Abstract
Earlier studies have shown that herpes simplex virus 1 (HSV-1) blocks the interferon response pathways, at least at two sites, by circumventing the effects of activation of protein kinase R by double-stranded RNA and interferon and through the degradation of promyelocytic leukemia protein (PML) since interferon has no antiviral effects in PML−/− cells. Here we report on two effects of viral genes on other sites of the interferon signaling pathway. (i) In infected cells, Jak1 kinase associated with interferon receptors and Stat2 associated with the interferon signaling pathway rapidly disappear from infected cells. The level of interferon alpha receptor is also reduced, albeit less drastically at times after 4 h postinfection. Other members of the Stat family of proteins were either decreased in amount or posttranslationally processed in a manner different from those of mock-infected cells. The decrease in the levels of Jak1 and Stat2 may account for the decrease in the formation of complexes consisting of Stat1 or ISGF3 and DNA sequences containing the interferon-stimulated response elements after exposure to interferon. (ii) The disappearance of Jak1 and Stat2 was related at least in part to the function of the virion host shutoff protein, the product of the viral UL41 gene. Consistent with this observation, a mutant lacking the UL41 gene and treated with interferon produced lesser amounts of a late protein (UL38) than the wild-type parent. We conclude that HSV-1 blocks the interferon signaling pathways at several sites.
The effect of interferons (IFNs) on the replication of herpes simplex virus 1 (HSV-1) can be readily demonstrated by appropriate adjustment of IFN dose (high) and multiplicity of infection (low). Although the virus appears to be resistant to IFN in cultured cells (29) and IFN production and viral replication are incompatible (3), the importance of IFNs in the pathogenesis associated with HSV-1 infections is readily apparent from studies of mice in which specific genes in the IFN pathway have been knocked out (6, 31, 32). In these studies HSV-1 mutants lacking specific genes and that were highly attenuated in wild-type mice became virulent in mice lacking an intact IFN signaling pathway. The number of viral genes whose function it is to block IFNs also adds credence to the importance of this host defense mechanism.
Alpha IFN (IFN-α) and IFN-β are the common responders of cells infected by viruses. Immune cells secrete class II or IFN-γ to activate the innate and adaptive immune response and also aid in upregulating the antiviral state in cells to limit viral infection. This defense pathway begins with induction of IFNs by stimulated cells. In the case of viral infection, the most common inducer of IFN is the accumulation of complementary viral RNAs capable of annealing to form double-stranded RNA (dsRNA) (25, 28). dsRNA activates a cytoplasmic protein, IFN regulatory factor 3 (IRF3), that then translocates into the nucleus and binds to the promoter of the gene for IFN-β (35). IFN-β is secreted and binds to the receptors of the same cell or neighboring cells. The interaction of IFN with its receptors leads to activation of signaling pathways initiated by the phosphorylation of signal transducers and activators of transcription (Stat) proteins. The Stat proteins are phosphorylated by kinases (e.g., Jak1, Tyk2, etc.) associated with IFN receptors. Stat proteins in turn activate other factors (e.g., IRF7 [38]). IFN-α/β and IFN-γ activate similar but distinct Jak/Stat pathways (14). In the presence of IFN-α, the IFN-α receptor chains dimerize. Upon dimerization of the IFN-α receptor chains 1 and 2, the associated Janus kinases (Jaks), Tyk2 and Jak1, are tyrosine phosphorylated. Activated Jak1 and Tyk2 phosphorylate Stat1 and Stat2. Stat1 and Stat2 heterodimerize and bind to p48 (also known as IRF9) to form the complex: IFN-stimulated growth factor 3 (ISGF3). ISGF3 complexes translocate into the nucleus and bind to IFN-stimulated response elements (ISRE) to promote transcription. In the presence of IFN-γ, the IFN-γ receptor chains dimerize and activate associated Jak1 and Jak2 kinases. Stat1 is tyrosine phosphorylated by Jak1, homodimerizes, and translocates into the nucleus to bind to gamma-activated sequences (GAS) to promote transcription. Serine phosphorylation of Stat1 enhances its transcriptional activity.
Studies on the mechanisms by which HSV-1 blocks IFN-induced response have led to the identification of at least two viral proteins with specific anti-IFN functions. Thus, in infected cells, dsRNA activates the preexisting, inactive protein kinase R (PKR) (60). Activated PKR phosphorylates the α subunit of the translation initiation factor 2 (eIF-2α). Phosphorylated eIF-2α shuts off all protein synthesis, and as a consequence, viral replication is terminated. Viruses that induce the synthesis of complementary RNAs have evolved elaborate mechanisms to block PKR. HSV has evolved a gene, γ134.5, whose product recruits protein phosphatase 1α and redirects it to dephosphorylate eIF-2α (22). In effect, γ134.5 protein acts as a phosphatase accessory factor (21). In other studies, the product of the α0 gene, infected cell protein 0 (ICP0), has been reported to play a role in the resistance of IFN inasmuch as Δα0 mutants are more sensitive to IFNs than the wild-type parent (42). One of the functions mediated by ICP0 is the dispersal of nuclear structures known as PODs, Kr bodies, or ND10, and the degradation of sumoylated forms of promyelocytic leukemia protein (PML) and other constituents of these structures. PML is considered to be the organizer of ND10 structures and has been ascribed numerous functions. The resistance to IFN attributed to ICP0 may well be due to its degradation of PML, since HSV-1 infection of PML−/− cells appears to be insensitive to IFN compared to the infection of wild-type, PML+/+ cells (9).
The objective of the present study was to determine whether HSV-1 blocks the IFN signaling pathway at other sites. We report here that in wild-type-virus-infected cells several members of the Stat protein family appear to be posttranslationally modified and that the levels of both Stat2 and Jak1 decrease after infection. Thus, HSV-1 blocks both the initiation of the signaling pathway and the activation of IFN response genes at the end of the pathway.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells, HeLa, and HEp-2 cells were obtained from American Type Culture Collection. HSV-1 strain F [HSV-1(F)], a limited extra human passage isolate, is the prototype HSV-1 strain used in this laboratory (16). Recombinant virus R2621 lacks the UL41 gene encoding the virion host shutoff (vhs) protein (47). The recombinant viruses derived by genetic engineering from HSV-1(F) and used in these studies were R3616 (Δγ134.5 gene [13]), R7802 (Δα22 gene [44]), R7041 (ΔUS3 gene [50]), R7036 (ΔUL13 gene [49]), R6016 (Δα47 gene [39]), R6047 (ΔUs9 gene [4]), and R7910 (Δα0 [26]). Titers of all virus stocks were determined in Vero cells.
Antibodies and reagents.
Monoclonal antibodies to Stat1, Stat2, Stat3, Stat5, Stat6, and Jak1 were purchased from Transduction Laboratories (catalog no. S87080). Phospho-epitope specific polyclonal antibodies against Stat1 phosphorylated at tyrosine 701 (06-657) and Stat1 phosphorylated at serine 727 (06-802) were purchased from Upstate Biotechnology. Monoclonal antibodies against IFN-α receptor (SC-7391) and rabbit polyclonal antibodies against p48 (catalog no. SC-496) were purchased from Santa Cruz Biotechnology. Rabbit polyclonal antisera against HSV-1(F) UL38 were described elsewhere (57). Recombinant human IFN-γ (catalog no. RDI-3002) and human IFN-α (catalog no. RDI-PB11101) were purchased from Research Diagnostics, Inc.
