Abstract
While liganded nuclear receptors are established to regulate Pol II-dependent transcription units, their role in regulating Pol III-transcribed DNA repeats remains largely unknown. Here, we report that ~2–3% of the ~100,000–200,000 human DR2 Alu repeats in proximity to activated Pol II transcription units are activated by retinoic acid receptor in human embryonic stem cells to generate Pol III-dependent RNAs. These transcripts are processed, initially in a DICER-dependent fashion, into small RNAs (~28–65nt.), referred to as riRNAs, that cause degradation of a subset of critical stem cell mRNAs, including Nanog, modulating exit from the proliferative stem cell state. This regulation requires AGO3-dependent accumulation of processed DR2 Alu transcripts and subsequent recruitment of AGO3-associated decapping complexes to the target mRNA. In this way, the RAR and Pol III-dependent DR2 Alu transcriptional events in stem cells functionally complement the Pol II-dependent neuronal transcriptional program.
Introduction
Nuclear receptors are a cohort of transcription factors involved in many critical biological processes, such as development, metabolism and tumorigenesis1,2. They are usually activated by respective ligands and subsequently regulate RNA Pol II-dependent transcription of target genes in conjunction with a group of cofactors called the coactivator–corepressor complex. The target genes usually harbor specific response elements in their regulatory regions served for recognition and binding by nuclear receptors.
Retinoic acid nuclear receptors (RARs) are essential for embryonic development, neural specification and homeostasis2,3. It has three homologs, alpha, beta and gamma-RAR, responding to both the all-trans retinoic acid (atRA) and 9-cis RA. RAR forms a heterodimer structure with retinoid X receptor (RXR). The ligand-activated RAR–RXR plays as an activator for transcription while an unliganded RAR as a transcription repressor, which is attributed to its interaction with the canonical Nuclear receptor CoRepressor (NCoR) and the closely-related Silencing Mediator for Retinoic acid and Thyroid hormone receptors (SMRT)4,5. Upon atRA binding to the receptor, NCoR and SMRT are dismissed, which is accompanied by recruitment of coactivators, hence switching RAR into an active state to regulate target gene transcription in a Pol II-dependent manner6.
Interestingly, in the human genome, protein-coding regions only account for ~2% of the DNA content7, while ~47% can be classified into numerous types of repetitive elements, long-interspersed element (LINE), short-interspersed element (SINE), SVA, LTR, satellite repeats, etc. For a long time, these DNAs had been referred to as "junk DNAs" in human cells8 since no definitive functions had been fully identified. However, many lines of evidence have started emerging to suggest that these DNA repeats could actually represent overlooked sequences that have tremendous influence on cells, including the integrity of human genome, gene expression regulation and development9. In particular, there are over one million copies of the ancient 7SL-derived retrotransposons, Alu elements, embedded in both intragenic and intergenic regions of the human genome. They first diverged from the 7SL elements around 65 million years ago, and have evolved into multiple subfamilies, AluJ, AluS and AluY10. These repeats are about 300 bp in length, and contain the A and B boxes of an internal promoter for RNA polymerase III11. Typically, due to a basal level of transcription, a human cell typically contains a few hundred to a thousand copies of Alu transcripts12, the biological significance of which has not been well appreciated. Recently, some bioinformatics analyses predicted that a great number of Alu repeats contain potential binding sites for specific transcription factors13,14, including one subset of Alu repeats harbors the characteristic direct repeat of 6 bp core RAR binding site spaced by two nucleotides-the DR2 element14. We refer to this subset as “DR2 Alu” in this paper. Indeed, the presence of DR2 signature sequence in some Alu repeats suggests a potential interaction of DNA repeats and nuclear receptors.
We therefore explored whether atRA could activate DR2 Alu repeats, characterizing the potential role of DR2 Alu transcripts in stem cell state maintenance. Here, we report that many DR2 Alu repeats located in proximity to active Pol II transcriptional units are generally bound and induced by RAR to generate Pol III-dependent transcripts in human embryonic stem cells (hESCs). The primary transcripts are processed to small 28–65nt. sequences, initially in a Dicer-dependent fashion. These small products, collectively referred to as DR2 Alu repeat induced small RNAs (riRNAs), are stabilized by binding with Ago3. They further exploit an Ago3-associated decapping machinery to cause degradation of a cohort of ESC-expressed mRNAs that contain the complementary sequences to riRNAs in their 3'-untranslated regions (3'UTRs), thus modulating exit from the stem cell proliferative state. Hence, RAR signaling coordinates both the Pol II- and Pol III-dependent programs in ES cells.
Results
Retinoic acid-induced DR2 Alu transcription in stem cells
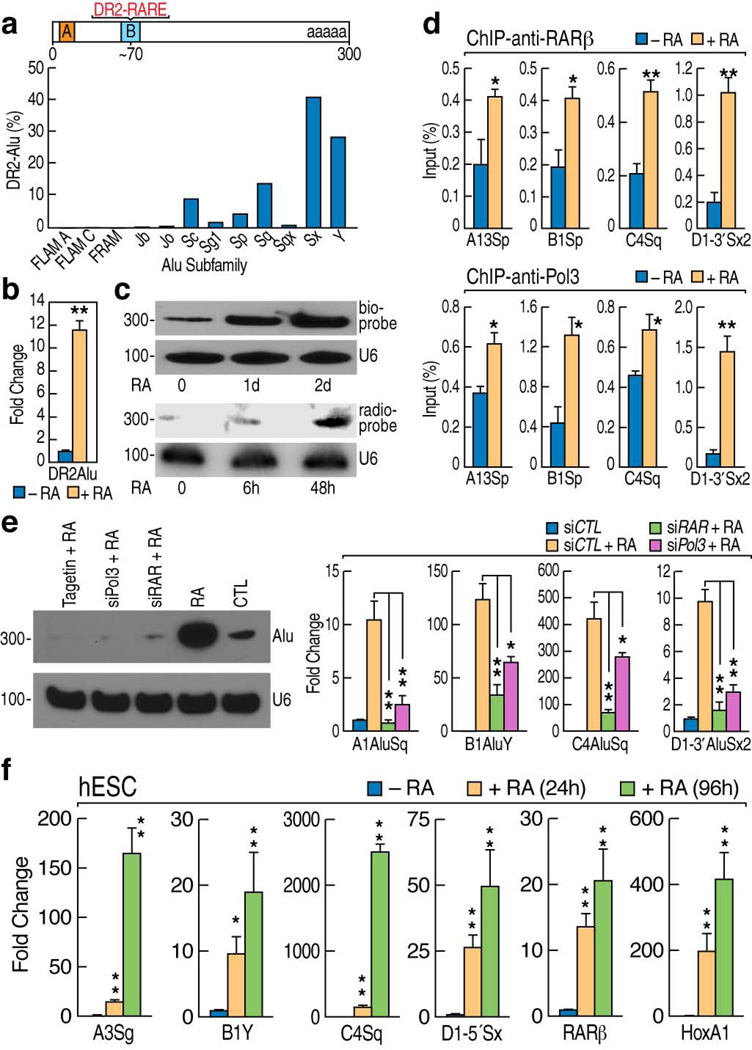
Bioinformatic predictions reveal that ~10% of the genomic Alu population contain DR2 elements14, referred to as DR2 Alu repeats, which are differentially distributed across the subclasses of Alu repeats (Fig. 1a). The presence of a canonical RAR response element raise an interesting possibility that atRA might also exert effects on at least a subset of these ancient Alu units15. Therefore, we utilized as an initial model the Ntera2 human embryonic carcinoma (EC) cells16 to determine whether atRA-induced neural developmental program might involve activation of DR2 Alu repeats. A significant atRA-induced increase of DR2 Alu repeat transcripts was observed in quantitative RT-PCR assays (RT-qPCR) (Fig. 1b). By using radio-labeled or biotin-conjugated oligonucleotide probes, Northern blot analysis confirmed that levels of DR2 Alu transcripts increased in response to atRA treatment (Fig. 1c). Consistent with the previous observation that ~75% of Pol II-regulated coding genes are flanked by Alu elements17, we noted that Hox clusters are flanked by DR2 Alu repeats (Supplementary Fig. 1a,b). Chromatin immunoprecipitation (ChIP) revealed a atRA-dependent increased binding of RARβ, Pol III and general transcription factor 3C (TF3C) to the DR2 Alu repeats flanking within ~15kb of Hox clusters (Fig. 1d; Supplementary Fig. 1c,d). Consistently, RAR knockdown, RNA Pol III knockdown or Tagetin, a specific inhibitor of Pol III18, abolished the induction of DR2 Alu repeats as determined by Northern blot and RT-qPCR (Fig. 1e), indicating that atRA-induced DR2 Alu activation is RAR and Pol III-dependent.
