Abstract
CBF/DREB1 (C-repeat-binding factor/dehydration responsive element-binding factor 1) genes encode a small family of transcriptional activators that have been described as playing an important role in freezing tolerance and cold acclimation in Arabidopsis. To specify this role, we used a reverse genetic approach and identified a mutant, cbf2, in which the CBF2/DREB1C gene was disrupted. Here, we show that cbf2 plants have higher capacity to tolerate freezing than WT ones before and after cold acclimation and are more tolerant to dehydration and salt stress. All these phenotypes correlate with a stronger and more sustained expression of CBF/DREB1-regulated genes, which results from an increased expression of CBF1/DREB1B and CBF3/DREB1A in the mutant. In addition, we show that the expression of CBF1/DREB1B and CBF3/DREB1A in response to low temperature precedes that of CBF2/DREB1C. These results indicate that CBF2/DREB1C negatively regulates CBF1/DREB1B and CBF3/DREB1A, ensuring that their expression is transient and tightly controlled, which, in turn, guarantees the proper induction of downstream genes and the accurate development of Arabidopsis tolerance to freezing and related stresses.
Freezing temperatures greatly limit the geographical distribution and growing season of plants and cause negative effects on crop quality and productivity. As a consequence, appreciable effort has been conducted to determine the adaptive mechanisms plants have evolved to survive this adverse environmental condition. Many plants, including Arabidopsis, increase their freezing tolerance in response to low, nonfreezing temperatures. This process, called cold acclimation (1), is complex and involves a number of biochemical and physiological changes, ranging from alterations in lipid composition to accumulation of sugars (2). Different studies have suggested that low-temperature-regulated gene expression is critical in plants for cold acclimation (2). Low-temperature-responsive genes encode a diverse number of proteins, including enzymes involved in respiration and metabolism of carbohydrates, lipids, phenylpropanoids and antioxidants, molecular chaperones, antifreeze proteins, among others, with a believed function in freezing tolerance (2).
During the past few years, substantial progress has been made toward understanding how low temperatures regulate gene expression. In particular, a family of transcription factors in Arabidopsis known either as C-repeat-binding factor (CBF)1, CBF2, and CBF3 (3, 4) or dehydration-responsive element-binding factor (DREB)1B, DREB1C, and DREB1A (5), respectively, has been identified. These factors belong to the Apetala 2/ethylene-responsive element-binding protein (AP2/EREBP) family of DNA-binding proteins (6) and bind to the cold- and dehydration-responsive DNA regulatory element (DRE) (7), also termed C-repeat (CRT) (8). CRT/DRE elements contain the conserved CCGAC core sequence, which is sufficient to induce gene transcription under cold stress (7, 8) and is present in the promoters of many cold-inducible genes (2). Interestingly, the CBF/DREB1 genes do not contain the CCGAC sequence in their promoters but are also induced by low temperature. This induction is transient and precedes that of cold-inducible genes with the CRT/DRE cis-element (4, 5, 9). Ectopic overexpression of CBF1/DREB1B and CBF3/DREB1A in Arabidopsis results in the constitutive expression of downstream cold-inducible genes, even at warm temperatures and in increased freezing tolerance (5, 10-12), suggesting that CBF/DREB1 genes may play an important role in cold acclimation. In addition, overexpression of CBF3/DREB1A also enhances drought and salt tolerance (5, 11). To our knowledge, overexpression of CBF2/DREB1C has not been reported.
Unfortunately, mutant plants in the CBF/DREB1 genes have not been so far identified, which has prevented the analysis of their actual contribution to the cold-acclimation response. In fact, despite the extensive investigations carried out, our understanding of CBF/DREB1 gene function(s) remains elusive, and a clear role of their requirement for stress tolerance has not still emerged. For example, whether all three CBF/DREB1 genes are required for freezing tolerance and cold acclimation and how the expression of CBF/DREB1 genes is regulated in response to low temperatures are essential questions that are still unanswered. To dissect the precise role of these genes and shed some light on these issues, we screened a transferred DNA (T-DNA) mutagenized population of Arabidopsis for plants containing T-DNA insertions in the CBF/DREB1 genes. Here, we report on the isolation and characterization for the first time of a mutant plant in which a CBF/DREB1 gene, namely CBF2/DREB1C, is disrupted. The results obtained indicate that CBF2/DREB1C plays a critical role in the development of Arabidopsis tolerance to freezing and other related stresses by controlling the precise expression of CBF1/DREB1B and CBF3/DREB1A and, hence, that of the downstream genes. On the basis of these results, a model for the function of CBF2/DREB1C in cold acclimation and the regulation of CBF/DREB1 gene expression in response to low temperature is proposed.
