Abstract
N-glycosylation of proteins is an essential, highly conserved modification reaction that occurs in all eukaryotes and some prokaryotes. This process is catalyzed by oligosaccharyltransferase (OT), a multisubunit enzyme localized in the endoplasmic reticulum. Complete loss of N-glycosylation is lethal in all organisms. In Saccharomyces cerevisiae, OT is composed of nine nonidentical membrane proteins. Here, we report the atomic structure of an OT subunit from S. cerevisiae, Ost4p. This unusually small membrane protein containing only 36 residues folds into a well formed, kinked helix in the model-membrane solvent system used in this study. The residues critical for the OT activity and the stability of Stt3p-Ost4p-Ost3p subcomplex are located in helix α2, the larger cytosolic half of this kinked helix. The residues known to disrupt Ost4p-Stt3p complex form a well defined ridge in the 3D structure. Taking together prior mutational studies and the NMR structure of Ost4p, we propose that in the OT complex Stt3p is packed against the α2-helix of Ost4p by using a “ridges-into-grooves” model, with Met-18, Leu-21, and Ile-24 as the packing interface on one face, whereas Ost3p is involved in interactions with Met-19, Thr-20, Ile-22, and Val-23 on the other face.
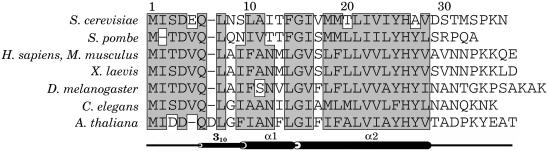
Oligosaccharyltransferase (OT) is a membrane-associated enzyme complex that catalyzes N-glycosylation, an essential and highly conserved protein modification reaction that occurs in some prokaryotic (1) and all eukaryotic (2, 3) organisms. This co- and posttranslational modification reaction affects a large number of both secretory and membrane proteins. OT catalyzes the transfer of a preassembled high mannose oligosaccharide (Glc3Man9GlcNAc2) from a lipid-linked dolichol pyrophosphate onto an asparagine residue in an Asn-X-Ser/Thr consensus sequence on nascent polypeptides as they are translocated into the lumen of the rough endoplasmic reticulum (ER). Although studies on N-glycosylation have been carried out for over three decades (4), it is less than a decade since it became clear that this enzyme is a complex consisting of multiple, nonidentical membrane proteins residing in the ER of both yeast and higher eukaryotes (3, 5). In the case of the yeast Saccharomyces cerevisiae, it has become clear that nine nonidentical membrane protein subunits (Swp1p, Wbp1p, Stt3p, Ost1p, Ost2p, Ost3p, Ost4p, Ost5p, and Ost6p) are components of the OT complex (3). The genes encoding Ost1p, Ost2p, Stt3p, Wbp1p, and Swp1p are essential for the viability of the cell. The OST4 gene is essential for growth at 37°C, but not at 25°C. OST3, OST5, and OST6 genes are not essential, but required for optimal OT activity (6-14). A high sequence similarity has been observed between most of the subunits of OT thus far identified in higher eukaryotes compared with those found in yeast. Homologs of Ost4p that also are minimembrane proteins have been found in Caenorhabditis elegans, mouse, and humans (Fig. 1).
Fig. 1.
Sequence alignment of Ost4p from the yeast S. cerevisiae and Ost4p analogs from other species: Schizosaccharomyces pombe (fission yeast), Homo sapiens (human), Mus musculus (house mouse), Xenopus laevis (clawed frog), Drosophila melanogaster (fruit fly), C. elegans (nematode), and Arabidopsis thaliana (thale cress). The secondary structure elements of Ost4p are shown below the sequence alignment.
All nine subunits of yeast OT have been cloned and sequenced. Genetic and biochemical studies have led to the identification of eight of the nine yeast OT proteins in three subcomplexes (Ost1p-Ost5p, Wbp1p-Swp1p-Ost2p, and Ost3p-Ost4p-Stt3p) in the ER membrane (15). Unlike most other membrane-associated glycosyl transferases that usually exist as monomers having one membrane-spanning segment, OT is far more complex. Although three of the subunits have single membrane-spanning segments, the others have multiple transmembrane (TM) domains ranging from 2 to 11. A major challenge for the future is to gain an understanding of the function of these membrane proteins in the N-glycosylation process and how they interact with each other in the ER.
