported in Table 2, we found that these mutations were not viable in combination with the rad27Δ mutation, indicating that partial Rad52p repair activities are not sufficient to compensate for the rad27Δ defect. There are several possibilities to account for this insufficiency: (i) these truncated proteins lack putative Rad51 binding sites, which have been mapped to the Rad52 COOH-terminal region; (ii) they may be unable to repair specific types of DNA lesion formed in the absence of Rad27 (such as single-strand gaps or DSBs); or (iii) they may be unable to process the excessive number of “potentially reparable” lesions that likely accumulate upstream of each Okazaki fragment during lagging strand synthesis in rad27Δ cells. Additional experiments, including the systematic screening of all known rad52 group mutations in this hypersensitive rad27Δ background, are required to distinguish among these hypotheses.
Synthetic Lethality of rad27Δ with Separation of Function Alleles of Rad50 and Mre11, and Deletion of the COM1/SAE2 Gene.
The heterotrimeric Mre11/Rad50/Xrs2 protein complex is a key element in various aspects of DNA metabolism (reviewed in ref. 53). Null mutation of each gene confers extreme hypersensitivity to ionizing radiation and MMS, enhances spontaneous recombination, retards the single-stranded resection of DSBs induced by the HO endonuclease, and abolishes DSB formation in meiosis. Moreover, the Mre11/Rad50/Xrs2 complex is involved in nonhomologous end joining and telomere maintenance. To gain further insight into the genetic interactions between this complex and Rad27, we examined the compatibility of the rad27Δ mutation with several mutations in RAD50 and MRE11.
Analysis of the RAD50 gene has led to the identification of several non-null mutations that define “separation-of-function” alleles (rad50S). The affected amino acids map in the N-terminal ATP binding domain of Rad50p (54). During vegetative growth, rad50S cells are substantially wild type with normal level of spontaneous recombination and only weak MMS sensitivity, but during meiosis, rad50S diploids have a more severe phenotype in that they are totally defective in recombination between homologues. In contrast to the rad50Δ null allele that abolishes meiotic DSB formation, the rad50S mutation allows DSB formation but blocks DSB processing by resection (54, 55). This is caused by a defect in the removal of the Spo11p transesterase (56, 57), which remains covalently attached to the 5′ end of DSBs (57). Table 1 shows the result of our analyses of the compatibility of the rad27Δ mutation with three representative rad50S mutations, rad50S-KE6, rad50S-KI81, and rad50S-QK99 (54). Surprisingly, we found that each of these mutations is synthetically lethal with rad27Δ; the mitotic lethality conferred by the combination of the two mutations contrast with the more subtle phenotype of rad50S RAD27 cells.
The above observation and the previous observation that mre11-H125N is synthetically lethal with rad27Δ (58) prompted us to examine additional separation-of-function mutations of the MRE11 gene, which confer a phenotype similar to that of rad50S mutants. In vitro and in vivo studies indicate that the Mre11 protein has two distinct functional domains: the N-terminal phosphoesterase domain, which is required for single-stranded processing of DSBs, and the C-terminal double-strand DNA binding domain, which is essential for DSB formation in meiosis (44, 45, 58–60). The presence of discrete functional domains correlates with the genetic observation of separation-of-function mutations. One class of mutants exhibits a deficiency in processing of DSBs and is represented by the mre11-D16A, mre11–6 (which results in a deletion of amino acid residues 410–421 from the mature protein) and mre11–58S (H213Y) mutations. The second class of mutants is represented by the mre11–5 (a deletion of 136-aa residues from the C terminus) and mre11ΔC49 (a deletion of 49-aa residues from the C terminus) mutations, which remove the DNA binding domain B required for meiotic DSB formation. As indicated in Table 2, we observed that as observed for a complete deletion of MRE11, the mre11-D16A, mre11–6, and mre11–58S alleles are synthetically lethal with rad27Δ, as is the mre11-H125N mutation (58). In contrast, mre11–5 rad27Δ and mre11ΔC49 rad27Δ strains are viable and do not exhibit a synergistic growth phenotype. Altogether, these results can be interpreted with respect to the distinct functional domains of Mre11 to indicate that the nuclease activity of Mre11 is essential for survival in the absence of Rad27, as previously discussed (58), and they shed further light on the role of Mre11 in homologous recombination. Notably, these results also show that the two mutants (mre11–5 and mre11ΔC49) that are as resistant as wild type to MMS exposure (44, 60), are also viable when they have the rad27Δ mutation, whereas the three mutants that are inviable when they also contain the rad27Δ mutation are more sensitive to MMS than wild type, although to various extents. That is, the mre11–58 strain is as sensitive as the mre11Δ mutant but the mre11D16A and mre11–6 strains exhibit an MMS sensibility intermediate between that of the wild-type and deletion strains (44, 60).