Figure 1.
RA induction of DR2 Alu transcription. (a) Bioinformatics analysis showed the distribution of the DR2 motif binding sites across the different subfamilies of Alu repeats, based on the binding profile shown in Fig.S1b. (b) RT-qPCR analysis of DR2 Alu transcript levels in RA-treated Ntera2 cells using the generic primer pairs for DR2 Alu repeats. RA: atRA in all figure labels. (c) Northern blot analysis of the total RNAs from untreated or RA-treated NT2 cells with biotin-conjugated or radio-labeled DR2 Alu probes. U6 snRNA was used as a control. (d) ChIP assays showed RA-induced recruitment of RARβ and Pol III to the DR2 Alu repeats flanking the Hox clusters. (e) Northern blot (left panel) and RT-qPCR (right panels) showed the requirement of RARβ and Pol III in activation of the DR2 Alu repeats. (f) RT-qPCR showed an RA-dependent activation of DR2 Alu repeats in human ES cells. The names of individual DR2 Alu elements were abbreviated (e.g. C4Sq denotes C4AluSq). (mean±SEM; *p<0.05, **p<0.01)
We next examined whether atRA-induced DR2 Alu transcription also occurred in human embryonic stem cells (hESCs). The RT-qPCR assays showed that DR2 Alu transcription was indeed induced by atRA in H9 hESCs (Fig. 1f). In contrast, we did not observe induced DR2 Alu transcription in Hela, U2OS, or human lung fibroblasts (NHLFs), even though the efficacy of the atRA–RAR pathway in each of these cell lines was confirmed with demonstration of RARβ expression (Supplementary Fig. 1e). These data indicated that the robust RA-dependent DR2 Alu activation was uniquely observed in stem cells, but not in terminally differentiated cells. To explain this specificity, we performed ChIP assays to assess the binding of RAR, Pol II and Pol III to targeted DR2 Alu repeats in U2OS cells. The data showed that atRA did not cause any significant recruitment of these factors to DR2 Alu elements in U2OS cells (Supplementary Fig. 1f).
To investigate the extent of the atRA-induced activation of DR2 Alu transcription, we performed genome-wide RNA sequencing at 50, 76 or 100 cycles to obtain sufficient resolution for assignment of repetitive sequences. These data sets suggest that at least ~2–3% of the estimated 100,000–200,000 DR2 Alu repeats are activated by atRA in Ntera2 cells, distributed amongst various classes of DR2 Alu repeats, mostly “middle-aged” classes of DR2 Alu repeats (Supplementary Fig. 2a). Our initial sequencing data also suggest that a key, correlated feature determining DR2 Alu activation might be proximity to an active Pol II transcription unit (Supplementary Fig. 2b), as similarly implied by other genome-wide profilings19,20. This was exemplified by the observation that DR2 Alu repeats <10 kb from HoxA1 could be activated by atRA, while those located ~20 kb from HoxA1 failed to exhibit RA-induced activation (Supplementary Fig. 2c). In the case of HoxA13 locus, the proximal (~18kb from HoxA13) DR2 Alu repeat was activated by atRA, while the distal ones (~28kb–31kb) were not (Supplementary Fig. 2c). These findings provoked a more generalized question whether active transcription of DR2 Alu repeats depends on activation of the adjacent Pol II transcription unit. We initiated investigation of this point by taking advantage of the heat shock 70 (Hsp70) locus that can be activated by the heat shock stimulation21. We found that the two DR2 Alu repeats located within 10kb of Hsp70 exhibited some detectable activation after heat shock, and also showed some activation in response to atRA treatment (Supplementary Fig. 2d), but when the cells were exposed to the atRA treatment followed by heat shock, these DR2 Alu repeats showed a significant, combinatorially enhanced induction of transcription. In contrast, for the HoxA1-AluSq repeat, no such effects were observed (Supplementary Fig. 2d). Given that heat shock did not trigger HoxA1 transcription, these data suggest that, while RAR binds directly onto the DR2 sequence embedded in the Alu repeats near Hsp70, their transcriptional activity is highly augmented by activation of the adjacent Pol II-dependent Hsp70 transcription unit.
DR2 Alu transcripts were further processed into small RNAs
To explore whether DR2 Alu transcripts are subsequently relocated to the cytoplasm, RNA FISH was performed by using a degenerate probe to DR2 Alu transcripts, and revealed accumulation of DR2 Alu transcripts "encircling" the nucleus (Supplementary Fig. 2e, left panels). Given potential distortion of cellular structures during the preparation of cells for RNA FISH, we designed specific molecular beacons22 containing a degenerate complementary sequence to DR214. By introducing the beacons into live cells pre-treated with atRA for 24 hours, we observed a prominent increase in DR2 Alu transcripts in the peri-nuclear and cytoplasmic regions (Supplementary Fig. 2e, right panels). In contrast, we did not observe detectable atRA-induced signals in Hela or U2OS cells, which confirmed a cell lineage specificity of induced DR2 Alu transcription (Supplementary Fig. 2f). Since the transported DR2 Alu transcripts appeared clustered in the cytoplasm (Supplementary Fig. 2e), a pattern reminiscent of cytoplasmic structures such as P bodies23, we used the same molecular beacons together with immunofluorescent staining of argonaute proteins. Strikingly, DR2 Alu RNAs substantially co-localized with argonaute proteins, including AGO3 in RA-treated Ntera2 cells (Supplementary Fig. 2g,h).
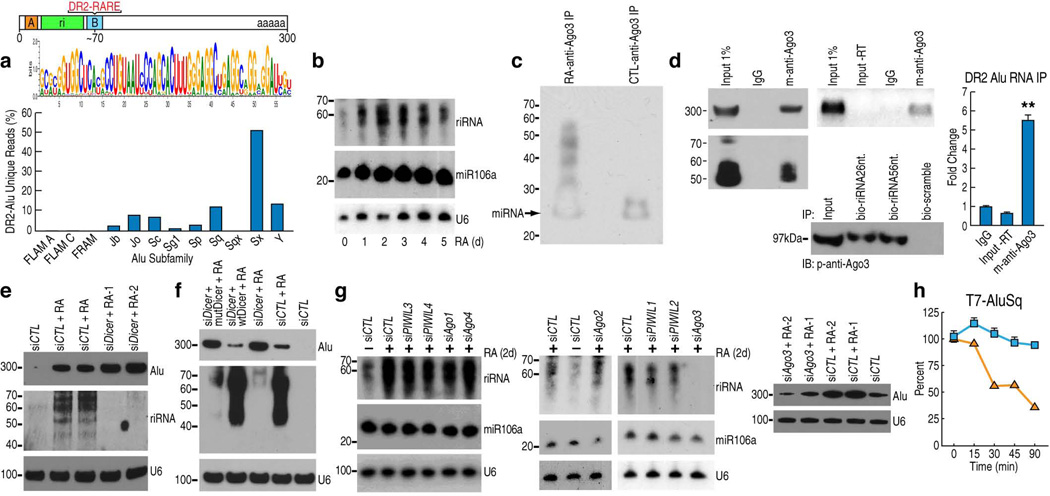
Given that P bodies serve as sites for RNA processing machinery, we next explored the possibility that the DR2 Alu transcripts might be processed into smaller RNA moieties. Bioinformatics analysis of small RNA databases24 revealed that some small RNAs might be derived from DR2 Alu repeats. Hence, we performed small RNA sequencing in atRA-treated Ntera2 cells with the Illumina platform. By aligning the sequencing data to all subclasses of the DR2 Alu repeats using the NOVOalign program, we found that atRA-treated Ntera2 cells contained DR2 Alu-derived small RNAs varying in length from ~28–65 nt., largely mapped to the 5’ region of the repeat, and we assigned these small RNAs to each subclass of DR2 Alu from which they were originated (Fig. 2a). While the Illumina Small RNA Kit introduced a strong artifactual bias towards RNAs <30 nt., Northern blot analysis revealed a much higher abundance of longer RNAs products (~40–65nt.) in atRA-treated cells (Fig. 2b). We refer to this heterogeneous population of small RNAs as riRNAs, with some ~30nt. sequences potentially classified as piRNAs25.