The yeast OT subunit Ost4p is an unusually small membrane protein, consisting of one TM domain and only 36 amino acid residues (12). This minimembrane protein serves as a bridge between Stt3p and Ost3p subunits in the Stt3p-Ost4p-Ost3p subcomplex (16). Recent mutagenesis studies have shown that introduction of a single ionizable amino acid in positions 18-24 of Ost4p results in impaired cell growth and in vitro OT activity (16, 17). These mutations were shown to cause the disruption of the interactions between Ost4p, Ost3p, and Stt3p (17). In contrast, no effect was observed when similar single mutations were made in residues 2-17 of Ost4p (17). These findings, suggesting the functional importance of Ost4p in OT activity, along with its extraordinarily small mass, made this minimembrane protein an attractive target to initiate structural analysis and to learn how it interacts with other OT subunits.
Despite the success of solution NMR and x-ray crystallography, structure analysis of membrane proteins remains a challenging task. For every 500 published high-resolution protein structures, only 1 is the structure of a membrane protein, despite the fact that one-third of the genome in any living organism encodes for membrane proteins (18). One of the limitations of membrane protein NMR is the fact that a protein associated with a membrane mimetic, such as a micelle or a bicelle, tumbles as a very large complex, which leads to poor NMR sensitivity and low resolution. This problem can be avoided by dissolving the membrane protein in a suitable aqueous-organic solvent mixture, such as chloroform:methanol:water. This mixture has been shown to serve as a membrane mimetic environment for a number of integral membrane proteins (19-22). It is possible that water, chloroform, and methanol organize around different regions of the protein to mimic a biphase system such as a lipid bilayer.
Here, we report the 3D structure of a eukaryotic OT subunit, Ost4p. We used NMR spectroscopy to characterize the solution structure of this minimembrane protein of S. cerevisiae. This small membrane protein containing only 36 residues folds into a well formed, kinked helix in the model-membrane solvent system used in this study and allowed us to identify the specific structural elements responsible for interaction with the other two OT subunits.
Materials and Methods
Sample Preparation. The Ost4p peptide was obtained as 95% pure powder from Genemed Synthesis (San Francisco). Peptide quality was assessed with HPLC and electron spray ionization mass spectroscopy. The NMR sample of Ost4p was prepared by dissolving 8 mg of dry peptide in 400 μl of the deuterated solvent mixture containing 4:4:1 of CHCl3, CH3OH, and H2O.
NMR Experiments. All NMR data were collected on a Bruker DMX 750 or a Bruker DMX 500 spectrometer (Bruker, Billerica, MA) and processed offline with nmrpipe (23) and nmrview (24). All of the 2D NMR experiments reported here were recorded at 25°C and 38°C. Quadrature detection was used in both dimensions with the carrier frequency placed on the water resonance for all experiments. A 2D phase-sensitive double quantum-filtered (DQF) COSY experiment (25) and a total correlation spectroscopy (TOCSY, ref. 26) experiment were acquired in the time proportional phase increments (TPPI) mode with standard phase cycling schemes. Two-dimensional NOESY spectra (27) were recorded with mixing times of 350, 400, and 500 ms. The spectral width was set to 8,993 Hz (12.0 ppm) in both dimensions. The data were collected with 1,024 complex points in the t2 dimension and 256 complex free induction decays (FIDs) in the t1 dimension with 32 transients per FID. Squared sine bell window functions with a skew factor of 0.95 and a phase shift of 32 deg were applied in both dimension. The first experiment in t1 dimension was multiplied by 0.5 to suppress t1 ridges in the spectra (28). The data in the t2 dimension were zero-filled to 2,048 complex points and in t1 dimension to 512 points before Fourier transform.