Strains with deletions of the COM1/SAE2 gene are phenotypically similar to rad50S and mre11S mutants (61, 62). During vegetative growth, com1/sae2Δ mutants exhibit a wild-type growth rate at all temperatures and are not sensitive to γ-rays, but they are weakly sensitive to MMS (like rad50S-KI81 mutants) and undergo a slightly elevated level of spontaneous mitotic intragenic recombination. During meiosis, the com1/sae2Δ mutation prevents the processing of DSB ends. We found that this mutation is synthetically lethal with rad27Δ at 30°C (Table 1).
In summary, these analyses indicate that the nuclease activity of the Rad50/Mre11/Xrs2 complex and the still unknown function of the Com1/Sae2 protein are essential for the survival of rad27Δ cells. Moreover, this experimental approach illustrates how the rad27Δ genetic background can exacerbate the effect of mutations, which otherwise confer only a weak phenotype—such as MMS sensitivity—in a RAD27 background. One interpretation of the hypersensitive rad27Δ background is that rad27 cells accumulate so many DNA interruptions or lesions that the capacity of alternative pathways, including the recombinational repair pathway, is saturated.
Synthetic Lethality of rad27Δ with Separation of Function Alleles of RPA.
To further define functions allowing survival of rad27Δ cells, we examined their requirement for the activity of the single-stranded DNA binding RPA, which is involved in various aspects of DNA replication, repair, and recombination (reviewed in ref. 63). This multisubunit complex contains three polypeptides encoded by the essential genes RFA1, RFA2, and RFA3. The large subunit (Rfa1) contains the major single-stranded DNA binding activity, and in vitro studies clearly indicate that RPA stimulates Rad51-dependent strand exchange during homologous recombination, similar to the way that the Escherichia coli single-strand binding protein stimulates RecA-mediated strand transfer activities (reviewed in refs. 64 and 65). Several genetic screens have yielded viable rfa1 mutants that have been characterized with respect to temperature sensitivity, UV and MMS sensitivity, mutability, and recombination efficiency (46, 66, 67). Among these are several separation-of-function mutants that have allowed the numerous functions of this protein complex to be analyzed. Two of these, the rfa1-t11 (Lys-45–Glu) and rfa1-t48 (Leu-221–Pro) mutants, are viable and proficient for DNA replication, but are strongly sensitive to MMS treatment (2,000-fold more sensitive than wild-type strains) and deficient for HO-induced DSB repair (46). Notably, the rfa1-t11 mutation has no effect on the kinetics of resection of HO-generated DSB ends, in contrast to the delay in processing conferred by mre11, rad50, or xrs2 null mutations (68). As shown in Table 2, we did not recover viable rfa1-t11 rad27Δ or rfa1-t48 rad27Δ double mutants. This result indicates that RPA activity is required for rad27Δ survival, most likely because of its role in recombinational repair, because these mutants are inefficient in DSB repair rather than in replication.
SRS2 but Not SGS1 Is Required for the Viability of rad27Δ.
The interpretation that the absence of the Rad27 function leads to the accumulation of unprocessed DNA replication intermediates that can be repaired by homologous recombination raises the question whether this repair may require the SRS2 gene, which encodes a 3′-5′ DNA helicase (69). The characterization of srs2Δ phenotypes observed in homozygous diploids (sensitivity to both ionizing and UV radiation, sensitivity to MMS, spontaneous hyperrecombination) and their suppression by several mutations in genes of the Rad52 pathway suggested that SRS2 has a role in reversing aberrant recombination structures (70, 71). We created rad27Δ srs2Δ heterozygotes but did not recover the double mutant among their haploid progeny (Table 1), indicating that Srs2p contributes to the survival of rad27 defective cells. One interpretation is that Srs2p channels aberrant replicative DNA structures formed in the absence of Rad27 toward the recombinational repair pathway. An alternative and not mutually exclusive hypothesis is that Srs2p participates in the reversal of these structures, possibly through its interaction with the Pol32 subunit of polymerase δ (72). The requirement for Srs2 also indicates that its function is not compensated for by the activity of a redundant endogeneous DNA helicase such as the Dna2 helicase, which is essential for replication (9, 34, 35). However, the synthetic lethality of the rad27Δ mutation with the temperature-sensitive dna2–1 allele, the suppression of the temperature-sensitive growth defect of rad27Δ strains by overexpression of the Dna2–1 protein (and vice versa) (9), and biochemical evidence for an interaction between the Rad27 and Dna2 proteins (9) strongly suggest that Dna2 is essential for the viability of rad27Δ cells. The role of Dna2, which remains to be elucidated, may depend on its helicase activity or its flap endonuclease activity (10, 36), the latter of which may biochemically compensate for the absence of Rad27 protein. We also examined whether the Sgs1 helicase (73) is required for the viability of rad27Δ cells and found that rad27Δ sgs1Δ haploids are viable and do not exibit a synergistic growth phenotype.
