Background: MAPK cascades are signaling modules that function as switch generators.
Results: In vitro phosphorylation in the p38 MAPK cascade tracked by LC-MS/MS revealed specific phosphorylation intermediates at each level.
Conclusion: The p38 MAPK cascade reactions occur through intermediates MEK6/ST* and p38α/TY*.
Significance: The precise order of reactions may contribute to the diverse kinetic outputs of the cascades, including those with large Hill coefficients.
Keywords: MAP Kinases (MAPKs), Mass Spectrometry (MS), p38 MAPK, Phosphorylation Enzymes, Protein Kinases, Signal Transduction, Cascade Signaling, Ordered Phosphorylation
Abstract
The MAP kinase cascades, composed of a MAP3K, a MAP2K, and a MAPK, control switch responses to extracellular stimuli and stress in eukaryotes. The most important feature of these modules is thought to be the two double phosphorylation reactions catalyzed by MAP3Ks and MAP2Ks. We addressed whether the reactions are sequential or random in the p38 MAP kinase module. Mass spectrometry was used to track the phosphorylation of the MAP2K MEK6 by two MAP3Ks, TAO2 and ASK1, and the subsequent phosphorylation of p38α by MEK6/S*T* (where S (Ser) and T (Thr) are the two phosphorylation sites and * denotes phosphorylation). Both double phosphorylation reactions are precisely ordered. MEK6 is phosphorylated first on Thr-211 and then on Ser-207 by both MAP3Ks. This is the first demonstration of a precise reaction order for a MAP2K. p38α is phosphorylated first on Tyr-182 and then on Thr-180, the same reaction order observed previously in ERK2. Thus, intermediates were MEK6/ST* and p38α/TY*. Similarly, the phosphorylation of the p38α transcription factor substrate ATF2 occurs in a precise sequence. Progress curves for the appearance of intermediates were fit to kinetic models. The models confirmed the reaction order, revealed processivity in the phosphorylation of MEK6 by ASK1, and suggested that the order of phosphorylation is dictated by both binding and catalysis rates.
Introduction
MAP2 kinase cascades mediate decision-making processes throughout eukaryotic life. Changes in cell fate leading to differentiation, proliferation, and apoptosis in response to hormones, as well as metabolic changes in response to stress, involve MAP kinase pathways (1–5). Each MAP kinase module has a canonical architecture of three kinases, a MAP3K, a MAP2K, and a MAPK. The cascades catalyze a sequence of phosphorylation reactions. MAP3Ks phosphorylate two Ser/Thr residues in MAP2Ks (6), and MAP2Ks phosphorylate MAPKs on a Tyr and a Thr residue (7), such that MAP2Ks are true dual (Tyr and Ser/Thr) specificity kinases. The doubly phosphorylated MAP kinase is the product of the cascade, a proline-directed Ser/Thr kinase (8). Four distinct MAP kinase cascades have been elucidated, each named for the MAPK-activated, ERK1/2 (extracellular signal-regulated kinase 1 and 2), p38 MAP kinase, JNK (c-Jun kinase), and ERK5 pathways (4). These modules are activated by a variety of inputs, including other kinases, referred to as MAP4Ks, interactions with small G-proteins (such as Ras), or scaffold proteins (such as JNK-interacting protein (JIP)). The modules also have numerous substrates, consisting primarily of transcription factors and other protein kinases. Thus, apparently, the three-tiered modules “plug in” to a variety of cellular contexts. The cascades are further capable of a variety of kinetic outputs as measured by the dose-response activity of the MAPK. In oocytes, the ERK2 MAP kinase module generates highly cooperative or sigmoid responses (9, 10). Responses observed in other cell types can be more graded (11–13). Signal response can further be modified by the strength and duration of inputs (12, 14).
Several recent studies have addressed the dose-response behavior of MAP kinase modules in cells (13–17). Although MAPK phosphorylation occurs through a specific intermediate (13, 18), the phosphorylation order in MAP2Ks has not been addressed. The strong sigmoid properties of the cascade occurring in oocytes is thought to be derived from the phosphorylation reactions catalyzed by MAP3Ks and MAP2Ks (19). There are two models (20). One is that both the phosphorylation reactions occur at random but both are required for the activity of the MAP2K (19). A second possibility is that the reactions occur in a precise sequence (as proposed in Fig. 1), and the activity is associated with the second phosphorylation event. In either case, sigmoid behavior should be possible when the reactions are distributive (nonprocessive) (19). Some support for the sequential model is in the literature. It has been established that the MAPK ERK2 is phosphorylated first on Tyr-185 to give ERK2/TY* and then on Thr-183 to give ERK2/T*Y* (18, 19, 21, 22) (where T (Thr) and Y (Tyr) are the two phosphorylation sites and * denotes phosphorylation). Engelberg and colleagues (23) have shown that in the yeast Hog1 pathway, Tyr phosphorylation is not required for activity, but is essential for the salt stress activation of Hog1 in cells (see also Refs. 24 and 25). The phosphorylation of MAP2Ks has not been well studied. Mutational analysis of the role of phosphorylation sites in the activity and activation of MAP2Ks gave mixed results (6, 26, 27), except that in vitro studies of MEK1 and MEK6 (MAP/ERK kinase 1 and 6) showed that one alanine mutant is phosphorylated more easily than the other (MEK1/AS more than MEK1/SA; MEK6/AT more than MEK6/SA) (27, 28). These data are suggestive that phosphorylation of MAP2Ks may be sequential.
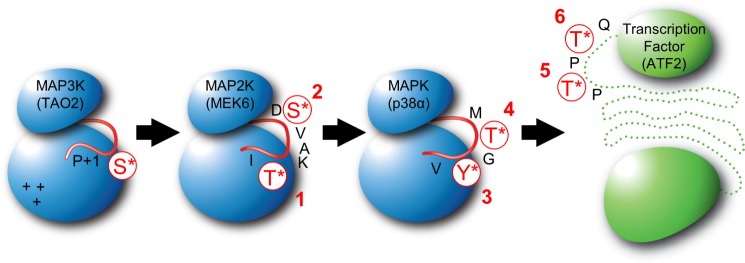
FIGURE 1.
