Abstract
Bacteriophage T4 uses two modes of replication initiation: origin-dependent replication early in infection and recombination-dependent replication at later times. The same relatively simple complex of T4 replication proteins is responsible for both modes of DNA synthesis. Thus the mechanism for loading the T4 41 helicase must be versatile enough to allow it to be loaded on R loops created by transcription at several origins, on D loops created by recombination, and on stalled replication forks. T4 59 helicase-loading protein is a small, basic, almost completely α-helical protein whose N-terminal domain has structural similarity to high mobility group family proteins. In this paper we review recent evidence that 59 protein recognizes specific structures rather than specific sequences. It binds and loads the helicase on replication forks and on three- and four-stranded (Holliday junction) recombination structures, without sequence specificity. We summarize our experiments showing that purified T4 enzymes catalyze complete unidirectional replication of a plasmid containing the T4 ori(uvsY) origin, with a preformed R loop at the position of the R loop identified at this origin in vivo. This replication depends on the 41 helicase and is strongly stimulated by 59 protein. Moreover, the helicase-loading protein helps to coordinate leading and lagging strand synthesis by blocking replication on the ori(uvsY) R loop plasmid until the helicase is loaded. The T4 enzymes also can replicate plasmids with R loops that do not have a T4 origin sequence, but only if the R loops are within an easily unwound DNA sequence.
The bacteriophage T4-infected cell is a replication factory designed for the rapid production of multiple copies of its genome. T4 replication does not need to be coordinated with cell division or a cell cycle. Instead, the T4 replication strategy is optimized for quickly producing and packaging its DNA. The phage uses two modes of replication initiation: origin-dependent replication early in infection and recombination-dependent replication at later times. Because the 168-kb linear phage genome is terminally redundant and circularly permuted, the end of one DNA molecule is homologous to the middle of another. Thus the products of early origin-dependent replication can invade each other to form D loops for recombination-initiated replication. The same complex of relatively few T4-encoded replication proteins is responsible for DNA synthesis in both modes of replication (reviewed in refs. 1–4).
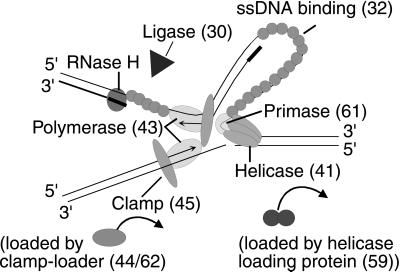
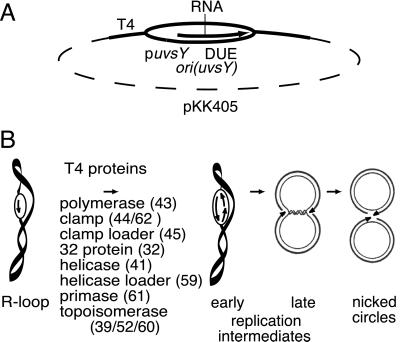
The genes for each of the T4 replication proteins have been cloned, and the functions of the proteins have been characterized by in vitro reactions on model templates (Fig. 1) (reviewed in ref. 5). T4 DNA polymerase (gene 43), which catalyzes DNA synthesis on both leading and lagging strands, is attached to a clamp protein (gene 45) that is loaded by the complex of the gene 44 and 62 proteins. In the presence of the T4 gene 32 single-stranded DNA binding protein, T4 DNA polymerase, the clamp, and the clamp loader are sufficient for slow strand displacement synthesis of the leading strand. The 5′ to 3′ gene 41 helicase unwinds DNA ahead of the fork and increases the elongation rate more than 10-fold to 400 nt/sec, comparable to that in vivo. Although the helicase can load on nicked and forked DNA by itself, its loading is greatly accelerated by the 59 helicase-loading protein. Lagging strand fragments are initiated by pentamer RNA primers, whose synthesis requires both the helicase and gene 61 primase. These primers ultimately are removed by a T4 encoded 5′ to 3′ nuclease (T4 RNase H), and after gap repair, the adjacent fragments are joined by T4 DNA ligase. T4 type II topoisomerase (genes 39, 52, and 60) is required for replication in vitro when the parental duplex is a covalently closed circle.
Figure 1.
Model of a replication fork with bacteriophage T4 proteins. Numbers indicate the name of the gene encoding each protein. See text for description of the functions of the proteins.
In tightly controlled replication systems like Escherichia coli oriC or yeast ARS I, sequence-specific origin binding proteins help to load the helicase at the origin (6). In contrast, the seven known T4 replication origins, identified by a variety of methods, do not share DNA sequences (1, 3). The four best-characterized origins, oriA, oriE, ori(uvsY)(F), and ori(34)(G), each have a transcription promoter, whose transcript is thought to act as the primer for leading strand synthesis. With the exception of the repEB and repEA proteins at oriE (7), there is no evidence for T4-encoded proteins that are required for initiation at a specific origin. Consequently, the mechanism for loading the T4 41 helicase must be versatile enough to allow it to load on R loops created by transcription at several origins, on D loops created by recombination, and on stalled replication forks.
In this paper we review recent evidence demonstrating that the T4 gene 59 helicase-loading protein recognizes specific structures rather than specific sequences. It binds and loads the helicase on replication forks and on three- and four-stranded (Holliday junction) recombination structures, without sequence specificity (8, 9). We summarize our experiments showing that the purified T4 enzymes catalyze complete unidirectional replication of a plasmid containing T4 ori(uvsY), with a preformed R loop at the position of the R loop identified in vivo (10). This replication depends on the helicase and is strongly stimulated by 59 protein. Moreover, the helicase-loading protein helps to coordinate leading and lagging strand synthesis by blocking replication on the ori(uvsY) R loop plasmid until the helicase is loaded.