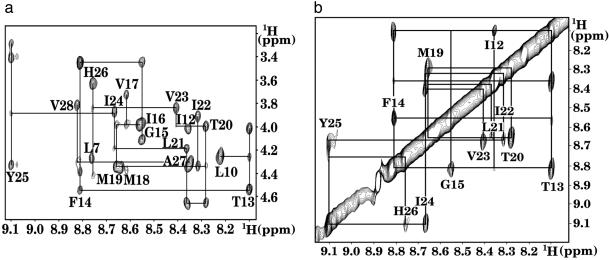
NMR Assignment and Structure Calculation. Ost4p was successfully assigned following the two-step procedure developed by Wüthrich (29), i.e., identification of the spin system followed by sequence-specific assignment of these spin systems to specific amino acid residues in the protein. The scalar-coupled spin systems were identified in DQF-COSY and TOCSY experiments. The Hα-Hβ cross peaks of Ala and Ser and Hβ-Hγ cross peaks of Thr residues were identified by using DQF-COSY. The side chains of Leu, Ile, Val, Pro, and Lys were identified by using TOCSY. The DQF-COSY spectrum was used to connect observable amide protons to their Hα protons. After identification of the few unique amino acid spin systems using DQF-COSY and TOCSY, the next step in the sequential assignment process was to assign each spin system to specific residues by combining the information of the TOCSY and NOESY spectra. Sequential assignments were made by using backbone HN-HN (dNN) and Hα-HN (dαN) nuclear Overhauser effect (NOE) connectivities with the unique aromatic residues Phe-14, Tyr-25, and His-26 used as starting points. Sequential connectivity in the HN-Hα region (Fig. 2a) and in the HN-HN region (Fig. 2b) could be observed only for the residues in the predicted TM domain. The residues outside the TM domain have weak or missing amide NOE cross peaks presumably due to increased mobility and fast exchange with the solvent. The sequential assignment of these residues was carried out by using Hα-Hα and Hα-Hβ connectivities, wherever possible. We assigned 90% of nonlabile protons in all residues with the exception of the N-terminal Met 1 and Ile 2. Temperature dependence of the chemical shifts (30) was measured for the slowly exchanging amide protons to determine hydrogen bonding (Fig. 3a). The 287 experimental upper distance restraints were derived from the intensities of the assigned NOE cross peaks with the program cyana (31) by using two calibration functions: d-6 for the backbone of the residues 8-30 and d-4 for the rest of the backbone and for all side chain resonances. A total of 58 backbone dihedral angle constraints were derived from the assigned chemical shifts by using the program talos (32). The structure of Ost4p was calculated by torsion angle dynamics simulated annealing as implemented in the program cyana (31). The length of the annealing procedure was increased to 10,000 steps. The calculation was carried out with 500 randomized starting conformations.
Fig. 2.
(a) Fingerprint region of the 2D NOESY spectrum of Ost4p showing sequential connectivities dαN(i,i + 1), dβN(i,i + 1), and dαN(i,i + 3). (b) Amide proton region of the 2D NOESY spectrum of Ost4p showing sequential connectivities dNN(i,i + 1) and dNN(i,i + 2).
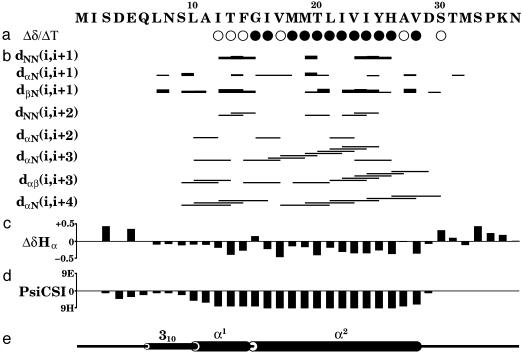
Fig. 3.
Ost4p secondary structure as determined by solution NMR spectroscopy. (a) Temperature coefficient for amide protons. Filled circles indicate coefficients more positive than -4.5 ppb/K (hydrogen bond present), and open circles indicate coefficients more negative than -4.5 ppb/K. (b) Summary of sequential and medium-range NOE connectivities. (c) Deviations of Hα chemical shifts from random coil values. (d) PsiCSI prediction of secondary structure. Negative bars correspond to helical conformation. (e) Secondary structure of Ost4p as observed in the 3D structure.
Solvent Refinement. Each of the 50 structures with the lowest cyana target function was placed in the center of a 40 × 40 × 28-Å octane slab, aligned with the z axis. The protein and the octane slab were immersed in an 8-Å shell of TIP3P (33) water molecules according to the solvent refinement procedure by Linge et al. (34). The refinement protocol consisted of three stages: a heating stage from 100 to 300 K in steps of 100 K with 200 steps of restrained molecular dynamics at each temperature, a refinement stage with 3,333 steps at 300 K, and a cooling stage from 300 K to 25 K in steps of 25 K with 500 steps of molecular dynamics at each temperature. After every 100 steps during the refinement stage, the solvent molecules leaving the simulation box were returned, and the excess energy was removed from the solvent as done in the refinement protocol by Spronk et al. (35) All solvent refinement was performed by using the program aria (36). The 20 structures with the lowest NOE violations were validated with procheck-nmr (37) and visualized with molmol (38). Secondary structure boundaries were defined by using the algorithm of Kabsch and Sander as implemented in molmol (39). The restraints used in calculation and statistical parameters of the refined structures are summarized in Table 1.
