Abstract
The final steps in butyrate synthesis by anaerobic bacteria can occur via butyrate kinase and phosphotransbutyrylase or via butyryl-coenzyme A (CoA):acetate CoA-transferase. Degenerate PCR and enzymatic assays were used to assess the presence of butyrate kinase among 38 anaerobic butyrate-producing bacterial isolates from human feces that represent three different clostridial clusters (IV, XIVa, and XVI). Only four strains were found to possess detectable butyrate kinase activity. These were also the only strains to give PCR products (verifiable by sequencing) with degenerate primer pairs designed within the butyrate kinase gene or between the linked butyrate kinase/phosphotransbutyrylase genes. Further analysis of the butyrate kinase/phosphotransbutyrylase genes of one isolate, L2-50, revealed similar organization to that described previously from different groups of clostridia, along with differences in flanking sequences and phylogenetic relationships. Butyryl-CoA:acetate CoA-transferase activity was detected in all 38 strains examined, suggesting that it, rather than butyrate kinase, provides the dominant route for butyrate formation in the human colonic ecosystem that contains a constantly high concentration of acetate.
The human colon harbors a diverse range of bacteria that form a complex anaerobic ecosystem. Apart from interacting with each other, for example by cross-feeding, colonic bacteria also interact with the human host and play a vital role in maintaining human health through mechanisms such as protecting against pathogenic bacteria, influencing the host's immune system, and supplying nutrients (23).
The recent development of molecular techniques for studying phylogenetic diversity of the colonic microflora is leading to a better understanding of its composition (9, 21). One group of bacteria, which has received relatively little previous attention because of their fastidious growth requirements, are the strictly anaerobic gram-positive bacteria of low mol% G+C-content clostridial groups. Several studies employing a variety of molecular approaches indicate that members of this group related to clostridial clusters IV and XIVa (14) might make up about 30 to 60% of the total bacterial population within the colon (22, 25, 38, 46, 50).
Bacteria have been identified within these groups that carry out a fermentative metabolism leading to the buildup of butyrate; lactate and formate are often being produced, while acetate can be either produced or consumed (18). Butyrate is considered to exert health-promoting effects on the colon. It serves as a major energy source for the colonocytes and has also been claimed to be protective against colon cancer and inflammatory bowel diseases via effects on gene expression and cellular development (reviewed in references 36 and 51).
The pathway for butyrate formation and its respective genes have been examined in the solventogenic bacterium Clostridium acetobutylicum. Two molecules of acetyl coenzyme A (acetyl-CoA) are condensed and subsequently reduced to butyryl-CoA in a sequence of reactions that resembles the reverse β-oxidation of fatty acids (7, 11, 32). For the final step of butyrate synthesis from butyryl-CoA, two alternative pathways are known: the enzymes phosphotransbutyrylase and butyrate kinase convert butyryl-CoA to butyrate with the intermediate formation of butyryl-phosphate. This pathway, which is found in C. acetobutylicum (52), is analogous to the formation of acetate from acetyl-CoA and leads to energy generation in the form of ATP. Alternatively, a butyryl-CoA:acetate CoA-transferase transfers the CoA moiety to external acetate, which leads to the formation of acetyl-CoA and butyrate (18). The extent to which the two pathways are used by the intestinal microflora is unclear. Several reports have indicated that butyrate kinase is the route used in intestinal ecosystems (27-29). A study of several strains of Butyrivibrio fibrisolvens from the rumen showed that they exhibited either butyrate kinase or butyryl-CoA:acetate CoA-transferase activities, but not both (17). Recently, Duncan et al. (18) revealed that human strains belonging to the Roseburia and Faecalibacterium genera (19, 20) possessed butyryl-CoA:acetate CoA-transferase activity only, whereas strain L2-50, which is remotely related to a Coprococcus sp. or to Eubacterium ruminantium, exhibited activities for both pathways.
The aim of this study was to screen a wider range of phylogenetically diverse butyrate-producing bacteria from the human large intestine to determine the extent to which the two routes of butyrate formation are employed within this ecosystem. The two different methodologies used were degenerate PCR and enzymatic activities.
MATERIALS AND METHODS
Bacterial strains and growth media.