Cell infection.
In all of the experiments described here replicate cell cultures grown in 25-cm2 flasks were either mock infected or exposed to virus in mixture 199 supplemented with 1% calf serum at the PFU/cell ratios given in Results. The cells were exposed to virus for 2 h at 37°C. The inoculum was then replaced with mixture 199 supplemented with 5% newborn calf serum, and the cultures were then incubated at 37°C for the intervals given in Results.
Harvesting and fractionation of cells.
To prepare whole-cell lysates, the cells were scraped, rinsed in phosphate-buffered saline (PBS), lysed in whole-cell lysis buffer (50 mM Tris, 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, 1 mM dithiothreitol [DTT], 10% glycerol, and protease inhibitors) or in PBS* (1% deoxycholate, 1% NP-40, and protease inhibitors in PBS), and centrifuged for 10 min at 13,000 rpm in Sorvall Biofuge to remove the insoluble fraction. For fractionation, the cells were scraped, rinsed with PBS, resuspended in hypotonic lysis buffer (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 0.1 mM sodium orthovanadate, 1 mM DTT, 0.2% NP-40, 10% glycerol, and protease inhibitors), lysed for 5 min on ice, and centrifuged at 13,000 rpm for 10 s to separate the cytoplasmic (supernatant) and nuclear (pellet) fractions. The cytoplasmic fraction was transferred into a new tube. The pelleted nuclei were gently rinsed with hypotonic buffer and pelleted as described above. The nuclear fractions were resuspended in hypertonic lysis buffer (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 0.1 mM sodium orthovanadate, 1 mM DTT, 0.2% NP-40, 20% glycerol, 420 mM NaCl, and protease inhibitors), lysed for 30 min in ice, and centrifuged at 13,000 rpm for 5 min to remove the insoluble fraction. Whole-cell lysates or the soluble nuclear and cytoplasmic fractions were used for electrophoretic mobility shift assays (EMSAs) with defined labeled probes or for immunoblotting after electrophoretic separation in denaturing gels.
Immunoblotting of electrophoretically separated proteins.
Protein concentrations of the cell lysates were measured by using the Bradford assay (Bio-Rad). Protein lysates (200 μg/lane for the detection of cellular proteins and 100 μg/lane for the detection of viral proteins) were denatured by boiling for 5 min in disruption buffer (final concentration of 2% sodium dodecyl sulfate, 50 mM Tris [pH 7.0], 2.75% sucrose, 5% β-mercaptoethanol, and bromophenol blue), electrophoretically separated on a denaturing 12, 10, or 8.5% polyacrylamide gels cross-linked with N,N′-diallyltartardiamide (DATD), and transferred to nitrocellulose membranes. Blots were blocked for 1 h with 5% milk buffer and then probed with the appropriate primary antibody for 2 h at room temperature or overnight at 4°C. Antibodies were diluted in PBS with 0.05% Tween 20 and 1% bovine serum albumin as follows: Stat1, 1:2,000; Stat2, 1:250; Stat3, 1:2,500; Stat5, 1:250; Stat6, 1:500; Jak1, 1:250; Stat1 P-tyr701, 1:500; Stat1P-ser727, 1:500; IFN-α receptor, 1:500; Stat1 P-ser727, 1:1,000; and UL38, 1:500. The immunoblots were incubated with secondary antibodies conjugated with peroxidase (Sigma) or conjugated with alkaline phosphatase (Bio-Rad) for 1 h. Peroxidase-conjugated antibodies were detected and quantified by using enhanced chemiluminescence (ECL Plus; Amersham) in a Storm 860 PhosphorImager (Molecular Dynamics). Alkaline phosphatase (AP)-conjugated antibodies were developed by using a colorimetric assay in AP buffer (100 mM Tris [pH 9.5], 100 mM NaCl, and 5 mM MgCl2) containing BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium.
Oligonucleotide labeling for EMSAs.
The oligonucleotides M67, which contains a mutant high-affinity Stat1 binding site from the c-fos promoter (58), sense strand (5′-CATTTCCCGTAAATCAT-3′), ISRE sense strand (5′-CTCGGGAAAGGGAAACCGAAACTGAAGCC-3′), and their antisense complements were end labeled with 32P. Three picomoles of oligonucleotide, 150 μCi of [γ-32P]ATP, 15 U of bacteriophage T4 polynucleotide kinase, and bacteriophage T4 polynucleotide kinase buffer were reacted in 20 μl at 37°C for 45 min. Heating the reaction mixture at 68°C for 10 min inactivated the T4 polynucleotide kinase. Oligonucleotides were incubated with their complements, denatured at 95°C for 5 min, and allowed to reanneal at room temperature. Labeled oligonucleotides were separated from free [γ32P]ATP by Sephadex G-50 spin column chromatography and then stored at −20°C.
EMSA and supershift assays.
Cytoplasmic and nuclear fractions (40 μg) were incubated with 3 μg of poly(dI-dC), 0.5 ng of 32P-labeled M67, or ISRE probe for 30 min on wet ice in binding buffer (final concentration of 20 mM Tris [pH 8.0], 12% glycerol, 2 mM MgCl2, and 0.6 mM DTT) at a total volume of 20 or 30 μl. For supershift assays, 1 μl of each antibody was added for an additional 30 min on wet ice (30). The complexes were electrophoretically separated on a 4% nondenaturing gel in Tris-borate-EDTA buffer for 75 min at 180 V. Gels were dried and analyzed in the Storm 860 PhosphorImager (Molecular Dynamics).
RESULTS
HSV-1(F) blocks activation of Stat1 by IFN-γ in infected HEp-2 cells.
Two series of experiments were done to test the effect of IFN-γ signaling in HEp-2 cells infected with HSV-1(F). In the first, replicate cultures of HEp-2 cells grown in 25-cm2 flasks were exposed to 10 PFU of HSV-1(F) per cell. After 2 h at 37°C, the inoculum was replaced with Dulbecco modified Eagle medium supplemented with 5% of newborn calf serum. At 7 h after infection the cells were exposed to 500 U of IFN-γ in 1 ml for 1 h and then harvested and probed for the presence of proteins binding to labeled M67 probe in EMSAs.
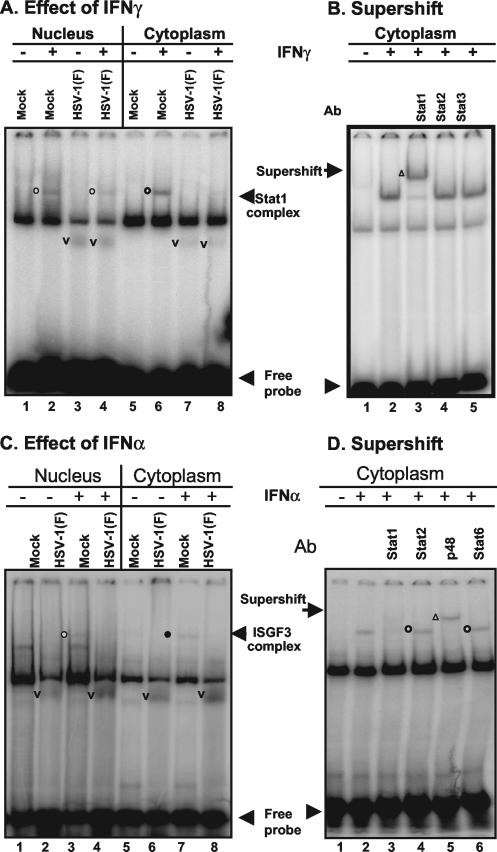
The results shown in Fig. 1A were as follows. (i) IFN-γ induced the appearance of a new M67 DNA-protein band that migrated more slowly than the band observed in untreated mock-infected cells. Lysates of IFN-γ-treated HSV-1-infected cells formed a similar band but much reduced in intensity. (ii) Lysates of infected cells, both IFN-γ treated and untreated, formed a M67-DNA band (Fig. 1A, v) that migrated faster than the protein band formed by lysates of untreated and treated mock-infected cells.