Table 1. Experimental restraints and structural statistics.
| Distance restraints | |
| Unambiguous | 287 |
| Intraresidue | 131 |
| Sequential | 57 |
| Medium range | 99 |
| Hydrogen bond restraints* | 24 |
| Dihedral angle restraints | 58 |
| Ideal geometry rmsd | |
| Bonds, Å | 0.012 |
| Angles, ° | 6.6 |
| Interresidue distance restraints violations | |
| Violations > 0.2 Å | 12 |
| Maximum violation, Å | 0.56 |
| rmsd to average structure, Å | |
| Backbone (residues 8-29) | 0.158 |
| Heavy atoms (residues 8-29) | 0.415 |
| Ramachandran plot statistics, % | |
| Most favored regions | 78.1 |
| Additionally allowed regions | 21.6 |
| Generously allowed regions | 0.3 |
| Disallowed regions | 0.0 |
Two restraints per bond, derived from the temperature dependence of amide chemical shifts (Fig. 3a). rmsd, rms deviation.
Results and Discussion
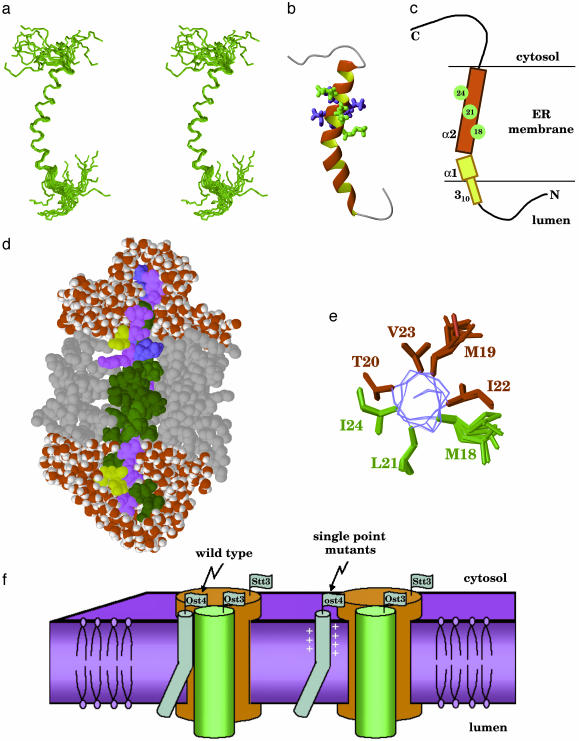
NMR Structure of Ost4p. The secondary structure of Ost4p, derived from the assigned chemical shifts by psicsi (40), suggests only a single α-helix encompassing residues 10-28 (Fig. 3d). However, according to the results of simulated annealing and solvent refinement calculations, this unusually small membrane protein consists of a kinked α-helix (residues 10-28) in the TM domain and a short 310-helical extension (residues 6-9) in the N-terminal domain proximal to the membrane as shown in Fig. 4 a and b. The N-terminal residues 1-5 and the entire C-terminal domain 29-36 are unstructured.
Fig. 4.
(a) Stereoview of the ensemble of the 20 lowest-energy NMR structures of Ost4p after solvent refinement. (b) Ribbon view of the structure of Ost4p. Residues 18, 21, and 24, expected to interact with Stt3p, are shown in green. Other residues found to be important for optimal protein activity are in blue. These residues may be interacting with Ost3p. (c) Schematic representation of Ost4p structure in membrane. Residues expected to interact with Stt3p are shown in green. (d) Structure of Ost4p from the restricted molecular dynamics calculation in a water/octane/water simulation cell. Octane molecules are shown in gray, and water molecules are shown in red and white. The hydrophobic residues V, I, L, A, F, M, and G are shown in green, hydrophilic S, T, N, Q, and Y are shown in purple, negatively charged D and E are shown in yellow, and positively charged K and H are shown in blue. (e) Location of the mutation-sensitive residues on the helix α2, viewed along the helix axis. (f) Model of the putative structure of Stt3p-Ost4p-Ost4p complex.