Figure 2.
DR2 Alu transcripts associate with AGO3 and produce riRNAs in a DICER-dependent manner. (a) Bioinformatics analysis of small RNA-seq datasets identified a consensus sequence of riRNAs derived from the 5’ end of DR2 Alu (the green box). The bar graph shows the distribution of riRNAs among Alu subfamilies. (b) Northern blot detected riRNAs in a time-course study. miR106a and U6 were used as controls. (c) Biotin-labeling of AGO3-immunoprecipitated RNAs from control and RA-treated cells and chemiluminescence detection. Due to the biotin conjugation, the putative miRNA appeared ~25nt. as pointed by the arrow. (d) AGO3-immunoprecipitated RNAs were analyzed by Northern blot (left gel), RT-PCR (middle upper gel) and RT-qPCR (mean±SEM; **p<0.01). RNA pull-down assays with biotin-conjugated riRNA oligonucleotides, both the 26nt. and 56nt. sequences, confirmed interaction with AGO3 (middle lower gel). (e) Northern blot examined the full-length DR2 Alu transcripts and riRNAs in Ntera2 transfected with two individual DICER siRNAs. siCTL: control siRNA. (f) Northern blot showed that overexpression of wild-type DICER (wtDicer), but not slicer-deficient mutant (mutDicer), in DICER knock-down cells, reduced the full length DR2 Alu transcripts and caused re-appearance of riRNAs. (g) Northern showed that among AGO1–4 and PIWIL1–4, only AGO3 knock-down reduced both riRNA (left two panels) and the full-length DR2 Alu RNA (right panel; two different AGO3 siRNAs were used). (h) RT-qPCR showed that in AGO3 knock-down Ntera2 cells, microinjected T7 DR2 Alu RNAs were rapidly decreased. Blue squares represent control siRNA and orange squares AGO3 siRNA.
Consistent with the observation that DR2 Alu transcripts were colocalized with AGO3 (Supplementary Fig. 2g), biotin labeling of AGO3-associated RNAs revealed that AGO3 was associated with an atRA-induced species of small RNAs sized ~30–60nt. suggestive of riRNAs, as well as with the putative miRNAs and siRNAs as indicated by the arrow (Fig. 2c). Northern blot, RT-PCR, RT-qPCR and biotinylated oligonucleotide pull-down assays confirmed interactions between AGO3 and DR2 Alu transcripts or riRNAs (Fig. 2d).
Accordingly, we also explored the role of DICER in potential processing of atRA-induced DR2 Alu transcripts into riRNAs. Using specific siRNAs, we found that in DICER knock-down Ntera2 cells, riRNA generation was abolished as determined by Northern blot analysis, and concurrently the accumulation of full-length DR2 Alu transcripts was increased (Fig. 2e). These effects were reversed by overexpression of wild-type, but not slicer-deficient, DICER constructs26 (Fig. 2f) in DICER-knockdown cells. Although this implied that the slicing activity of DICER is initially involved in DR2 Alu processing, given the current knowledge on DICER enzymology and structure, we cannot exclude the interpretation that these observations could reflect an indirect effect. Interestingly, we observed partial colocalization of DR2 Alu RNAs and DICER in the cytoplasm of atRA-treated cells (Supplementary Fig. 2i). Indeed, two recent articles reported an essential role of DICER in preventing cytotoxic accumulation of Alu transcripts, which leads to geographic atrophy27,28.
We next examined the potential roles of AGO1–4 and PIWIL1–4 which are expressed in Ntera2 cells (Supplementary Fig. 3a), in the processing of full-length DR2 Alu RNAs. We knocked down the eight individual argonaute homologs with specific individual siRNAs and, surprisingly, found that only AGO3 knock-down blocked the generation of riRNAs by Northern blot (Fig. 2g, left two panels). Remarkably, the levels of full length DR2 Alu RNAs also decreased substantially in AGO3 knock-down cells (Fig. 2g, right panel), indicating that AGO3 is involved in maintaining the levels of full length DR2 Alu and riRNAs once they are generated in a Dicer-dependent manner. This observation is consistent with previous reports that AGO3 lacks intrinsic enzymatic activity29,30.
To better understand the rather surprising role of AGO3 in accumulating and maintaining levels of DR2 Alu transcripts, we adopted a microinjection strategy, as described previously31. Ntera2 cells were first transfected with either control siRNA or siRNA against AGO3, and then micro-injected with either full length DR2 Alu or a 68-nt synthesized riRNA oligonucleotide. The cells were then harvested at different time points and subjected to RT-qPCR analysis. We noticed that the injected full length DR2 Alu or riRNAs remained stable at time points examined in control siRNA-transfected cells, while in AGO3-knockdown cells, the injected RNAs and oligonucleotides exhibited rapid loss and degradation, indicating that AGO3 is indeed involved in maintaining and stabilizing the level of Alu RNAs in cells (Fig. 2h; Supplementary Fig. 3b). Indeed, a recent study described a similar stabilizing role of AGO2 in maintaining miRNA levels32.
To confirm this, Ntera2 cells were first transfected with siRNA against AGO or DICER, and 48 hours post-transfection, the cells were further transfected with full length DR2 Alu or the 68-nt riRNA oligonucleotide for another 4 hours. The cells were then immediately harvested and RNAs analyzed in RT-qPCR. Again, we found that in AGO3-knockdown cells, the introduced full length DR2 Alu or riRNA oligonucleotides exhibited much lower levels than those observed in mock-transfected cells (Supplementary Fig. 3c). In contrast, the introduced full length DR2 Alu were remained largely intact in DICER-knockdown cells (Supplementary Fig. 3c), consistent with the data that DICER is involved in the initial processing of full length DR2 Alu transcripts.