Phosphorylation sites in MAP3K TAO2, MAP2K MEK6, MAPK p38α, and substrate ATF2. Schematics of the kinases (blue) and a transcription factor substrate (green) of the p38 cascade showing phosphorylation sites and the order of phosphorylation tested in this study are displayed. Graphic representations of the activation loops of each kinase and specific phosphorylation sites are indicated in red. A cluster of positive changes and the P+1 site of TAO2 (indicated on the MAP3K) may interact with the Asp-206 and Ile-212 in the MEK6 activation loop sequence 206DSVAKTI212 (underlining indicates phosphorylation sites) and potentially dictate Thr-first phosphorylation (28).
Here we phosphorylated MEK6 in vitro, using two different activators, the MAP3K ASK1 (apoptosis signaling kinase-1) (29) and TAO2 (thousand and one kinase-2) (30), tracking the appearance and disappearance of all MEK6 phosphorylation states by LC-MS/MS. We found that MEK6 phosphorylation goes through a precise intermediate, MEK6/ST* (Fig. 1). We also found that p38α MAP kinase, the MEK6 substrate, is phosphorylated through p38α/TY*, as anticipated from studies of ERK2. We further confirmed that p38α phosphorylates the transcription factor ATF2 in a specific order (31). Progress curves for the appearance of phosphorylation intermediates and products were fit to kinetic models for the double phosphorylation reactions, which validated the apparent sequence of phosphorylation at each level. The double phosphorylation of MEK6 by TAO2 occurred processively, whereas the rest of the reactions were distributive.
EXPERIMENTAL PROCEDURES
Protein Reagents
MEK6, TAO2, ASK1, p38α, and ATF2 proteins were expressed and purified following standard procedures (32). A rat cDNA encoding MEK6 was subcloned into a modified pET29b (Novagen) expression vector possessing a C-terminal tobacco etch virus protease recognition sequence and a His tag. This expression vector was modified to encode the kinase-dead MEK6/K82M. Full-length MEK6 and MEK6/K82M expression and purification were conducted essentially as described in Ref. 33. Viral stock harboring TAO2 (residues 1–451) cDNA was transfected into Sf9 insect cells for protein expression as described previously (28). Human ASK1 (residues 659–951) was synthesized in Escherichia coli with an N-terminal His tag using the expression vector pLIC (obtained from Stefan Knapp, Structural Genomics Consortium at Oxford (34)). A pET14b plasmid encoding p38α described previously (35) was modified to produce kinase-dead p38α/K53M using the Stratagene QuikChange kit. Wild-type p38α and p38α/K53M proteins were expressed and purified as described in Ref. 36. A pGEX(KG) plasmid encoding GST-tagged human ATF2 (residues 33–254) was obtained from Melanie Cobb, University of Texas Southwestern (UT Southwestern) Medical Center, expressed in bacteria, and purified on a glutathione-Sepharose column (GE Healthcare) following the manufacturer's protocol.
Phosphorylated Active Kinases
TAO2 and ASK1 are constitutively phosphorylated as expressed in either insect cells (TAO2) or bacteria (ASK1). Doubly phosphorylated p38α (p38α/T*Y*) was obtained by coexpression of p38α with MEK6/DD (MEK6/S207D,T211D) in E. coli as described previously (37). MEK6/S*T* was obtained by incubation of ASK1 and MEK6 (in the molar ratio of 1 to 100) at 30 °C for 30 min using standard kinase reaction conditions (see below). ASK1 was removed using nickel-Sepharose (GE Healthcare). The phosphorylation states of these proteins were verified by LC-MS/MS (see below).
Phosphorylation Assays
All phosphorylation assays were carried out under the standard conditions of 20 mm HEPES, pH 7.4, 20 mm MgCl2, 2.5 mm ATP at 30 °C. Reactions were initiated by the addition of the enzyme TAO2/S* (TAO2/Ser(P)-181, where “P” denotes phosphorylation), ASK1/T* (ASK1/Thr(P)-838), MEK6/S*T* (MEK6 Ser(P)-207/Thr(P)-211), and p38α/T*Y* (p38α Thr(P)-180/Tyr(P)-182) to their respective substrates (MEK6/ST, p38α/TY, and ATF2/TT (ATF2 Thr-69/Thr-71)). In general, initial substrate proteins concentrations were 20 μm. Reactions were run over a 30-min time course, with aliquots removed at each time point. ASK1:MEK6 and TAO2:MEK6 reactions (molar ratios of 1:100 and 1:3; respectively) were terminated in 1 m guanidine-HCl. MEK6:p38α and p38α:ATF2 (molar ratios of 1:300 and 1:1000, respectively) reactions were stopped with urea to a final concentration of 1 m. Stopping the reaction in high denaturant promoted protein solubility throughout the experiment, thus reducing errors from time point to time point. Peptides were separated from denaturant on an RP-C18 column by diverting early eluents.
Peptide Standards for Mass Spectrometry
MEK6 and p38α peptide standards corresponding to all activation loop phosphorylated species of proteolysis-derived peptides (supplemental Table S1, entries for MEK6 and p38α) were synthesized (Haydn Ball, UT Southwestern Medical Center). Standards were used to optimize HPLC peptide separation, to determine elution times, and to optimize MS ionization and detection protocols. Peptide total ion current signal and total absorption (λ = 190–400 nm) as measured by an in-line diode array detector (Agilent) were compared. Phosphorylation had minimal effect on peptide ionization (0–10% variation) (data not shown). Thus, it was possible to use the assumption that MS responses for each phospho-species were similar for both MEK6 and p38α activation loop peptides.
Protein Digestion and HPLC Peptide Separation
Chymotrypsin protease reaction mix (containing 100 mm Tris-HCl, pH 6.0, 25 mm CaCl2, 50 nm sequencing grade chymotrypsin (Roche Applied Science)) was used to digest MEK6. p38α reactions were digested using trypsin protease reaction mix (50 mm NH4HCO3, 50 mm Tris-HCl, pH 8.0, and 10 nm trypsin (Promega)). All proteolysis reactions were conducted at an 80:1 protein:protease molar ratio. Activation loop peptides (supplemental Table S1) were separated by an Agilent 1100 series LC system (Agilent Technologies, Palo Alto, CA) with an RP-C18 microbore HPLC column (Phenomenex Aeris WIDEPORE 150 × 2.1 mm, 3-μm particle size, 200 Å pore diameter). Peptides were eluted using a water/acetonitrile gradient with 0.2% formic acid. MEK6 peptides of interest eluted at ∼24–26% acetonitrile, whereas p38α peptides eluted at ∼16–19% acetonitrile. ATF2 was proteolyzed by trypsin as described previously (21, 31), giving the peptides listed in supplemental Table S1, which were separated by HPLC as above, with all activation loop fragments eluting at 14–17% acetonitrile.