Human fecal bacteria used in this study are described in Table 1. The bacterial isolations were made from human fecal samples from one healthy adult male and three healthy adult females, all of whom were consuming an omnivorous diet and had not taken any antibiotics or other medication known to influence human colonic microflora for a period of more than 3 months. Ethical approval for this work was granted by the Grampian Research Ethics Committee (project number 00/00133). The human fecal bacterial strains were isolated on rumen fluid-based M2 medium (24) roll tubes containing a range of added carbon sources (0.5% [wt/vol]) (Table 1), on M2GSC (31), or on medium previously described for the isolation of Selenomonas species (2). Serial 10-fold dilutions were prepared from 1 g (wet weight) of freshly voided feces mixed in 9 ml of anaerobic diluent, and 0.5-ml aliquots were inoculated into roll tubes. All bacterial isolates were repurified following a second passage on roll tubes. Strains Clostridium lituseburense (DSM 797), Clostridium propionicum (DSM 1682), Clostridium sporosphaeroides (DSM 1294), and Eubacterium barkeri (DSM 1223) were obtained from the German Collection of Microorganisms and Cell Cultures. C. acetobutylicum NCIMB 8052 was obtained from the National Collection of Industrial and Marine Bacteria (United Kingdom), and E. ruminantium GA195 was obtained from John Wallace (Rowett Research Institute, Aberdeen, United Kingdom).
TABLE 1.
Human colonic isolates examined in this studya
| Strain | 16SrRNA sequence relationship (% identity BlastN) for:
|
Isolation mediumc | SCFA concn (mM) ofd:
|
Reference | ||
|---|---|---|---|---|---|---|
| Closest speciesb | Best match | But. | Ac. | |||
| A2-194 | Roseburia intestinalis (94%) | Clone HuCB37 (99%) | M2GSC | 7.3 | −3.5 | 5 |
| B16/4 | B. fibrisolvens | Closest species | NSM | 18.8 | −3.9 | 42 |
| M72/1 | Roseburia intestinalis (97%) | Clone HuCA13 (99%) | M2WS | 13.9 | −5.4 | This study |
| M6/1 | Roseburia intestinalis (97%) | Clone HuCA13 (99%) | M2GSC | 17.0 | −3.6 | This study |
| M88/1 | Roseburia intestinalis (97%) | Clone HuCA13 (99%) | M2WS | 24.0 | −8.3 | This study |
| M91/1 | Roseburia intestinalis (97%) | Clone HuCA13 (99%) | M2WS | 23.6 | −7.4 | This study |
| M101/3 | Roseburia intestinalis (97%) | Clone HuCA13 (99%) | M2CS | 18.6 | −1.6 | This study |
| A2-183 | Roseburia intestinalis (97%) | Clone adhufec225 (99%) | M2GSC | 20.0 | −8.2 | 5, 18 |
| A2-181 | Roseburia intestinalis (96%) | Strain A2-183 (98%) | M2GSC | 16.8 | −11.5 | 4, 18 |
| L1-82 | Roseburia intestinalis | Closest species | M2GSC | 18.5 | −9.1 | 4, 18, 19 |
| L1-952 | Roseburia intestinalis (99%) | Closest species | M2GSC | 21.3 | −10.0 | 5, 18, 19 |
| L1-8151 | Roseburia intestinalis | Closest species | M2GSC | 16.0 | −12.3 | 4, 18, 19 |
| M50/1 | Roseburia intestinalis (99%) | Strain L1-93 (99%) | M2GSC | 17.5 | −10.9 | This study |
| T1-815 | E. rectale (99%) | Strain A1-86 (99%) | M2GSC | 5.3 | 0.8 | 5 |
| A1-86 | E. rectale (99%) | Closest species | M2GSC | 21.7 | −9.3 | 5 |
| M104/1 | E. rectale (99%) | Strain A1-86 (99%) | M2WS | 10.8 | 1.1 | This study |
| A2-232 | C. nexile (98%) | Strain A2-231 (99%) | M2GSC | 3.8 | 2.1 | 4 |
| SL7/1 | C. nexile (98%) | Strain A2-231 (99%) | M2L | 4.2 | 2.1 | This study |
| M62/1 | C. saccharolyticum (95%) | Closest species | M2GSC | 9.7 | 4.8 | This study |
| SM4/1 | C. sp. DR7 (95%) | Closest species | M2M | 3.6 | −1.0 | This study |
| SS3/4 | C. hathewayi (91%) | Clone HuCA19 (94%) | SS | 13.7 | 10.3 | This study |
| GM2/1 | Ruminococcus gnavus (92%) | Clone HuCA19 (94%) | M2M | 10.6 | 2.3 | This study |
| SR1/1 | Ruminococcus obeum (95%) | Clone HuCB12 (97%) | SR | 5.5 | 18.6 | This study |
| SR1/5 | Ruminococcus obeum (94%) | Clone HuCB12 (97%) | SR | ND | ND | This study |
| SSC/2 | C. indolis (94%) | Clone adhufec25 (99%) | SS | 22.8 | 0.1 | This study |
| SS2/1 | C. indolis (94%) | Clone adhufec25 (99%) | SS | 9.4 | −7.0 | This study |
| L1-92 | A. caccae | Closest species | M2GSC | 20.4 | −16.9 | 5, 44 |
| SM6/1 | E. hallii (98%) | Clone p-2001-s959-5 (99%) | M2M | 14.9 | −1.4 | This study |
| L2-7 | E. hallii (96%) | CloneHuCA15 (98%) | M2GSC | 11.9 | 2.9 | 5 |
| SL6/1/1 | E. hallii (97%) | CloneHuCA15 (99%) | M2L | 11.0 | −4.4 | This study |
| ART55/1 | E. ruminantium (92%) | Clone A19 (99%) | SSL | 1.9 | 0.7 | This study |
| L2-50 | E. ruminantium (93%) | Clone A19 (96%) | M2GSC | 6.2 | 3.1 | 5, 18 |
| T2-87 | E. cylindroides (99%) | Closest species | M2GSC | 5.7 | 2.3 | 4 |
| SM7/11 | E. cylindroides (99%) | Closest species | M2M | 6.0 | 0.3 | This study |
| A2-165 | F. prausnitzii | Closest species | M2GSC | 14.4 | −7.4 | 5, 18, 20 |
| M21/2 | F. prausnitzii (99%) | Closest species | M2CS | 16.0 | −10.0 | This study |
| SL3/3 | F. prausnitzii (98%) | Strain A2-165 (98%) | M2L | 19.5 | −14.5 | This study |
| SL4/4 | F. prausnitzii (98%) | Closest species | M2L | 17.6 | −9.4 | This study |
Shown are data for 16S rRNA relationships, isolation media, and net production and utilization of butyrate and acetate.