Synergisitic Growth Defect of the rad27Δ rad59Δ Double Mutant at 23°C.
The Rad59 protein shares partial sequence homology with Rad52p and is required for Rad51-independent mitotic recombination (74). A previous study using strains of the W303 background (37) reported the surprising result that rad27Δ is not viable in combination with a deletion of the RAD59 gene at 30°C, although rad59Δ single mutants are more resistant to ionizing radiation than are other mutants of the Rad52 epistasis group. We also examined genetic interactions between the rad27Δ and rad59Δ mutations and confirmed that the vast majority of double mutant cells are inviable at 30°C. In one case, a single spore (of 24 presumed to have the rad27Δ rad59Δ genotype) gave rise to a slowly growing haploid colony. However, at 23°C, all putative double mutant spores germinated and grew very slowly. Therefore, our results indicate that Rad59 is not strictly required for survival of rad27Δ mutants, in contrast to what is seen for all other members of the Rad52 group, at least in the S288C background. In summary, both studies strongly suggest that the process that allows the Rad27 requirement to be bypassed requires the activity of Rad59. The current view that Rad59p is involved in sister chromatid recombination is consistent with the hypothesis that lesions that may accumulate during replication might be repaired by homologous recombination involving the replicated sister chromatid in a Rad52- and Rad59-dependent pathway.
Concluding Remarks
The Rad27 protein plays an important role in DNA replication for Okazaki fragment maturation, in particular by removing the RNA–DNA primer that initiates lagging strand synthesis. Indeed, in vivo analyses demonstrated that single-stranded DNA fragments accumulate in rad27Δ cells during replication. The extensive synthetic lethality assays reported here and in other studies favor two classes of bypass process: (i) the RNA primers may be removed by the similar biochemical activities of other proteins that act at the replication fork [RNase H(35), Exo1, Dna2, Polδ], or (ii) the improperly processed Okazaki fragments may be channeled into the Rad52 recombinational repair pathway. This pathway must be fully intact to compensate for rad27Δ defects, as shown here by synthetic lethality conferred by rad27Δ and mutations such as rad52-Δ251, rad52-Δ327, rfa-t11, rad50S, mre11S, or com1/sge2. Those mutations confer only a leaky or weak phenotype in RAD27 cells. The mechanism by which the Rad52 pathway functions to rescue mutants with defects in DNA replication remains to be elucidated, but the current data are consistent with the hypothesis that postreplicative recombinational repair activity is necessary to repair any single-stranded nicks, gaps, or secondarily formed DSBs (12, 75), which may arise (by replication and/or base-excision repair defects) and therefore to allow complete replication of the genome. This hypothesis is also consistent with the previous observation of the colethality of rad27Δ with mutations in genes required for DNA damage checkpoint pathways, including RAD9, RAD17, RAD24, and MEC3 (13, 38). These checkpoint functions ensure the orderly progression of the cell cycle in response to replication blocks or to the presence of unprocessed DNA lesions. Those data also emphasize that the hypersensitive background created by the rad27Δ defect allows us to easily screen for mutations in genes that function in DNA metabolism. Such mutations may confer an imperceptible phenotype in a RAD27 background. Future studies should further describe the overlapping and complementary role of the various events that allow the absence of the Rad27/FEN1 flap endonuclease during replication to be bypassed.
Acknowledgments
We thank B. de Massy, F. Fabre, E. Friedberg, S. Gangloff, S. Huang, D. Livingston, R. Kolodner, N. Kleckner, H. Ogawa, T. Ogawa, K. Ohta, M. Reagan, V. Rocco, C. Soustelle, L. Symington, and members of the EUROFAN and Yeast International Consortium for providing plasmids and yeast strains used in this study. We thank all members of our laboratory for helpful discussion, C. Mézard for critical reading of the manuscript, and K. Smith for English corrections. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS UMR144), the Institut Curie, the Association pour la Recherche sur le Cancer, and the Ministère de la Défense. H.D. was successively supported by a postdoctoral fellowship from the Ministère Education Nationale de la Recherche et de la Technologie and the Association pour la Recherche sur le Cancer. J.L. is supported by a postdoctoral fellowship from the Ligue Nationale contre le Cancer.