The presence of the N-terminal helix extension is also supported by the chemical shift difference of residues 7-9 (Fig. 3c). The Hα chemical shifts of these residues are less than their random coil values, which indicates helical secondary structure. Such small 310-helices have been shown to appear at the N termini of many α-helices (41). The 310-helix in Ost4p is not well defined in the NMR structure and exhibits increased flexibility as can be seen from the weak or missing NOE contacts (Fig. 3b).
The TM α-helix kinks between residues Phe 14 and Gly 15 with a kink angle of 37°, resulting in a smaller luminal part (α1, residues 10-14) and a larger cytosolic part (α2, residues 15-28). That the helix has a kink is also supported by the Hα chemical shift difference of Gly-15, which is positive, indicating a nonhelical conformation. Kinks, i.e., changes in direction of the helical axis, are a functionally important feature of many TM helices, typically induced by proline residues (42, 43). The only proline in Ost4p is located in the unstructured C-terminal segment, outside the TM domain. However, kinks in TM helices have also been associated with residues other than proline, particularly hydrophobic residues Ala, Leu, Ile, and Val and aromatic residues located near glycines (43). In case of Ost4p, aromatic Phe-14 is followed by Gly-15, similar to the proline-less kink motif described above. Surprisingly, Gly-15 is strictly conserved through Ost4p analogs in all eukaryotes (Fig. 1). However, it has not been identified as important for Ost4p function based on the mutagenesis studies (17). Because kinks in TM helices usually result from cooperative participation of several residues in up to two helical turns (43), it is possible that single point mutations did not significantly alter the kink in Ost4p.
TM Domain of Ost4p. The boundaries of the TM domain were determined from the results of water/octane/water solvent refinement (Fig. 4d). The octane box system used for solvent refinement has been shown (44-46) to act as a simplified model of a phospholipid bilayer that has a significantly lower viscosity and enables substantial changes in protein structure and orientation in nanosecond timescale, if they are energetically favorable. The solvent refinement of the NMR structures of Ost4p did not result in any significant changes in geometry, with the root-mean-square difference between the average structures before and after refinement not exceeding 0.5 Å. The major result of the solvent refinement was a substantially improved local geometry of the 310-helix. In the refined structure (Fig. 4d), all residues from Leu-10 to Val-28 are found inside the octane layer, which is in perfect agreement with the hydrophobicity profile (47). The boundaries defined in this study differ from the earlier estimations of the TM domain of Ost4p, which has been predicted on the basis of various programs to span the residues 8-28 (17) or 10-25 (16).
Functional Implications of Ost4p Structure. It is very interesting that the 3D structure of Ost4p in chloroform:methanol:water mixture explains and rationalizes the results of comprehensive mutagenesis study on Ost4p (17). Single point mutations of the residues 18-24 have a negative effect on OT activity and disrupt the complex formation between Ost4p, Ost3p, and Stt3p (17). In the NMR structure determined in this study (Fig. 4 a and b), these residues form the central part of the α2 helix (residues 15-28). Mutations in the α1 helix (residues 10-14) have no effect on OT activity, which implies that the helix α2 is the structural element necessary to form a functional subcomplex between the three membrane proteins.
It is of interest to note that mutation of residue Met-18, Leu-21, or Ile-24 disrupts the Ost4p and Stt3p complex (17) whereas mutation of any residue from Met-18 through Ile-24 disrupts the Ost4p-Ost3p complex (17). In the NMR structure, residues Met-18, Leu-21, and Ile-24 are located on the same side of the Ost4p TM helix (Fig. 4 b and e) and form a well defined i + 4 ridge (Fig. 4c). Such ridges are the key elements in the classical “ridges-into-grooves” helix packing model (48). This finding leads us to believe that, in the Stt3p-Ost4p-Ost3p complex, one of the TM domains of Stt3p is packed against Ost4p in ridges-into-grooves manner, with the Ost4p residues 18, 21, and 24 forming the packing interface. Thus, in the model of Ost4p serving as the bridge molecule between Stt3p and Ost3p, residues Met-18, Leu-21, and Ile-24 on one side of the helix (Fig. 4e) are involved in interactions with Stt3p whereas residues Met-19, Thr-20, Ile-22, and Val-23 on the other side of the helix are crucial for the interactions with Ost3p (Fig. 4f). These interactions may include interhelical hydrogen bonding of the side chain of Thr-20, as often found in other membrane proteins (49, 50).