Materials and Methods
Plant Materials, Growth Conditions, and Treatments. Seeds from Arabidopsis thaliana (L.) Heynh, ecotype Columbia, were purchased from Lehle Seeds (Round Rock, TX). Plants were grown in pots containing a mixture of organic substrate and vermiculite (3:1, vol/vol) and irrigated with mineral nutrient solution (13) once a week. Plants for dehydration and salt tolerance assays were grown under sterile conditions in Petri dishes containing GM medium (Murashige and Skoog medium (14) supplemented with 1% sucrose) solidified with 0.8% (wt/vol) agar. In all cases, plants were developed at 20°C under a long-day photoperiod (16 h of cool-white fluorescent light, photon flux of 70 μMm-2·s-1). All treatments were performed on 3-week-old plants.
Low-temperature treatments were performed by transferring plants to a growth chamber set to 4°C for different periods of time under the light and photoperiodic conditions described above. Freezing assays were carried out in a temperature programmable freezer. Nonacclimated or cold-acclimated (7 days at 4°C) plants were exposed to 4°C for 30 min in darkness and subsequently temperature was lowered by 2°C per h. The final desired freezing temperature was maintained for 6 h, and then the temperature was increased again to 4°C at the same rate. After thawing at 4°C for 4 h in the dark, plants were returned to their original growth conditions (see above). Tolerance to freezing was determined as the capacity of plants to resume growth after 7 days of recovery under control conditions. Dehydration was induced by removing plants from the medium, placing them on a dry filter paper, and allowing them to develop for 2 days without watering. The rate of dehydration was estimated as the percentage of initial fresh weight (FW) that remains after treatment. Salt stress was accomplished by transferring plants to new Petri dishes containing the agar medium plus 100 mM NaCl. Tolerance was estimated by determining the root elongation and the FW of the plants after 7 days of treatment. After low-temperature treatment, plants used for RNA-blot hybridizations were immediately frozen in liquid N2 and stored at -80°C until their use.
Identification of the cbf2 Mutant. The cbf2 mutant was identified by PCR screening of ≈30,000 Arabidopsis T-DNA insertion lines (J.M.A. and J.R.E., unpublished data), by using specific oligonucleotides for the CBF2/DREB1C gene (5′-TCCGGTTTCCTCAGGCGGTGATTACA-3′ and 5′-TAAGGACACGTCATCATCTCCCTGAC-3′) and the T-DNA (5′-GCTCATGA-TCAGATTGTCGTTTCCCGCCTT-3′ and 5′-GGCAAT-CAGCTGTTGCCCGTCTCACTGGTG-3′). DNA sequencing showed that the T-DNA insertion was 179 bp upstream of the start codon of CBF2/DREB1C.
Cosegregation Analysis. Cosegregation of the T-DNA insertion with the mutant (stress-tolerant) phenotypes was determined by crossing homozygous cbf2 plants to WT Arabidopsis of the same ecotype (Columbia). For cosegregation analysis, the genotype of segregating F2 plants was analyzed by PCR with CBF2/DREB1C and T-DNA-specific primers. More than 95% of homozygous cbf2 plants showed tolerant phenotypes, and 96% of plants that were homozygous WT displayed WT phenotypes. Several homozygous WT and mutant F2 plants were self-crossed and their descendents analyzed. Ninety percent of the progeny from the cbf2 homozygous parents were phenotypically mutant, and 91% of the progeny from WT parents were phenotypically WT, demonstrating that the mutant phenotypes are genetically linked to the cbf2 locus.
Genetic Complementation Analysis. A 1,810-bp DNA fragment containing the CBF2/DREB1C gene and 1,138 bp of its native promoter was obtained by PCR. The fragment was then cloned upstream of the nopaline synthase terminator in pCAMBIA1381 binary vector. The resulting plasmid was introduced into Agrobacterium C58C1 and used for transformation of cbf2 mutant. T1 plants were selected by using hygromycin.
Molecular Biology Methods. Total RNA was isolated as described (15). Restriction digestions, cloning, and RNA-blot hybridizations were performed by following standard protocols (16). Specific probes for the CBF/DREB1 genes have been described (9). The probe for LTI78 was a 1.0-kb genomic fragment produced by using the primers 5′-CGGGATTTGACGGAGAACCA-3′ and 5′-ACCATAATACATCAAAGACG-3′. The probe for KIN-1 was a 700-bp genomic fragment obtained by using the primers 5′-GGCACCACACTCCCTTTAGC-3′ and 5′-GAATATAAGTTTGGCTCGTC-3′. The COR47 and COR15A probes were 400-bp and 1.0-kb fragments, respectively, obtained from the corresponding cDNAs (17, 18). The RCI1A probe consisted of a 200-bp DNA fragment corresponding to the 3′ untranslated region (19). The RCI2A probe consisted of a 300-bp DNA fragment from the 3′ noncoding region (20). The probe for DREB2A was a 300-bp genomic fragment produced by using the primers: 5′-GATGTGGATCAGAGTCACTT-3′ and 5′-CAACAGTCGTTGTGGGATTAAGG-3′. The L18 probe was a 791-bp cDNA fragment (21). Equal RNA loading in the experiments was monitored by rRNA staining. RNA samples for each experiment were analyzed in at least two independent blots, and each experiment was repeated at least twice.