FIG. 1.
HSV-1 blocks the formation of DNA protein complexes induced by IFN. Replicate HEp2 cells (A and B) or HeLa cells (C and D) were mock infected or exposed to10 PFU of HSV-1(F) per cell as described in Materials and Methods After 7 h of incubation, the cells were treated (+) or not treated (−) with 500 U of IFN-γ/ml for 1 h (A and B) or 5,000 U of IFN-α/ml for 15 min (C and D). Cells were harvested, fractionated, and processed as described in Materials and Methods. (A and C) EMSAs. Nuclear (lanes 1 to 4) or cytoplasmic fractions (40 μg/reaction) were reacted with either labeled M67 probe (A) or ISRE probe (C) in total volumes of 30 μl per lane. Complexes containing Stat1 (circles, A) or ISGF3 (C) are identified by open circles to the left of the bands. Bands containing virus-induced proteins are identified with a “v” placed to the left of the bands. Although not specifically identified as such here, HSV-1 induces proteins capable of binding DNA, and this could account for the virus-specific bands. (B and D) Supershift assays. To the reaction mixtures of untreated and IFN-treated cells described above were added 1 μl of Stat1 (lane 3), Stat2 (lane 4), or Stat3 (lane 5) antibody (A) or 1 μl of Stat1 (lane 3), Stat2 (lane 4), ISGF3 p48 (lane 5), or Stat6 (lane 6) antibody (D). The reaction mixtures were incubated for an additional 30 min and then subjected to electrophoresis in denaturing gels. The bands supershifted by antibodies are marked by an open triangle to the left of the band.
We conclude from this experiment that HSV-1(F) blocked the induction or activation of Stat DNA-binding proteins by IFN-γ. The faster-migrating band formed in infected cells may represent a DNA-viral protein complex or a complex from which one or more proteins present in slower-migrating complex formed by lysates of mock-infected cells were missing. The components of this complex were not further identified. This experiment was also done with extracts of mock-infected and infected HeLa cells with similar results (data not shown).
The second series of experiments was designed to identify the Stat component of the M67 DNA-protein complex. In this series of experiments, 40 μg of mock-infected cytoplasmic protein were reacted with M67 labeled DNA probe for 30 min. Antibody to Stat1, Stat2, or Stat3 was then added and allowed to react for an additional 30 min. The reaction mixture was next subjected to electrophoresis on a nondenaturing gel. The results (Fig. 1B) indicate that Stat1 was the predominant component of the M67 DNA-protein complex, whose formation was induced by IFN-γ in HEp-2 cells. We conclude that in the system analyzed in the present study IFN either induced or activated Stat1 and that HSV-1 blocked the formation of Stat1-DNA complex.
The predominant response to the IFN-α is activation of Stat1, Stat2, and p48 DNA-binding complex: a step also blocked by infection with HSV-1.
The experiments described above were repeated with HeLa cells except that at 7 h after infection, the cells were treated with 5,000 U of IFN-α in 1 ml of medium for 15 min and that the cell lysates were probed for the presence of proteins binding to labeled ISRE probe. As shown in Fig. 1C, IFN-α induced the formation of novel ISRE probe-protein complex that migrated more slowly than those formed by lysates of untreated mock-infected cells. These complexes were not formed by lysates of infected cells either untreated or treated with IFN-α. At the same time, a new, faster-migrating ISRE-protein complex was formed by lysates of both infected, IFN-α-treated cells and untreated cells. In this instance, these DNA-protein complexes were more abundant in mixtures containing lysates from IFN-α-treated cells than those containing lysates of untreated, infected cells. The results presented in Fig. 1D showed that the antibody to p48 supershifted the complex, whereas antibody to Stat1 disrupted the complex. The results suggest that the ISRE DNA-protein complex contained both p48 and Stat1 proteins. The monoclonal antibody against Stat2 did not supershift the complex. The epitope for Stat2 in this complex may not have been accessible to the antibody.
The Stat family of proteins is modified in HeLa cells infected with HSV-1.
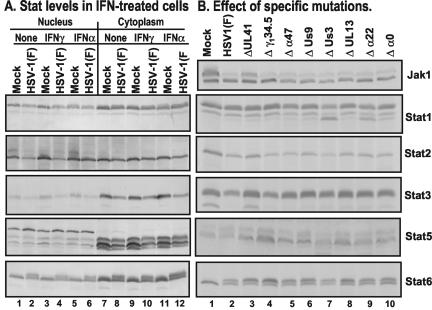
In the preceding section we showed that HSV-1(F) infection was able to block both IFN-α- and IFN-γ-induced pathways. The question addressed in the following experiments was to determine whether inhibition of the Jak/Stat pathway by wild-type virus correlated with destabilization or modification of the Stat proteins. In this series of experiments (Fig. 2A), replicate HeLa cell cultures in 25-cm2 flask were mock infected or exposed for 2 h to 10 PFU of HSV-1(F) per cell. At 7 h after infection the cultures were either left untreated or treated with 500 U of IFN-γ in 1 ml for 1 h or to 5,000 U of IFN-α in 1 ml of medium for 15 min. After treatment, the cells were harvested, fractionated into nuclear and cytoplasmic fractions, and subjected to electrophoresis (200 μg/lane) and then reacted with appropriate antibodies as described in Materials and Methods.
FIG. 2.
Effect of HSV-1 on members of the Stat family of proteins. (A) Replicate HeLa cells were mock infected or inoculated with 10 PFU of HSV-1(F) per cell as described in Materials and Methods. After 7 h of incubation, the cells were either not treated or treated with 500 U of IFN-γ/ml for 1 h or with 5,000 U of IFN-α/ml for 15 min. The cells were harvested and fractionated, and nuclear and cytoplasmic fractions (200 μg/lane) were electrophoretically separated on denaturing polyacrylamide gels, transferred to nitrocellulose membranes, and reacted with antibodies to Stat1, Stat2, Stat3, Stat5, or Stat6 (Transduction Laboratories) and then with secondary antibodies conjugated to AP. AP-conjugated antibodies were detected by using a colorimetric reaction (Bio-Rad). (B) Replicate HeLa cells were mock infected or exposed to 5 PFU of wild-type virus per cell or 5 PFU of mutant viruses lacking UL41, γ134.5, α47, US9, US3, UL13, α22, or α0 genes per cell. The cells were harvested at 18 h after infection and processed as described in Materials and Methods. Whole-cell lysates were subjected to electrophoresis in denaturing gels (200 μg/lane) and reacted with antibodies as described above. Bound peroxidase-conjugated antibodies were detected with the aid of enhanced chemiluminescence.