Post-transcriptional regulation of target mRNAs by riRNAs
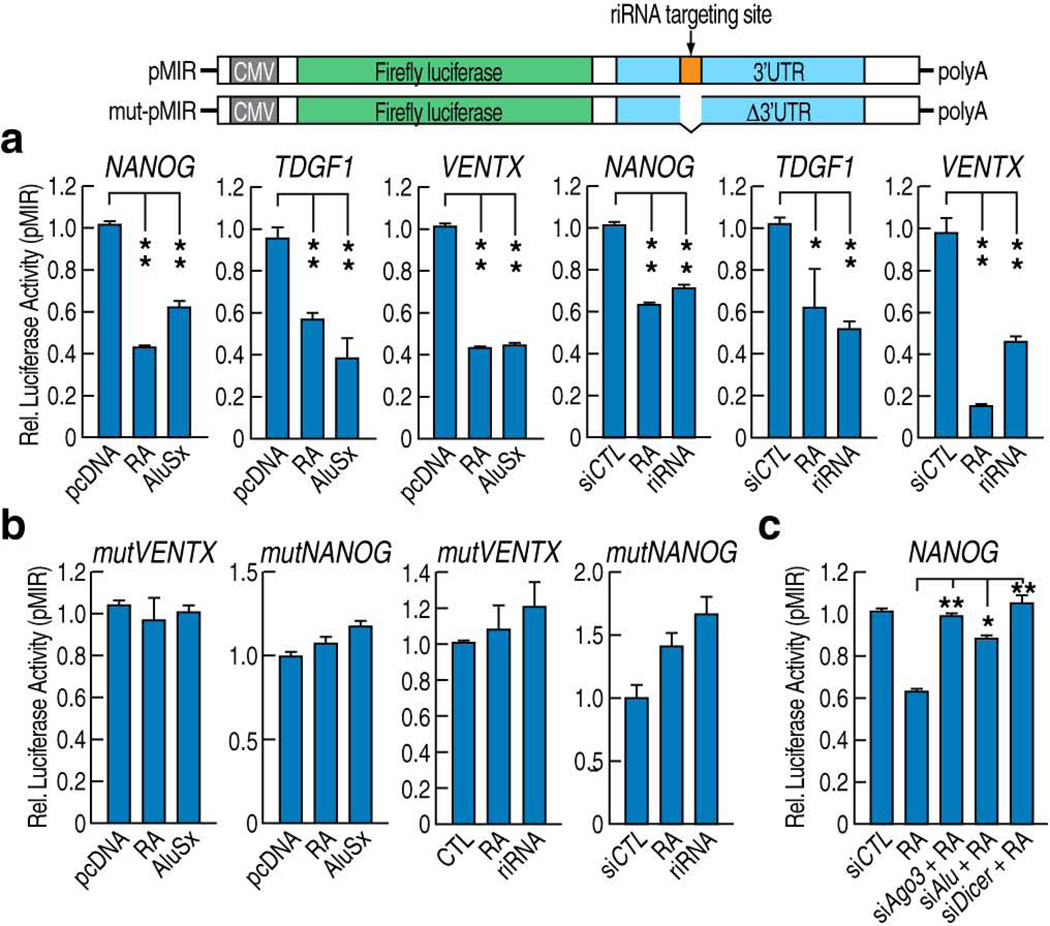
Given that riRNAs contained a consensus sequence, after BLAST analysis and Transpogene database mining, we hypothesized that they might exert functions somewhat comparable to those of miRNAs, possibly targeting some messenger RNAs by recognition of complementary sequences. Indeed, bioinformatics analyses of the RNA-sequencing data further revealed that among genes down-regulated by atRA, ~79 mRNAs contain complementary sequences to riRNAs in their 3′UTRs. The list includes many genes with known functional roles in embryogenesis, regulation of programmed cell death and cancer (Supplementary Fig. 3d). 20 genes particularly down-regulated by atRA were selected and 18 were confirmed to be down-regulated by atRA in RT-qPCR (Supplementary Fig. 3e). Five key genes involved in stem cell maintenance, cell proliferation regulation and apoptosis- Nanog33, Tdgf134, Ventx35, Lzts136, and Phlda137-were chosen for further study; schematic structures depicting potential riRNA targeting sites at the 3’ UTRs are shown (Fig. 3a; Supplementary Fig. 3f). RNA immunoprecipitation confirmed that AGO3 was recruited to the 3’UTRs of NANOG and TDGF1 mRNAs in an atRA- and Alu-dependent manner (Fig. 3b).
Figure 3.
riRNAs recognize complementary sequences in the 3'UTRs of target mRNAs. (a) Schematic structure of selected RA negatively-regulated transcripts, Tdgf1 and Nanog. The wave line denotes the 3’UTR. (b) RNA immunoprecipitation showed RA-dependent and DR2 Alu-dependent interaction of AGO3 with the 3’UTRs of Nanog and Tdgf1 mRNAs. (c) RT-qPCR showed that RA treatment decreased the transcript levels of examined genes and the specific siRNA against AGO3 reversed the effect. Over-expression of full-length AluSx transcripts (d) or synthesized 56 nt. riRNA (e) in Ntera2 cells mimicked the RA action in decreasing transcript levels of those genes. (f) Two siRNAs against DR2 Alu transcripts abolished RA-mediated downregulation of those transcripts. (mean±SEM; *p<0.05, **p<0.01)
To further explore how riRNAs may potentially regulate mRNAs, we first examined the effect of DICER knockdown on the expression of these genes. As expected, atRA-mediated down-regulation of these transcripts was either inhibited or abolished (Supplementary Fig. 3g). Similarly, in cells transfected with the AGO3 siRNA, down-regulation of these transcripts was also reversed (Fig. 3c; Supplementary Fig. 4a). The observation that AGO3 knockdown only partially, although statistically significantly, restored the Nanog mRNA level, can be reasonably explained by observing that the Nanog transcription unit surprisingly also exhibits an NCoR-dependent, putatively transcriptional, repression in response to atRA in ChIP assays (Supplementary Fig. 4b). The dual regulation of Nanog transcript levels was further supported by decreased reads of the H3K36me3 mark, a marker for actively transcribed region, across Nanog in ChIP-Seq (Supplementary Fig. 4c). In contrast, some transcription units, such as Phlda1, was not regulated at the transcriptional level, as shown by unaltered tag reads in either ChIP-Seq for H3K36me3 or global nuclear run-on-sequencing (GRO-Seq) assays38 (Supplementary Fig. 4c). Remarkably, AGO1, 2, 4 or PIWIL1–4 knockdown had no effects on atRA-repression of these transcripts (Supplementary Fig. 5a–d), implying a unique role of AGO3 in atRA-mediated down-regulation of target mRNAs.
We then examined the effects of over-expression of DR2 Alu repeats on the levels of the selected transcripts. Interestingly, overexpression of a specific DR2 Alu repeat (AluSx; chr7: 27,093,354–27,093,650; hg18) in Ntera2 cells mimicked the actions of atRA in reducing the levels of the selected transcripts (Fig. 3d), and introduction of synthetic riRNAs, either a long sequence (56nt; Fig. 3e) or a short species (26nt; Supplementary Fig. 5e), into Ntera2 cells produced similar effects. However, when riRNAs or AluSx-expressing constructs were co-transfected with siRNAs against AGO3, the repression on these genes was reversed (Supplementary Fig. 6a,b). In contrast, DICER knockdown did not affect the riRNA-mediated transcript down-regulation (Supplementary Fig. 6c). These data again imply that, while DICER exerts its function on initial processing of full length DR2 Alu, AGO3 potentially helps maintain the level of riRNAs that post-transcriptionally regulate target genes. As expected, depletion of DR2 Alu transcripts by two generic siRNAs abolished RA-induced down-regulation of these genes (Fig. 3f). Interestingly, we further noticed that knock-down of DROSHA1 and DGCR8 did not affect the transcript levels of these genes (Supplementary Fig. 6d,e), indicating that biogenesis of riRNAs follows a distinct mechanistic pathway from that of miRNAs.
To confirm that riRNAs mediate gene expression at post-transcriptional levels, we cloned the 3’UTRs of Tdgf1, Ventx and Nanog mRNAs into the pMIR luciferase vector and performed luciferase assays in Ntera2 cells. We noticed that over-expression of full length DR2 Alu or riRNAs decreased the luciferase activities of these constructed reporters (Fig. 4a). To assure that targeting of the 3’UTRs by riRNAs was essential for the observed effects, we deleted the 50 nt. complementary sequences to riRNAs from the 3’UTRs of Ventx or Nanog and performed the luciferase assays. In this case, we found that the DR2 Alu or riRNA-dependent decrease in luciferase activities was abolished (Fig. 4b). Interestingly, co-transfection of siAGO3, siAlu or siDICER with the wild-type Nanog luciferase construct abolished atRA-induced decrease in luciferase activities (Fig. 4c). These data confirmed that full length DR2 Alu and riRNAs can act post-transcriptionally to decrease the levels of RNAs that harbor complementary sequences in their 3’UTRs. In a reciprocal assay, we “neutralized” the post-transcriptional function of riRNA by co-transfecting both riRNA and an antisense oligonucleotide of riRNA into Ntera2 cells. As expected, the riRNA-mediated down-regulation of target mRNAs was abolished (Supplementary Fig. 6f). Together, these observations support that riRNA and its complementary sequence in the 3’UTR potentially play a role in the post-transcriptional regulation.
Figure 4.
