Abstract
There is an increasing availability of complete or draft genome sequences for microbial organisms. These data form a potentially valuable resource for genotype–phenotype association and gene function prediction, provided that phenotypes are consistently annotated for all the sequenced strains. In this review, we address the requirements for successful gene-trait matching. We outline a basic protocol for microbial functional genomics, including genome assembly, annotation of genotypes (including single nucleotide polymorphisms, orthologous groups and prophages), data pre-processing, genotype–phenotype association, visualization and interpretation of results. The methodologies for association described herein can be applied to other data types, opening up possibilities to analyze transcriptome–phenotype associations, and correlate microbial population structure or activity, as measured by metagenomics, to environmental parameters.
Keywords: genotype–phenotype association, genome-wide association studies, functional genomics, microbial genomics, random forest
INTRODUCTION
The ‘function’ of a gene or protein is a complex concept that consists of several layers, such as molecular function, cellular component and biological process (phenotypic function) [1,2]. Deciphering the function of microbial genes often involves the use of molecular biological techniques to establish function at the molecular level, or knock-out studies to specify cellular or phenotypic annotations. Since (near) complete microbial genomes started becoming available, comparative genomics has been used to prioritize candidate genes for further laboratory testing [1], minimizing the costs of experimental research. The best-known example of functional annotation by comparative analysis is the transfer of functional annotations between orthologous genes, but many other associative methods have been developed [3] including conservation of gene order [4,5] and phylogenetic profiling [6,7]. The recent drop in the cost of genome sequencing has enabled an increase in the scale of comparative genomic analyses. Several thousand bacterial genomes have been sequenced thus far, opening up the potential for microbial genome-wide association studies (GWAS).
In human genomics, GWAS investigate the correlation of genetic variants with phenotypic traits across different individuals [8,9]. These genetic variants mostly consist of simple nucleotide polymorphisms (SNPs) [10], i.e. either point mutations or small insertions or deletions (indels) in the genome sequence. In the first GWAS study to be published, 1133 affected individuals versus 1006 controls were tested for SNPs linked to myocardial infarction [11]. A total of 92 788 SNPs were genotyped, revealing one SNP that was strongly associated to the disease. Because such a correlation does not prove causality, further experimental testing confirmed that the SNP induced several cell-adhesion molecules and enhanced the transcription of lymphotoxin-α [11]. Thus, GWAS can be a useful tool for bioinformatic prioritization of candidate SNPs for further research.
A comparable approach is possible for the association of bacterial genes to phenotypes. When the first complete microbial genomes emerged, ‘differential genome analysis’ of the gene content of related microbes shortlisted gene candidates responsible for certain phenotypes [12,13]. In that study, candidate genes were selected based on a genome-wide comparison of Helicobacter pylori to its close relatives Haemophilus influenzae and Escherichia coli. It was proposed that the genes that were differentially present or absent might be responsible for H. pylori’s species-specific features, including the ability to colonize host cells in highly acidic environments. Currently, gene-trait matching or genotype–phenotype association is a more commonly used term for similar analyses.
The concept of correlating phenotypes to the presence or absence of genes in bacterial genomes was scaled up with the introduction of comparative genome hybridization (CGH) microarrays that measured genotypes with high throughput [14,15]. These microarrays were spotted with probes that map to the genes present in a single genome (CGH arrays) or in a number of strains of the same species [16,17], coined pan-genome arrays [18]. The arrays could then be hybridized with fluorescently labeled genomic cDNA to determine the gene content of a query strain [19,20]. For both single genome CGH arrays and the pan-genome arrays, the genes in the query genome can only be determined in terms of the reference sequences, provided that they are sufficiently similar to the probes on the array [20,21]. There are only a few studies describing CGH-based gene-trait matching. In part, this is because consistent trait annotations have been lacking. To correlate the gene content of the selected strains to their phenotypes, consistent phenotyping is required, i.e. the phenotypes should be determined under the same conditions for all the strains that are genotyped.
Today, sequencing has surpassed microarrays for accurately determining the gene content of an organism. Advantages of sequencing include the direct determination of transcripts by RNA sequencing, and the fact that sequencing allows the discovery of new genes, while the genomic diversity that can be determined by microarrays is limited to the reference sequences used for array probe design. Moreover, sequencing increases the resolution of comparative genomic analyses from gene content to individual SNPs. Several high-impact studies have provided draft genome sequences of many strains of the same species [22–25], where genome alignments allow the discovery of single nucleotide differences.
We argue that the greatest bottleneck for applying GWAS to microbial datasets is the availability of experimentally consistent phenotypes or other annotations, such as environmental parameters (metadata). Both types of genotyping data introduced above, CGH arrays and complete genome sequences, are potentially of high value for gene-trait matching, provided that the phenotypes of the strains are also consistently annotated. There are some promising examples, including a publication where 12 phenotypes were determined for 42 Lactobacillus plantarum strains [26], however, such datasets are scarce, in part because consistent culturing is crucial to reduce experimental noise. Thus, culturing studies and underpinning microbial physiology are crucial for making best use of the wealth of information that will be available.
In this review, we will focus on genotype–phenotype associations for groups of bacterial strains of the same species. First, it may be more likely that similar traits have been annotated for strains, than for phylogenetically more divergent organisms. Second, the genome sequences of different species are not easily alignable due to the greater differences in gene content and in genome structure. As a result, we would not be able to illustrate the use of SNPs as genotypic characters. Nevertheless, the methods outlined herein are equally suitable for genotype–phenotype associations across different species. We conclude with an outlook of the application of these methods to other data types, including the use of transcriptomic data across different experimental conditions for linking genes to functions within a single species, and the use of functional or taxonomic profiles across metagenomes to link functions or taxa to environmental parameters.
ASSEMBLY AND ANNOTATION
Understanding the functional potential encoded by a given genome starts with an accurate genome sequence and gene annotation. Next-generation sequencing techniques are increasingly being used to sequence the genomes of new microbial isolates [27–30]. As read lengths of most sequencing platforms are in the hundreds of nucleotides, it is imperative to assemble reads into larger contiguous sequences (contigs) and to order and orient contigs into larger scaffolds [31]. These larger DNA fragments allow better prediction of open reading frames (ORFs) and facilitate gene context analyses with comparative genomic approaches. For SNP typing of bacterial strains, the sequence quality of the assembly is very important and there are several strategies to correct the assembly for sequencing errors, including the detection of frameshifts by comparative genomics, and the correction of SNPs in an assembly using Illumina reads [32,33].
Genome annotations often start with submitting a genome sequence to an online annotation service [34,35]. This results in ab initio predicted ORFs consisting of start and stop positions, as well as a predicted function. Start and stop codon prediction is usually performed by ORF calling software implemented in these annotation engines, such as GLIMMER [36], GeneMark [37,38] or Prodigal [39]. It is crucial to use the same ORF prediction method for the different strains of interest, as differences in the ORF predictions could influence downstream analyses, including determining orthologs (see below). It should be mentioned that sequencing of transcripts now enables direct measurement of ORFs, which may be more accurate than automated ORF predictions.