Results
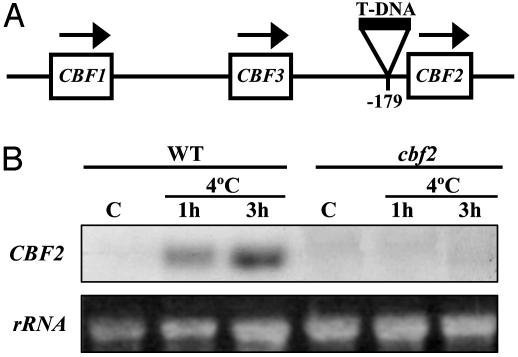
Isolation and Characterization of the cbf2 Mutant. A T-DNA mutagenized population of Arabidopsis was screened by PCR for plants containing an insertion in the CBF/DREB1 genes. From a population of ≈30,000 individual T-DNA insertion lines, we identified a single mutant plant (cbf2) bearing a disruption mutation in CBF2/DREB1C. Sequence analysis revealed that the insertion was within the putative TATA box (9), 179 bases upstream of the start codon (Fig. 1A). The progeny of heterozygous plants showed a segregation of the cbf2 phenotypes (described below) of ≈1:3 between mutant and WT (data not shown), indicating that cbf2 is a recessive mutation in a single nuclear gene. RNA-blot analysis confirmed that the CBF2/DREB1C mRNA did not accumulate in the mutant after cold stress treatment (Fig. 1B), suggesting that the cbf2 is a null or severely hypomorphic allele. Compared to the WT under control growth conditions, cbf2 mutant plants did not exhibit any obvious morphological or developmental abnormality.
Fig. 1.
Structure of CBF/DREB1 cluster and T-DNA insertion in the CBF2/DREB1C gene. (A) Schematic representation of the CBF/DREB1 cluster with the T-DNA insertion. The site of insertion in the cbf2 mutant, 179 bp upstream of the CBF2/DREB1C start codon, is represented. Arrows indicate the direction of transcription in the CBF/DREB1 genes. The CBF locus is not drawn to scale. (B) RNA-blot hybridization by using a specific probe for CBF2/DREB1C and total RNA (20 μg) prepared from 3-week-old rosette leaves of WT and cbf2 mutant plants grown under control conditions (C) or exposed to 4°C for 1 h and 3 h. Equal amounts of RNA were present in each sample as confirmed by ethidium bromide staining of rRNAs.
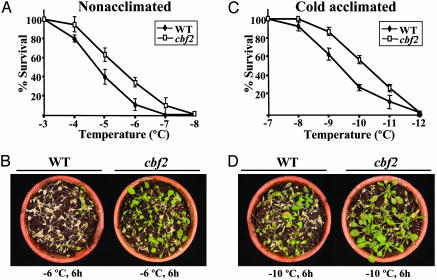
The cbf2 Mutation Enhances Constitutive Freezing Tolerance and Cold Acclimation. To understand the precise role of CBF2/DREB1C in cold acclimation, we examined the freezing tolerance of the cbf2 mutant. Plants, with or without cold acclimation for 7 days at 4°C, were exposed for 6 h to different freezing temperatures. Plant survival was scored after 7 days of recovery under control growth conditions. Unexpectedly, both nonacclimated and cold-acclimated mutant plants were significantly more tolerant to freezing than the corresponding WT plants (Fig. 2). The enhancement of freezing tolerance in cbf2 plants compared with WT was very similar before and after cold acclimation. In fact, a comparison of LT50 (temperature that causes 50% lethality) values between nonacclimated mutant and WT plants showed that cbf2 plants had LT50 values of -5.7°C, whereas WT plants had a LT50 of -4.8°C (Fig. 2A). In the case of cold-acclimated plants, the LT50 values of cbf2 and WT plants were -10.4°C and -9.4°C, respectively (Fig. 2C). The increased freezing tolerance manifested by the mutant with respect to the WT was very apparent in both nonacclimated (Fig. 2B) and cold-acclimated (Fig. 2D) plants. Therefore, the cbf2 mutation does not impair cold acclimation. Rather, it actually enhances both the constitutive freezing tolerance and the freezing tolerance of Arabidopsis after cold acclimation. These results indicate that CBF2/DREB1C acts as a negative regulator of freezing tolerance in Arabidopsis.
Fig. 2.