LC-MS/MS Analysis
HPLC-MS/MS analysis was performed on an LCQ DECA XP ion trap mass spectrometer (ThermoFinnigan) in which the HPLC was coupled in-line to an orthogonal electrospray ionization source. Integration under ion traces corresponding to mass ranges for activation loop peptides were used to acquire raw MS detector responses (see Fig. 2). Time courses for the appearance of peptides were conducted in triplicate, and the different experiments were scaled by the total ion current over an elution window encompassing peptides from the digestion (38).
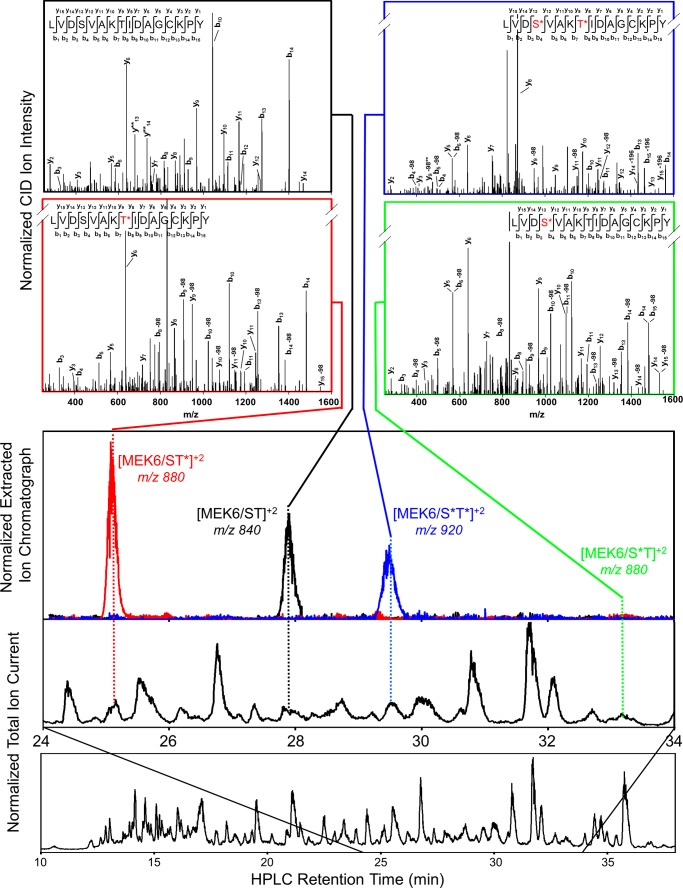
FIGURE 2.
LC-MS/MS identification of MEK6/K82M phosphorylation intermediates. MEK6/K82M was phosphorylated by TAO2 for 10 min and then proteolyzed with chymotrypsin and separated on an RP-C18 column (see “Experimental Procedures”). The total ion trace shows separation of individual peptides (bottom). Extracted ion chromatographs of masses correspond to doubly charged activation loop peptides (middle). MASCOT analysis of collision-induced dissociation-generated peptide fragments provide MS/MS peptide identification (top).
MS/MS spectra were acquired in a data-dependent mode (recording the masses and fragmentation patterns of the strongest to weakest ions) in each MS scan (ThermoFinnigan). MASCOT software (Matrix Science Ltd., London, UK) (39) was used for identifying peptides from their MS spectrum. Better than 80% of the expected proteolytically generated peptides for MEK6 and p38α were identified by the software and included the activation loop-derived peptides.
Cascade Modeling
Kinetic models for the phosphorylation reactions were defined in DynaFit (40, 41). The DynaFit software uses the Levenberg-Marquardt algorithm to perform nonlinear least squares regression of progress curve data. Global minimization was achieved by a differential evolution of trial parameter sets (41). Trial parameter values were taken from those obtained for cellular assays of the ERK2 cascade (15). The program outputs standard error for each variable and a correlation matrix for the model parameters. All likely kinetic steps were included in the initial models, including separate binding steps leading to different chemical outcomes (supplemental Fig. S1). However, backward dissociation reactions were eliminated because they were too slow to affect the model, leading to an irreversible mechanism throughout (41, 42). Other steps that were too fast to be kinetically relevant were eliminated. Due to correlation between variables, one constant was fixed at each level (based on values from the literature), thus improving the fit and the calculated standard error. Equations used are listed in supplemental Table S2. For ASK1 phosphorylation of MEK6, 224 data points were used to model eight variable parameters, giving a 28:1 data/parameter ratio. For MEK6 phosphorylation of p38α, 156 data points were used to model five variable parameters, giving a 31:1 data/parameter ratio. In general, all reactions were assumed to be distributive (with dissociation between phosphorylation reactions) (43).
RESULTS
To determine the order of phosphorylation reactions at each level of the p38 MAP kinase cascade, the phosphorylation of MEK6 by two MAP3Ks and the phosphorylation of p38α by MEK6 were studied separately. The appearance of phosphorylation at each site of either MEK6 or p38α was measured as a function of time of incubation with its activating enzyme. The position and extent of phosphorylation was determined by HPLC-MS/MS of proteolysis-derived peptides. The phosphorylation of ATF2 by p38α was measured similarly.
Phosphorylation Intermediates in the Dual Phosphorylation of the MAP2K MEK6 Were Identified by LC-MS/MS
MEK6 was phosphorylated by the MAP3K TAO2. MS/MS spectra of synthetic peptides corresponding to expected chymotrypsin-derived activation loop peptides (LVDSVAKTIDAGKPY, supplemental Table S1) in alternative states of phosphorylation were measured. Then chymotrypsin-derived MEK6 activation loop peptides were separated to base line by reverse phase HPLC and identified by tandem mass spectrometry using the MASCOT software (Matrix Science, Inc.) (39) (Fig. 2). After 10 min, substantial quantities of three phosphorylation states were observed (MEK6/ST, MEK6/ST*, MEK6/S*T*), with MS/MS spectra corresponding to those obtained for standard peptides. A small amount of the disfavored intermediate, MEK6/S*T, was also seen. Standard peptides run under similar conditions were used for approximating peptide concentrations (see “Experimental Procedures” and the legend for Fig. 3).