Functional annotation of the predicted ORFs may involve many steps including homology searches to annotated databases, such as RefSeq [40], Genbank [41] and SwissProt [42] using BLAST [43], or hidden Markov model screenings with Pfam [44]. Annotation engines generally provide reasonably accurate automated function annotations for proteins, although they may show deficiencies in genotype–phenotype extrapolation [45–47]. Specifically, they are suited for annotating core metabolic genes, while for genes that are not widely conserved, manual curation remains an important step in identifying function [48]. The time necessary for the curation of gene functions can be reduced by (i) performing the function curation for a representative member of an orthologous group (OG) (see below) instead of for all members; (ii) concentrating curation efforts on the molecular functions of interest and (iii) by examining gene function predictions for targets resulting from the genotype–phenotype matching. The DNA sequences with putative ORFs and their annotations are then ready for comparative genomics and determining structural variations (SVs), single nucleotide polymorphisms (SNPs) and small insertions or deletions (indels).
ORTHOLOGOUS GROUPS OF GENES
Comparing the genes in a selection of genome sequences depends on a reliable annotation of orthologs. Coined by Fitch in 1970, orthology is an evolutionary concept that describes the relationship between genes that diverged following a speciation event [49]. Conversely, paralogy refers to genes that diverged following a gene duplication (Figure 1). A frequent misinterpretation of the concept of orthology is the idea that it signifies functional equivalence. Indeed, orthologs may be likely to represent functional equivalents because of their evolutionary definition, but the original definition per se contains no statement about conservation of function [49].
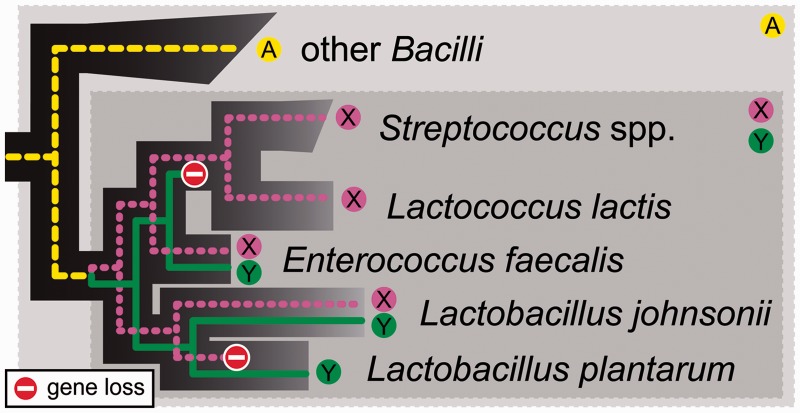
Figure 1:
The resolution of an OG depends on the age of the LCA for the studied species. The dark background tree indicates the evolutionary history of the included Bacilli; colored lines indicate the evolutionary history of the genes. Gene family A in the Bacilli duplicated in the LCA of the Lactobacillales to form the paralogs X and Y. When constructing OGs for all Bacilli, all the homologs A, X and Y will be united in one OG, where X and Y are called ‘in-paralogs’. If only Lactobacillales are taken into account, X and Y are placed in separate OGs because they had different ancestral genes in the more recent LCA. Note that when species are compared in pairs, paralogs may be mistaken for orthologs due to differential loss of paralogs, e.g. the Lactococcus lactis gene X and the L. plantarum gene Y. Orthology can be inferred at different levels of resolution by analyzing speciation events and gene duplication events in phylogenetic trees [119].
It is relatively straightforward to identify the orthologous genes or proteins for pairs of species by reciprocal homology searches [50]. Comparative genomics of more than two genomes requires an annotation of group orthology [51,52]. OGs of genes can be interpreted as gene families, and consist of all the genes that have descended from a single ancestral gene in the last common ancestor (LCA) of the species considered. Thus, the resolution of a set of OGs depends on which group of species is considered (Figure 1). If the group shares an ancient LCA, say the ancestor of all bacteria (or Bacilli in Figure 1), the single ancestral genes will have had ample time to evolve and diverge, resulting in large and inclusive OGs that may have many representatives in each genome (paralogs). Conversely, a very recent LCA, say for a group of strains of one bacterial species (or the order Lactobacillales in Figure 1) leads to smaller, higher resolved OGs with one or a few paralogs in each of the genomes.
There are several methods and tools available that assign genes to OGs for a given set of genomes. These can be divided into methods that construct de novo OGs and methods that map proteins to existing OGs by using sequence similarity (both approaches are elaborated in the paragraphs below). Methods that map proteins to existing OGs are more practical to use than de novo reconstruction of OGs, but have several disadvantages. For example, the resolution of the existing OGs may not be optimal for the selection of species under consideration: the resolution may either be too low (if OGs were reconstructed for a more ancient LCA), or too high, leading to arbitrary subdivision of proteins into distinct OGs, that actually belong in the same OG because they share a single ancestral gene. Moreover, genes that are specific to the newly sequenced genome will not be mapped.
For construction of new OGs, it is imperative that complete genome sequences are available with accurate gene predictions, so that any paralogy relationships can be identified. Given that we are currently within the era of short-read shotgun sequencing, it should be noted that paralogs may accidentally be collapsed by short-read assemblers if they are very similar, e.g. when creating a genome assembly by mapping the sequencing reads to a closely related reference genome [53]. Established tools for de novo OG reconstruction include InParanoid/Multiparanoid [50,54] and OrthoMCL [55]. These programs do not rely on phylogenetic reconstruction, which is computationally expensive and becomes increasingly prohibitive with larger datasets. Finally, orthologs may not necessarily be more similar in sequence [56], and additional information including gene neighborhood (operon structure) can be used to refine their definition.
As sequence databases become more comprehensive, the mapping methods gain in resolution and form a viable alternative to de novo OG reconstruction methods. A simple approach is to provide the proteins in the database with OG annotations, and after searching this database with the query genes, assign each to the OG of its highest scoring hit. This works well if the query genome is a close relative of genomes in the database, and the speed of the RAST server (Rapid Annotations using Subsystems Technology [35]) in part depends on this approach. For genomes that do not have reliably sequenced close relatives, the top hit approach might be more likely to spuriously yield distant hits from a different OG. In these cases, Cognitor [57] provides an extension of the simple top-hit rule, by assigning genes to an OG if the majority of the top scoring proteins map to the same OG. Note that proteins belonging to different OGs may occur as fused genes, in which case the Cognitor rule should be applied separately to each of the fused regions. The Signature webserver [58] automatically performs these steps, assigning proteins or protein regions to the OGs from the STRING database [59]. Alternatively, the EggNOG webserver [60] also allows the assignment of protein sequences to OGs.
PHAGES
Prophages and mobile elements are the most variable fraction of microbial genomes and often encode functions related to interaction with the changing environment [61], including pathogenicity genes [62]. Moreover, they may contain genes that help bacteria cope with adverse environmental conditions like sub-lethal concentrations of antibiotics or withstanding osmotic, oxidative and acid stresses, as well as increasing growth and influencing biofilm formation [63]. Prophages often contribute the biggest difference between strains and may account for many kilobases of divergent sequence. For example, Sodalis glossinidius strain ‘morsitans’ contains 26 prophages that together cover 956 349 bp of this 5MB genome [64]. In general, prophages encode three types of genes: (i) genes required for the phage cell cycle including phage entry, DNA integration and excision, DNA replication, packaging, and cell lysis, and the expression regulation of those genes; (ii) genes that inhibit the cell cycle of other phages (super-infection exclusion) and (iii) auxiliary metabolic genes that generate energy in the cell and promote viral replication. Thus, we expect prophage and mobile element content to be important for explaining phenotypic differences between closely related strains.