Freezing tolerance of cbf2 mutant plants. Three-week-old WT and cbf2 plants grown under long-day photoperiods at 20°C were exposed to different freezing temperatures for 6 h. Freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 days of recovery under unstressed conditions. (A) Tolerance of nonacclimated plants. (B) Representative nonacclimated WT and cbf2 plants 7 days after being exposed to -6°C for 6 h. (C) Tolerance of cold-acclimated (7 days at 4°C) plants. (D) Representative cold-acclimated WT and cbf2 plants 7 days after being exposed to -10°C for 6 h. In A and C, data are expressed as means of three independent experiments with 50 plants each. Bars indicate SE.
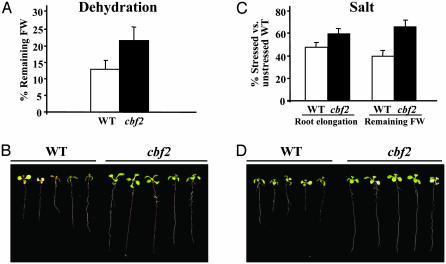
cbf2 Mutant Plants Show Increased Tolerance to Dehydration and Salt Stress. To characterize the capacity of the cbf2 mutant to respond to other types of stresses, we examined its tolerance to dehydration and high salt. Dehydration was induced by maintaining plants on a dry filter paper for 2 days without watering. The rate of dehydration was determined as the percentage of initial FW remaining after treatment. Mutant and WT plants did not show significant differences in their initial FW values (data not shown). After dehydration, cbf2 plants maintained an average of 22% of their initial FW, whereas WT plants maintained only 13.5% (Fig. 3A). No differences in stomatal closure were found in any case (data not shown). Correspondingly, cbf2 plants had a much less severe dehydration phenotype than that observed in WT plants (Fig. 3B). This indicates that the mutation at the CBF2/DREB1C gene significantly increases the tolerance of Arabidopsis to dehydration. The tolerance to salt stress was estimated by determining the root elongation in cbf2 and WT plants after growing for 7 days in a medium containing 100 mM NaCl. WT and cbf2 mutant plants showed identical root elongation under control conditions. The FW of the plants after the stress treatment also provided to be an estimate of their salt tolerance. Mutant plants subjected to salt stress displayed 60% and 66% of the root elongation and FW shown by unstressed WT plants. In turn, WT plants only showed 48% and 40% of the root elongation and FW that they displayed under control conditions (Fig. 3C). These significant differences among cbf2 and WT plants were clearly apparent at the phenotypic level (Fig. 3D) and demonstrate that the cbf2 mutant also has an increased tolerance to salt stress. Taken together, these data reveal that, in addition to freezing tolerance, CBF2/DREB1C acts as a negative regulator of dehydration and salt tolerance in Arabidopsis. Importantly, the freezing, dehydration, and salt tolerance phenotypes shown by cbf2 were found to be genetically linked to the T-DNA insertion, as demonstrated by cosegregation analysis (see Materials and Methods).
Fig. 3.
Tolerance to dehydration and salt stress of cbf2 mutant plants. (A) Dehydration tolerance of 3-week-old WT and cbf2 plants. Tolerance was estimated as the percentage of initial FW that remains after transferring plants to a dry filter paper and allowing them to develop for 2 days without watering. (B) Representative WT and cbf2 plants after dehydration treatment. (C) Salt tolerance of 3-week-old WT and cbf2 plants. Tolerance was estimated by determining the root elongation and FW of plants transferred to a medium containing 100 mM NaCl for 7 days. These values are represented as a percentage of root elongation and FW of WT unstressed plants. (D) Representative WT and cbf2 plants after salt treatment. In A and C, data are expressed as means of three independent experiments with 20 plants each. Bars indicate SE. Values obtained from WT and cbf2 were in all cases significantly different (P < 0.05) as determined by Student's t test.
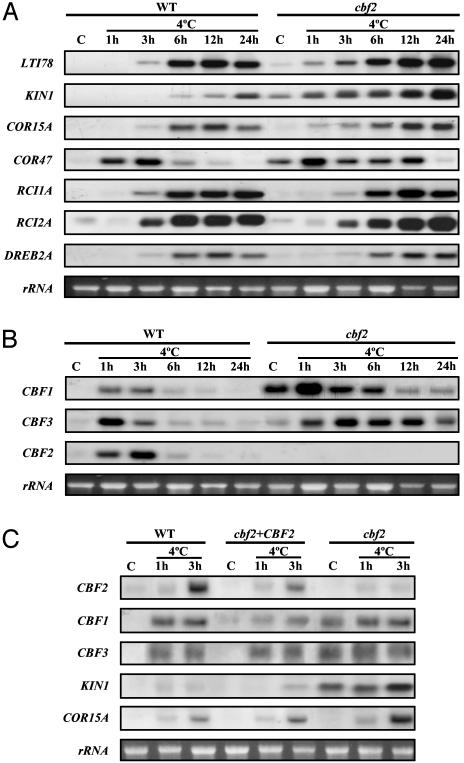
The cbf2 Mutation Enhances Transcription of CBF1/DREB1B, CBF3/DREB1A, and Downstream Genes. In an attempt to reconcile the unexpected results described above with earlier predictions on the role of CBF2/DREB1C, we examined the impact of the cbf2 mutation on the transcript levels of several cold-inducible genes. The LTI78, KIN1, COR47, and COR15A genes have been described as CBF/DREB1 target genes and to be part of the CBF/DREB1 regulon (22, 23). Interestingly, mRNAs for these cold-inducible genes were found to be constitutively expressed at low levels in the cbf2 mutant under unstressed control conditions (Fig. 4A). After cold treatment, the levels of these mRNAs were further induced and more sustained than in the WT (Fig. 4A). Moreover, the effects of the cbf2 mutation on the expression of CBF/DREB1 target genes seem to be specific because the expression of RCI1A, RCI2A, and DREB2A cold-inducible genes (19, 20, 24), which do not contain CRT/DRE elements in their promoters (ref. 25; GenBank accession nos. AL391145 and AB010692), was unaffected in control and cold-treated cbf2 plants (Fig. 4A). We conclude from these results that CBF2/DREB1C negatively regulates the transcription of downstream CBF/DREB1 target genes and is essential for the accurate expression of these genes in response to low temperature.