T4 Gene 59 Helicase-Loading Protein
T4 mutants in gene 59 are UV-sensitive and defective in repair, recombination, and the recombination-initiated replication predominant in the late stage of infection (11, 12). The initial characterization of purified 59 protein showed that it was a small, monomeric, and basic protein that was capable of binding both single- and double-stranded DNA. 59 Protein also was shown to interact specifically with 41 helicase and gene 32 single-stranded DNA binding protein in the presence and absence of DNA (13–16). Barry and Alberts (16) showed that 59 protein increased the rate of loading of the 41 helicase on both single-stranded and nicked duplex templates. Its ability to load the helicase on single-stranded DNA covered with 32 protein was particularly striking because 32 protein strongly inhibits DNA unwinding by the helicase without 59 protein.
Structure of 59 Protein.
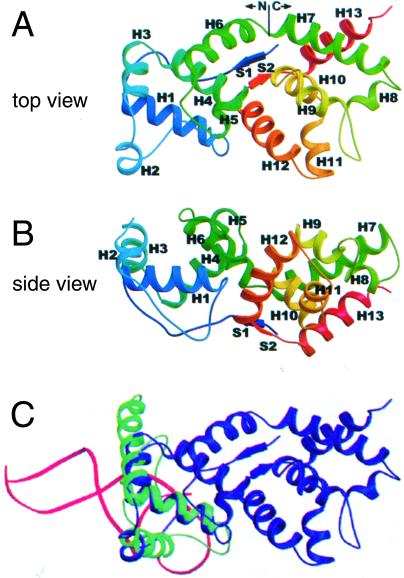
The crystal structure of the full-length (217 residues) 59 helicase-loading protein [Protein Data Bank (PDB) code 1C1K; ref. 8] revealed a novel, almost entirely α-helical protein, with no similarity to the structures of other single-stranded DNA binding proteins (Fig. 2). Its 13 α-helices are divided into N and C domains of similar size. The single short β-sheet connects N-terminal residues 2–4 with residues 197–199 near the C terminus. There is a narrow groove between the two domains on the “top” of the protein. The surface of the protein is notable for the high density of basic and hydrophobic residues, which may be important for its DNA and protein interactions.
Figure 2.
Ribbon diagrams of the crystal structure of the T4 gene 59 helicase-loading protein showing its structural similarity with HMG proteins. (A) and (B) are, respectively, “top” and “side” view rainbow-colored ribbon diagrams of 59 protein (PDB 1C1K, ref. 8) from the blue N terminus to the red C terminus. S1–2 are β-sheets and H1–13 are α-helices. The line between H6 and H7 indicates the junction between the N and C domains of the protein. (C) Superposition of the H1, H2, and H3 α-helices of T4 59 helicase-loading protein (blue) and rat HMG1 protein (green, PDB 1AAB, ref. 20) and with the DNA (red) from the model of lymphoid enhancer-binding factor LEF1-DNA, (PDB 2LEF, ref. 21). Adapted from Mueser et al. (8).
The sequence of 59 protein has only limited similarity to the sequences of functionally related proteins. However, the N domain of 59 protein has strong structural similarity to several members of the high mobility group (HMG) family proteins, including rat HMG1A and the LEF-1 lymphoid enhancer-binding factor (8). The HMG domain has been found in structure-specific nonhistone chromatin-binding proteins and sequence-specific transcription factors. Proteins with the HMG domain have been shown to bind to the minor groove of DNA, bending and partially unwinding the duplex. HMG1 and other family members bind and unstack cruciform DNA (reviewed in refs. 17–19). The structural alignment on the bottom of Fig. 2 shows the close superposition of 59 protein helices H1, H2, and H3 (residues 11–67) (blue) with residues 9–65 of the rat HMG1 structure (green) (PDB code 1AAB; ref. 20). The superposition of the duplex DNA from the LEF1-DNA complex (red) (PDB code 2LEF; ref. 21) suggests a possible orientation for duplex DNA on 59 protein.
59 Protein Binds Preferentially to Forked DNA.
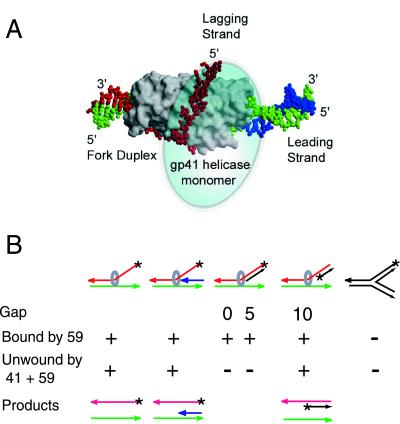
Although 59 protein binds to both single- and double-stranded DNA, it shows a clear preference for binding to DNA structures related to replication forks and recombination intermediates (8, 9). Our binding data, summarized in Fig. 3B, indicate that 59 protein binds strongly to fork DNA with either single- or double-stranded arms. These arms needed to be at least 6 nt long, but binding affinity was not increased by lengthening the arms from 12 to 18 nt. Whereas 59 protein bound to forks with completely duplex arms, it could not load the helicase unless there was a single-stranded gap of 10 nt on the fork arm corresponding to the lagging strand template. There was no requirement for a gap on the leading strand template. Three-way junction branched DNA, which is less flexible than fork and flap structures (22), was not bound by 59 protein. These observations are consistent with the role of the 41 helicase during DNA replication. The helicase moves 5′ to 3′ on the lagging strand template, opening the duplex ahead of the leading strand polymerase, and interacting with the primase to allow it to make the pentanucleotide primers that initiate lagging strand synthesis.
Figure 3.