Phage genes can be identified by similarity to known homologs in a relevant database of viral proteins, such as Phantome (http://www.phantome.org/). However, because viruses mutate rapidly [65] and are under-represented in the databases [66], this may not always be sufficient to identify all prophages in a newly sequenced genome. Moreover, it should be noted that prophages often contain homologous sequences inserted at different loci around the genome, hindering sequence assembly efforts. Prophages have specific characteristics, including protein length, transcription strand directionality, AT/GC skews, phage-specific oligonucleotides and phage insertion points (even though different strains can have different prophages inserted at the same chromosomal integration site), that can be used for identification. Recently, the tool PhiSpy [64] has been developed that uses combinations of these characteristics, as well as homology, to identify prophages in bacterial genome sequences. Once phages have been identified, they can be mapped between strains using orthology and gene content.
MUTATIONS
It is possible that the differential presence of OGs, functions, mobile elements or phages cannot explain a given phenotype, if mutations such as SNPs or indels are responsible for gain- or loss-of-function. One famous example is the presence of point mutations in the E. coli fimH gene that alters its host specificity [67,68]. Another recent example in fungi is that of very recently diverged Aspergillus fumigatus strains that have acquired resistance to toxic azole compounds by a mutation in the transcription factor subunit HapE [69], but have an otherwise identical genetic background. Such small mutations can also be included into the list of genotypic features, and associated to phenotypes in the same way as presence/absence patterns of OGs or phages (above). For brevity we will refer to these mutations as SNPs, which could stand for simple (instead of single) nucleotide polymorphisms [10]. In microbial genomics, the state of the art for SNP detection between strain variants consists of mapping the reads to a reference sequence and identifying variants in the conserved regions [23]. Although this will miss larger SV, it will suffice for our purpose, as differences in OG or phage content are already identified with the approaches above.
PHENOTYPES
Phenotypes are here defined as the observable characteristics of an organism. For bacteria, these phenotypes often consist of growth on specific media, for example, containing different carbon sources [26]. Phenotype microarrays [70,71] are a relatively straightforward way of assessing many conditions in parallel, as exemplified by a study of the soil bacterium Sinorhizobium meliloti [72]. Another example of a phenotype is the resistance to certain antibiotics, such as measured for the progeny of sexual crossing experiments of A. fumigatus [69], or for different bacteria isolated from patients at intensive care units [73]. In the latter study, the observed resistance correlates with geography or other differences in the patient populations. Other phenotypes include: (i) survival/growth under different experimental conditions or presence in ecological niches [74–81]; (ii) the ability to perform a specific molecular function [26,76,82,83] and (iii) metabolomic fingerprints [84].
To perform comparative studies encompassing many strains, it is imperative that phenotypic annotations are consistently performed across the strains. A standard in the description of the environment (metadata including sampling point and habitat) and the actual nucleic acid sequence of individual strains has been launched by the Genomic Standards Consortium, coined the Minimum Information about a Genome Sequence [85]. We recommend that this minimal standard be followed, also when large collections of strains are sequenced simultaneously. Moreover, we imagine that extensions to this standard, that describes the environment and the sequence, may be formulated for the further phenotypic description of specific taxonomic clades or species. An example could be running a standard phenotype microarray for every sequenced strain and one that measures specific properties for the taxonomic group in question. Optimally, the availability of consistent phenotypic information about sequenced microbial strains will allow the application of gene-trait matching approaches, such as those described below, even to bacterial strains sequenced and phenotyped by different research groups. It should be noted that this is not always possible. For example, genome sequences are increasingly being produced from organisms that are not grown in pure culture or even from single uncultured cells.
Phenotype data may be noisy due to several factors, including inaccuracies in the measurements. Moreover, phenotypic measurements assessed on a continuous scale are often converted to categorical values such as ‘Yes’ or ‘No’. However, it might be difficult to categorize some measurements accurately, resulting in ambiguous categories (or phenotypes) such as ‘Maybe’ or ‘Mild’. In association analysis, samples with ambiguous phenotypes are preferably discarded to increase statistical power. For example, some cases that are annotated as ‘Maybe’ may in fact be a ‘No’ or a ‘Yes’, and to prevent noise from obscuring the association signal we recommend excluding such cases from the analysis.
ASSOCIATION ANALYSIS
Association analysis is a multi-step process that links the individual elements of the genotype (e.g. genes, phages, SNPs) to specific phenotypes (e.g. growth on different media, antibiotic resistance; see Figure 2). Depending on the experiment and the association approach, different pre-processing steps will be necessary for the genotype and phenotype data. Below we will describe these steps in more detail.
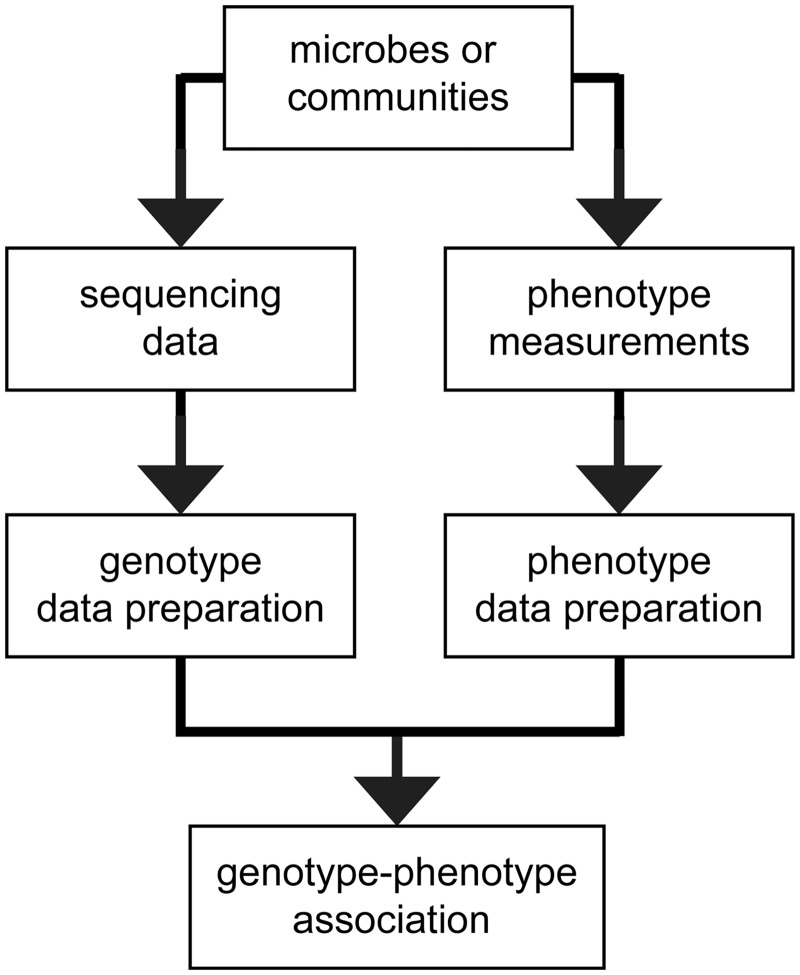
Figure 2:
Flow diagram for genotype–phenotype association analysis. Genomic and phenotypic data are collected for microbial strains. Phenotypes can be determined by, e.g. phenotype microarrays or analytical profile indices. Both the genotypic and phenotypic data are then preprocessed before genotype–phenotype association analysis. In the association analysis, correlations between genotype and phenotype are determined and visualized.