Fig. 4.
Transcript levels of cold-induced genes in the cbf2 mutant and in the complemented cbf2 mutant. RNA-blot hybridizations were performed with total RNA (20 μg) isolated from 3-week-old rosette leaves of WT, cbf2, and complemented cbf2 (cbf2+CBF2) plants grown under control conditions (C) or exposed to 4°C for the indicated times. In all cases, gene-specific probes were used for the hybridizations. (A) Transcript levels of LTI78, KIN1, COR15A, COR47, RCI1A, RCI2A, and DREB2A cold-inducible genes in WT and cbf2. LTI78, KIN1, COR15A, and COR47 contain the CRT/DRE element in their promoters, whereas RCI1A, RCI2A, and DREB2A do not. (B) Transcript levels of CBF/DREB1 genes in WT and cbf2. (C) Transcript levels of CBF/DREB1, KIN1, and COR15A genes in WT, cbf2, and cbf2+CBF2. Equal amounts of RNA were present in each sample as confirmed by ethidium bromide staining of rRNAs.
Overexpression of CBF1/DREB1B and CBF3/DREB1A in transgenic Arabidopsis has been reported to induce constitutive expression of the CRT/DRE-containing genes, which in turn would promote freezing, dehydration and salt tolerance (5, 10-12). Furthermore, a positive correlation was found between the expression levels of CBF1/DREB1B and CBF3/DREB1A, the level of expression of the CBF/DREB1-target genes, and the level of tolerance to stress conditions (5, 10-12). One hypothesis to explain the intriguing physiological and molecular phenotypes shown by the cbf2 mutant is that the level of expression of CBF1/DREB1B and/or CBF3/DREB1A under control conditions and/or in response to low temperature may be higher than in the WT. RNA-blot analysis revealed that under control conditions, the transcript levels of both CBF1/DREB1B and CBF3/DREB1A were higher in cbf2 plants than in the WT. After cold treatment, the level of expression of CBF1/DREB1B and CBF3/DREB1A was more sustained in the cbf2 mutant compared to the WT (Fig. 4B). These data indicate that CBF2/DREB1C acts to negatively regulate the expression of CBF1/DREB1B and CBF3/DREB1A. Nevertheless, because the three CBF/DREB1 genes are arranged in tandem, the possibility exists that the T-DNA insertion in the cbf2 mutant could induce directly the expression of CBF1/DREB1B and CBF3/DREB1A. Complementation of the mutant with the CBF2/DREB1C gene driven by its native promoter reestablished the WT expression patterns of CBF1/DREB1B and CBF3/DREB1A and those of their target genes (Fig. 4C). This complementation confirms that CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A. As expected from these results, the expression of CBF2/DREB1C in cbf2 mutant also reestablished the WT phenotypes to freezing, dehydration, and salt tolerance (data not shown).
Contrary to what was described in CBF3/DREB1A-overexpressing plants (10-12), cbf2 mutants did not display a growth-retardation phenotype. The degree to which overexpressing plants were stunted in growth was shown to positively correlate with the level of CBF3/DREB1A expression and, therefore, with the level of expression of CBF/DREB1 target genes (11, 12). It seems likely that the lack of effect on growth and development observed in cbf2 plants is due to their low levels of CBF3/DREB1A expression compared with the overexpressing plants.