Fork DNA structures bound by T4 59 helicase-loading protein and unwound by the 41 helicase loaded by 59 protein. (A) A speculative model of T4 gene 59 helicase-loading protein bound on a DNA replication fork. This model is taken from ref. 9 and was adapted from Mueser et al. (8). It is based on the distribution of hydrophobic and basic residues on the surface of 59 protein and on the assumption that the HMG-like region of its N domain binds and unstacks the duplex (red and green) ahead of the fork, as shown for HMG proteins (17, 19). The leading strand duplex [template (green)/primer (blue)] is docked on the bottom surface of the C domain. A long segment of single-stranded DNA, representing the lagging strand (red), traverses the shallow groove between the N and C domains. A helicase monomer (light blue oval) is proposed to bind to 59 protein between the lagging and leading strand arms (8). The other subunits of the hexameric helicase (not shown) would surround the lagging strand. (B) 59 protein binds to forks with single or duplex arms, but cannot load the helicase without a single-stranded gap of more than 5 nt on the lagging strand template. The fork DNA structures are drawn in the same orientation as the fork DNA on the model of 59 protein in A. The hexameric helicase (circle) is placed on the strand that would occupy the position of the fork lagging strand, if each of these DNAs binds to 59 protein as predicted by the model in A. Arrowheads represent the 3′ end of each strand; * marks the position of 32P label in the substrates tested. [Reproduced with permission from ref. 9 (Copyright 2000, American Society for Biochemistry and Molecular Biology).]
Because the helicase-loading protein has much greater affinity for fork DNA with single- or double-stranded arms than for either single-stranded or simple duplex DNA, this small protein must have sites for the duplex region of the fork, as well as two arm sites that can accommodate either single- or double-stranded arms. Unfortunately, we do not yet have a structure for 59 protein bound to DNA. Fig. 3A shows a speculative model for 59 protein on a replication fork DNA that is based on the assumption that the “HMG-like” N domain binds the duplex ahead of the fork and holds the beginning of the fork arms in an open configuration (8). The leading strand duplex then could bind to a hydrophobic region on the bottom surface of the C domain, whereas the lagging strand template occupies the groove between the two domains on the top surface of the protein. If 59 protein binds as proposed in this model, it would separate the two fork strands and provide a docking area for 41 protein, which has been shown to surround the lagging strand (23). It also would be in position to bind to 32 protein coating the template for the lagging strand.
Binding and Unwinding on Forks within Recombination Structures.
Both the 59 helicase-loading protein and the 41 helicase play vital roles in recombination. Gene 59 mutants and some gene 41 mutants have a recombination-defective phenotype (12, 24–26). Salinas and Kodadek (27) showed that after homologous pairing in vitro by the T4 uvsX, uvsY, and 32 proteins, both the 41 helicase and 59 helicase-loading protein were needed for extensive polar branch migration. Branch migration from circular single-stranded DNA annealed on a single-stranded extension of a complementary duplex is carried out by the combination of 41 helicase and the 59 and 32 proteins (28).
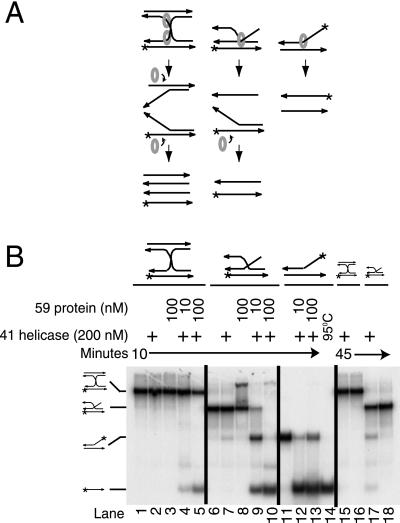
As anticipated because of the role of 41 helicase and 59 loading protein in recombination, 59 protein bound a three-stranded DNA strand invasion structure and four-stranded Holliday junction DNA (9) (Fig. 4). However, as indicated above (Fig. 3B), it did not bind three-way junction branch DNA. The Holliday junction and strand invasion structures were unwound by the helicase if 59 protein was present. In Fig. 4A, the strand invasion and cruciform structures are drawn in the same orientation as the fork DNA on the model of 59 protein in Fig. 3A. The hexameric helicase (circle) is shown on the strand that would occupy the position of the lagging strand, if these DNAs bind to 59 protein as predicted by the fork model.
Figure 4.
T4 59 protein stimulates unwinding of cruciform DNA substrates by 41 DNA helicase. (A) Diagram showing the expected unwinding products and positions of the helicase (circle) on a four-stranded cruciform and three-stranded invasion structure, assuming that 59 protein binds to the fork portions of these structures and loads the helicase as predicted by the speculative model in Fig. 3A. Arrowheads represent the 3′ end of each strand; * marks the position of 32P label in the substrates tested. Strand exchange was not possible because the arms of the structures were not homologous. (B) Unwinding of cruciform and strand invasion structures. Helicase was present at a final concentration of 200 nM (monomer) where indicated. DNA was present at 10 nM. Adapted from ref. 9.
In the three-stranded structure, the 3′ end of the invading strand has annealed to a complementary region of the duplex. 41 Helicase would be expected to bind to the 5′ end of this invading strand, because it is the only 5′-ended single strand in the structure (Fig. 4A). The duplex that includes the 3′ end of this strand would be predicted by the model to be bound by the HMG-like N-terminal domain of 59 protein. In the experiment shown, the P32 label (indicated by * on Fig. 4A) was not on this invading strand. Thus helicase moving 5′ to 3′ on the invading strand would give unlabeled single-strand and labeled fork as the initial products. The labeled fork would be rapidly unwound to single strands by the helicase. In agreement with this prediction, both labeled fork and single strand were observed as products in the reaction with the lower concentration of 59 protein, whereas at the higher 59 protein concentration there was only labeled single-stranded product (Fig. 4B). This is the helicase reaction required for conservative recombination-initiated DNA replication and bubble-branch migration (see Fig. 7).
Figure 7.