In a parallel experiment, replicate HeLa cell cultures were mock infected or exposed to 5 PFU of wild-type virus or mutants lacking US9, US3, UL13, α22, α47, γ134.5, or α0 genes. The cells were harvested at 18 h after incubation at 37°C and processed as described in Materials and Methods for whole-cell lysates. These lysates were probed for the levels of Stat proteins and also for Jak1. The results of the studies on Stat proteins are shown in Fig. 2B. The results of analyses of Jak1 levels are discussed in a later section. The results of the analyses of Stat levels shown in Fig. 2 can be summarized as follows.
(i) Stat1 was detected in both nucleus and cytoplasm (Fig. 2A). IFN-γ has a slight stimulatory effect on the accumulation of nuclear Stat1 in mock-infected cells but not in infected cells. No significant differences were observed in the accumulation or electrophoretic mobility of Stat1 in untreated or IFN-α-treated infected cells and in uninfected cells. A faster-migrating band of protein reacting with the anti-Stat1 antibody accumulated at higher levels in cells infected with the Δα22 or ΔUS3 mutant viruses (Fig. 2B). This observation suggests that HSV mediates or suppresses a modification of Stat1 that is dependent on α22 and US3 genes. Stat1 has two known isoforms, Stat1α and Stat1β (24, 51). The mobility of the fastest migrating is consistent with that of Stat1β, although it could be a novel isoform found in infected cells that has been observed in other human cell lines (data not shown).
(ii) Stat2 formed two bands in nuclear extracts of mock-infected cells that were either untreated or treated with IFN-α or IFN-γ. The slower-migrating band was reduced in amount or absent in the lysates of nuclei of untreated infected cells. The band formed by the cytoplasmic fractions was unaffected. IFN-α had a slight stimulatory effect on the accumulation of Stat2 in nuclei of both infected and uninfected cells. Neither IFN-α nor IFN-γ affected the electrophoretic mobility of Stat2. The results shown in Fig. 2B (compare lane 3 with lanes 4 to 10) suggest that the decrease in Stat2 was mediated at least in part by vhs, the product of the UL41 gene, although other factors may also play a role.
(iii) Stat3 accumulated predominantly in the cytoplasm (Fig. 2A). Neither IFN-α nor IFN-γ had an effect on the localization of Stat3. However, the amounts of Stat3 were reduced in infected cells compared to those of uninfected cells. Stat3 protein levels were similar in cells infected with all of the mutant viruses. A faster-migrating band was present in mock-infected and in ΔUL41 mutant-infected cells but not in cells infected with wild-type virus or other mutants (Fig. 2B). Other isoforms of Stat3 with a faster mobility has been observed in other studies (5, 8).
(iv) Cytoplasmic and nuclear fractions formed multiple bands reactive with anti-Stat5 antibody. These bands have been observed in other cell lines (36, 56). The cytoplasmic bands were more intense and migrated faster than the corresponding nuclear bands. In addition, Stat5 accumulating in the infected cell lysates appeared to be slightly less abundant than those of uninfected cell lysates (Fig. 2A). A significantly higher level of Stat5 accumulated in cells infected with Δγ134.5 mutant than in cells mock infected or infected with wild-type virus (Fig. 2B). The increased accumulation of Stat5 in ΔUL41-infected cells suggests the possibility that Stat5 synthesis was induced in cells infected with some mutants but not in wild-type-virus-infected cells.
(v) Stat6 was more abundant in the cytoplasmic than in the nuclear fractions. In both the nucleus and the cytoplasm, Stat6 accumulating in infected cell lysates formed a second, slower-migrating band (Fig. 2A). The levels of the slow-migrating band were higher in cells infected with several mutants, but particularly in cells infected with Δγ134.5 or ΔUL41 rather than in cells infected with wild-type virus (Fig. 2B). In this instance as well, the results suggest the possibility that the synthesis of Stat6 was induced after infection.
We conclude from the studies that the Stat proteins were not modified in infected cells in a uniform fashion. After infection there was a discernible decrease in the accumulation of Stat2 in the nuclear compartment and of Stat3 in the cytoplasmic compartment. The decrease in the accumulation of Stat2 is to a large extent mediated by the product of the UL41 gene. Stat1 and Stat3 appear to undergo posttranslational modification mediated by HSV. In the case of Stat5 and Stat6, the results suggest the possibility of de novo synthesis of posttranslationally modified proteins. The key conclusion to be drawn from these studies is that the Stat proteins are extensively modified after infection and this, to a large extent, may account for the insensitivity of HSV for IFN.
Accumulation of Stat1 and Stat2 proteins in the course of infection with HSV-1(F).
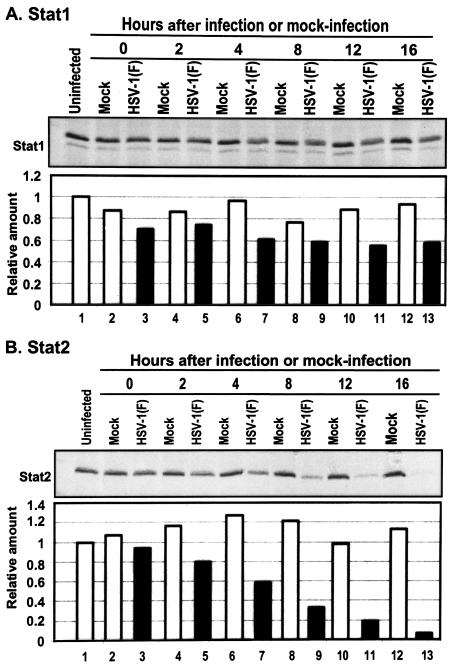
In this series of experiments replicate 25-cm2 cultures of HeLa cells were mock infected or exposed to 10 PFU of HSV-1(F) per cell for 2 h as described above and in Materials and Methods. The cells were harvested at 2, 4, 6, 10, 14 or 18 h after infection or mock infection, solubilized, subjected to electrophoresis in denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, reacted with anti-Stat1 or anti-Stat2 antibodies, and processed as described in Materials and Methods. The bound antibody was quantified with the aid of enhanced chemiluminescence in the Storm 860 PhosphorImager. The results were normalized with respect to the Stat1 or Stat2 levels to those measured in the uninfected cell sample.
As shown in Fig. 3A, there was a 20 to 40% decrease in Stat1 levels detected by the phosphorimager in cells after infection by wild-type virus. Stat1, at this lower level, was maintained throughout infection. In contrast (Fig. 3B), the levels of Stat2 gradually decreased throughout the replicative cycle. At 16 h after infection the amount of Stat2 present in infected cells was ∼10% of that present in mock-infected cells throughout the interval of study.
FIG. 3.
Effect of HSV-1 on the accumulation of Stat1 and Stat2 protein levels. Replicate HeLa cell cultures were either mock infected or exposed to 10 PFU of HSV-1(F) per cell and then harvested at 2, 4, 8, 12, or 16 h after infection, processed, subjected to electrophoresis in denaturing gels (200 μg of protein per lane), transferred to a nitrocellulose sheet, and reacted with anti-Stat1 (A) or Stat2 (B) antibodies and then with secondary antibodies conjugated to peroxidase. Bound antibodies were detected by using enhanced chemiluminescence and quantified with the aid of the Storm 860 PhosphorImager. The results were normalized to the Stat protein levels in the uninfected cell samples.
We conclude that HSV-1(F) infection mediates the reduction in the levels of Stat2. Downregulation of Stat2 during infection may play a significant role in the inhibition of the Jak/Stat pathway that is activated by IFN-α.