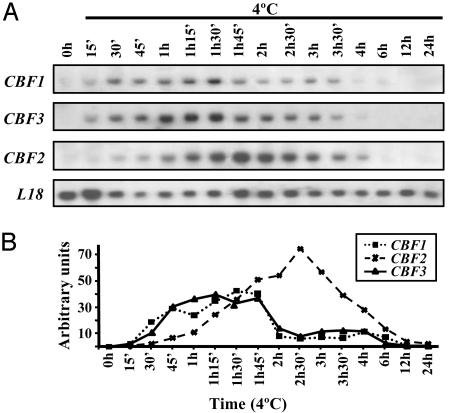
The Expression of CBF1/DREB1B and CBF3/DREB1A in Response to Cold Precedes That of CBF2/DREB1C. The three CBF/DREB1 genes are generally assumed to be induced at the same time in response to low temperature. Nevertheless, because the induction is transient and the results described above demonstrate that CBF2/DREB1C represses CBF1/DREB1B and CBF3/DREB1A, this might not be the case. To test this possibility, we performed a detailed analysis of the induction of each individual CBF/DREB1 gene by exposing Arabidopsis plants to 4°C for various periods of time. RNA-blot hybridizations revealed that CBF1/DREB1B and CBF3/DREB1A transcripts have very similar patterns of accumulation. The transcripts started to accumulate after 15 min of cold treatment, and they increased over the next 90 min. Subsequently, transcript levels decreased rapidly (Fig. 5). CBF2/DREB1C transcripts accumulated at a slower rate, reaching maximal levels after 2.5 h of cold exposure and then gradually declining (Fig. 5). From these results, we conclude that the three CBF/DREB1 genes are not induced at the same time in response to low temperature, CBF1/DREB1B and CBF3/DREB1A preceding CBF2/DREB1C.
Fig. 5.
Transcript levels of CBF/DREB1 genes in response to low temperature. (A) RNA-blot hybridizations were performed with total RNA (20 μg) isolated from 3-week-old rosette leaves of Columbia plants grown under control conditions (C) or exposed to 4°C for the indicated times. Specific probes were used for the hybridizations. Hybridization with a probe representing L18, a ribosomal gene, was used to normalize signals from the CBF/DREB1 genes. (B) Quantitative representation of the relative expression of CBF/DREB1 genes (CBF/L18) in response to low temperature.
Discussion
Low temperature triggers the expression of the CBF/DREB1 family of transcription factors (4, 5, 9). It has been suggested that these factors have an important role in cold acclimation from the observation that ectopic expression of CBF1/DREB1B and CBF3/DREB1A induces the transcription of genes containing the CRT/DRE promoter element and enhances freezing tolerance of nonacclimated Arabidopsis transgenic plants (5, 10-12). Nevertheless, the individual contribution of each CBF/DREB1 gene to that adaptive response has not been examined by loss-of-function analysis and important points on their function(s) remain uncertain. In this study, we have identified and characterized cbf2, a mutant with a T-DNA disrupting the CBF2/DREB1C gene. Our results show that cbf2 mutants have higher capacity to tolerate freezing than WT plants before and after cold acclimation and are more tolerant to dehydration and salt stress. Interestingly, these unexpected phenotypes correlate with an increased expression of CBF1/DREB1B, CBF3/DREB1A, and, hence, downstream-regulated genes. The CBF/DREB1 regulon has been described as including genes that act in concert to improve freezing tolerance (10, 26). Furthermore, high expression levels of CBF/DREB1 target genes have been shown to result in improved tolerance to dehydration and salt stress (5, 11). Therefore, the increased tolerance to freezing, dehydration, and salt stress exhibited by cbf2 mutant plants is likely a consequence of their high levels of CBF/DREB1-regulated gene expression. These data indicate that CBF2/DREB1C negatively regulates CBF1/DREB1B and CBF3/DREB1A expression and plays a critical role not only in cold acclimation but also in the proper development of constitutive tolerance to freezing and related stresses in Arabidopsis.
The mechanism(s) whereby the expression of CBF/DREB1 genes is regulated by low temperature is unknown at present. Analysis of transgenic Arabidopsis-expressing reporter genes fused to CBF/DREB1 promoters has revealed that the induction of these genes in response to cold is regulated at the transcriptional level (ref. 4; F.N. and J.S., unpublished data). On the other hand, available evidence has suggested that CBF/DREB1 genes are not subjected to autoregulation, because the CRT/DRE core motif, CCGAC, is not present in their promoters (4, 9) and overexpression of CBF1/DREB1B does not result in accumulation of CBF3/DREB1A transcripts (4). Thus, it was proposed (4) that induction of CBF/DREB1 genes in response to low temperature could involve the modification of either a CBF/DREB1 activator, named ICE (inducer of CBF expression), that would be inactive at warm temperatures or an associated protein that would allow the activator to induce the expression of these genes. Recently, the identification of an Arabidopsis MYC-like basic helix-loop-helix transcriptional activator, called ICE1, that binds the CBF3/DREB1A promoter has been reported (27). ICE1 is expressed constitutively, and its overexpression enhances the expression of the CBF/DREB1 regulon in the cold and improves the freezing tolerance of the Arabidopsis transgenic plants (27). The expression of the CBF/DREB1 genes has also been suggested to be feedback inhibited by their gene products or by products of their downstream target genes (24). Our results provide direct evidence that CBF/DREB1 gene expression can be controlled, at least in part, by the CBF/DREB1 factors themselves. These results are consistent with the role of CBF2/DREB1C as a negative regulator of the expression of CBF1/DREB1B and CBF3/DREB1A, and, therefore, also of their downstream-target genes. In fact, the cbf2 mutation leads to an accumulation of CBF1/DREB1B and CBF3/DREB1A transcripts that results in the induction of target genes and subsequent increase in freezing, dehydration, and salt tolerance.