Role of riRNA complementary sequence in post-transcriptional regulation. (a) Luciferase assays using constructs harboring 3’UTRs of Nanog, Tdgf1 or Ventx showed RA-dependent reduction in luciferase activity in Ntera2 cells. Expression of either full-length AluSx transcripts or synthesized short riRNAs gave similar results. (b) Luciferase assays showed that when the putative ~50bp riRNA targeting sequence was deleted from the Ventx or Nanog 3'UTR luciferase reporters, they no longer responded to RA treatment or over-expression of full-length AluSx or riRNAs. (c) In luciferase assays using the wild-type Nanog 3’UTR luciferase reporter, knockdown of AGO3 and DR2 Alu restored the luciferase activity in the presence of RA. (mean±SEM; *p<0.05, **p<0.01)
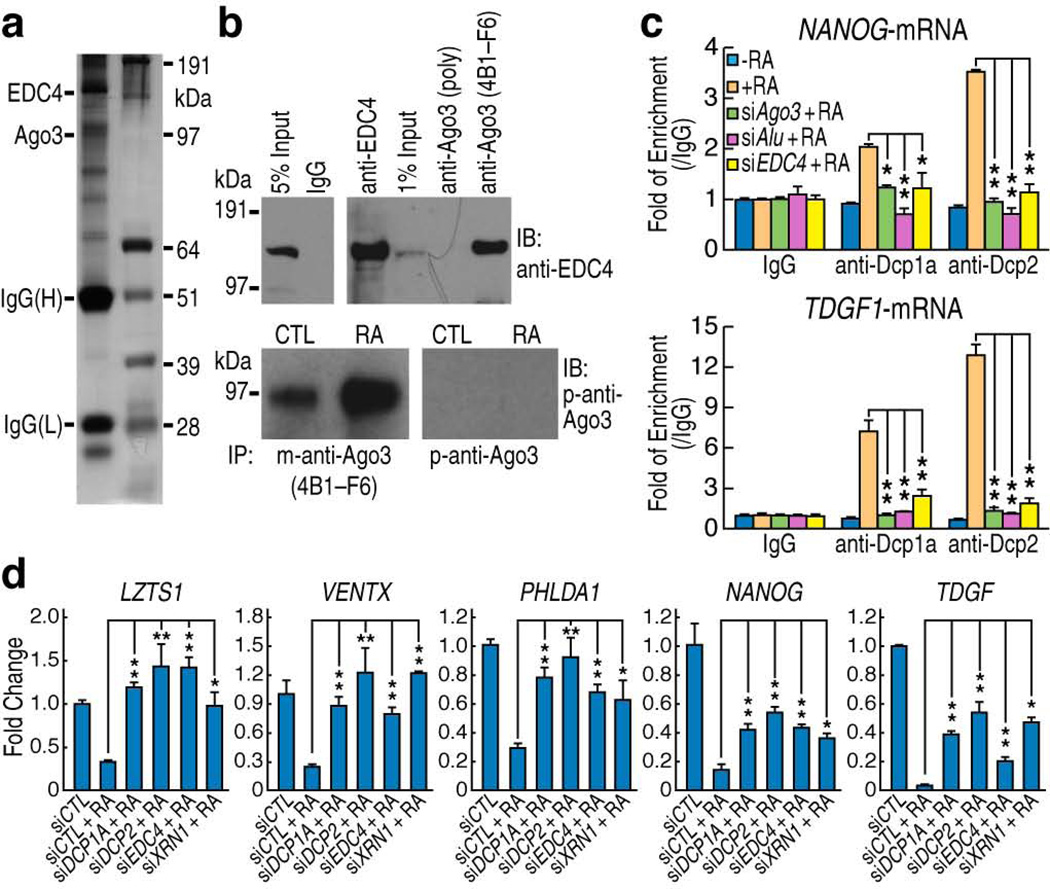
riRNA downregulated mRNAs through the decapping complex
To further clarify the mechanism of AGO3-dependent riRNA-mediated post-transcriptional regulation, we performed immunoprecipitation using a specific mono-clonal antibody against AGO3 (clone 4B1-F6) and analyzed the precipitates by mass spectrometry (Fig. 5a). Among the proteins found to associate with AGO3, RCD8 (also known as Enhancer of Decapping Complexes 4,EDC4) was of particular interest because it helps recruit DCP2 and stabilizes the DCP1a–DCP2 complex to remove the 5’-capped structure of target mRNAs39, a prerequisite for subsequent degradation by the exonuclease XRN140. The interaction between AGO3 and EDC4 was then confirmed by co-immunoprecipitation (Fig. 5b). The specificity of this protein interaction was corroborated by using another AGO3 antibody, characterized to be useful only for immunoblotting and immunofluorescence purposes, but not for immunoprecipitation. When used in the immunoprecipitation assay as a control, the protein interaction was not detected (Fig. 5b). Hence, these findings suggested a decapping mechanism for riRNA-mediated transcript down-regulation. To confirm a physical interaction or recruitment of the decapping complex to the target mRNAs, we performed RNA immunoprecipitation using specific antibodies against DCP1a and DCP2. RT-qPCR showed that these decapping components were indeed recruited to the target mRNAs in an atRA-, AGO3-, DR2 Alu-, and EDC4-dependent manner (Fig. 5c). We therefore tested whether knockdown of the decapping complexes and the exonuclease, XRN1, would interfere with the riRNA-mediated post-transcriptional regulation. As expected, transfection of the specific siRNAs against these components significantly restored the atRA-reduced transcript levels of the five genes tested (Fig. 5d). These data indicate that the AGO3-associated decapping complexes, at least in part, account for the riRNA-mediated target mRNA degradation.
Figure 5.
Role of the AGO3-associated decapping complex in post-transcriptional regulation. (a) Silver-staining of anti-AGO3 immunoprecipitates in Ntera2 cells. The same precipitates were subsequently analyzed by mass spectrometry. (b) Co-immunoprecipitation with the AGO3 antibody (4B1-F6) confirmed the interaction between AGO3 and EDC-4 (right upper panel). 4B1-F6 proved to be applicable in the immunoprecipitation assays while another AGO3 polyclonal IgG (p-anti-Ago3) manufactured for immunoblotting purposes effectively served only as a negative control (right lower panel). (c) RNA immunoprecipitation revealed RA-dependent and Alu-dependent recruitment of DCP1a and DCP2 proteins to the target mRNAs. (d) RT-qPCR showed that Ntera2siRNAs knock-down of DCP1a, DCP2, EDC4 and XRN1 restored the transcript levels of indicted genes in Ntera2 cells. (mean±SEM; *p<0.05, **p<0.01)
DR2 Alu RNAs-riRNAs regulate progenitor cell proliferation
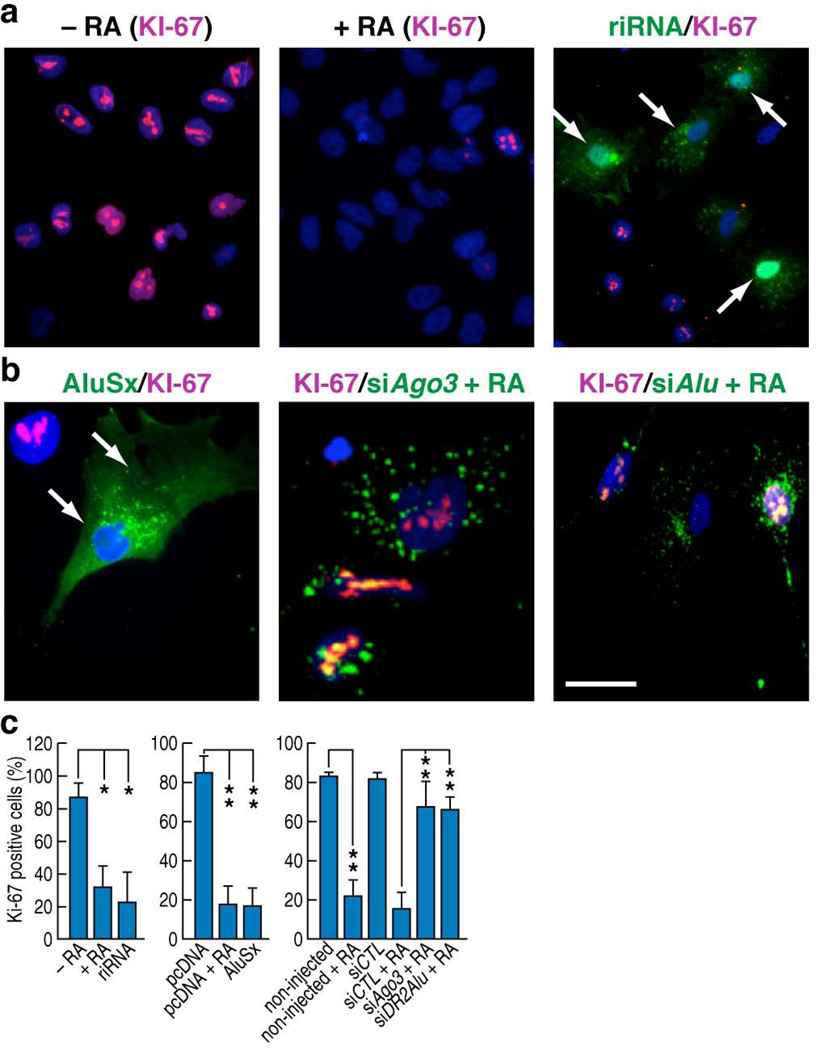
In accordance with previous reports41,42, we noticed proliferation inhibition in atRA-treated Ntera2 cells, and in full length DR2 Alu-transfected cells. Hence, we proceeded to evaluate the effects of riRNAs on Ntera2 proliferation using single cell microinjection technique, finding that the injected oligonucleotides caused a similar reduction in KI-67 staining as the atRA treatment (Fig. 6a). Cell proliferation arrest was also observed when AluSx expression vectors were injected into Ntera2 cells (Fig. 6b). Intriguingly, when cells were first microinjected with siRNAs against AGO3 or DR2 Alu, they sustained the proliferative capacity even in the presence of atRA (Fig. 6b,c), suggesting that atRA-induced DR2 Alu–riRNAs are an intrinsic component of the machinery regulating proliferation in stem cells.