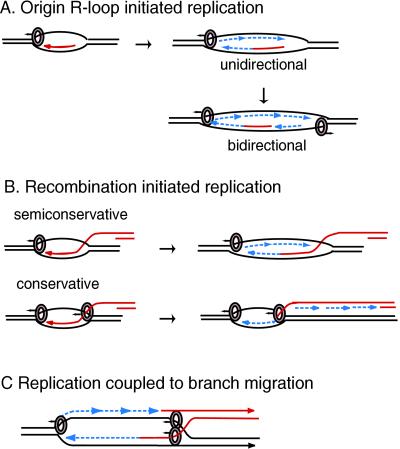
Models for the role of 41 helicase in origin-initiated and recombination-initiated replication. Helicase hexamers (circles) are shown on the strands they surround. Arrows above the circles show the direction of helicase movement. Arrowheads indicate the 3′ strand ends. RNA and invading DNA molecules are in red. Newly synthesized DNA is a dashed line in blue. (A) One or two helicases are required, respectively, for unidirectional or bidirectional replication from an origin. The helicase at each fork opens the duplex ahead of the polymerase and enables primer synthesis by the primase. (B) Semiconservative and conservative replication at forks initiated by recombination. In semiconservative replication, the displaced strand serves as the lagging strand template, and the helicase role is like that at an origin. In conservative replication, the invading strand serves as the lagging strand template. The helicase at the right catalyzes the branch migration needed for reannealing the duplex behind the leading strand and also interacts with the primase on the invading strand. (C) Semiconservative replication coupled to branch migration at a four-way junction. Helicase at the fork opens the duplex and interacts with primase. Branch migration is catalyzed by 5′ to 3′ movement of helicase on the four-way junction behind the leading strand polymerase. Two helicase hexamers are shown on the junction because the products of unwinding a cruciform with nonhomologous arms are most easily explained by two 41 helicases unwinding the cruciform simultaneously (see Fig. 4). However, it remains to be determined whether a single hexamer is sufficient for strand exchange at a Holliday junction. See text for further discussion. Adapted from ref. 9.
The observation that helicase can unwind the cruciform, which lacks single-stranded arms, suggests that there are enough accessible unpaired nucleotides at the cruciform junction to permit slow loading of the helicase (9). This would be expected if the HMG-like domain of 59 protein, like other HMG proteins, binds the cruciform in an unstacked open configuration.
The arms of the cruciform substrate in Fig. 4 do not share common sequences, so strand exchange was not possible in this experiment. If two helicases were loaded on the cruciform as shown on the diagram in Fig. 4A, the initial products would be two forks. Additional 41 helicase would be quickly loaded on these forks by 59 protein. Thus, the finding of single-stranded DNA as the only product (Fig. 4B) is not surprising, because the rate of unwinding of forks to single strands is so much faster than unwinding of the cruciform. If only one of the two helicases shown on the cruciform in Fig. 4A was present, no unwound product would have been detected. The single helicase would move 5′ to 3′ on the strand it encircles, opening the duplex ahead. Because the other ends of the strands forming this duplex are still bound in the four-stranded structure, the unwound duplex would reanneal rapidly. However, it remains to be determined whether unwinding of one arm of a cruciform by a single gene 41 helicase would be enough to facilitate strand exchange between homologous arms of a cruciform.
Role of the Helicase and Helicase-Loading Protein During Origin-Dependent Replication
Gene 41 helicase is needed for both early origin-dependent T4 phage replication and replication of plasmids with the cloned T4 origins ori(uvsY) and ori(34). The role of gene 59 helicase-loading protein in origin-dependent replication is less clear because amber mutations in gene 59 did not reduce early T4 phage DNA synthesis (29), or replication of ori(uvsY) plasmids (30), but did abolish plasmid replication via the recombination-dependent mode. Thus either 59 protein is necessary to load the helicase onto recombination intermediates, but not onto replication origins, or the gene 59 amber mutations are leaky, and a small amount of read-through 59 protein is sufficient for origin- but not recombination-initiated replication.
Two T4 origins, ori(uvsY) and ori(34), were found to have a minimal sequence of about 100 bp that is sufficient for maximal levels of replication of plasmids containing these origins in T4-infected cells (31). Each of these minimal origins consists of a phage middle-mode promoter just upstream of a DNA-unwinding element (DUE) (32). T4 middle-mode promoters are transcribed by E. coli RNA polymerase that has been modified by addition of the T4 AsiA protein and must be activated by T4 MotA protein (33). Analysis of replicative intermediates during phage infection in vivo indicated that the RNA within a stable R loop at ori(uvsY) is used to prime leading-strand replication in one direction from the origin, and that assembly of a functional replication complex for the second direction is delayed significantly (34).
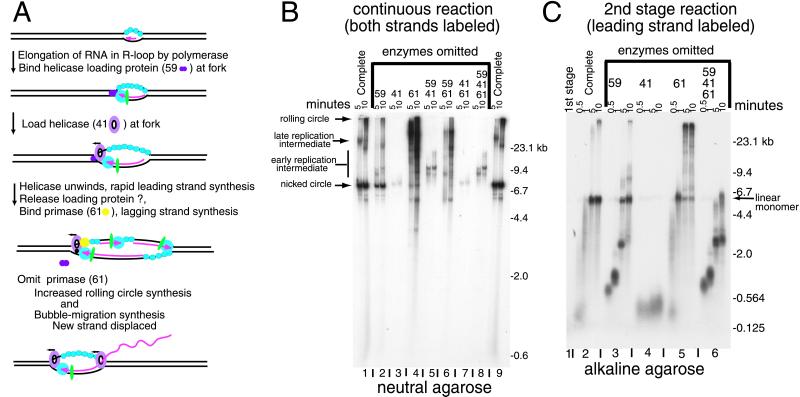
As an initial step toward reconstructing origin-initiated replication, we recently have shown that bacteriophage T4 proteins catalyze complete unidirectional replication of a plasmid, pKK405, with a preformed R loop at the T4 ori(uvsY) replication origin in vitro (10) (Fig. 5A). The 104-b RNA in the R loop serves as the primer for leading strand synthesis. The origin R loop plasmid was converted to two nicked circles in a reaction with T4 DNA polymerase, clamp, clamp loader, 32 protein, 41 helicase, 61 primase, 59 helicase-loading protein, and topoisomerase (Fig. 5B, and reactions 1 and 9 on the neutral gel, Fig. 6B). There were also some slower moving products, labeled late replication intermediates, which moved in the position expected for almost completed theta structures (35), as well as products just entering the gel, which are likely to be the result of rolling circle synthesis from the nicked circles. We showed that the nicked circular products were sealed if T4 RNaseH and DNA ligase are added to the reaction (10). As expected, leading strand synthesis stalled prematurely in the absence of T4 DNA topoisomerase.