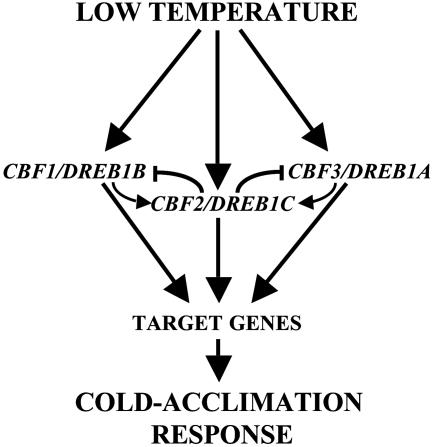
Because CBF/DREB1 transcripts are not usually detected in RNA-blot hybridizations under control conditions, it is generally presumed that CBF/DREB1 genes are not transcribed at normal growth temperatures. Therefore, if CBF2/DREB1C is required to repress the expression of CBF1/DREB1B and CBF3/DREB1A, it could be expected that these genes were expressed at those temperatures. A detailed analysis of the quantitative data available from different microarray experiments (26-28) reveals that, under nonstressed conditions, the transcript levels of CBF2/DREB1C are 5- to 8-fold higher than those of CBF1/DREB1B and CBF3/DREB1A, which is consistent with the function that we propose for CBF2/DREB1C. Another general assumption concerning the expression of CBF/DREB1 genes is that they have a similar pattern of induction in response to low temperature. Our results demonstrate that this assumption is not the case, the expression of CBF1/DREB1B and CBF3/DREB1A preceding that of CBF2/DREB1C. This asynchronism, together with the negative regulation of CBF1/DREB1B and CBF3/DREB1A by CBF2/DREB1C, may be important to ensure that CBF/DREB1 gene expression is transient and tightly controlled. Based on the results described here, a hypothetical model for CBF2/DREB1C function during cold acclimation is presented in Fig. 6. Under control conditions, steady-state levels of CBF2/DREB1C would be repressing CBF1/DREB1B and CBF3/DREB1A expression. When plants are exposed to low temperature, the activation of different, perhaps specific, regulators, such as ICE1 (27), would induce very rapidly the expression of these genes escaping from CBF2/DREB1C repression. Later on, probably once CBF1/DREB1B and CBF3/DREB1A products attain certain levels, CBF2/DREB1C is induced, which in turn would provoke the suppression of CBF1/DREB1B and CBF3/DREB1A transcription. We propose that CBF1/DREB1B and CBF3/DREB1A proteins, likely in the presence of low temperature, could contribute to activate CBF2/DREB1C expression. Finally, CBF1/DREB1B and CBF3/DREB1A would activate the transcription of downstream target genes and, subsequently, the development of the cold-acclimation response (5, 10-12). The possibility remains that, in addition to repressing CBF1/DREB1B and CBF3/DREB1A, CBF2/DREB1C might also activate cold-induced gene expression. Unfortunately, such a function cannot be revealed by studying the cbf2 mutation alone, given that CBF1/DREB1B and CBF3/DREB1A likely have a redundant role in this regard and thus mask the effect of the mutation on cold-induced gene expression. Needless to say, this model does not rule out the existence of additional regulatory mechanisms involved in controlling the precise expression of CBF/DREB1 genes (24, 29-31).
Fig. 6.
Proposed model for the function of CBF2/DREB1C in cold acclimation and the regulation of CBF/DREB1 gene expression in response to low temperature. Arrowheads and end lines indicate positive and negative regulation, respectively.
As mentioned before, freezing temperature is a common stress condition that adversely affects plant growth and agricultural production. Even modest increases (1-2°C) in the freezing tolerance of crop species would have a dramatic impact on agricultural productivity and profitability (32). Determining the molecular mechanisms that plants have evolved to survive freezing would not only increase our fundamental knowledge of how plants adapt to changes in the environment but would also contribute to the development of new strategies to improve the tolerance of crop species to this adverse situation. Here, we demonstrate that CBF2/DREB1C plays a crucial role in the development of Arabidopsis tolerance to freezing and other related stresses by ensuring the accurate expression of CBF1/DREB1B and CBF3/DREB1A and, hence, that of the CBF/DREB1-target genes. Although the mechanisms by which CBF2/DREB1C regulates CBF1/DREB1B and CBF3/DREB1A expression are currently unknown and need to be investigated, these findings provide a new perspective on the genetic control of freezing tolerance and further our understanding of the molecular basis of plant responses to adverse environmental conditions.