Large p small n
Many statistical and classification methods are suited for datasets including few measurements (e.g. genotypic variables) for many samples (e.g. strains). Conversely, the number of variables measured in high-throughput experiments is often much larger than the number of samples (genomes). This is referred to as the large p small n problem. With few samples, each individual sample has a relatively high importance, and there is a high risk of overtraining the data to individual samples. In such situations, machine learning methods are more suitable than classical statistical methods [86]. Accuracy of a machine learning algorithm can be improved by pre-processing input data, which could also decrease the number of variables significantly. A valid question is how many strains are necessary for a good signal in genotype–phenotype association studies. In general, more samples yield a better statistical power. While we have been able to extract meaningful links with as few as five positive and five negative cases (see also the section on ‘Visualization’ below), it has been stated that statistical issues can arise when sample sizes drop below 30 subjects. Such a small sample lacks heterogeneity (i.e. diversity) and does not approximate the normal distribution [87]. If possible, we recommend using at least 30 genomes in the association analysis, provided that matching phenotypic data is available.
Data filtering
When using data from high-throughput platforms that simultaneously assess many variables for many samples (e.g. next-generation sequencing technologies or phenotype microarrays), it is often necessary to apply some kind of pre-processing to filter for intrinsic noise [70,88]. Moreover, separately observed variables may contain redundant information for association analysis, for example, if two genes are in the same operon they will likely show the same presence/absence pattern across strains, and it might be better to collapse them. The most straightforward way to remove redundant variables is by using a correlation metric and combining strongly correlated features [26]. Finally, variables could be non-informative: i.e. a variable has the same or similar values across all samples, for example, a housekeeping gene that is present in every genome. Because these noisy, redundant or non-informative observations will not add relevant information about the different phenotypes, they need to be excluded from the association analysis [89,90].
Biased sampling of phenotypes
In addition to noise, the number of samples for each phenotype also affects the association analysis. In particular, an imbalanced distribution of samples across phenotypes decreases the accuracy of classification algorithms [91]. Due to overtraining of the algorithm toward the largest phenotypic group, strains from relatively rare phenotypic groups will often be classified into the more frequent phenotypic groups, as this is a ‘safe bet’ for the classifier. The effect of this phenotype imbalance can be decreased by bagging, where each bag contains an equal number of randomly selected samples, or by assigning reversely proportional weights to phenotypes [91,92].
Genotype–phenotype association
Here, we describe three approaches for genotype–phenotype association (Figure 3): statistical tests, correlation analysis and machine learning. Note that although these methods are often used, they are merely examples that illustrate a range of possibilities.
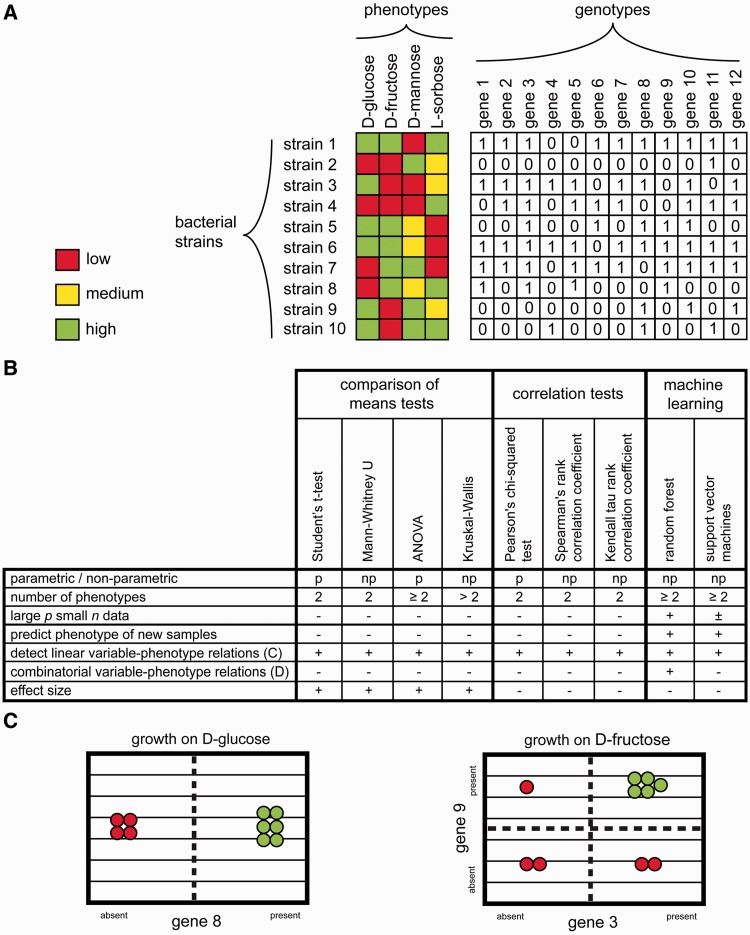
Figure 3:
Choosing an approach for genotype–phenotype association. (A) Dataset consisting of phenotypes (e.g. growth rates on different carbon sources) and genotypes (e.g. gene content) for 10 bacterial strains (rows). (B) Nine possible methods (four comparison of means statistical tests, three correlation analyses and two machine learning methods) for detecting genotype–phenotype associations. The compatibility with specific data types and applicability in microbial GWAS is shown. (C) Hypothetical example of a linear genotype–phenotype relation. Green strains grow on d-glucose; red strains do not. The presence of gene 8 is predictive of the growth on d-glucose. (D) Hypothetical example of a combinatorial genotype–phenotype relation. All six strains that grow on d-fructose contain gene 9 and gene 3. In other words: the interaction between gene 9 and gene 3 is predictive of the growth on d-fructose.
Comparison of means
One of the most straightforward ways of detecting genotype–phenotype associations is by using a statistical test to compare the mean values of the genotypic variables between different phenotypes. It is important to know the distribution of the data, because it determines which tests can be used. For normally distributed data, (parametric) the Student’s t-test (two phenotypes) [93] and ANOVA (two or more phenotypes) [94] are used, for other data the respective non-parametric (rank based) counterparts can be used, i.e. the Mann–Whitney U (two phenotypes) [93] and Kruskal–Wallis tests (two or more phenotypes) [95]. After multiple testing correction, these statistical tests supply the user with a P-value for each variable, which indicates the probability that the distributions of the different phenotypes do not differ significantly. As each genotypic variable is tested individually, no combinatorial dependencies between variables can be detected.
Correlation tests
Thus far, we have only discussed discrete genotypic variables (e.g. presence/absence of OGs, SNPs or phages). However, in some cases, it may be possible to assign continuous values (e.g. gene copy number or percent sequence identity). Similarly, some phenotypes may also be better represented as continuous values rather than dividing them into groups. In such cases, it is more appropriate to correlate the genotypic and phenotypic values. Similarly as for the comparison of means tests, the distribution of the data is an important factor in determining which method to use. For normally distributed data, the Pearson’s chi-squared test [96] can be applied. The other two methods listed in Figure 3, the Kendall tau rank correlation coefficient (tau) [97] and Spearman’s rank correlation coefficient (rho) [98] are both non-parametric, which means that they do not rely on a specific distribution in the data. On average, rho gives a higher correlation value than tau, while more severely penalizing individual samples that correlate badly [99]. As for the comparison of means tests, each genotypic variable is tested individually, and no combinatorial dependencies can be detected.