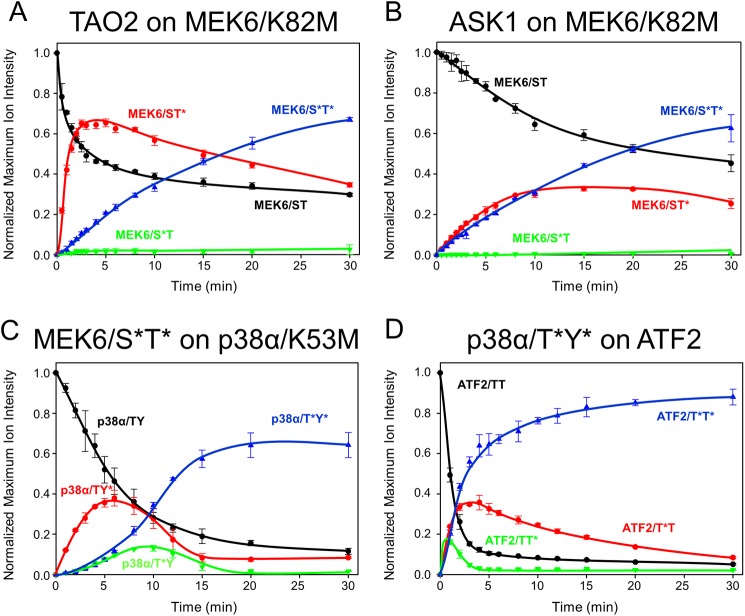
FIGURE 3.
Phosphorylation progress curves in the p38 cascade. A–D, time courses show MEK6/K82M phosphorylation by TAO2 (A) and ASK1 (B), p38α/K53M phosphorylation by MEK6/S*T* (C), and GST-ATF2 phosphorylation by p38α/T*Y* (D). Traces represent total ion current normalized selected ion response curves for each proteolytically derived activation loop phosphopeptide. Experiments were performed in triplicate with the error bars representing one standard deviation. Unphosphorylated (black), monophosphorylated (red and green), and doubly phosphorylated (blue) activation loop peptides were tracked for 30 min. In A and B, the sum of peptide signal apparently exceeds the starting value, probably due to errors in the total ion current normalization (38) and matrix ionization effects (55). In C and D, these effects were minimal.
Dual Phosphorylation of MEK6/K82M Occurs through a Single Intermediate, MEK6/K82M/ST*
MEK6/K82M phosphorylation by TAO2 and ASK1 was tracked over 30 min (Fig. 3, A and B). Using TAO2, the intermediate MEK6/K82M/ST* appeared immediately and peaked at about 2 min before declining over 10 min. The apparent rate of the disappearance of MEK6 and appearance of MEK6/ST* mirror each other in the first 2 min. Further, the rates of MEK6/ST* consumption and MEK6/S*T* formation appear equivalent. Apparently, the flux of phosphorylation goes through MEK6/ST*. Both the substrate MEK6 and the intermediate MEK6/ST* persisted over the time course. Kinetic modeling, discussed below, was used to discover the potential origins of this behavior. ASK1 phosphorylation of MEK6 generated the same MEK6 intermediate as observed with TAO2. However, using ASK1, the concentration of MEK6/ST* does not dramatically exceed that of MEK6/S*T*, and the rate of formation of MEK6/S*T* mirrors the disappearance of MEK6. Both of these observations are suggestive of processivity in this reaction.
Dual Phosphorylation of p38α by MEK6 Occurs Primarily through the Intermediate p38α/TY*
MEK6/S*T* was used to phosphorylate kinase-dead p38α (p38α/K53M). HPLC-MS of trypsinized p38α was recorded as above to generate progress curves for p38α activation loop phosphorylation. The p38α activation loop peptides (supplemental Table S1) derived from p38α/TY, p38α/TY*, p38α/T*Y, and p38α/T*Y* were observed (supplemental Fig. S2). The time course (Fig. 3C) reveals a single dominant intermediate, p38α/TY*, with small amounts of the alternate intermediate, p38α/T*Y, also present. This order of phosphorylation, through p38α/TY*, is the same as that observed in the MAPK ERK2 (13, 18). Over the first 2 min, the appearance of p38α/TY* mirrors the disappearance of p38α/TY. Further, the appearance of p38α/T*Y* occurs with the same apparent rate as the disappearance of p38α/TY*. These data indicate that the flux of phosphorylation occurs through the intermediate p38α/TY*. In contrast to the phosphorylation of MEK6, there is a very strong lag in the formation of p38α/T*Y*, indicative of a distributive mechanism (little or no processivity). The substrate p38α approaches the base line over the time course, in contrast with the MEK6 phosphorylation.
p38α Phosphorylates ATF2 through ATF2/TT* in Vitro
We tested whether ordered phosphorylation occurs at the level of MAPK pathway output. Wild-type p38α phosphorylates a native substrate, ATF2, on two threonine residues, Thr-69 and Thr-71 (44). Using the same methods as for MEK6 and p38α phosphorylation, we tracked tryptic peptides encompassing Thr-69 and Thr-71 (supplemental Table S1) of GST-ATF2 and observed both monophosphorylated intermediates, ATF2/T*T and ATF2/TT* (Fig. 3D). ATF2/TT* is the dominant intermediate observed over the time course, although the alternative intermediate is also present. There is a strong processive component, as indicated by the simultaneous appearance of ATF2/TT* and ATF2/T*T* as seen previously through in vitro (31) and cellular ATF2 phosphorylation assays (45). The rate of phosphorylation of ATF2 by p38α is significantly faster than the rate of phosphorylation of p38α by MEK6.