Figure 5.
Replication of a plasmid with a preformed R loop at the T4 ori(uvsY) replication origin by T4 replication proteins in vitro. (A) Diagram of the pKK405 plasmid (57), which contains a 1.35-kb fragment of T4 DNA (solid line) with the ori(uvsY) replication origin in a pBR322-based vector (dashed line). As indicated in the text, this origin contains the PuvsY transcription promoter adjoining a DUE. The R loop in the plasmid was formed by annealing a 104-b RNA corresponding to −5 to +99 relative to the transcription start site (10). (B) T4 replication proteins required for unidirectional replication from the pKK405 R loop plasmid, giving nicked circular products. [B, Reproduced with permission from ref. 10 (Copyright 2001, Elsevier Science).]
Figure 6.
Replication of the R loop plasmid with the T4 ori(uvsY) origin requires 41 helicase and is strongly stimulated by the 59 helicase-loading protein. (A) Model showing the roles of the 59 helicase-loading protein, 41 helicase, and 61 primase in unidirectional replication from T4 ori(uvsY). 59 Protein binds ahead of the polymerase holoenzyme elongating the RNA in the R loop and loads the helicase essential for rapid leading strand synthesis. 59 Protein inhibits leading strand synthesis when there is no helicase. (B) Neutral agarose gel of the products of unidirectional continuous replication of the pKK405 ori(uvsY) R loop plasmid. The complete reactions contained the T4 replication proteins listed in Fig. 5B. 59 Helicase-loading protein, 41 helicase, and 61 primase were omitted as indicated. (C) Alkaline agarose gel of the replication of the pKK405 R loop plasmid in a two-stage reaction in which only the leading strand is labeled. The RNA was 3′ end-labeled by limited synthesis with dATP, dTTP, 32P-dCTP in a first-stage reaction that required T4 polymerase, clamp, clamp loader, and 32 protein. 41 Helicase, 59 helicase loader, 61 primase, and topoisomerase then were added along with dGTP and enough unlabeled dCTP to prevent further 32P-dCTP incorporation. The indicated enzymes were omitted in the second stage. T4 topoisomerase was present in all reactions. See text for description of the inhibition by 59 helicase-loading protein when 41 helicase is omitted and of the aberrant products made without the 61 primase. [B and C, Reproduced with permission from ref. 10 (Copyright 2001, Elsevier Science).]
Helicase was required for the synthesis of the nicked circular products from the R loop plasmid (10). When helicase activity was eliminated by omitting both the helicase and the loading protein, the products migrated as partially completed theta structures (early replication intermediates) (reaction 5, Fig. 6B). This limited synthesis depended on polymerase, clamp, clamp loader, and 32 protein, in agreement with the protein requirements for strand displacement synthesis (10). When the helicase-loading protein was omitted but the helicase was present (reaction 2), the DNA products were similar to those in the complete reaction, but the extent of replication was reduced about 4-fold. Thus helicase by itself can load on the replication bubble formed at the R loop, but loading is much more efficient with the loading protein. This is consistent with previous findings that 59 protein greatly stimulates, but is not essential for, strand displacement synthesis on nicked and forked duplex DNA (16, 36), and that there is some origin-initiated replication with T4 gene 59 mutants (1, 29).
When 61 primase was omitted (reaction 4 in Fig. 6B), there was a large increase in the synthesis of products with long single strands (10). The longest of these are likely formed by rolling circle synthesis from nicked circles. There also may be bubble-branch migration synthesis in which the leading strand polymerase moves continuously around the circle without enlarging the bubble because the single-stranded side of the bubble branch migrates to displace the newly synthesized leading strand. Bubble branch migration was first shown in replication reactions with the T4 polymerase holoenzyme, UvsX recombination protein, and Dda helicase (37). It also would be expected in these reactions with the 41 helicase and 59 protein, but without primase, because 41 helicase can unwind the invading strand from a three-stranded invasion structure (Fig. 4B) (9), and the combination of 59 helicase-loading protein, 41 helicase, and 32 single-stranded DNA binding protein catalyzes strand exchange (27, 28).
59 Protein Helps to Coordinate Leading and Lagging Strand Synthesis by Blocking Replication in the Absence of the Helicase.