Machine learning
Machine learning methods, including random forest (RF) [100] and support vector machines (SVMs) [101] use training data to create a classifier (predictor) to classify phenotypes of new samples. These methods can generate an importance score for each of the genotypic variables for distinguishing the phenotypes (this is a native feature of RF; SVM requires an additional module such as the R module caret). Using this importance, variables can be removed that do not significantly contribute to the classification or improve the prediction accuracy [26,102,103]. The advantage of machine learning methods is that they build decision models that encompass multiple variables at once, allowing the prediction of phenotypes based on combinations of genotypic variables, i.e. two variables that contain no information on their own, but are predictive when assessed together (see Figure 3D) [104–106]. These interactions are implicitly modeled in current implementations of RF and SVM. However, there is no explicit importance measure for interactions between variables. Using machine learning methods allows the selection of the most important variables for further visualization (see below), interpretation and follow-up experiments in the laboratory.
We note that the steps outlined earlier can be applied to answer many different types of research questions that aim to find relations between large-scale datasets. Advanced users may choose to perform all the data processing steps using R and a set of specific modules. As an alternative, web tools are available such as PhenoLink [26] that uses RFs, and automatically performs many of the steps outlined above. Such automated pipelines allow for rapid, in-depth analysis of large amounts of data.
VISUALIZATION
Mining very large datasets, like the collections of genotypic variables described above, may yield many significant associations. Although these genotype–phenotype associations are selected on the basis of their statistical significance, some associations might make more biological sense than others. For instance, the absence of a gene could have a high correlation with growth on a given carbon source, but if there is no function known for that gene (e.g. the gene could be a regulator repressing an operon required for growth on that sugar) a meaningful explanation for the statistical association might be difficult to predict. The integration of additional biological descriptors can help to formulate a meaningful interpretation of the results and to select the most promising associations for follow-up experiments.
Visualization allows us to integrate multiple information sources, including biological descriptors, and facilitate distinguishing relevant from irrelevant links. An optimal visualization would allow a general overview of the data, while simultaneously providing the possibility for in-depth examination of specific associations. To allow the inclusion of different sources of information, the visualization may require using multiple dimensions, each dimension representing a different source of information. Network graphs are an often used visualization that allows the incorporation of data from different sources [107]. However, such graphs can quickly become complex and often very large for many associations. Moreover, incorporating information from different sources is cumbersome when they are inter-dependent. For instance, visualizing a link between a gene and a phenotype based on their co-occurrence across a selection of strains could illustrate why this specific gene is predicted to be important. While visualization of such a three-way relationship (gene-strain-phenotype) is straightforward for a single gene, it is not easy for hundreds of genes and multiple phenotypes. Thus, color-coded tables (Figure 4) can be used to represent information using two different views: (i) a general figure that shows the relationships between all selected variables and their related classes and (ii) a detailed figure that shows the relationships between variables and samples for only a few classes. Similarly, multiple scales can be achieved in graph-based visualizations by interactive visualization of associations, initially showing a general view and also allowing interactive browsing of specific relations [108].
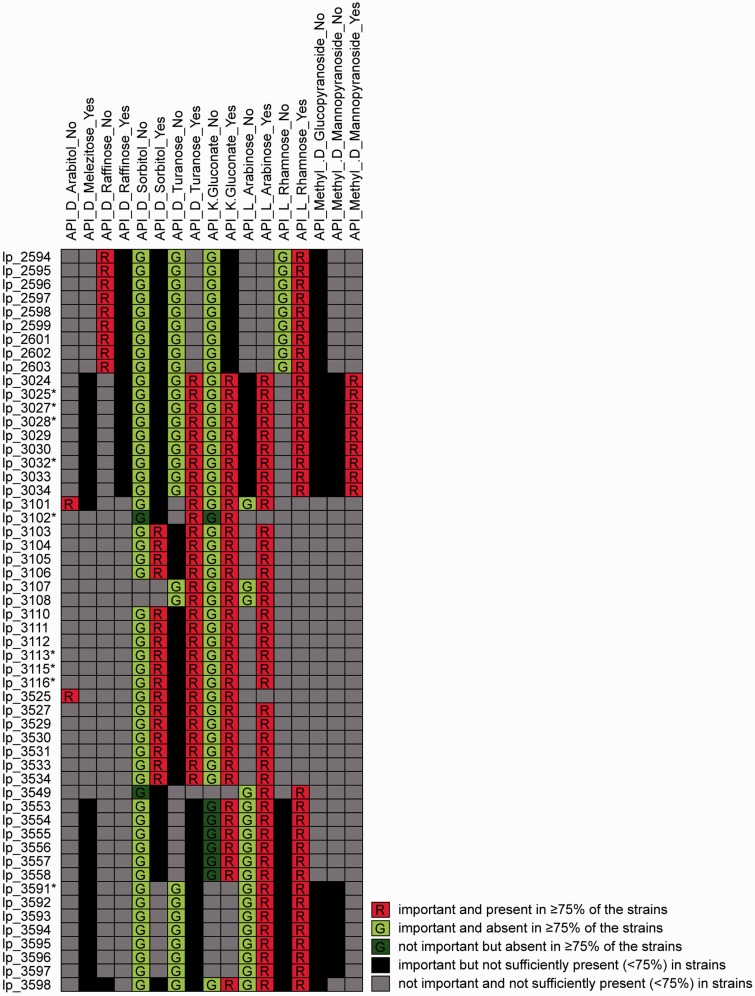
Figure 4:
Different ways to visualize L. plantarum genes that were found to be important to predict growth or non-growth on multiple sugars using PhenoLink [26]. Color-coded table of links between the 54 selected genes and growth on different sugars using from PhenoLink. ‘Yes’ or ‘No’ suffixes in column names indicate growth and non-growth, respectively. Asterisks (*) besides gene names (rows) indicate that the gene could not be mapped to COGs (see Figure 6). The color scheme integrates the importance of genes to predict phenotypes, and their occurrence in strains with that phenotype: bright red/green indicates genes that are important to a phenotype and present/absent in ≥75% of the strains with this phenotype; dim red/green indicates genes that are not important to a phenotype but are present/absent in ≥75% of the strains with this phenotype; black indicates genes that are important to a phenotype but are not sufficiently present/absent (<75%) in strains with this phenotype; gray indicates genes that are not important to a phenotype and are not sufficiently present/absent (<75%) in strains with this phenotype.