Phosphorylation in the p38α MAPK Module Follows a Sequential Mechanism
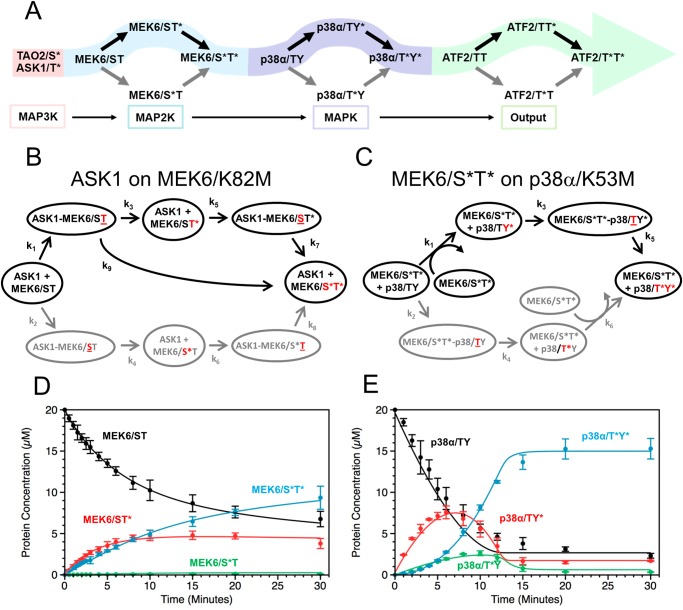
Given the above data, it appears that the phosphorylation events through the cascade composed of ASK1 (or TAO2), MEK6, and p38α follow a sequential mechanism. ASK1 or TAO2 phosphorylates MEK6 through MEK6/ST* (in the sequence 206DSVAKTI212); MEK6/S*T* phosphorylates p38α through p38α/TY* (in the sequence 183TGY185). Finally, p38α/T*Y* phosphorylates ATF2 through ATF2/TT* (in the sequence 67DQTPTTR73). The sequential mechanism is summarized in Fig. 4A.
FIGURE 4.
The sequence of cascade phosphorylation events. A, signal propagation through the p38 MAP kinase cascade follows double phosphorylation reactions that apparently occur in a preferred sequence, highlighted in colored arrows. The MAP2K MEK6 is phosphorylated first on Thr-211 to give MEK6/ST* and then on Ser-207 to give MEK6/S*T*. p38α is phosphorylated first on Tyr-182 to give p38α/TY* and then on Thr-180 to give p38α/T*Y*. B and C, kinetic models were generated for ASK1 phosphorylation of MEK6 (B) and MEK6 phosphorylation of p38α (C). The phosphorylation site involved at each step is shown in red, and underlining indicates binding in the active site. Protein complexes leading to catalytically competent interactions were defined. This is followed by an irreversible phosphorylation step and rebinding of the monophosphorylated substrate. A second phosphorylation step is similarly modeled, leading to a final, double phosphorylated product. In both models, one set of reactions, initiated by binding MEK6/ST in (B) and p38α/TY in (C), is unfavored and shown in gray. Only reactions contributing kinetically to the model are included, to reduce the number of variables. In B, a processive component was required (k9) to account for the rapid appearance of the doubly phosphorylated MEK6/S*T*. In C, it was not necessary to separate binding and catalysis in the formation of p38α/TY* to fit the phosphorylation data. Models were evaluated using the DynaFit software (40, 56). Calculated progress curves from the models for the ASK1 phosphorylation of MEK6 (D) and MEK6 phosphorylation of p38α (E) are shown superimposed on the experimental data.
Kinetic Models Describe MEK6 and p38α Phosphorylation
Several questions arise from the shapes of the progress curves. Does the flux of the reactions track through the apparent observed phospho-intermediates? Further, what is the origin of the distinctive shapes of the curves at the two levels? Does this arise from the existence of different kinetic steps, or is there just a variation in the rates of the same steps? Also, is the reaction specificity due to rates of binding or catalytic events? To gain qualitative answers to these questions, kinetic models were fit to the phosphorylation progress curves. As described under “Experimental Procedures,” all potential binding and catalytic steps were initially considered; then the reactions that contributed kinetically were retained in the final model. At the MAP2K level, we focused on the ASK1 phosphorylation of MEK6. At both levels of the cascade, complexes leading to different phosphorylation reactions were distinguished by underlining the amino acid in the active site (for example, the complex ASK1-MEK6/ST in Fig. 4B leads to chemistry on Thr). Further, catalysis and dissociation steps were held separate. In general, the back reactions were too slow to affect the model. At the MAPK level, the persistence of p38α and p38α/TY* was modeled as a slow loss of competence of all p38α proteins to be phosphorylated over the duration of the time course experiment (see Fig. 4E legend for p38α and see also supplemental Table S2). At the MAP2K level, no such additional terms were required to obtain a fit. The final models are given in Fig. 4, B and C, and the overlays of the calculated curves to the data are given in Fig. 4, D and E. Reactions leading to the unfavored intermediates, MEK6/S*T and p38α/T*Y, were much slower (shown in gray in Fig. 4, B and C). The derived kinetic constants for both MEK6 and p38α phosphorylation are given in supplemental Tables S3 and S4, respectively. The catalytic constants from our data fit approximately with those presented by Fujioka et al. (15) (11 min−1) for phosphorylation of MEK and ERK in cells.
The final reaction schemes are slightly different. At the MAP2K level, it was necessary to include a term for direct conversion of ASK1-MEK6/ST* to MEK6/S*T* (k9), thus accounting for the apparent processivity in Fig. 3B, where the intermediate and product appear simultaneously. In the phosphorylation of p38α, the tyrosine phosphorylation was too fast to affect the model, allowing the binding and catalytic steps to be conflated.
Kinetic Modeling Confirms Flux through Specific Intermediates
Fitting confirms that the flux to doubly phosphorylated MEK6 occurs primarily through MEK6/ST* (with k2 much smaller than k1, supplemental Table S2). Likewise, the phosphorylation of p38α by MEK6/S*T* was fit to the model shown in Fig. 4C, with the resulting calculated curves validating p38α/TY* as the predominant intermediates, again with k2 < k1 (supplemental Table S2).