The 59 helicase-loading protein strongly inhibited replication when it was present in reactions without the helicase (10). There was much less synthesis in the reaction with 59 protein but no helicase (reaction 3, neutral gel, Fig. 6B), than in reaction 5 in which both the helicase and loading protein were omitted. The most likely explanation for this inhibition was that 59 protein bound tightly to the fork ahead of the leading strand polymerase, and inhibited polymerase progression (Fig. 6A). To test this hypothesis, we carried out the replication of the ori(uvsY) R loop plasmid in a two-stage reaction in which only the leading strand is labeled (Fig. 6C). The RNA was 3′ end-labeled by limited synthesis with dATP, dTTP, and P32-dCTP in a first-stage reaction that required T4 polymerase, clamp, clamp loader, and 32 protein (10). 41 Helicase, 59 helicase loader, 61 primase, and topoisomerase then were added along with dGTP and enough unlabeled dCTP to prevent further 32P-dCTP incorporation. In the complete reaction most of the leading strand was full-length linear monomer on an alkaline gel (reaction 2, Fig. 6C). When 41 helicase, 59 helicase-loading protein, and primase were omitted, the remaining enzymes (polymerase, clamp, clamp loader, 32 protein, and topoisomerase) were sufficient for limited extension of the leading strand to about 2.5 kb (reaction 6). Helicase was loaded slowly without the loading protein (reaction 3). The length of the leading strand at early times was the same as that in reaction 6, in which helicase, helicase loader, and primase all were omitted. However, by 5 min there was some full-length linear monomer in the reaction with the helicase but no 59 protein (reaction 3). Dramatically, the leading strand elongation stopped at about 300–500 b, when 59 protein was present without the helicase (reaction 4). This result suggests that 59 protein, which as indicated above is a tight fork DNA binding protein (8, 9), binds ahead of polymerase at the replication fork and remains there until the helicase is loaded (Fig. 6A). From this position, the helicase-loading protein could help to coordinate leading and lagging strand synthesis by slowing the leading strand until helicase was available to work with primase to begin the lagging strand. It should be noted that in this experiment polymerase began elongating the RNA in the R loop in the first-stage reaction, and 59 protein was not added until the second stage. Thus, some or all of the 300–500 nt added when 59 protein was present without the helicase (reaction 4) may have been added by polymerase before 59 protein bound to the DNA.
The strong replication inhibition by 59 protein without the helicase with the supercoiled ori(uvsY) plasmid was much greater than its inhibition of rolling circle synthesis (16) (C. E. Jones and N.G.N., unpublished experiments). Possibly, the helicase-loading protein binds with higher affinity to a fork in a replication bubble on a supercoiled plasmid than to a fork on relaxed DNA.
The question of whether 59 protein blocks phage replication in vivo in the absence of the helicase has not yet been addressed directly. However, it has been shown that wild-type T4 growth is inhibited in a host expressing 59 protein from a plasmid (36). This inhibition may be the result of replication blockage by 59 protein present at high levels at the time of infection.
A DUE Is Required for Replication from an R Loop.
Because T4 41 helicase and 59 helicase-loading protein work on both replication forks and recombination intermediates, they must be able to bind at many sequences. Our recent experiments with R loop plasmids demonstrate that the requirement for loading the T4 replication enzymes, including the helicase and 59 helicase-loading protein, is not a specific T4 sequence, but rather a specific structure, an R loop within an easily unwound DNA sequence (DUE). The minimal T4 origins ori(uvsY) and ori(34) each contain AT-rich DUE sequences downstream of a T4 middle promoter. Deletion analysis showed that plasmid replication in a T4-infected cell gradually decreased as more of the DUE sequence was removed (31). A plasmid whose only T4 DNA was the ori(uvsY) middle promoter (pY074Δ) replicated poorly. However, replication could be restored by replacing the T4 DUE with a well characterized DUE from the pBR322 plasmid (32). Consistent with these results we found that although a stable R loop could be formed downstream of the T4 middle promoter on pY074Δ, this R loop was not extended by the T4 replication proteins in vitro (10), because of the absence of a DUE sequence. Furthermore, although pBR322 cannot be replicated in a T4-infected cell because it lacks the T4 promoter necessary to make RNA (32), pBR322 with a preformed R loop in the DUE region was replicated in vitro as well as the plasmid pKK405 with the R loop at T4 ori(uvsY) (10). These experiments explain earlier observations that origin function was retained when the T4 middle promoter at ori(uvsY) was replaced by other T4 middle promoter sequences, but middle promoters without an adjacent DUE are not origins (32).
The Versatility of T4 41 Helicase and 59 Helicase-Loading Protein Couples Replication and Recombination
T4 59 protein can bind and load the 41 helicase on forked DNA without sequence specificity as long as there are 10 single-stranded nt on the lagging strand template at the fork. It also can load the helicase on R loops if they are within easily unwound sequences, and on D loops, cruciforms, and strand invasion structures. This versatility allows T4 replication forks to be established on R loops created by transcription and at D loops created by recombination (Fig. 7).
In a T4-infected cell, the origin R loops are formed by transcription using E. coli RNA polymerase modified for middle transcription in the case of ori(uvsY), ori(34), and ori A and unmodified host polymerase at ori E (3, 7, 31). Once the R loops are formed within DUEs, polymerase can be clamped at the 3′ end of the RNA by the gene 45 clamp and 44/62 clamp loader. 59 Protein can bind at the fork in front of the RNA and position the helicase on the 32 protein-covered displaced strand, without additional phage or host proteins (Fig. 7A). From this position helicase unwinds the duplex ahead of the leading strand polymerase and binds primase to begin lagging strand synthesis. Addition of topoisomerase is then sufficient for unidirectional replication from the origin. Bidirectional replication is established when helicase is loaded on the strand displaced by elongation of the first lagging strand fragment.
D loops and strand invasion structures are formed at double-stranded breaks and at the ends of the T4 linear genome by the T4-encoded uvsX recombinase, loaded with the help of the uvsY and 32 proteins (see refs. 2, 4, and 38 for recent reviews) (Fig. 7 B and C). Unidirectional semiconservative replication from a D loop requires a single helicase and is mechanistically similar to unidirectional replication from an origin R loop. At present there is no evidence indicating whether assembly of the T4 replication enzymes at these D loops is enhanced by an easily unwound sequence, as has been shown for replication from R loops.