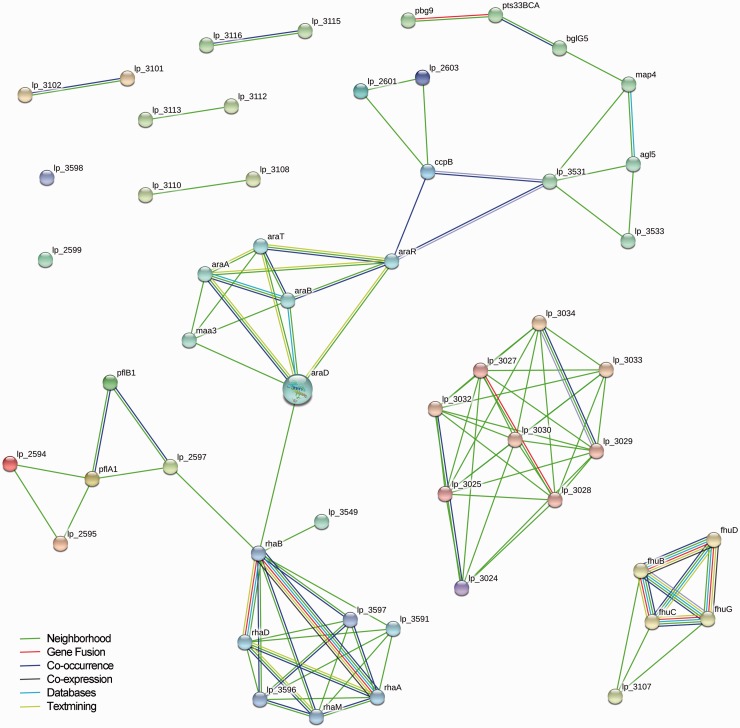
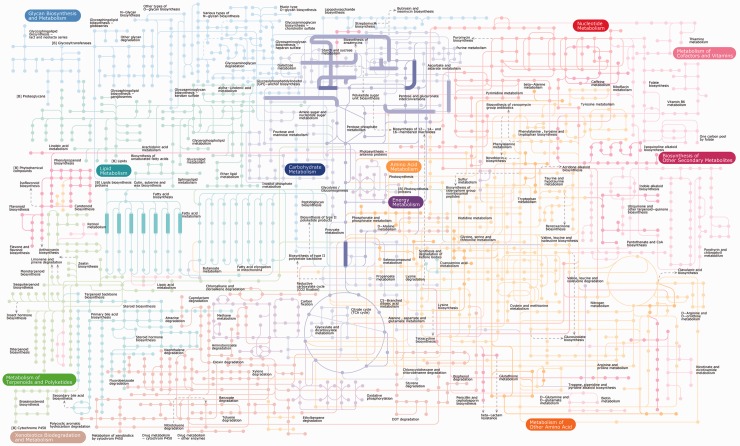
To illustrate the different possible visualizations, we show the results of a genotype–phenotype association analysis of 42 L. plantarum strains performed using PhenoLink [26]. All three visualizations show the same genes (genotype) selected for their importance in predicting one of the phenotypes (growth or non-growth on several sugars). The first visualization of these data comes from PhenoLink, using color-coding as an additional dimension of information to visualize the relations between 54 genes and 17 phenotypes (Figure 4). This figure highlights: (i) relations between genes that are relevant to similar phenotypes; (ii) relations between phenotypes that share a similar set of related genes and (iii) relations between individual genes and phenotypes. The second visualization, made with the graph-based visualization tool STRING [3], shows proteins as nodes and predicted pairwise interactions as edges (Figure 5). This graph shows additional links that have been mined from publicly available literature and experimental datasets. The third visualization (Figure 6) was made with iPath [109], and highlights the proteins in a global map of metabolic pathways, placing the selected genes in the context of their biological system.
Figure 5:
Different ways to visualize L. plantarum genes that were found to be important to predict growth or non-growth on multiple sugars using PhenoLink [26]. STRING evidence graph [3] of all 53 genes important for growth or non-growth on multiple sugars (all phenotypes combined). The gene lp_3111 did not encode a protein and was omitted from this figure.
Figure 6:
Different ways to visualize L. plantarum genes that were found to be important to predict growth or non-growth on multiple sugars using PhenoLink [26]. iPath global metabolic map [109] of the same genes mapped to COGs (47 unique COGs), where reactions with at least one mapped COG are indicated with a thick line.
CONCLUSIONS AND FUTURE PERSPECTIVES
Recent technological advances, combined with large-scale genotype–phenotype association studies, hold a great promise for microbial functional genomics. Specifically, next-generation sequencing technologies have made DNA sequencing orders of magnitude faster and cheaper [27], and advances including the phenotype microarray [70,71] allow high-throughput measurement of microbial phenotypes. However, while bacterial genome sequences are appearing faster than they can be analyzed, the consistent measurement of phenotypes across the sequenced strains is often still lacking. Some examples of available datasets involving lactic acid bacteria include growth on different carbon sources [26,74,78]] organic acid production (measured in a single L. plantarum strain grown under different fermentation conditions [110]), adhesion to eukaryotic cells (Saccharomyces cerevisiae [111]) and IL-10 and IL-12 response in human cell lines (measured for 42 L. plantarum strains [112,113]).
The association methods described herein for discovering genotype–phenotype associations can also be applied to other types of data. For example, it may be valuable to link metagenomic entities including functions or taxa (operational taxonomic units) observed across metagenomic samples to clinical [114] or environmental [115] metadata. Indeed, discovery approaches have been published for such ‘metagenome-wide’ association of environmental parameters to metagenomic entities [115–117].
One of the disadvantages of the static measures of genotype described in this review (e.g. presence of OGs, phages and SNPs on the genome) is that they do not take into account other levels of cellular regulation, such as gene expression and protein abundance. Although the presence of OGs, phages or SNPs on a genome may have important consequences for the functioning of an organism, the question remains whether the gene product is actually present and functional. Addressing this issue, the first transcriptome-trait matching studies have recently been published [110,118], which are a particularly attractive way of comparing genotype and phenotype, and designing testable hypotheses. These studies determined the transcriptomes of a single strain grown under different conditions. Moreover, several phenotypes were measured in each of these cultures, including the production of organic acids, and the survival in the gastrointestinal tract. These phenotypes were then linked to the genes identified in the transcriptomes to discover which gene products correlated best with each specific phenotype.
Future microbial genotype–phenotype association studies will require the integration of consistent genome annotations with consistent phenotypic datasets, available in a computer readable format. The challenge does not lie in the generation of sequence data or in the development of novel statistical techniques, but rather in the generation, annotation and databasing of phenotypic data about the genomes and metagenomes that are being studied. Novel computational techniques, see for example [102], applied to these large datasets will allow determining interacting genes or SNPs that govern currently not understood complex phenotypes. These findings in turn will fuel the prediction of novel microbial properties for newly sequenced strains.
Key points.
The growing number of (draft) genome sequences constitutes a rich resource for microbial functional genomics.
Phenotypes should be consistently measured and documented for the sequenced strains, so that computational tools can be readily applied.
Datasets of consistently measured phenotypes across a collection of sequenced strains are still rare.
Visualization can turn the sometimes abundant statistically significant genotype–phenotype associations into biological interpretations.
Funding
B.E.D. is supported by the Netherlands Organisation for Scientific Research (NWO) Veni grant (016.111.075). R.A.E. is supported by NSF grants DBI: 0850356 from the Division of Biological Infrastructure and DEB: 1046413 from the Division of Environmental Biology. J.R.B. is supported by the Dutch e-Science Grid (BiG Grid), which is financially supported by NWO.
Biographies
Bas E. Dutilh uses comparative genomics and other approaches to answer questions about the functioning and evolution of genomes and microbial communities.
Lennart Backus a PhD student and develops phylogenomics techniques for sequence-based prediction of microbial interactions.
Robert A. Edwards develops computational tools for genome and metagenome analysis and data mining.
Michiel Wels is group leader bioinformatics at NIZO food research and is involved in applying bioinformatics approaches to different food-related research questions.
Jumamurat R. Bayjanov is involved in analyzing next-generation sequence data and developing machine-learning tools.