Complete understanding of the chemistry and function of the OT complex will require the combination of many approaches. In the present study, NMR spectroscopy identified the 3D structure of an OT subunit, which clarified the results of mutagenesis studies and allowed us to deduce a structural model of the interactions within three proteins in an OT subcomplex. This result represents an important step toward understanding the mechanism of N-glycosylation in eukaryotes, which, in the long term, will provide us with a new basis for treatment of congenital disorders of glycosylation.
Note Added in Proof. While this article was in press, a very interesting work on the evolution of transmembrane helix kinks was reported by Yohannan et al. (51) describing a strong correlation between the frequency of prolines at a position and a kink at that site in the helix, even if the helix in the known protein does not contain a proline. These nonproline kinks have arisen during evolution through compensatory mutations that help to stabilize the bent conformation without the need of proline.
Acknowledgments
We thank undergraduate student Bina Farooqi and graduate student Arun Ramasamy for initial NMR data processing. This research was supported by U.S. Department of Agriculture Grants 99-35302-8106 and 2003-35302-12930 (Presidential Early Career Award program), National Science Foundation Grant IBN-0074591 (to S.M.), and National Institutes of Health Grant GM 33185 (to W.J.L.).
Abbreviations: OT, oligosaccharyltransferase; ER, endoplasmic reticulum; TM, transmembrane; NOE, nuclear Overhauser effect; DQF, double quantum-filtered; TOCSY, total correlation spectroscopy.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1RKL). The assigned chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (BMRB accession no. 6056).
References
- 1.Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W., et al. (2002) Science 298, 1790-1793. [DOI] [PubMed] [Google Scholar]
- 2.Kornfeld, R. & Kornfeld, S. (1985) Annu. Rev. Biochem. 54, 631-664. [DOI] [PubMed] [Google Scholar]
- 3.Knauer, R. & Lehle, L. (1999) Biochim. Biophys. Acta 1426, 256-273. [DOI] [PubMed] [Google Scholar]
- 4.Marshall, R. D. (1972) Annu. Rev. Biochem. 41, 673-702. [DOI] [PubMed] [Google Scholar]
- 5.Kelleher, D. J. & Gilmore, R. (1997) Proc. Natl. Acad. Sci. USA 94, 4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pathak, R., Parker, C. S. & Imperiali, B. (1995) FEBS Lett. 362, 229-234. [DOI] [PubMed] [Google Scholar]
- 7.te Heesen, S., Janetzky, B., Lehle, L. & Aebi, M. (1992) EMBO J. 11, 2071-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.te Heesen, S., Knauer, R., Lehle, L. & Aebi, M. (1993) EMBO J. 12, 279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silberstein, S., Collins, P. G., Kelleher, D. J. & Gilmore, R. (1995) J. Cell Biol. 131, 371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zufferey, R., Knauer, R., Burda, P., Stagljar, I., te Heesen, S., Lehle, L. & Aebi, M. (1995) EMBO J. 14, 4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karaoglu, D., Kelleher, D. J. & Gilmore, R. (1995) J. Cell Biol. 130, 567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi, J. H., Roos, J. & Dean, N. (1996) J. Biol. Chem. 271, 3132-3140. [DOI] [PubMed] [Google Scholar]
- 13.Reiss, G., te Heesen, S., Gilmore, R., Zufferey, R. & Aebi, M. (1997) EMBO J. 16, 1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knauer, R. & Lehle, L. (1999) J. Biol. Chem. 274, 17249-17256. [DOI] [PubMed] [Google Scholar]
- 15.Karaoglu, D., Kelleher, D. J. & Gilmore, R. (1997) J. Biol. Chem. 272, 32513-32520. [DOI] [PubMed] [Google Scholar]
- 16.Kim, H., Yan, G., Heijne, G., Caputo, G. A. & Lennarz, W. J. (2003) Proc. Natl. Acad. Sci. USA 100, 7460-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H., Park, H., Montalvo, L. & Lennarz, W. J. (2000) Proc. Natl. Acad. Sci. USA 97, 1516-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001) J. Mol. Biol. 305, 567-580. [DOI] [PubMed] [Google Scholar]