A second helicase would be required if conservative replication followed strand invasion (Fig. 7B). While the helicase at the left opened the duplex ahead of the leading strand polymerase elongating the invading strand, the helicase at the right would move on the invading strand in the same direction as the polymerase, unwinding the newly synthesized strand. Interaction between the second 41 helicase and 61 primase would allow synthesis of the primers needed to initiate discontinuous synthesis on the unwound portion of the newly synthesized strand. In the absence of primase, helicase traveling behind polymerase on the newly synthesized strand leads to bubble branch migration, as first observed in the T4 system with the Dda helicase by Formosa and Alberts (37). Although 59 protein can bind and load helicase on an invading strand (Fig. 4 and ref. 9), at least two factors favor semiconservative rather than conservative replication after strand invasion. First, the helicase on the invading strand would be less stable than the helicase ahead of the polymerase, because there is evidence suggesting that interaction between 41 helicase and T4 DNA polymerase helps to keep the helicase on the fork (36, 39). Second, uvsX protein coating the invading strand would inhibit binding by the 59 helicase-loading protein (13, 38). Conservative replication with the T4 proteins has not been demonstrated in vivo or in vitro.
T4 59 helicase-loading protein, 41 helicase, and 32 protein together catalyze strand exchange (27, 28) and unwind four-stranded Holliday junctions (9). Recombination-initiated semiconservative replication can be coupled to branch migration if 41 helicase binds to the four-way junction behind the leading strand polymerase (Fig. 7C) and moves the junction toward the replication fork. The T4 helicase responsible for semiconservative replication must be 41 helicase, because it is the only helicase that interacts with the primase. T4 Dda helicase could replace 41 helicase at the four-way junction. These coupled replication and recombination reactions would result in coordinated extension of the two strands of the invading DNA, leaving the donor DNA unchanged, as recently proposed for the break-induced replication model for double-strand break repair (40, 41). This replication mechanism with helicase at the fork coordinating leading and lagging strand synthesis, and helicase behind the fork catalyzing strand exchange, is well suited to the T4-infected cell where most replication begins with recombination. It also may account for the extensive chromosomal replication (termed ECR) that has been shown to accompany double-strand break repair in T4 phage DNA (2, 42).
Comparison with Other Helicases and Loading Proteins
T4 gene 59 helicase-loading protein differs from other proteins with this function in being able to load a helicase on origins, on replication forks remote from origins, and on three- and four-stranded replication intermediates. Separate sets of proteins are required for these multiple functions in E. coli. Like T4 gene 41 helicase, E. coli dnaB helicase can slowly load on forked DNA by itself (43). However, both the dnaA protein, which binds to dnaA box sequences on oriC, and dnaC protein, which forms a hexameric complex with the dnaB hexamer, are required to load the dnaB helicase at this origin. The O and P proteins of phage lambda are required to load the dnaB helicase at the phage origin (reviewed in ref. 44). In contrast to 59 protein, the single-stranded DNA binding activities of the lambda P and E. coli dnaC proteins are only evident when these proteins form complexes with the dnaB helicase (45).
E. coli Pri A protein shares with T4 59 protein the ability to bind with high affinity to forked DNA and to the three-stranded junction at the 3′ end of an invading strand in D loops, but Pri A does not bind to four-stranded Holliday junctions. PriA is needed to load the dnaB helicase on stalled replication forks (46–49), on D loops at specialized origins during inducible stable replication, on R loops during constitutive stable replication in E. coli RNase HI mutants (50, 51), and on the phage Mu strand transfer complex (52, 53). Thus PriA resembles T4 59 in recognizing forks and D loops without sequence specificity, but it does not bind or load dnaB helicase on four-stranded Holliday junctions.
In E. coli a separate helicase (RuvB) and helicase-loading protein (RuvA) are responsible for branch migration on recombination structures. The RuvAB complex operates by a mechanism distinct from that of the replicative helicases, and it has no role in leading or lagging strand synthesis (54–56). In the simpler bacteriophage T4 system, 41 helicase, loaded by 59 protein, can catalyze branch migration and is essential for both origin- and recombination-initiated replication.
Acknowledgments
We thank Deborah Hinton for helpful comments on the manuscript. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases intramural program (to N.G.N.), National Institutes of Health Grant GM34622 (to K.N.K.), and National Institutes of Health Fellowship F32 GM19000 (to K.C.D.).