HSV-1 infection decreases the tyrosine phosphorylation but not the serine phosphorylation of Stat1.
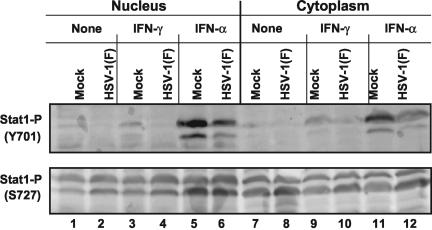
Since Stat1 protein levels were maintained throughout infection, the question arose whether the virus affects the tyrosine or serine phosphorylation of Stat1. Inhibition of tyrosine phosphorylation of Stat1 would inhibit its activation, homodimerization, and translocation into the nucleus, whereas the inhibition of serine phosphorylation of Stat1 would decrease its transcriptional activity (59). In this series of experiments, replicate 25-cm2 HeLa cell flask cultures were mock infected or exposed for 2 h to 10 PFU of HSV-1(F) per cell as described in Materials and Methods. At 7 h after incubation at 37°C, the cells were either left untreated or exposed to 500 U of IFN-γ for 1 h or to 5,000 U of IFN-α for 15 min in 1 ml of medium. After treatment, the cells were harvested, fractionated into nuclear and cytoplasmic fractions, subjected to electrophoresis in denaturing polyacrylamide gels (200 μg/lane), transferred to a nitrocellulose sheet, and reacted with phospho-specific primary antibodies against tyrosine phosphorylated Stat1 or serine phosphorylated Stat1.
The results (Fig. 4) were as follows. The levels of tyrosine phosphorylated Stat1 were too low to be detected in either nuclear or cytoplasmic fraction of untreated cells. After treatment with IFN-γ and especially after treatment with IFN-α, tyrosine phosphorylated Stat1 was detected in both nuclear and cytoplasmic fractions. In addition, there was a decrease in tyrosine phosphorylated Stat1 in infected cells treated with IFN compared to that of mock-infected IFN-treated cells. As shown in the lower panel of Fig. 4, there was no apparent inhibition of serine phosphorylation of Stat1 in wild-type virus-infected cells or an increase in the amounts of serine phosphorylation of Stat1 in IFN-treated cells.
FIG. 4.
HSV-1 infection can inhibit tyrosine phosphorylation but not serine phosphorylation of Stat1 in HeLa cells. Replicate HeLa cell cultures were mock infected or exposed to 10 PFU of HSV-1(F) per cell. At 7 h after mock infection or infection the cells were mock treated or exposed to 500 U of IFN-γ/ml for 1 h or 5,000 U of IFN-α/ml for 15 min. At the conclusion of the treatment the cells were harvested and fractionated, and nuclear and cytoplasmic fractions were subjected to electrophoresis in denaturing gels (200 μg/lane). The electrophoretically separated proteins were transferred to a nitrocellulose sheet and reacted with primary antibodies to phospho-epitope-specific polyclonal antibodies tyrosine 701 or serine 727 of Stat1 (Upstate Biotechnology) and then with a secondary antibody conjugated to peroxidase. Bound peroxidase-conjugated antibodies were detected by using enhanced chemiluminescence.
These results indicate that tyrosine phosphorylation of Stat1 was partially inhibited during infection. Since Stat1 is necessary for IFN-α- or IFN-γ-induced Jak/Stat pathways, reduction in the tyrosine phosphorylation of Stat1, even in the absence of any reduction of the amounts of the protein, would have a negative effect on the activation of the cellular IFN defense pathway.
HSV-1 inhibits Stat1 tyrosine phosphorylation by decreasing the levels of Jak1.
Stat1 tyrosine is phosphorylated by activated Jak1 protein kinase. Inhibition of tyrosine phosphorylation of Stat1 during infection could result from the inhibition of Jak1 kinase activity or the reduction of Jak1 protein level. In preliminary experiments we found that, whereas the Jak1 sample provided by the manufacturer of the anti-Jak1 antibody formed one band in denaturing gels, the antibody reacted with three bands in lysates of mock-infected cells. To test this hypothesis presented above three series of experiments were carried out.
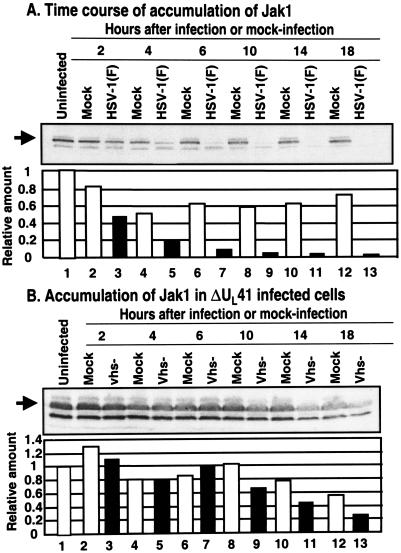
In the first series of experiments, we examined the levels of Jak1 in HeLa cells harvested at different times after mock infection or a 2-h exposure to 10 PFU of HSV-1(F) per cell. The cells were processed as described in Materials and Methods, and the amount of Jak1 present in the cells was quantified with the aid of the Storm 860 PhosphorImager. As shown in Fig. 5A, Jak1 decreased by ca. 50% at 2 h after infection and to nearly 10% of the amount recovered from mock-infected cells by 18 h after infection.
FIG. 5.
Downregulation of Jak1 protein levels is delayed in HeLa cells infected with UL41 mutant virus. Replicate HeLa cell cultures were mock infected or were exposed to 10 PFU of the parent, HSV-1(F) virus (A) or the UL41 mutant virus (B) per cell and then harvested at 2, 4, 6, 10, 14, or 18 h postinfection. The harvested cells were solubilized, subjected to electrophoresis (200 μg/lane), transferred to a nitrocellulose sheet, and reacted first with primary antibodies to Jak1 and then with secondary conjugated antibodies. The bound antibodies were detected by enhanced luminescence and were quantified by using a Storm 860 PhosphorImager and then normalized with respect to the protein level detected in the uninfected samples.
To determine whether a specific viral gene product was responsible for the rapid disappearance of Jak1, we carried out the experiments described above and in the legend to the Fig. 2B. As illustrated in that figure, Jak1 levels accumulated in cells infected with the ΔUL41 mutant to the same level as in mock-infected cells but at a higher level than in cells infected with wild-type virus or various other mutants.
The objective of the third series of experiments was to determine the contribution of the UL41 gene to the rapid disappearance of the Jak1 from infected cells. In these experiments, replicate HeLa cell flask cultures were mock infected or exposed to 10 PFU of ΔUL41 mutant virus per cell. The cells were harvested at 2, 4, 6, 10, 14, or 18 h after infection or mock infection and processed as described above for the experiment illustrated in Fig. 5A. The results (Fig. 5B) were as follows. There was little or no significant difference between Jak1 levels in mock-infected and infected cells until at least 6 h after infection. At between 6 and 18 h the Jak1 levels gradually decreased to ca. 40% of the amount detected in mock-infected cells at those times.
We conclude from these experiments that the initial decreased in the levels of Jak1, i.e., between the time of exposure of the virus to cells to ca. 6 h after infection, is due the action of the product of UL41 gene. At later times, additional factors may play a role in curbing the replenishment of Jak1 in infected cells.