Figure 6.
DR2 Alu transcripts and riRNAs regulate stem cell proliferation. (a,b) Immunostaining showed that RA treatment suppressed cell proliferation as marked by KI-67. Microinjection of synthesized riRNAs or the full-length AluSx overexpression construct similarly suppressed proliferation of Ntera2 cells. However, microinjection of AGO3 or DR2 Alu siRNAs prevented the RA-mediated proliferation arrest. Scale bar: 20 µm for the middle image in panel b; 60 µm for the rest of a, b panels. (c) Bar graphs showed the counting results from three independent injection experiments. (mean±SEM; *p<0.05, **p<0.01)
Discussion
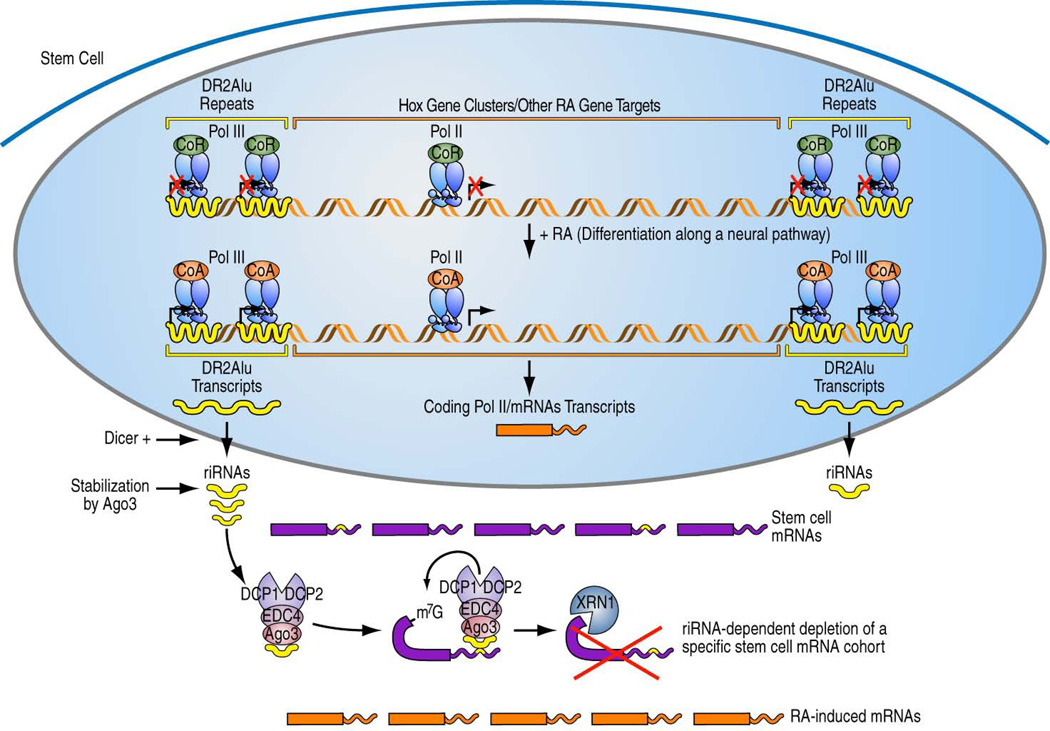
Together these data have begun to uncover an unexpected aspect of ligand-induced stem cell developmental regulatory program, whereby liganded RAR not only induces its long-recognized Pol II transcriptional program, encoding functionally-important proteins critical for differentiation and exit from the stem cell state, but also subsequently activates Pol III-dependent transcription of a larger number of DR2-containing ancient Alu repeats that flank these coding transcription units. These transcripts are processed in an as yet incompletely delineated pathway into small riRNAs in the cytoplasm. While a recent paper reported potential generation of miRNAs from downstream of some Alu elements43, our findings reveal that some novel species of small RNAs can be generated from within the Alu repeat. The stable accumulation of DR2 Alu transcripts and their processed small RNA derivatives requires the presence of AGO3, and they appear to cause the degradation of a subset of Pol II-transcribed coding transcripts, including Nanog and Tdgf1, by targeting the complementary sequences in their 3’UTRs, causing AGO3-dependent recruitment of EDC4, an important component of the decapping machinery. The EDC4-dependent assembly of the decapping complex and XRN1-dependent digestion of the target mRNAs explain, at least in part, the RA-induced stem cell exiting from the pluripotency stage (Fig. 7).
Figure 7.
Working model. Retinoic acid induced Pol II and Pol III transcription programs in stem cells, resulting in generation of DR2 Alu-derived riRNAs that act on a subset of mRNAs containing complementary sequences and exert developmental effects. Dicer+ indicates the actions of Dicer and other putative additional nucleases.
These findings reveal a previously unsuspected aspect of retinoic acid-mediated differentiation. In addition, we have noticed that by depletion of AGO3 or DR2 Alu RNAs, the ability to observe atRA-induced loss of stem cell proliferation is significantly impaired. Our study has, for the first time, identified an important function of the processed products of Alu RNAs in stem cell maintenance and proliferation. The murine genome also appears to harbor 7SL-derived retrotransposons, referred to as SINE B1 repeats7, and bioinformatic analysis suggest that these repeats are, similarly, capable of serving as precursors of riRNAs, indicating a utilization of these retrotransposon-dependent strategies in vertebrate evolution.
Because subclasses of Alu repeats also contain consensus sequences for other nuclear receptors and additional classes of regulatory DNA-binding transcription factors, including TR and ERα hormone receptors13, we are tempted to speculate that analogous co-regulated Pol II and Pol III DNA repeat transcriptional programs will be elucidated for many classes of transcription factors, providing a key aspect of regulatory pathways that perturbate diverse aspects of development, homeostasis, aging and disease.