- 19.Rastogi, V. K. & Girvin, M. E. (1999) Nature 402, 263-268. [DOI] [PubMed] [Google Scholar]
- 20.Girvin, M. E., Rastogi, V. K., Abildgaard, F., Markley, J. L. & Fillingame, R. H. (1998) Biochemistry 37, 8817-8824. [DOI] [PubMed] [Google Scholar]
- 21.Schwaiger, M., Lebendiker, M., Yerushalmi, H., Coles, M., Gröger, A., Schwarz, C., Schuldiner, S. & Kessler, H. (1998) Eur. J. Biochem. 254, 610-619. [DOI] [PubMed] [Google Scholar]
- 22.Dmitriev, O. Y., Altendorf, K. & Fillingame, R. H. (2004) FEBS Lett. 556, 35-38. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277-293. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, B. A. & Blevins, R. A. (1994) J. Biomol. NMR 4, 603-614. [DOI] [PubMed] [Google Scholar]
- 25.Piantini, U., Sørensen, O. W. & Ernst, R. R. (1982) J. Am. Chem. Soc. 104, 6800-6801. [Google Scholar]
- 26.Bax, A. & Davis, D. G. (1985) J. Magn. Reson. 65, 355-360. [Google Scholar]
- 27.Kumar, A., Ernst, R. R. & Wüthrich, K. (1980) Biochem. Biophys. Res. Commun. 95, 1-6. [DOI] [PubMed] [Google Scholar]
- 28.Otting, G., Widmer, H., Wagner, G. & Wüthrich, K. (1986) J. Magn. Reson. 66, 187-193. [Google Scholar]
- 29.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 30.Baxter, N. J. & Williamson, M. P. (1997) J. Biomol. NMR 9, 359-369. [DOI] [PubMed] [Google Scholar]
- 31.Güntert, P., Mumenthaler, C. & Wüthrich, K. (1997) J. Mol. Biol. 273, 283-298. [DOI] [PubMed] [Google Scholar]
- 32.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289-302. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. (1983) J. Chem. Phys. 79, 926-935. [Google Scholar]
- 34.Linge, J. P., Williams, M. A., Spronk, C. A., Bonvin, A. M. & Nilges, M. (2003) Proteins 50, 496-506. [DOI] [PubMed] [Google Scholar]
- 35.Spronk, C. A., Linge, J. P., Hilbers, C. W. & Vuister, G. W. (2002) J. Biomol. NMR 22, 281-289. [DOI] [PubMed] [Google Scholar]
- 36.Linge, J. P., Habeck, M., Rieping, W. & Nilges, M. (2003) Bioinformatics 19, 315-316. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996) J. Biomol. NMR 8, 477-486. [DOI] [PubMed] [Google Scholar]
- 38.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-55. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577-2637. [DOI] [PubMed] [Google Scholar]
- 40.Hung, L.-H. & Samudrala, R. (2003) Protein Sci. 12, 288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doig, A. J., MacArthur, M. W., Stapley, B. J. & Thornton, J. M. (1997) Protein Sci. 6, 147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordes, F. S., Bright, J. N. & Sansom, M. S. P. (2002) J. Mol. Biol. 323, 951-960. [DOI] [PubMed] [Google Scholar]
- 43.Riek, R. P., Rigoutsos, I., Novotny, J. & Graham, R. M. (2001) J. Mol. Biol. 306, 349-362. [DOI] [PubMed] [Google Scholar]
- 44.Guidoni, L., Torre, V. & Carloni, P. (2000) FEBS Lett. 477, 37-42. [DOI] [PubMed] [Google Scholar]
- 45.Tieleman, D. P., Berendsen, H. J. C. & Sansom, M. S. P. (2001) Biophys. J. 80, 331-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell, J. D., Biggin, P. C., Baaden, M. & Sansom, M. S. P. (2003) Biochemistry, 42, 3666-3673. [DOI] [PubMed] [Google Scholar]
- 47.Kyte, J. & Doolittle, R. (1982) J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]
- 48.Chothia, C., Levitt, M. & Richardson, D. (1981) J. Mol. Biol. 145, 215-250. [DOI] [PubMed] [Google Scholar]
- 49.Smith, S. O., Eilers, M., Song, D., Crocker, E., Ying, W., Groesbeek, M., Metz, G., Ziliox, M. & Aimoto, S. (2002) Biophys. J. 82, 2476-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eilers, M., Shekar, S. C., Shieh, T., Smith, S. O. & Fleming, P. J. (2000) Proc. Natl. Acad. Sci. USA 97, 5796-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yohannan, S., Faham, S., Yang, D., Whitelegge, J. P. & Bowie, J. U. (2004) Proc. Natl. Acad. Sci. USA 101, 959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]