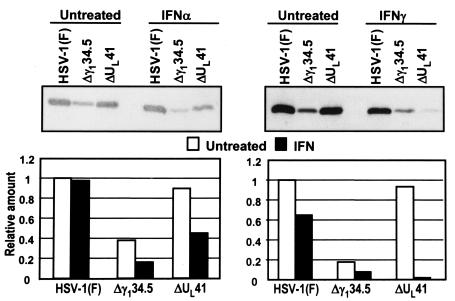
The ΔUL41 mutant virus is more sensitive to IFN than wild-type virus.
The results presented above showed that Jak1 levels were maintained at least for several hours after infection with the ΔUL41 mutant. This observation allows the prediction that ΔUL41 mutant virus would be more sensitive than the parent virus to IFNs. To test this prediction, we measured the effect of IFN on the accumulation of UL38, a γ2 protein in HeLa cells infected with wild-type or ΔUL41 mutant viruses and which were either mock treated or exposed to IFN. A late protein was selected in the expectation that IFN would reduce viral gene expression at each step of the replicative cycle, and the effects on the early events would be additive and exaggerate the effects on late steps in viral replication. As a known positive control, we also tested cells infected with the Δγ134.5 mutant virus. In cells infected with wild-type virus, PKR is activated, the α subunit of the translation initiation factor (eIF-2α) is phosphorylated and, were it not for the γ134.5 protein, all protein synthesis would cease. The γ134.5 protein binds protein phosphatase 1 and diverts it to dephosphorylate eIF-2α. Because of the rapid shutoff of protein synthesis in Δγ134.5-infected cells, late or γ2 proteins are produced in grossly diminished amounts (12, 22).
In this experiment, replicate 25-cm2 flask cultures of HeLa cells were treated with 1,000 U of IFN-γ/ml or with 5,000 U of IFN-α/ml for 24 h and then exposed to 10 PFU of HSV-1(F), ΔUL41, or Δγ134.5 mutant virus per cell. Cells were harvested at 12 h after infection and processed as described in Materials and Methods. The electrophoretically separated proteins were reacted first with polyclonal antibodies against UL38 and then with peroxidase-conjugated anti-rabbit immunoglobulin G antibodies. The bound antibody was quantified with the aid of enhanced chemiluminescence and the Storm 860 PhosphorImager. All values were normalized with respect to the levels of viral proteins were detected in the untreated, wild-type virus-infected cells. These experiments (Fig. 6) showed that IFN-α had no effect on the accumulation of UL38 protein in wild-type virus-infected cells but reduced by ca. 50% the accumulation of UL38 protein in mutant-infected cells compared to the untreated infected cells. Consistent with earlier results (12), there was significantly less UL38 protein in untreated cells infected with Δγ134.5 mutant. IFN-γ caused a reduction in the accumulation of UL38 protein both wild-type virus-infected cells and mutant virus-infected cells. In this instance, the reduction in accumulation of UL38 protein was ∼2-fold in wild-type-virus- or Δγ134.5 mutant virus-infected cells but >10-fold in ΔUL41-infected cells. The dramatic decrease in the accumulation of UL38 protein in IFN-γ-treated cells infected with ΔUL41 mutant is consistent with the expectation that it would be more sensitive to IFN because of the maintenance of Jak/Stat signaling pathway in these cells.
FIG. 6.
IFN has a stronger inhibitory effect on the accumulation of late viral proteins in cells infected by UL41 mutant virus than in cells infected with the wild-type virus. Replicate HeLa cell cultures were exposed to either 1,000 U of IFN-γ or to 5,000 U of IFN-α per ml of medium for 24 h and then exposed to 10 PFU of parent HSV-1(F) virus to ΔUL41 or Δγ134.5 mutant viruses per cell. After 2 h, the inoculum was replaced with medium supplemented with 5% newborn calf serum with 1,000 U of IFN-γ per ml or with 5,000 U of IFN-α per ml. The cells were harvested at 12 h postinfection, solubilized, electrophoretically separated on denaturing 12% polyacrylamide gels, transferred to nitrocellulose membranes, and reacted first with polyclonal antibody to UL38 and then to anti-rabbit immunoglobulin G conjugated to peroxidase. Bound antibody was detected by using enhanced chemiluminescence, quantified with the aid of a Storm 860 PhosphorImager, and normalized with respect to the levels of UL38 protein levels detected in untreated HeLa cells infected by wild-type virus.
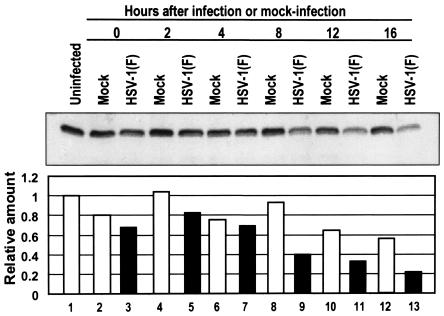
HSV-1 decreases the levels of the IFN-α receptor late in infection.
Jak1 kinases are associated with IFN receptors. To determine whether the IFN receptor is also targeted for destruction, along with Jak1, we examined the protein levels of the IFN-α receptor at different times after infection. In this experiment, replicate 25-cm2 cultures of HeLa cells were mock infected or exposed to 10 PFU of HSV-1(F) per cell. The cells were harvested at 0, 2, 4, 8, 12, or 16 h after infection or mock infection and then processed as described in Materials and Methods. The electrophoretically separated proteins were reacted with anti-IFN-α receptor antibody, and the bound antibodies were quantified as described above. The results shown in Fig. 7 indicate that IFN-α receptor was maintained with minimal reduction in quantity until past 4 h postinfection. The lowest level, at 16 h postinfection, represented ca. 40% of the amount present in mock-infected cells harvested at the same time.
FIG. 7.
IFN-α receptor protein levels decrease late in HSV-1 infection. Replicate HeLa cell cultures were mock infected or exposed to 10 PFU of HSV-1(F) per cell and then at 0, 2, 4, 8, 12, or 16 h after infection. The harvested cells were processed as described in Materials and Methods, subjected to electrophoresis in denaturing polyacrylamide gels (200 μg/lane), transferred to a nitrocellulose sheet, and reacted with primary antibody to IFN-α receptor and a secondary antibody conjugated to peroxidase. Bound antibodies were detected by enhanced chemiluminescence, quantified with the aid of a Storm 860 PhosphorImager, and normalized to the protein level detected in mock-infected cells.
DISCUSSION
In this report we show that HSV also precludes the maintenance of normal levels of Jak1 and Stat2 and, to a lesser extent, the level of the IFN-α receptor. Another study reported a similar downregulation of Jak1 and Stat2 protein levels by HSV but at later time points (62). Analyses of viral mutants suggest that at least one gene product associated with the decrease in the levels of Jak1 and Stat2 is vhs. The effect of vhs on Jak1 is readily evident during the first 6 h after infection. Other factors may play a role in the decline of Jak1 at later times after infection. Stat2 was less intensively investigated in the present study, but the accumulated results suggest that the levels of this protein are also affected by vhs and viral gene products. The impact of the vhs gene can be deduced from the observation that ΔUL41 mutant was more sensitive to IFN than the parent virus. The working hypothesis entertained at this time is that these proteins have a relatively short half-life and that vhs precludes their replenishment through the translation of newly synthesized mRNA. It is likely, although we have not directly tested this hypothesis, that ICP27 contributes to the decline of this and other cellular proteins that turn over with a half-life significantly smaller than the viral replicative cycle.