Reactions at the MAP2K but Not MAPK Level Are Partially Processive
At the MAP2K level, the reaction is partially processive, without dissociation between catalytic steps. The processivity was modeled as an additional kinetic term for the conversion from ASK1-MEK6/ST* to ASK1-MEK6/S*T* without complex dissociation (k9, supplemental Table S2, Fig. 4B). Models lacking a processive step resulted in a much poorer fit (data not shown). In p38α phosphorylation, the reaction is distributive (nonprocessive). This is apparent from the sudden appearance p38α/TY* relative to p38α/T*Y*. The slowest reaction in p38α/T*Y* formation is the last step, and the first step, k1, is much faster (k1/k5 ∼ 20), further explaining the buildup of intermediates. No improvement in the model fit was obtained by including a processive reaction term (data not shown). Thus, the difference in shapes of the two different sets of curves is significantly due to an added processive step in the ASK1 phosphorylation of MEK6.
Specific Intermediates Result from kcat as well as Binding Effects
In the ASK1 phosphorylation of MEK6, the binding constant k1 is greater than k2 (2 > 0.04 μm−1 min−1). Similarly, the kcat constant k3 is greater than k4 (11 > 0.7 min−1). Apparently, both binding and catalysis contribute to the reaction specificity. In the phosphorylation of p38α, k1 > k2; kcat for the conversion of p38α/TY to p38a/TY* is too fast to model, giving rise to the same conclusion, that both binding and catalysis contribute to the reaction specificity.
The Rate of Reaction Increases through the Cascade
The progress curve data allow us to consider overall rates for each step in the full MAPK cascade. As a first approximation, the slowest step leading to doubly phosphorylated product is the rate-limiting step. If we only consider the dominant phosphorylation pathways, the slowest step in the formation of MEK6/S*T* is k1 at about 2 μm−1 min−1. The slowest step to form p38α/T*Y* is k5 at about 7 min−1. Phosphorylation of ATF2 is fastest of all, at about 500 min−1, as estimated from the initial rate of phosphorylation (Fig. 3).
DISCUSSION
Phosphorylation order in MAP kinase modules could theoretically be either random or sequential (20). Here we show the p38 MAP kinase module clearly follows the sequential model. The order of reactions is well defined at both levels of the cascade. The MAP2K MEK6 is phosphorylated primarily through the intermediate MEK6/ST*. The phosphorylation of p38α goes through a dominant intermediate, p38α/TY*. This is the same order that occurs in the MAPK ERK2 (18, 21). At both levels, the order is apparently determined by a difference in initial rates leading to different chemical outcomes (k1/k2 > 1). We also confirmed that ATF2 is phosphorylated through the intermediate ATF2/TT* by p38α as reported by Waas et al. (31).
The basis for the observed order of phosphorylation events at either level of the cascade is not understood in molecular detail. Our earlier studies of MEK6-TAO2 interaction showed that mutation of either Ile-212 or Asp-206 in the activation loop of MEK6 (in the sequence 206DSVAKTI212) reduced phosphorylation by TAO2 (28). Mutational analysis of TAO2 revealed a cluster of basic residues (Lys-120, Arg-221, and Lys-222) that is important for activity (marked as + in Fig. 1). This led to a model in which the isoleucine (Ile-212) in MEK6 binds to the P+1 pocket (46) of TAO2, and the aspartic acid (Asp-206) binds the basic cluster, thus dictating the reaction order.
No structures or molecular models are available to indicate why tyrosine in the MAPK activation loop sequence TXY is phosphorylated first by MAP2Ks. However, the progress curves and model fitting presented here show that MEK6 binding to p38α at Tyr-185 leads to rapid catalysis in comparison with p38α Thr-183 binding. This is also the case in MAP kinase phosphatases, which dephosphorylate first on Tyr (47, 48). It is also unknown how ATF2 is phosphorylated in a preferred order. Further data will be required to fully understand the underlying mechanisms driving the reaction orders established here.
At the MAPK level, the role of a phosphotyrosine intermediate has been in dispute, with the suggestion that either it is required for activity (49) or it is a stepping stone in MAPK activation (23). Here we have shown that the p38α phosphorylation occurs through p38α/TY*. This result offers support to the idea that phospho-tyrosine formation is a stepping stone in MAPK activation. The stepping stone idea gains strength from the lack of Tyr residues in the ERK2 homolog ERK3 (4), suggesting that the ERK3/S* (occupying the homologous position to Thr in ERK2 and p38α) is all that is required for activity (50) (supplemental Fig. S3). It is interesting that the overall design of the cascade represents an excursion from Ser/Thr to Tyr chemistry. The cascade starts with a Ser/Thr MAP3K and ends as a Ser/Thr MAPK but requires Tyr kinase activity in the intermediate MAP2K.
We further observe both distributive (dissociative) and processive reactions. We observe processivity at the MAP2K level, with the appearance of the intermediate and product occurring simultaneously. The shapes of the progress curves for the appearance of MEK6/ST* and MEK6/S*T* are reminiscent of processive phosphorylation reactions induced by high viscosity reported by Aoki et al. (13) for ERK2. The phosphorylation of p38α is clearly distributive, with significant amounts of the intermediate forming before the final product. The kinetic modeling suggests that the last step to form p38a/T*Y* is slow, thus contributing to the buildup of intermediates. The presence of intermediates should contribute to the sigmoid properties of the cascade by slowing the formation of final product (43) and by creating competing binding interactions for the upstream enzyme (51).
Because of the differences in experimental approaches, the ability to compare rate constants with published cellular assays is limited. Nevertheless, the kcat values at the MAP2K level are comparable. We derived a kcat of 11 min−1, which is in close agreement to the results of Fujioka et al. (15). On the other hand, they report significantly faster on rates than we observed at the MAP2K level. This could be due to our use of MAP3K kinase domains instead of full-length proteins and the lack of other cellular scaffolds.
Many multiple phosphorylation events in signal transduction are being discovered, and in a very few cases, the order of reactions has been shown to occur in a precise sequence (31, 52). It is interesting that ATF2 is also phosphorylated in a specific order, and further, this is an integrator of signals from multiple MAP kinases (53, 54). Additional analysis is required to determine whether the reaction order is maintained across multiple MAPKs acting on ATF2.
This study represents the first in vitro analysis of a complete MAP kinase cascade using native kinase substrates and their upstream activators. This approach, in conjunction with mass spectrometry, generates high fidelity data for modeling cascade reaction kinetics. It is clear from the reaction schemes in Figs. 1 and 4A that maximally switch-like responses should be available if intermediates are inactive. Future work will focus on comparisons of the activities of phosphorylation intermediates across different MAP kinase cascades and studies of reconstituted cascades in vitro.