Abbreviations
- MMS
methyl methanesulfonate
- DSB
double-strand break
- RPA
replication protein A
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Jacquier A, Legrain P, Dujon B. Yeast. 1992;8:121–132. doi: 10.1002/yea.320080207. [DOI] [PubMed] [Google Scholar]
- 2.Reagan M S, Pittenberg C, Siede W, Friedberg E C. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieber M R. BioEssays. 1997;19:233–239. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 4.Bambara R A, Murante R S, Henricksen L A. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 5.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 6.Turchi J J, Bambara R A. J Biol Chem. 1993;268:15136–15141. [PubMed] [Google Scholar]
- 7.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 8.Waga S, Bauer G, Stillman B. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 9.Budd M E, Campbell J L. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae S-H, Choi E, Lee K-H, Park J S, Lee S-H, Seo Y-S. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 11.Sommers C H, Miller E J, Dujon B, Parakash S, Prakash L. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 12.Tishkoff D X, Filosi N, Gaida G M, Kolodner R. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 13.Vallen E A, Cross F R. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R E, Kovvali G K, Prakash L, Prakash S. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 15.Freudenreich C H, Kantrow S M, Zakian V A. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 16.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweitzer J K, Livingston D M. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 18.Richard G-F, Dujon B, Haber J E. Mol Gen Genet. 1999;261:871–882. doi: 10.1007/s004380050031. [DOI] [PubMed] [Google Scholar]
- 19.White P J, Borts R H, Hirst M C. Mol Cell Biol. 1999;19:5675–5684. doi: 10.1128/mcb.19.8.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parenteau J, Wellinger R J. Mol Cell Biol. 1999;19:4243–4252. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Wilson T E, Lieber M R. Proc Natl Acad Sci USA. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson R E, Kovvali G K, Prakash L, Prakash S. Curr Genet. 1998;34:21–29. doi: 10.1007/s002940050362. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Wang Z. Nucleic Acids Res. 1999;27:956–962. doi: 10.1093/nar/27.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merill J B, Holm C. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szankasi P, Smith G R. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 27.Huang K N, Symington L S. Mol Cell Biol. 1993;13:3125–3134. doi: 10.1128/mcb.13.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu J, Qian Y, Chen V, Guan M-X, Shen B. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 29.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington J J, Lieber M R. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 31.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fikus M U, Mieczkowski P A, Koprowski P, Rytka J, Sledziewska G E, Ciesla Z. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mieczkowski P A, Fikus M U, Ciesla Z. Mol Gen Genet. 1997;253:655–665. doi: 10.1007/s004380050369. [DOI] [PubMed] [Google Scholar]
- 34.Budd M E, Chloe W-C, Campbell J L. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 35.Bae S, Seo Y. J Biol Chem. 2000;48:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 36.Gary R, Park M S, Nolan J P, Cornelius H L, Kozyreva O G, Tran H T, Lobachev K S, Resnick M A, Gordenin D A. Mol Cell Biol. 1999;19:5373–5382. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Symington L S. Nucleic Acids Res. 1998;26:5589–5595. doi: 10.1093/nar/26.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulovich A G, Armour C D, Hartwell L H. Genetics. 1998;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Massy B, Nicolas A. EMBO J. 1993;12:1459–1466. doi: 10.1002/j.1460-2075.1993.tb05789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winston F, Dollard C, Ricupero-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 41.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 42.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 43.Rocco V, de Massy B, Nicolas A. Proc Natl Acad Sci USA. 1992;89:12068–12072. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 46.Umezu K, Suguwara N, Chen C, Haber J, Kolodner R. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 48.Shinohara A, Ogawa T. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 49.Game J C. In: Yeast Genetics: Fundamental and Applied Aspects. Spencer J F T, Spencer D M, Smith A R M, editors. New York: Springer; 1983. pp. 109–137. [Google Scholar]
- 50.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 52.Asleson E N, Okagaki R J, Livingston D M. Genetics. 1999;153:681–692. doi: 10.1093/genetics/153.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haber J E. Trends Genet. 1998;14:317–321. doi: 10.1016/s0168-9525(98)01501-7. [DOI] [PubMed] [Google Scholar]
- 54.Alani E, Padmore R, Kleckner N. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 55.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 56.Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. Nature (London) 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 57.Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 58.Nairz K, Klein F. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsubouchi H, Ogawa H. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreau S, Ferguson J R, Symington L S. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prinz S, Amon A, Klein F. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKee A H Z, Kleckner N. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 64.Radding C M. Curr Biol. 1993;3:358–360. doi: 10.1016/0960-9822(93)90200-8. [DOI] [PubMed] [Google Scholar]
- 65.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith J, Rothstein R. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hays S L, Firmenich A A, Massey P, Banerjee R, Berg P. Mol Cell Biol. 1998;18:4400–4406. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R, Haber J E. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 69.Rong L, Klein H L. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- 70.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chanet R, Heude M, Adjiri A, Maloisel L, Fabre F. Mol Cell Biol. 1996;16:4782–4789. doi: 10.1128/mcb.16.9.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang M-E, de Calignon A, Nicolas A, Galibert F. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 73.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bai Y, Symington L S. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 75.Flores-Rozas H, Kolodner R D. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]