The net effect of the HSV-1 gene expression on the IFN signaling pathway is readily apparent from the formation of DNA-protein complexes activated by the IFN. Thus, complexes containing gamma-activated sequences of DNA-Stat1 homodimers were readily apparent in cells treated with IFN-γ at 7 h after mock infection but not in infected cells treated for a similar interval (Fig. 1). Similarly, complexes containing IFN-stimulated response elements DNA-Stat1, Stat2, and p48 were readily apparent in cells treated with IFN-α at 7 h after mock infection but not in cells treated with IFN-α after 7 h of infection with HSV-1. The effects are also apparent from examination of the role of vhs protein in blocking the antiviral activity of IFN-α and IFN-γ. This effect is readily demonstrable by comparison of late viral protein accumulation in wild-type and ΔUL41 virus-infected cells (Fig. 6). It should be noted the vhs protein also blocks the accumulation of other cytokines with potential antiviral effects in experimental animal systems (52).
Virtually all viruses have evolved mechanisms designed to inhibit or evade the IFN-based host defenses to infection, and viruses frequently target the Jak/Stat pathway. For example, adenovirus E1A can cause a decrease in Stat1 and p48 protein levels. Simian virus 5 infection targets Stat1 for proteasome-mediated degradation (15). Mumps virus also ubiquitylates and downregulates Stat1 protein levels (61). Human parainfluenza virus type 2 downregulates and polyubiquitylates Stat2 protein (45, 55). Varicella-zoster virus infection reduces the protein levels of Stat1α and Jak2 (1). Human cytomegalovirus encodes mechanisms that can reduce the protein levels of both Jak1 and p48 (40, 41). Sendai virus and human papillomavirus have mechanisms that can inhibit activation of Tyk2 (27, 34). Hepatitis C virus can also impair the IFN-α/β-induced signaling pathway (23). Although the mechanisms and target components of the Jak/Stat signaling pathway may differ between these viruses and HSV-1, the results are the same: prevention of the upregulation of IFN-stimulated genes and enhancement of viral production.
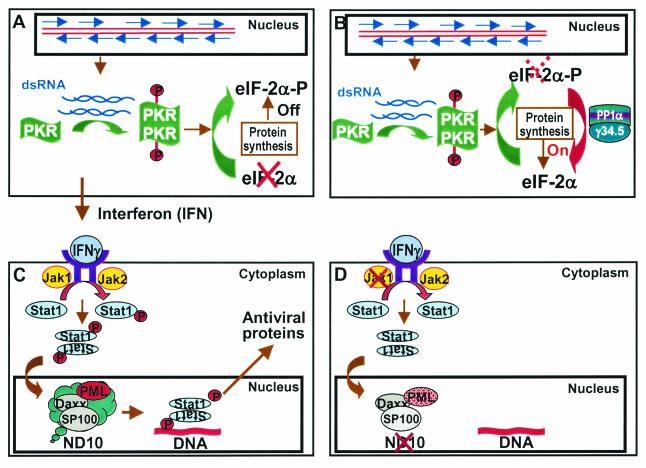
The striking features of the studies of HSV-1 viral gene function are the large number of genes targeting the IFN-dependent host defense systems. These are described below and schematically illustrated in Fig. 8.
FIG. 8.
Schematic representation of the multiple mechanisms by which HSV-1 blocks cellular IFN-based defense systems. (A) Accumulation of complementary RNA capable of annealing induces the synthesis of IFN and activates PKR (25, 28). Activated PKR dimerizes, phosphorylates eIF-2α and, in the absence of viral anti-IFN gene function, shuts off protein synthesis. (B) In cells infected with wild-type virus the γ134.5 protein binds and redirects phosphatase 1α to dephosphorylate eIF-2α (22). Protein synthesis is not affected by activated PKR. (C) Secreted IFN binds IFN receptor to activate the signaling pathway. In the case of IFN-γ, the receptor-associated kinases Jak1 and Jak2 are activated and phosphorylate Stat1 (14). These and associated proteins are translocated into the nucleus, where they induce antiviral proteins. The activation of antiviral response is mediated by PML (9). (D) In wild-type-virus-infected cells, Jak1 disappears, PML is degraded, and the constituents of ND10 are dispersed (17). As a result, the infected cell does not respond to exogenous IFN to curtail viral replication.
(i) For at least several hours, vhs, the product of the UL41 gene, mediates the degradation of cellular mRNAs. Although vhs causes the degradation of both cellular and viral mRNAs, the more vigorous transcription of viral genes enables viral protein synthesis but effectively shuts off the expression of a large number of cellular genes. Recent studies (16a, 53) indicate that only a small proportion of the transcripts of genes upregulated after infection are actually translated. An additional impediment to the translation of cellular mRNAs is that for the most part they arise by splicing. ICP27, a regulatory α protein, blocks splicing but enables the transport to the cytoplasm of unspliced RNAs, with the consequence that translation of the products does not ensue (11, 16a, 53). In the present study we have shown that the decrease in the levels of Jak1 and Stat2 proteins in infected cells is specifically related to vhs protein.
(ii) HSV-1 also targets preexisting proteins. Some, such as PML, Sp100, and CENP-A and -C, are degraded (10, 17, 18). In recent studies it has become apparent that ICP0 acts as a ubiquitin ligase. Thus, a ubiquitin ligase site encoded in exon III targets cdc34, whereas a ubiquitin ligase encoded in exon II uses the UbcH5a ubiquitin-conjugating enzyme to degrade at least PML and Sp100 proteins (20). As noted earlier, overexpression of PML has no effect on viral replication (37). PML, however, appears to mediate the antiviral signaling by IFNs, and destruction of PML is designed to block the exogenous IFN from affecting viral replication (9).
(iii) As noted in the introduction, HSV-1 infection of competent cells results in the activation of PKR. The accumulation of phosphorylated eIF-2α is precluded by the sequestration and diversion of phosphatase 1α to dephosphorylate eIF-2α (12, 22). The activation of other cellular proteins may be simply ignored; as described above, HSV-1 sequesters and redirects protein phosphatase 1α to dephosphorylate eIF-2α, thus ignoring the activated PKR.
(iv) Not targeting directly to the IFN defense pathways but related generally to host defenses are two sets of viral gene products. The products of at least three HSV-1 genes (α47, γ134.5, and UL41) have been reported to block presentation of antigenic peptides by major histocompatibility complex classes I and II (54, 63). A larger number (e.g., US3, gD, gJ, α27, and UL39) block programmed cell death induced by either exogenous agents or by viral gene products (2, 19, 33, 43, 46, 64). This list is likely to grow and may include at least ICP27 inasmuch as, as noted above, the protein blocks RNA splicing but enables the transport of unspliced RNA.
The studies on the role of viral gene products in blocking IFN-dependent host defenses leave a number of questions unresolved. For example, although the emphasis of our studies was on Jak1, Stat1, and Stat2, other members of the Stat family of proteins were also affected by viral infection. We have noted, but have no appreciation of the impact of, decreased levels of IFN-α receptor at times after 4 h postinfection. Although it is clear that ND10 and its organizer, PML, mediate the effects of IFN-α or IFN-γ, the precise mechanism by which these structures enable activation of the immune response also remains to be elucidated. Not considered in detail in this summary of anti-IFN effects is the role of US11. Although several lines of evidence indicate that US11 expressed early in infection also acts to preclude the accumulation of phosphorylated eIF-2α, the question remains as to whether US11 has any anti-IFN role in cells infected with wild-type virus in which this gene is expressed at late times (7, 48).