Supplementary Material
Acknowledgments
We thank Melanie Cobb for valuable discussions and several protein expression vectors. We also thank Elliott Ross, Hamid Mirzaei, and Xuewu Zhang for critical reading of this manuscript and Saurabh Mendiratta for protein expression and purification.
This work was supported, in whole or in part, by National Institutes of Health Grant DK46993 (to E. J. G.). This work was also supported by Welch Foundation Grant I1128.

This article contains supplemental Figs. S1–S3 and Tables S1–S4.
- MAP
- mitogen-activated protein
- ASK1
- apoptosis signaling kinase-1
- TAO2
- thousand and one kinase-2
- ATF2
- activating transcription factor 2
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1. Avruch J. (2007) MAP kinase pathways: the first twenty years. Biochim. Biophys. Acta 1773, 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lawrence M. C., Jivan A., Shao C., Duan L., Goad D., Zaganjor E., Osborne J., McGlynn K., Stippec S., Earnest S., Chen W., Cobb M. H. (2008) The roles of MAPKs in disease. Cell Res 18, 436–442 [DOI] [PubMed] [Google Scholar]
- 3. Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C., Cobb M. H. (2001) MAP kinases. Chem. Rev. 101, 2449–2476 [DOI] [PubMed] [Google Scholar]
- 4. Keshet Y., Seger R. (2010) The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol. Biol. 661, 3–38 [DOI] [PubMed] [Google Scholar]
- 5. Yang S. H., Sharrocks A. D., Whitmarsh A. J. (2013) MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13 [DOI] [PubMed] [Google Scholar]
- 6. Mansour S. J., Resing K. A., Candi J. M., Hermann A. S., Gloor J. W., Herskind K. R., Wartmann M., Davis R. J., Ahn N. G. (1994) Mitogen-activated protein (MAP) kinase phosphorylation of MAP kinase kinase: determination of phosphorylation sites by mass spectrometry and site-directed mutagenesis. J. Biochem. 116, 304–314 [DOI] [PubMed] [Google Scholar]
- 7. Ahn N. G., Seger R., Bratlien R. L., Diltz C. D., Tonks N. K., Krebs E. G. (1991) Multiple components in an epidermal growth factor-stimulated protein kinase cascade: in vitro activation of a myelin basic protein/microtubule-associated protein 2 kinase. J. Biol. Chem. 266, 4220–4227 [PubMed] [Google Scholar]
- 8. Gonzalez F. A., Raden D. L., Davis R. J. (1991) Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266, 22159–22163 [PubMed] [Google Scholar]
- 9. Ferrell J. E., Jr., Machleder E. M. (1998) The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 280, 895–898 [DOI] [PubMed] [Google Scholar]
- 10. Bagowski C. P., Besser J., Frey C. R., Ferrell J. E., Jr. (2003) The JNK cascade as a biochemical switch in mammalian cells: ultrasensitive and all-or-none responses. Curr. Biol. 13, 315–320 [DOI] [PubMed] [Google Scholar]
- 11. Whitehurst A., Cobb M. H., White M. A. (2004) Stimulus-coupled spatial restriction of extracellular signal-regulated kinase 1/2 activity contributes to the specificity of signal-response pathways. Mol. Cell. Biol. 24, 10145–10150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mackeigan J. P., Murphy L. O., Dimitri C. A., Blenis J. (2005) Graded mitogen-activated protein kinase activity precedes switch-like c-Fos induction in mammalian cells. Mol. Cell. Biol. 25, 4676–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aoki K., Yamada M., Kunida K., Yasuda S., Matsuda M. (2011) Processive phosphorylation of ERK MAP kinase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 108, 12675–12680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cirit M., Wang C. C., Haugh J. M. (2010) Systematic quantification of negative feedback mechanisms in the extracellular signal-regulated kinase (ERK) signaling network. J. Biol. Chem. 285, 36736–36744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujioka A., Terai K., Itoh R. E., Aoki K., Nakamura T., Kuroda S., Nishida E., Matsuda M. (2006) Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 281, 8917–8926 [DOI] [PubMed] [Google Scholar]
- 16. Malleshaiah M. K., Shahrezaei V., Swain P. S., Michnick S. W. (2010) The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature 465, 101–105 [DOI] [PubMed] [Google Scholar]
- 17. O'Shaughnessy E. C., Palani S., Collins J. J., Sarkar C. A. (2011) Tunable signal processing in synthetic MAP kinase cascades. Cell 144, 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haystead T. A., Dent P., Wu J., Haystead C. M., Sturgill T. W. (1992) Ordered phosphorylation of p42mapk by MAP kinase kinase. FEBS Lett. 306, 17–22 [DOI] [PubMed] [Google Scholar]
- 19. Ferrell J. E., Jr., Bhatt R. R. (1997) Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 272, 19008–19016 [DOI] [PubMed] [Google Scholar]
- 20. Salazar C., Höfer T. (2007) Versatile regulation of multisite protein phosphorylation by the order of phosphate processing and protein-protein interactions. FEBS J 274, 1046–1061 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y. Y., Mei Z. Q., Wu J. W., Wang Z. X. (2008) Enzymatic activity and substrate specificity of mitogen-activated protein kinase p38α in different phosphorylation states. J. Biol. Chem. 283, 26591–26601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi M. Y., Kang G. Y., Hur J. Y., Jung J. W., Kim K. P., Park S. H. (2008) Analysis of dual phosphorylation of Hog1 MAP kinase in Saccharomyces cerevisiae using quantitative mass spectrometry. Mol. Cells 26, 200–205 [PubMed] [Google Scholar]
- 23. Bell M., Engelberg D. (2003) Phosphorylation of Tyr-176 of the yeast MAPK Hog1/p38 is not vital for Hog1 biological activity. J. Biol. Chem. 278, 14603–14606 [DOI] [PubMed] [Google Scholar]
- 24. Robbins D. J., Zhen E., Cheng M., Xu S., Vanderbilt C. A., Ebert D., Garcia C., Dang A., Cobb M. H. (1993) Regulation and properties of extracellular signal-regulated protein kinase-1, kinase-2, and kinase-3. J. Am. Soc. Nephrol. 4, 1104–1110 [DOI] [PubMed] [Google Scholar]