Sacha A.F.T. van Hijum is a senior scientist bioinformatics at NIZO food research and head of the CMBI bacterial genomics group. Research at the CMBI group focuses on establishing the relation between microbial consortia and health.
References
- 1.Bork P, Dandekar T, Diaz-Lazcoz Y, et al. Predicting function: from genes to genomes and back. J Mol Biol. 1998;283:707–25. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 2.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–33. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korbel JO, Jensen LJ, von Mering C, et al. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22:911–7. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 5.Kensche PR, Oti M, Dutilh BE, et al. Conservation of divergent transcription in fungi. Trends Genet. 2008;24:207–11. doi: 10.1016/j.tig.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Kensche PR, van Noort V, Dutilh BE, et al. Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. J R Soc Interface. 2008;5:151–70. doi: 10.1098/rsif.2007.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorci L, Blaby I, De Ingeniis J, et al. Genomics-driven reconstruction of acinetobacter NAD metabolism: insights for antibacterial target selection. J Biol Chem. 2010;285:39490–9. doi: 10.1074/jbc.M110.185629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–83. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 10.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–9. [PubMed] [Google Scholar]
- 11.Ozaki K, Ohnishi Y, Iida A, et al. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32:650–4. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- 12.Huynen M, Dandekar T, Bork P. Differential genome analysis applied to the species-specific features of Helicobacter pylori. FEBS Lett. 1998;426:1–5. doi: 10.1016/s0014-5793(98)00276-2. [DOI] [PubMed] [Google Scholar]
- 13.Huynen MA, Diaz-Lazcoz Y, Bork P. Differential genome display. Trends Genet. 1997;13:389–90. doi: 10.1016/s0168-9525(97)01255-9. [DOI] [PubMed] [Google Scholar]
- 14.Salama N, Guillemin K, McDaniel TK, et al. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci U S A. 2000;97:14668–73. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malloff CA, Fernandez RC, Lam WL. Bacterial comparative genomic hybridization: a method for directly identifying lateral gene transfer. J Mol Biol. 2001;312:1–5. doi: 10.1006/jmbi.2001.4925. [DOI] [PubMed] [Google Scholar]
- 16.Willenbrock H, Petersen A, Sekse C, et al. Design of a seven-genome Escherichia coli microarray for comparative genomic profiling. J Bacteriol. 2006;188:7713–21. doi: 10.1128/JB.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayjanov JR, Wels M, Starrenburg M, et al. PanCGH: a genotype-calling algorithm for pangenome CGH data. Bioinformatics. 2009;25:309–14. doi: 10.1093/bioinformatics/btn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medini D, Donati C, Tettelin H, et al. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–94. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Dorrell N, Mangan JA, Laing KG, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–15. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gresham D, Dunham MJ, Botstein D. Comparing whole genomes using DNA microarrays. Nat Rev Genet. 2008;9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrenreich A. DNA microarray technology for the microbiologist: an overview. Appl Microbiol Biotechnol. 2006;73:255–73. doi: 10.1007/s00253-006-0584-2. [DOI] [PubMed] [Google Scholar]
- 22.Harris SR, Clarke IN, Seth-Smith HM, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 2012;44:413–9, S1. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris SR, Feil EJ, Holden MT, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutreja A, Kim DW, Thomson NR, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt KE, Baker S, Weill FX, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44:1056–9. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayjanov JR, Molenaar D, Tzeneva V, et al. PhenoLink—a web-tool for linking phenotype to ∼omics data for bacteria: application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics. 2012;13:170. doi: 10.1186/1471-2164-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean D, Jones JD, Studholme DJ. Application of ‘next-generation' sequencing technologies to microbial genetics. Nat Rev Microbiol. 2009;7:287–96. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- 28.Loman NJ, Constantinidou C, Chan JZ, et al. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol. 2012;10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 29.Wu D, Hugenholtz P, Mavromatis K, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–60. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavromatis K, Land ML, Brettin TS, et al. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS One. 2012;7:e48837. doi: 10.1371/journal.pone.0048837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6–12. doi: 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
- 32.Klassen JL, Currie CR. Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics. 2012;13:14. doi: 10.1186/1471-2164-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carneiro AR, Ramos RT, Barbosa HP, et al. Quality of prokaryote genome assembly: indispensable issues of factors affecting prokaryote genome assembly quality. Gene. 2012;505:365–7. doi: 10.1016/j.gene.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Siezen RJ, van Hijum SA. Genome (re-)annotation and open-source annotation pipelines. Microb Biotechnol. 2010;3:362–9. doi: 10.1111/j.1751-7915.2010.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delcher AL, Bratke KA, Powers EC, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–4. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyatt D, Chen GL, Locascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruitt KD, Tatusova T, Brown GR, et al. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–5. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2011;39:D32–7. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bairoch A, Apweiler R, Wu CH, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–9. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson DA, Tetu SG, Phillippy K, et al. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet. 2008;4:e1000211. doi: 10.1371/journal.pgen.1000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsiao TL, Revelles O, Chen L, et al. Automatic policing of biochemical annotations using genomic correlations. Nat Chem Biol. 2010;6:34–40. doi: 10.1038/nchembio.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnoes AM, Brown SD, Dodevski I, et al. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson EJ, Watson M. The automatic annotation of bacterial genomes. Brief Bioinform. 2013;14:1–12. doi: 10.1093/bib/bbs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 50.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–52. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 51.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 52.Dutilh BE, van Noort V, van der Heijden RT, et al. Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics. 2007;23:815–24. doi: 10.1093/bioinformatics/btm015. [DOI] [PubMed] [Google Scholar]
- 53.Dutilh BE, Huynen MA, Strous M. Increasing the coverage of a metapopulation consensus genome by iterative read mapping and assembly. Bioinformatics. 2009;25:2878–81. doi: 10.1093/bioinformatics/btp377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexeyenko A, Tamas I, Liu G, et al. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dutilh BE, Huynen MA, Snel B. A global definition of expression context is conserved between orthologs, but does not correlate with sequence conservation. BMC Genomics. 2006;7:10. doi: 10.1186/1471-2164-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatusov RL, Galperin MY, Natale DA, et al. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutilh BE, He Y, Hekkelman ML, et al. Signature, a web server for taxonomic characterization of sequence samples using signature genes. Nucleic Acids Res. 2008;36:W470–4. doi: 10.1093/nar/gkn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen LJ, Kuhn M, Stark M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Powell S, Szklarczyk D, Trachana K, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–9. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nogueira T, Rankin DJ, Touchon M, et al. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr Biol. 2009;19:1683–91. doi: 10.1016/j.cub.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brussow H. Bacteria between protists and phages: from antipredation strategies to the evolution of pathogenicity. Mol Microbiol. 2007;65:583–9. doi: 10.1111/j.1365-2958.2007.05826.x. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Kim Y, Ma Q, et al. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akhter S, Aziz RK, Edwards RA. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012;40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonhoeffer S, Sniegowski P. Virus evolution: the importance of being erroneous. Nature. 2002;420:367, 369. doi: 10.1038/420367a. [DOI] [PubMed] [Google Scholar]