The striking feature of the results of these and earlier studies is the number and range of function of viral anti-IFN genes. The results argue that the virus has met its nemesis and evolved the armamentarium necessary to overcome it.
Acknowledgments
These studies were aided by grants from the National Cancer Institute (CA87661, CA83939, CA71933, CA78766, and CA88860), U.S. Public Health Service.
REFERENCES
- 1.Abendroth, A., B. Slobedman, E. Lee, E. Mellins, M. Wallace, and A. M. Arvin. 2000. Modulation of major histocompatibility class II protein expression by varicella-zoster virus. J. Virol. 74:1900-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, M., S. A. Rice, and J. A. Blaho. 2001. Accumulation of herpes simplex virus type 1 early and leaky-late proteins correlates with apoptosis prevention in infected human HEp-2 cells. J. Virol. 75:1013-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aurelian, L., and B. Roizman. 1965. Abortive infection of canine cells by herpes simplex virus. II. The alternative suppression of synthesis of interferon and viral constituents. J. Mol. Biol. 11:539-548. [DOI] [PubMed] [Google Scholar]
- 4.Brandimarti, R., and B. Roizman. 1997. US9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proc. Natl. Acad. Sci. USA 94:13973-13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldenhoven, E., T. B. van Dijk, R. Solari, J. Armstrong, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and R. P. de Groot. 1996. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 271:13221-13227. [DOI] [PubMed] [Google Scholar]
- 6.Cantin, E., B. Tanamachi, H. Openshaw, J. Mann, and K. Clarke. 1999. Gamma interferon (IFN-γ) receptor null-mutant mice are more susceptible to herpes simplex virus type 1 infection than IFN-γ ligand null-mutant mice. J. Virol. 73:5196-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceresa, B. P., and J. E. Pessin. 1996. Insulin stimulates the serine phosphorylation of the signal transducer and activator of transcription (STAT3) isoform. J. Biol. Chem. 271:12121-12124. [DOI] [PubMed] [Google Scholar]
- 9.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, P., K. S. Ellison, R. Verity, and J. R. Smiley. 2000. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated α-globin pre-mRNA in infected HeLa cells. J. Virol. 74:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of an Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 α and premature shutoff of protein synthesis after infection with γ134.5 mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou, J., A. P. Poon, J. Johnson, and B. Roizman. 1994. Differential response of human cells to deletions and stop codons in the γ134.5 gene of herpes simplex virus. J. Virol. 68:8304-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 15.Didock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 16a.Esclatine, A., B. Taddeo, L. Evans, and B. Roizman. The herpes simplex virus UL41 gene dependent destabilization of cellular RNAs is selective, and may be sequence specific. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 17.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, H., and B. Roizman. 2003. The degradation of PML and Sp100 proteins by herpes simplex virus 1 is mediated by the interaction of ubiquitin ligase activity of the ICP0 ring finger domain and the ubiquitin conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 100:8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, B., M. Gross, and B. Roizman. 1998. The γ134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J. Biol. Chem. 273:20737-20743. [DOI] [PubMed] [Google Scholar]
- 22.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Improta, T., C. Schindler, C. M. Horvath, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1994. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl. Acad. Sci. USA 91:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquemont, B., and B. Roizman. 1975. Ribonucleic acid synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and double-stranded RNA prepared from them. J. Virol. 15:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks α interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak, M., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. IX. Evidence for accumulation of abundant symmetric transcripts in nuclei. J. Virol. 15:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer, M. J., R. Dennin, C. Kramer, G. Jones, E. Connell, N. Rolon, A. Gruarin, R. Kale, and P. W. Trown. 1983. Cell and virus sensitivity studies with recombinant human α interferons. J. Interferon Res. 3:425-435. [DOI] [PubMed] [Google Scholar]
- 30.Kristie, T. M., and B. Roizman. 1984. Separation of sequences defining basal expression from those conferring α gene recognition within the regulatory domains of herpes simplex virus 1 α genes. Proc. Natl. Acad. Sci. USA 81:4065-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 35.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lokuta, M. A., M. A. McDowell, and D. M. Paulnock. 1998. Identification of an additional isoform of STAT5 expressed in immature macrophages. J. Immunol. 161:1594-1597. [PubMed] [Google Scholar]
- 37.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meignier, B., R. Longnecker, P. Mavromara-Nazos, A. E. Sears, and B. Roizman. 1988. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162:251-254. [DOI] [PubMed] [Google Scholar]
- 40.Miller, D. M., B. M. Rahill, J. M. Boss, M. D. Lairmore, J. E. Durbin, J. W. Waldman, and D. D. Sedmak. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, D. M., Y. Zhang, B. M. Rahill, W. J. Waldman, and D. D. Sedmak. 1999. Human cytomegalovirus inhibits IFN-α-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-α signal transduction. J. Immunol. 162:6107-6113. [PubMed] [Google Scholar]
- 42.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munger, J., A. V. Chee, and B. Roizman. 2001. The U(S)3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogle, W. O., and B. Roizman. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 73:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perkins, D., E. F. Pereira, M. Gober, P. J. Yarowsky, and L. Aurelian. 2002. The herpes simplex virus type 2 R1 protein kinase (ICP10 PK) blocks apoptosis in hippocampal neurons, involving activation of the MEK/MAPK survival pathway. J. Virol. 76:1435-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon, A. P., and B. Roizman. 1997. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant γ134.5 genes of herpes simplex virus 1. Virology 229:98-105. [DOI] [PubMed] [Google Scholar]
- 48.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 US11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 52.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of UL41 gene of herpes simplex virus type 1 in evasion of nonspecific host defense mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 53.Taddeo, B., A. Esclatine, W. Zhang, and B. Roizman. 2003. The stress-inducible immediate-early responsive gene IEX-1 is activated in cells infected with herpes simplex virus 1, but several viral mechanisms, including 3′ degradation of its RNA, preclude expression of the gene. J. Virol. 77:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trgovcich, J., D. Johnson, and B. Roizman. 2002. Cell surface major histocompatibility complex class II proteins are regulated by the products of the γ134.5 and U L41 genes of herpes simplex virus 1. J. Virol. 76:6974-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 56.Wang, D., D. Stravopodis, S. Teglund, J. Kitazawa, and J. N. Ihle. 1996. Naturally occurring dominant negative variants of Stat5. Mol. Cell. Biol. 16:6141-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner, B. J., T. E. Hayes, C. J. Hoban, and B. H. Cochran. 1990. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 9:4477-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 60.Williams, B. R., C. S. Gilbert, and I. M. Kerr. 1979. The respective roles of the protein kinase and pppA2′ p5′ A2′ p5 A-activated endonuclease in the inhibition of protein synthesis by double stranded RNA in rabbit reticulocyte lysates. Nucleic Acids Res. 6:1335-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yokosawa, N., S. Yokota, T. Kubota, and N. Fujii. 2002. C-terminal region of STAT-1α is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 76:12683-12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yokota, S., N. Yokosawa, T. Kubota, T. Suzutani, I. Yoshida, S. Miura, K. Jimbow, and N. Fujii. 2001. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and Janus kinases during an early infection stage. Virology 286:119-124. [DOI] [PubMed] [Google Scholar]
- 63.York, I. A., C. Roop, D. W. Andrews, S. R. Riddell, F. L. Graham, and D. C. Johnson. 1994. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 77:525-535. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]