Online Methods
Antibodies and siRNAs
Monoclonal antibodies: anti-AGO3 (4B1-F6; Active Motif), anti-DICER (13D6; Abcam) and anti-KI67 (BD Pharmingen). Rabbit polyclonal antibodies: anti-TF3C, anti-H3K36me3, anti-EDC4, anti-AGO3, anti-HP1 (Abcam); anti-Pol III, anti-RARβ, mouse and rabbit IgG (Santa Cruz); anti-AGO (Upstate); anti-DCP1a, anti-DCP2 (Sigma Aldrich); anti-NCoR and SMRT44. For immunoprecipitation assays, the antibodies were used at 2ug/ml. For immunoblotting, the antibodies were diluted 1:500. For immunostaining, the antibodies were diluted 1:200.
siRNAs: Pol III, AGO1,2,4, DROSHA1, DGCR8 from Qiagen; DCP1a, DCP2, EDC-4, XRN1 from Sigma; AGO3 from Qiagen and Dharmacon; DICER from Qiagen and Sigma; DR2 Alu1–4, PIWIL1–4 from Dharmacon. The customized siDR2 Alu1-4 sequences are summarized in Supplementary Table 1.
Cell Culture
Ntera2/D1 cells (ATCC) were kept in MEMα (Invitrogen) plus 10% FBS. For induction, the cells were treated with 1 µM atRA (Sigma) for two days. Transfection was performed at 50% confluence using Lipofectamine 2000 (Invitrogen). The RNA polymerase III inhibitor, tagetin (Epicentre), was added to the medium at 100 units/ml (3µM). For heat shock, the cells were treated with atRA for two days and exposed to 43°C for 90 minutes. The cells were recovered under normal culture conditions for another 90 minutes.
RT-qPCR
Total RNAs were extracted using Trizol (Invitrogen), cleaned up by RNeasy Mini columns (Qiagen) and treated with the On-column DNase kit (Qiagen). The cDNA was prepared using the First-strand Superscript III kit (Invitrogen). RT-qPCR was performed with the SYBR Green reagent (Stratagene) on the StepOne Plus machine (Applied Biosystems). GAPDH was used as the internal control. Expression data from at least three independent experiments were used to determine the significance by the Student’s t-test. The full primer sequences are listed in Supplementary Table 2.
mRNA Sequencing
10 µg total RNAs were processed with the mRNA Sequencing Sample Preparation Kit (Illumina). Sequencing was performed on the Illumina platform. SeqMap45 and rSeq46 were used to align the sequencing reads to the UCSC transcript database and to quantify their expression levels by RPKM values. The significance of differential expression of the genes was assessed by DEGseq47 in MeV48.
DR2-Alus were identified by scanning Alu sequences with a DR2 PWM (PAZAR database49), by using PATSER50 at distinct mapping scores. The potential induced DR2 Alu were identified based on the difference of normalized read counts (−RA vs. +RA). To quantify the DR2 Alu expression levels, the 72nt sequencing reads were aligned to the hg18 assembly using Bowtie51. Both uniquely aligned reads and those aligned to less than 10, 100, 1000 or 10000 locations were analyzed. For reads aligned to multiple regions, a single location was chosen based on a Gibbs sampling algorithm52, using 5 or 10 iterations. The sequencing reads count on DR2 Alu was computed using intersectBed tool in BedTools53, and were normalized to the sequencing depth. The list of potentially expressed DR2 Alu is presented at http://rosenfeldlab.ucsd.edu/qidong/.
Small RNA Sequencing
10 µg total RNAs were subjected to the small RNA preparation procedure (v1.5) (Illumina), and the libraries sequenced on the Illumina platform. For the analysis, the 3'-adaptor sequence was removed using FASTX, and the clipped reads were aligned to the DR2 Alu sequence database and DeepBase53 using BLAT and NOVOALIGN. The mRNAs with Alu elements in their 3'UTRs were identified by aligning Alu sequence fragments to transcripts databases, or by intersecting the genes in the Transpogene database54, with RA-regulated genes. We also downloaded the genome coordinates of 3'UTRs from hg18 and intersected them with the locations of Alu repeats.
Chromatin Immunoprecipitation (ChIP)
Ntera2 cells were treated with atRA for 2 hours, crosslinked with 1% formaldehyde and resuspended in the lysis buffer containing 1% SDS and Complete Proteinase Inhibitor Cocktail (Roche). After centrifugation, the supernatant was collected and diluted. After pre-clearing, the sample was incubated with 1µg of control IgG or antibodies overnight at 4°C. 50 µl Protein A beads were added to the mixture for another 6 hours. After de-crosslinking at 65°C overnight, the DNA was recovered using PB and Quickspin column (Qiagen). The primer sequences are summarized in Supplementary Table 3.
The ChIP-seq reads of H3K36me3 were aligned to hg18 by using Bowtie51. The sequencing reads count on UCSC genes was computed using intersectBed tool in BedTools55, and were normalized to the sequencing depth of each sample. (13 million for each sample by randomly extracting the corresponding number of reads)
GRO-Seq
The GRO-Seq was performed as described56. The sequencing reads were aligned to hg18 by using BFAST57. An equal number of reads (4 million) was randomly extracted from each sample and the sequencing reads were counted over the entire gene body on the sense strand in respect to the gene orientation by using BedTools55.
Northern Blots
Northern blot was performed as previously described58. RNA was resolved on 6% or 15% TBE-Urea gels (Invitrogen) and transferred onto HyBond N+® (Amersham) membranes. The membranes were incubated overnight with radio-labeled or biotin-conjugated DNA oligonucleotide probes in ExpressHyb™ (Clontech) or UltraHyb™ (Ambion), respectively at 42°C. After hybridization, membranes were washed with 2X SSC containing 0.05% SDS and with 0.1X SSC containing 0.1% SDS. The blot was detected with the Chemiluminescent Nucleic Acid Detection Module Kit (Fisher). The same blot was stripped using boiling 0.5% SDS and re-probed with the control U6 snRNA or miR-106a probes. RNA Century Plus Marker™ was from Ambion and SRA 10bp RNA ladder was from Illumina. The probe sequences are summarized in Supplementary Table 4.
RNA FISH
The RNA FISH was performed as described at http://www.epigenesys.eu/index.php/en/protocols/fluorescence-microscopy. Ntera2 cells were treated with atRA for two days, before subject to the RNA FISH procedure. Hybridization was performed overnight at 42°C with the probes generated by using the Alexa 546 ULYSIS Nucleic Acid Labeling Kit (Invitrogen). The template was a specific DR2 Alu repeat (AluSx; chr7: 27,093,354–27,093,650; hg18).
Molecular beacon and immunofluorescence
Ntera2 cells treated with atRA for two days were transfected with molecular beacons overnight using Lipofectamine 2000. The cells were then fixed, blocked and immunostained with the antibodies against Dicer and Ago3, followed by detection with Alexa-conjugated secondary IgGs (Invitrogen). The molecular beacon sequences are listed in Supplementary Table 1.
Ribonucleoprotein complexes immunoprecipitation (RIP)
RIP was performed as described at http://www.epigenome-noe.net/researchtools/protocol.php?protid=28. 2×108 Ntera2 cells were sequentially lysed in Buffer A (5mM PIPES[pH8.0], 85mM KCl, 0.5%NP-40, Complete Protease Inhibitor Cocktail, 50U/ml SUPERaseIn) and Buffer B (1%SDS, 10mM EDTA, 50Mm Tris-HCl[pH8.1], Complete Protease Inhibitor Cocktail, 50U/ml SUPERaseIn). The lysates were pooled and diluted 1:10 with IP Buffer (0.01%SDS, 1.1%TritonX-100, 1.2mM EDTA, 16.7mM Tris-HCl[pH8.1], 167 mM NaCl, Complete Protease Inhibitor Cocktail, 50U/ml SUPERaseIn). The diluted lysates were incubated with 40 µg anti-AGO3 (clone 4B1-F6), anti-DCP1a or anti-DCP2 IgGs for overnight, and precipitated with Protein G-Sepharose (Sigma-Aldrich). RNAs were extracted from both input (1%) and immunocomplexes using Trizol LS (Invitrogen). The RNAs were analyzed by Northern blot, RT-PCR and RT-qPCR. The primer and probe sequences are detailed in Supplementary Table 3 and 4.