- 66.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weissman SJ, Moseley SL, Dykhuizen DE, et al. Enterobacterial adhesins and the case for studying SNPs in bacteria. Trends Microbiol. 2003;11:115–7. doi: 10.1016/s0966-842x(03)00010-6. [DOI] [PubMed] [Google Scholar]
- 68.Sokurenko EV, Courtney HS, Ohman DE, et al. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J Bacteriol. 1994;176:748–55. doi: 10.1128/jb.176.3.748-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camps SMT, Dutilh BE, Arendrup MC, et al. Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One. 2012;7(11):e50034. doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–55. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biondi EG, Tatti E, Comparini D, et al. Metabolic capacity of Sinorhizobium (Ensifer) meliloti strains as determined by phenotype MicroArray analysis. Appl Environ Microbiol. 2009;75:5396–404. doi: 10.1128/AEM.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akulian JA, Metersky ML. Antibiotic resistance patterns in medical and surgical patients in a combined medical-surgical intensive care unit. J Crit Care. 2012 doi: 10.1016/j.jcrc.2012.02.006. doi:10.1016/j.jcrc.2012.02.006 (Advance Access publication 27 March) [DOI] [PubMed] [Google Scholar]
- 74.Siezen RJ, Tzeneva VA, Castioni A, et al. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol. 2010;12:758–73. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- 75.Burall LS, Laksanalamai P, Datta AR. Listeria monocytogenes mutants with altered growth phenotypes at refrigeration temperature and high salt concentrations. Appl Environ Microbiol. 2012;78:1265–72. doi: 10.1128/AEM.06576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marti S, Nait Chabane Y, Alexandre S, et al. Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS One. 2011;6:e26030. doi: 10.1371/journal.pone.0026030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nannapaneni P, Hertwig F, Depke M, et al. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology. 2012;158:696–707. doi: 10.1099/mic.0.055434-0. [DOI] [PubMed] [Google Scholar]
- 78.Siezen RJ, Bayjanov JR, Felis GE, et al. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb Biotechnol. 2011;4:383–402. doi: 10.1111/j.1751-7915.2011.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filocamo A, Nueno-Palop C, Bisignano C, et al. Effect of garlic powder on the growth of commensal bacteria from the gastrointestinal tract. Phytomedicine. 2012;19:707–11. doi: 10.1016/j.phymed.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 80.Habib F, Johnson AD, Bundschuh R, et al. Large scale genotype-phenotype correlation analysis based on phylogenetic trees. Bioinformatics. 2007;23:785–8. doi: 10.1093/bioinformatics/btm003. [DOI] [PubMed] [Google Scholar]
- 81.Fuste E, Galisteo GJ, Jover L, et al. Comparison of antibiotic susceptibility of old and current Serratia. Future Microbiol. 2012;7:781–6. doi: 10.2217/fmb.12.40. [DOI] [PubMed] [Google Scholar]
- 82.Siezen RJ, Starrenburg MJ, Boekhorst J, et al. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl Environ Microbiol. 2008;74:424–36. doi: 10.1128/AEM.01850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y, Li J, Sam L, et al. An integrative genomic approach to uncover molecular mechanisms of prokaryotic traits. PLoS Comput Biol. 2006;2:e159. doi: 10.1371/journal.pcbi.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–71. [PubMed] [Google Scholar]
- 85.Field D, Garrity G, Gray T, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang M, Zhang D, Wells MT. Variable selection for large p small n regression models with incomplete data: mapping QTL with epistases. BMC Bioinformatics. 2008;9:251. doi: 10.1186/1471-2105-9-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fitzner K, Heckinger E. Sample size calculation and power analysis: a quick review. Diabetes Educ. 2010;36:701–7. doi: 10.1177/0145721710380791. [DOI] [PubMed] [Google Scholar]
- 88.Quince C, Lanzen A, Curtis TP, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–41. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 89.Strobl C, Boulesteix AL, Zeileis A, et al. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinformatics. 2007;8:25. doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nicodemus KK, Malley JD. Predictor correlation impacts machine learning algorithms: implications for genomic studies. Bioinformatics. 2009;25:1884–90. doi: 10.1093/bioinformatics/btp331. [DOI] [PubMed] [Google Scholar]
- 91.Blagus R, Lusa L. Class prediction for high-dimensional class-imbalanced data. BMC Bioinformatics. 2010;11:523. doi: 10.1186/1471-2105-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen C, Liaw A, Breiman L. Using Random Forest to Learn Imbalanced Data, in Statistics Technical Reports. Berkeley, CA: Department of Statistics, UC Berkeley; 2004. [Google Scholar]
- 93.Fay MP, Proschan MA. Wilcoxon-Mann-Whitney or t-test? On assumptions for hypothesis tests and multiple interpretations of decision rules. Stat Surv. 2010;4:1–39. doi: 10.1214/09-SS051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gelman A. Analysis of variance—Why it is more important than ever. Ann Stat. 2005;33:1–31. [Google Scholar]
- 95.Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621. [Google Scholar]
- 96.Plackett RL. Pearson, Karl and the chi-squared test. Int Stat Rev. 1983;51:59–72. [Google Scholar]
- 97.Kendall MG. A new measure of rank correlation. Biometrika. 1938;30:81–93. [Google Scholar]
- 98.Spearman C. The proof and measurement of association between two things. Int J Epidemiol. 2010;39:1137–50. doi: 10.1093/ije/dyq191. [DOI] [PubMed] [Google Scholar]
- 99.Gibbons JD. Nonparametric methods for quantitative analysis. Columbus, Ohio: American Sciences Press; 1997. [Google Scholar]
- 100.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 101.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. [Google Scholar]
- 102.Touw WG, Bayjanov JR, Overmars L, et al. Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform. 2012 doi: 10.1093/bib/bbs034. doi: 10.1093/bib/bbs034 (Advance Access publication 10 July) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roshan U, Chikkagoudar S, Wei Z, et al. Ranking causal variants and associated regions in genome-wide association studies by the support vector machine and random forest. Nucleic Acids Res. 2011;39:e62. doi: 10.1093/nar/gkr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De Lobel L, Geurts P, Baele G, et al. A screening methodology based on Random Forests to improve the detection of gene-gene interactions. Eur J Hum Genet. 2010;18:1127–32. doi: 10.1038/ejhg.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lunetta KL, Hayward LB, Segal J, et al. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang P, Wu H, Wang W, et al. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–44. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamada T, Letunic I, Okuda S, et al. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–5. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bron PA, Wels M, Bongers RS, et al. Transcriptomes reveal genetic signatures underlying physiological variations imposed by different fermentation conditions in Lactobacillus plantarum. PLoS One. 2012;7:e38720. doi: 10.1371/journal.pone.0038720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pretzer G, Snel J, Molenaar D, et al. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J Bacteriol. 2005;187:6128–36. doi: 10.1128/JB.187.17.6128-6136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Hemert S, Meijerink M, Molenaar D, et al. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol. 2010;10:293. doi: 10.1186/1471-2180-10-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meijerink M, van Hemert S, Taverne N, et al. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One. 2010;5:e10632. doi: 10.1371/journal.pone.0010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gianoulis TA, Raes J, Patel PV, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci U S A. 2009;106:1374–9. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baran Y, Halperin E. Joint analysis of multiple metagenomic samples. PLoS Comput Biol. 2012;8:e1002373. doi: 10.1371/journal.pcbi.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Bokhorst-van de Veen H, Lee IC, Marco ML, et al. Modulation of Lactobacillus plantarum gastrointestinal robustness by fermentation conditions enables identification of bacterial robustness markers. PLoS One. 2012;7:e39053. doi: 10.1371/journal.pone.0039053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van der Heijden RT, Snel B, van Noort V, et al. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]