- 25. Diskin R., Lebendiker M., Engelberg D., Livnah O. (2007) Structures of p38α active mutants reveal conformational changes in L16 loop that induce autophosphorylation and activation. J. Mol. Biol. 365, 66–76 [DOI] [PubMed] [Google Scholar]
- 26. Zheng C. F., Guan K. L. (1994) Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 13, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alessi D. R., Saito Y., Campbell D. G., Cohen P., Sithanandam G., Rapp U., Ashworth A., Marshall C. J., Cowley S. (1994) Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 13, 1610–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou T., Raman M., Gao Y., Earnest S., Chen Z., Machius M., Cobb M. H., Goldsmith E. J. (2004) Crystal structure of the TAO2 kinase domain: activation and specificity of a Ste20p MAP3K. Structure 12, 1891–1900 [DOI] [PubMed] [Google Scholar]
- 29. Matsukawa J., Matsuzawa A., Takeda K., Ichijo H. (2004) The ASK1-MAP kinase cascades in mammalian stress response. J. Biochem. 136, 261–265 [DOI] [PubMed] [Google Scholar]
- 30. Hutchison M., Berman K. S., Cobb M. H. (1998) Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 273, 28625–28632 [DOI] [PubMed] [Google Scholar]
- 31. Waas W. F., Lo H. H., Dalby K. N. (2001) The kinetic mechanism of the dual phosphorylation of the ATF2 transcription factor by p38 mitogen-activated protein (MAP) kinase α: implications for signal/response profiles of MAP kinase pathways. J. Biol. Chem. 276, 5676–5684 [DOI] [PubMed] [Google Scholar]
- 32. Goldsmith E. J., Min X., He H., Zhou T. (2010) Structural studies of MAP Kinase cascade components. Methods Mol. Biol. 661, 223–237 [DOI] [PubMed] [Google Scholar]
- 33. Min X., Akella R., He H., Humphreys J. M., Tsutakawa S. E., Lee S. J., Tainer J. A., Cobb M. H., Goldsmith E. J. (2009) The structure of the MAP2K MEK6 reveals an autoinhibitory dimer. Structure 17, 96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bunkoczi G., Salah E., Filippakopoulos P., Fedorov O., Müller S., Sobott F., Parker S. A., Zhang H., Min W., Turk B. E., Knapp S. (2007) Structural and functional characterization of the human protein kinase ASK1. Structure 15, 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Z., Harkins P. C., Ulevitch R. J., Han J., Cobb M. H., Goldsmith E. J. (1997) The structure of mitogen-activated protein kinase p38 at 2.1-Ä resolution. Proc. Natl. Acad. Sci. U.S.A. 94, 2327–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bukhtiyarova M., Northrop K., Chai X., Casper D., Karpusas M., Springman E. (2004) Improved expression, purification, and crystallization of p38α MAP kinase. Protein Expr. Purif. 37, 154–161 [DOI] [PubMed] [Google Scholar]
- 37. Akella R., Min X., Wu Q., Gardner K. H., Goldsmith E. J. (2010) The third conformation of p38α MAP kinase observed in phosphorylated p38α and in solution. Structure 18, 1571–1578 [DOI] [PubMed] [Google Scholar]
- 38. Deininger S. O., Cornett D. S., Paape R., Becker M., Pineau C., Rauser S., Walch A., Wolski E. (2011) Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal. Bioanal. Chem. 401, 167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 40. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 41. Kuzmic P. (2009) Application of the Van Slyke-Cullen irreversible mechanism in the analysis of enzymatic progress curves. Anal. Biochem. 394, 287–289 [DOI] [PubMed] [Google Scholar]
- 42. Van Slyke D. D., Cullen G. E. (1914) The mode of action of urease and of enzymes in general. J. Biol. Chem. 19, 141–180 [Google Scholar]
- 43. Huang C. Y., Ferrell J. E., Jr. (1996) Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A. 93, 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gupta S., Campbell D., Derijard B., Davis R. J. (1995) Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267, 389–393 [DOI] [PubMed] [Google Scholar]
- 45. Baan B., van der Zon G. C., Maassen J. A., Ouwens D. M. (2009) The nuclear appearance of ERK1/2 and p38 determines the sequential induction of ATF2-Thr71 and ATF2-Thr69 phosphorylation by serum in JNK-deficient cells. Mol. Cell. Endocrinol. 311, 94–100 [DOI] [PubMed] [Google Scholar]
- 46. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 47. Farooq A., Zhou M. M. (2004) Structure and regulation of MAPK phosphatases. Cell. Signal. 16, 769–779 [DOI] [PubMed] [Google Scholar]
- 48. Zhao Y., Zhang Z. Y. (2001) The mechanism of dephosphorylation of extracellular signal-regulated kinase 2 by mitogen-activated protein kinase phosphatase 3. J. Biol. Chem. 276, 32382–32391 [DOI] [PubMed] [Google Scholar]
- 49. Zhou B., Wang Z. X., Zhao Y., Brautigan D. L., Zhang Z. Y. (2002) The specificity of extracellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. J. Biol. Chem. 277, 31818–31825 [DOI] [PubMed] [Google Scholar]
- 50. Cheng M., Zhen E., Robinson M. J., Ebert D., Goldsmith E., Cobb M. H. (1996) Characterization of a protein kinase that phosphorylates serine 189 of the mitogen-activated protein kinase homolog ERK3. J. Biol. Chem. 271, 12057–12062 [DOI] [PubMed] [Google Scholar]
- 51. Kim S. Y., Ferrell J. E., Jr. (2007) Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell 128, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 52. Furdui C. M., Lew E. D., Schlessinger J., Anderson K. S. (2006) Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21, 711–717 [DOI] [PubMed] [Google Scholar]
- 53. Lau E., Ronai Z. A. (2012) ATF2 – at the crossroad of nuclear and cytosolic functions. J. Cell Sci. 125, 2815–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wagner E. F., Nebreda A. R. (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 [DOI] [PubMed] [Google Scholar]
- 55. Annesley T. M. (2003) Ion suppression in mass spectrometry. Clin. Chem. 49, 1041–1044 [DOI] [PubMed] [Google Scholar]
- 56. Kuzmic P. (2009) DynaFit–a software package for enzymology. Methods Enzymol. 467, 247–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.