Biotin-labeling of AGO3-immunoprecipitated RNAs
The AGO3-associated RNAs were labeled with the Psoralen-Biotin Kit (Ambion). To minimize multiple labeling, 0.2 µl labeling reagent was used per reaction and 365 nm UV irradiation lasted for only 5 minutes. The labeled RNAs were resolved in the 15% TBE-Urea gel and transferred to the nylon membrane. The Chemiluminescent Nucleic Acid Detection Module Kit was used to detect the labeled RNAs.
Single-cell Microinjection
We followed a previously described procedure59. Briefly, ~300 Ntera2 cells seeded on coverslips were transfected with the siRNA against Ago3. 48 hours post-transfection, the cells were micro-injected with the in-vitro transcribed DR2 Alu RNA (HoxA1-AluSq; chr7: 27,092,966–27,093,267; Ambion T7 MegaScript Kit) or the synthesized riRNA oligonucleotide (68nt.). Microinjection assays were carried out as previously described60. Each experiment was performed on four independent coverslips. The cells were then lyzed with Trizol at different time points. The injected RNAs were quantified by RT-qPCR.
Mass Spectrometry
1×108 Ntera2 cells were lysed in 50Mm Tris-HCl[pH8.0], 150mM NaCl, 1%NP-40. The lysates were immunoprecipitated with 10 µg anti-AGO3 (clone 4B1-F6) and Protein G-Sepharose (Sigma-Aldrich). The beads were washed five times with the lysis buffer containing 500 mM NaCl. The quality of the precipitates was checked by silver staining (Fermentas) and the complexes were analyzed by mass spectrometry using the MASCOT engine at the UCSD Biomolecular/Proteomics Mass Spectrometry Facility.
RNA Pull-down
The pull-down was performed as described at http://www.epigenome-noe.net/researchtools/protocol.php?protid=32). Briefly, 2×108 Ntera2 cells were lysed in 10mM Tris-HCl[pH7.4], 10mM NaCl, 3mM MgCl2, 0.1mM DTT, 0.5% NP-40, Complete Protease inhibitor, 20U/ml SUPERaseIn. The lysates were incubated with 500nM 5’Biotin-conjugated oligonucleotides for 3 hours. Coupled complexes were captured by Dynabeads™ M-280 Streptavidin magnetic beads for 1 hour. After washing with 10mM Tris-HCl[pH8.0], 1mM EDTA, 0.5M NaCl for five times, the samples were analyzed by Western blot.
Luciferase assays
Wild-type or deletion-mutant 3’UTRs of select transcripts were cloned into pMIR (Promega). Ntera2 cells seeded in 12-well plates were transfected with 0.2 µg plasmids (including 0.1 µg Renilla) using Lipofectamine 2000, or with 0.1 µg plasmids using Effectene (Qiagen) and simultaneously treated with atRA. 48 hours post-transfection, the cells were lysed in the passive lysis buffer (Promega) and the luciferase activity from at least three independent experiments was measured with the DualGlo kit (Promega) and analyzed by the Student's t-test.
Cell Proliferation and KI-67 staining
Ntera2 cells were microinjected with siRNAs (treated with 10 µM atRA concurrently), Alu overexpression vectors, or riRNA oligonucleotide for two days. Then this procedure was repeated once. The cells were immunostained with the KI-67 antibody.
Supplementary Material
Acknowledgements
We thank M. Mercola (Sanford Burnham Medical Research Institute, La Jolla, CA) and S. Ding (The Scripps Research Institute, La Jolla, CA) for providing H9 human embryonic stem cells; N. Kim (Seoul National University, Gwanak-gu, South Korea) for Dicer expression constructs; J. Nand for assistance with the RNA-Seq data analysis; C. Nelson for cell culture assistance; J. Hightower for assistance with figure and manuscript preparation; M. Ghassemian from the UCSD Biomolecular/Proteomics Mass Spectrometry Facility FOR assistance in mass-spectrometry. We also want to acknowledge the UCSD Cancer Center Specialized Support Grant P30 CA23100 for confocal microscopy. Q.H. is a Cancer Research Institute Postdoctoral Fellow. M.G.R. is an Investigator with the Howard Hughes Medical Institute. This work was supported by grants from NIH and NCI to M.G.R. and by awards from DOD and PCF to M.G.R.
Footnotes
Author Contributions
Q.D.H. performed the majority of experiments. B.T. contributed the bioinformatics analysis, making the initial finding that the 3’UTRs of specific stem cell mRNAs harbored sequences complementary to DR2 Alu-derived riRNAs. M.T. contributed part of the Northern blot data. W.L contributed GRO-seq. J.Z. provided sequencing and microinjection assistance. K.A.O provided sequencing and vector construction assistance. D.W.R. contributed microinjection. C.K.G helped design experiments and critically read the manuscript. M.G.R. designed the experiments and evaluated the data. Q.D.H. and M.G.R wrote the manuscript.
References
- 1.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 3.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 4.Hörlein AJ, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 6.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 9.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;35:501–538. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 11.Häsler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu WM, Maraia RJ, Rubin CM, Schmid CW. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umylny B, Presting G, Ward WS. Evidence of Alu and B1 expression in dbEST. Arch. Androl. 2007;53:207–218. doi: 10.1080/01485010701426422. [DOI] [PubMed] [Google Scholar]
- 16.Pleasure SJ, Lee VM. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J. Neurosci. Res. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- 17.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 18.Steinberg TH, Mathews DE, Durbin RD, Burgess RR. Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 1990;265:499–505. [PubMed] [Google Scholar]
- 19.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moqtaderi Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 23.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell. Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell. Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, et al. Dicer recognizes the 59 end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarallo V, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 30.Azuma-Mukai A, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heyse G, Jönsson F, Chang WJ, Lipps HJ. RNA-dependent control of gene amplification. Proc. Natl. Acad. Sci. USA. 2010;107:22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 33.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya B, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 35.Gao H, et al. VentX, a novel lymphoid-enhancing factor/T-cell factor-associated transcription repressor, is a putative tumor suppressor. Cancer Res. 2010;70:202–211. doi: 10.1158/0008-5472.CAN-09-2668. [DOI] [PubMed] [Google Scholar]
- 36.Vecchione A, et al. Fez1/Lzts1 absence impairs Cdk1/Cdc25C interaction during mitosis and predisposes mice to cancer development. Cancer Cell. 2007;11:275–289. doi: 10.1016/j.ccr.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neef R, Kuske MA, Pröls E, Johnson JP. Identification of the human PHLDA1/TDAG51 gene: down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 2002;62:5920–5929. [PubMed] [Google Scholar]
- 38.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tritschler F, et al. DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc Natl Acad Sci USA. 2009;106:21591–21596. doi: 10.1073/pnas.0909871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde EA. crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YH, et al. Growth inhibition of a human myeloma cell line by all-trans retinoic acid is not mediated through downregulation of interleukin-6 receptors but through upregulation of p21(WAF1) Blood. 1999;94:251–259. [PubMed] [Google Scholar]
- 42.Yu Z, et al. p21 is required for atRA-mediated growth inhibition of MEPM cells, which involves RAR. J. Cell. Biochem. 2008;104:2185–2192. doi: 10.1002/jcb.21773. [DOI] [PubMed] [Google Scholar]
- 43.Gu TJ, Yi X, Zhao XW, Zhao Y, Yin JQ. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10:563. doi: 10.1186/1471-2164-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 44.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 45.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 48.Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 49.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8(10):R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999 Jul-Aug;15(7–8):563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Huda A, Lunyak VV, Jordan IK. A Gibbs sampling strategy applied to the mapping of ambiguous short-sequence tags. Bioinformatics. 2010;26:2501–2508. doi: 10.1093/bioinformatics/btq460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy A, Sela N, Ast G. TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008;36:D47–D52. doi: 10.1093/nar/gkm949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heyse G, Jönsson F, Chang WJ, Lipps HJ. RNA-dependent control of gene amplification. Proc. Natl. Acad. Sci. USA. 2010;107:22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.