Acknowledgments
We thank D. Bartels and J. J. Sánchez-Serrano for suggestions on the manuscript and E. Rodríguez and A. Redondo for technical assistance. This work was supported by European Union Grant QLK3-CT00328 (to J.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CRT, C-repeat; CBF, CRT-binding factor; DREB, dehydration-responsive element-binding factor; FW, fresh weight; T-DNA, transferred DNA; DRE, dehydration-responsive DNA-regulatory element.
References
- 1.Levitt, J. (1980) in Responses of Plants to Environmental Stresses (Academic, New York), 2nd. Ed.
- 2.Thomashow, M. F. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571-599. [DOI] [PubMed] [Google Scholar]
- 3.Stockinger, E. J., Gilmour, S. J. & Thomashow, M. F. (1997) Proc. Natl. Acad. Sci. USA 94, 1035-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M. & Thomashow, M. F. (1998) Plant J. 16, 433-442. [DOI] [PubMed] [Google Scholar]
- 5.Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1998) Plant Cell 10, 1391-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riechmann, J. L. & Meyerowitz, E. M. (1998) Biol. Chem. 379, 633-646. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi-Shinozaki, K. & Shinozaki, K. (1994) Plant Cell 6, 251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker, S. S., Wilhelm, K. S. & Thomashow, M. F. (1994) Plant Mol. Biol. 24, 701-713. [DOI] [PubMed] [Google Scholar]
- 9.Medina, J., Bargues, J., Perol, M., Pérez-Alonso, J. & Salinas, J. (1999) Plant Physiol. 119, 463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O. & Thomashow, M. F. (1998) Science 280, 104-106. [DOI] [PubMed] [Google Scholar]
- 11.Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1999) Nat. Biotechnol. 17, 287-291. [DOI] [PubMed] [Google Scholar]
- 12.Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D. & Thomashow, M. F. (2000) Plant Physiol. 124, 1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haughn, G. & Somerville, C. (1986) Mol. General Genet. 204, 430-434. [Google Scholar]
- 14.Murashige, Y. & Skoog, F. (1962) Plant Physiol. 15, 473-497. [Google Scholar]
- 15.Logeman, J., Schell, J. & Willmitzer, L. (1987) Anal. Biochem. 163, 26-20. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) in Cloning: A Laboratory Manual, (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 17.Gilmour, S. J., Artus, N. N. & Thomashow, M. F. (1992) Plant Mol. Biol. 18, 13-21. [DOI] [PubMed] [Google Scholar]
- 18.Gilmour, S. J., Lin, C. & Thomashow, M. F. (1996) Plant Physiol. 111, 293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarillo, J. A., Capel, J., Leyva, A., Martínez-Zapater, J. M. & Salinas, J. (1994) Plant Mol. Biol. 25, 693-704. [DOI] [PubMed] [Google Scholar]
- 20.Capel, J., Jarillo, J. A., Salinas, J. & Martínez-Zapater, J. M. (1997) Plant Physiol. 115, 569-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baima, S., Sessa, G., Ruberti, I. & Morelli, G. (1995) Gene 153, 171-174. [DOI] [PubMed] [Google Scholar]
- 22.Thomashow, M. F. (2001) Plant Physiol. 125, 89-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y. & Shinozaki, K. (2001) Plant Cell 13, 61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, Y., Xion, L., Ishitani, M. & Zhu, J. K. (2002) Proc. Natl. Acad. Sci. USA 99, 7786-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina, J., Catalá, R. & Salinas, J. (2001) Plant Physiol. 125, 1655-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowler, S. & Thomashow, M. F. (2002) Plant Cell 14, 1675-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B.-H., Hong, X., Agarwal, M. & Zhu, J. K. (2003) Genes Dev. 17, 1043-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, W., Provart, N. J., Glazebrook, J., Katagiri, F., Chang, H. S., Eulgem, T., Mauch, F., Luan, S., Zou, G., Whitham, S. A., et al. (2002) Plant Cell 14, 559-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B. & Zhu, J. K. (2001) Genes Dev. 15, 912-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, H., Guo, Y., Ohta, M., Xiong, L., Stevenson, B. & Zhu, J. K. (2002) EMBO J. 21, 2692-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong, Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B. & Zhu, J. K. (2002) Proc. Natl. Acad. Sci. USA 99, 11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steponkus, P. L., Uemura, M., Joseph, R. A., Gilmour, S. J. & Thomashow, M. F. (1998) Proc. Natl. Acad. Sci. USA 95, 14570-14575. [DOI] [PMC free article] [PubMed] [Google Scholar]