Abbreviations
- HMG
high mobility group
- PDB
Protein Data Bank
- DUE
DNA-unwinding element
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Kreuzer K N, Morrical S W. In: Molecular Biology of Bacteriophage T4. Karem J, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 28–42. [Google Scholar]
- 2.Kreuzer K N. Trends Biochem Sci. 2000;25:165–173. doi: 10.1016/s0968-0004(00)01559-0. [DOI] [PubMed] [Google Scholar]
- 3.Mosig G, Colowick N, Gruidl M E, Chang A, Harvey A J. FEMS Microbiol Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 4.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 5.Nossal N G. In: Molecular Biology of Bacteriophage T4. Karem J, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 43–53. [Google Scholar]
- 6.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 7.Vaiskunaite R, Miller A, Davenport L, Mosig G. J Bacteriol. 1999;181:7115–7125. doi: 10.1128/jb.181.22.7115-7125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueser T C, Jones C E, Nossal N G, Hyde C C. J Mol Biol. 2000;296:597–612. doi: 10.1006/jmbi.1999.3438. [DOI] [PubMed] [Google Scholar]
- 9.Jones C E, Mueser T C, Nossal N G. J Biol Chem. 2000;275:27145–27154. doi: 10.1074/jbc.M003808200. [DOI] [PubMed] [Google Scholar]
- 10.Nossal N G, Dudas K C, Kreuzer K N. Mol Cell. 2001;7:31–41. doi: 10.1016/s1097-2765(01)00152-6. [DOI] [PubMed] [Google Scholar]
- 11.Wu R, Wu J L, Yeh Y C. J Virol. 1975;16:5–16. doi: 10.1128/jvi.16.1.5-16.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah D B. J Virol. 1976;17:175–182. doi: 10.1128/jvi.17.1.175-182.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonesaki T. J Biol Chem. 1994;269:1284–1289. [PubMed] [Google Scholar]
- 14.Morrical S W, Hempstead K, Morrical M D. J Biol Chem. 1994;269:33069–33081. [PubMed] [Google Scholar]
- 15.Morrical S W, Beernink H T H, Dash A, Hempstead K. J Biol Chem. 1996;271:20198–20207. doi: 10.1074/jbc.271.33.20198. [DOI] [PubMed] [Google Scholar]
- 16.Barry J, Alberts B. J Biol Chem. 1994;269:33049–33062. [PubMed] [Google Scholar]
- 17.Zlatanova J, van Holde K. FASEB J. 1998;12:421–431. [PubMed] [Google Scholar]
- 18.Pöhler J R, Norman D G, Bramham J, Bianchi M E, Lilley D M. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill D A, Pedulla M L, Reeves R. Nucleic Acids Res. 1999;27:2135–2144. doi: 10.1093/nar/27.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardman C H, Broadhurst R W, Raine A R, Grasser K D, Thomas J O, Laue E D. Biochemistry. 1995;34:16596–16607. doi: 10.1021/bi00051a007. [DOI] [PubMed] [Google Scholar]
- 21.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Nature (London) 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 22.Fenley M O, Manning G S, Marky N L, Olson W K. Biophys Chem. 1998;74:135–152. doi: 10.1016/s0301-4622(98)00171-9. [DOI] [PubMed] [Google Scholar]
- 23.Raney K D, Carver T E, Benkovic S J. J Biol Chem. 1996;271:14074–14081. doi: 10.1074/jbc.271.24.14074. [DOI] [PubMed] [Google Scholar]
- 24.Cunningham R P, Berger H. Virology. 1978;88:62–70. doi: 10.1016/0042-6822(78)90110-1. [DOI] [PubMed] [Google Scholar]
- 25.Yonesaki T. Genetics. 1994;138:247–252. doi: 10.1093/genetics/138.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroki E, Yonesaki T. Mol Gen Genet. 1999;262:525–533. doi: 10.1007/s004380051114. [DOI] [PubMed] [Google Scholar]
- 27.Salinas F, Kodadek T. Cell. 1995;82:111–119. doi: 10.1016/0092-8674(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 28.Kong D, Nossal N G, Richardson C C. J Biol Chem. 1997;272:8380–8387. doi: 10.1074/jbc.272.13.8380. [DOI] [PubMed] [Google Scholar]
- 29.Mosig G, Colowick N. Methods Enzymol. 1995;262:587–604. doi: 10.1016/0076-6879(95)62046-x. [DOI] [PubMed] [Google Scholar]
- 30.Kreuzer K N, Yap W Y, Menkens A E, Engman H W. J Biol Chem. 1988;263:11366–11373. [PubMed] [Google Scholar]
- 31.Menkens A E, Kreuzer K N. J Biol Chem. 1988;263:11358–11365. [PubMed] [Google Scholar]
- 32.Carles-Kinch K, Kreuzer K N. J Mol Biol. 1997;266:915–926. doi: 10.1006/jmbi.1996.0844. [DOI] [PubMed] [Google Scholar]
- 33.Stitt B, Hinton D. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 142–160. [Google Scholar]
- 34.Belanger K G, Kreuzer K N. Mol Cell. 1998;2:693–701. doi: 10.1016/s1097-2765(00)80167-7. [DOI] [PubMed] [Google Scholar]
- 35.Minden J S, Marians K J. J Biol Chem. 1986;261:11906–11917. [PubMed] [Google Scholar]
- 36.Spacciapoli P, Nossal N G. J Biol Chem. 1994;269:447–455. [PubMed] [Google Scholar]
- 37.Formosa T, Alberts B M. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 38.Beernink H T, Morrical S W. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 39.Dong F, Weitzel S E, von Hippel P H. Proc Natl Acad Sci USA. 1996;93:14456–14461. doi: 10.1073/pnas.93.25.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes A M, Haber J E. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 41.Haber J E. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 42.George J W, Kreuzer K N. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens K M, McMacken R. J Biol Chem. 1997;272:28800–28813. doi: 10.1074/jbc.272.45.28800. [DOI] [PubMed] [Google Scholar]
- 44.Baker T A, Wickner S H. Annu Rev Genet. 1992;26:447–477. doi: 10.1146/annurev.ge.26.120192.002311. [DOI] [PubMed] [Google Scholar]
- 45.Learn B A, Um S J, Huang L, McMacken R. Proc Natl Acad Sci USA. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Marians K J. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 47.Sandler S J, Marians K J. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marians K J. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 49.Marians K J. Curr Opin Genet Dev. 2000;10:151–156. doi: 10.1016/s0959-437x(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 50.Masai H, Asai T, Kubota Y, Arai K, Kogoma T. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones J M, Nakai H. J Mol Biol. 1999;289:503–516. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- 53.Jones J M, Nakai H. Mol Microbiol. 2000;36:519–527. doi: 10.1046/j.1365-2958.2000.01888.x. [DOI] [PubMed] [Google Scholar]
- 54.Parsons C A, Tsaneva I, Lloyd R G, West S C. Proc Natl Acad Sci USA. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, West S C, Egelman E H. J Mol Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 57.Kreuzer K N, Alberts B M. Proc Natl Acad Sci USA. 1985;82:3345–3349. doi: 10.1073/pnas.82.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]