A., Anaerostipes; B., Butyrivibrio; C., Clostridium; E., Eubacterium; F., Faecalibacterium.
M2GSC (31); M2CS, M2 (24) plus corn starch; M2WS, M2 plus wheat starch; M2M, M2 plus mannitol; M2L, M2 plus lactate; NSM (42); SS, Selenomonas selective medium; SR, Selenomonas ruminantium medium (2).
SCFA, short-chain fatty acid; But, butyrate; Ac, acetate; ND, not determined.
PCR amplification and sequencing.
PCR amplifications were performed according to standard protocols (3). Templates used for PCR were either genomic DNA purified with a Promega Wizard genomic DNA purification kit according to the manufacturer's instructions or bacterial cell pellets from 1 ml of 24-h-old cultures grown on M2GSC medium and resuspended in 50 μl of sterile water (0.5 μl per 50 μl of PCR). PCR products and DNA bands excised from agarose gels were purified with a QIAquick Kit (Qiagen) according to the manufacturer's instructions. PCR products were cloned into pGEM-T-Easy (Promega) according to the manufacturer's instructions. Sequencing was performed on a Beckman capillary sequencer. Deoxynucleotide triphosphates and DNA polymerases were from Bioline (BIOTAQ DNA polymerase for general PCRs and BIO-X-ACT DNA polymerase for genome walking). Primers (listed in Table 2) were obtained from MWG Biotech.
TABLE 2.
Primers used in this study
| Primer | Sequencea (position) |
|---|---|
| Degenerate PCR primers | |
| BUKfor1 | GTATAGATTACTIRYIATHAAYCCNGG |
| BUKrev1 | CAAGCTCRTCIACIACIACNGGRTCNAC |
| PTBfor2 | CAAATTAAAGGITGYRTIRTIGAYGGNCC |
| Genome-walking primers | |
| UFWL250buk1 | CAATCCGGGTTCAACATCTAC |
| UFWL250buk2 | GTATGTGTAGGCCGTGGTGGNNNNNNNNNN |
| UFWL250buk3 | CAACAGAGGAACTTGCAGGC |
| UFWL250buk4 | AGATCGGTAAGCAGGGACAG |
| UFWL250buk5 | CCAACCTTGGTGGTATCCTTG |
| UFWL250ptb1 | TGCTCGTTCTTGATTGCCTC |
| UFWL250ptb2 | ATGTCAGGATAACAGGAGCCNNNNNNNNNN |
| UFWL250ptb3 | CTTCAAAGCTGTCTGATCTGG |
| UFWL250ptb4 | ATACGAGGTTTCCTGCATGG |
| UFWL250ptb5 | ATCTTACGGTCTTCTGCTCC |
| Sequencing primers | |
| M13F | GGTTTTCCCAGTCACGAC |
| M13R | GGAAACAGCTATGACCATG |
| BUKfor2 | TCGTTCAGGAAGGTGATGAG |
| BUKrev2 | ATGGAATTCCCAACTCATCG |
| BUKrev3 | TCATCACCTTCCTGAACGAG |
| LuxSfor1 | CTCGCAGCTACTTATCTCAG |
| LuxSfor2 | CGAAGATCGCCTGAACTATCC |
| LuxSrev1 | CTGAGATAAGTAGCTGCGAG |
| PFSfor1 | GCGTTGTAAGCGGAGATCAG |
| PTBfor3 | CTTGACCTTGCAATCGATCC |
| PTBfor4 | TAAGGCTGTTGAGCTGGTAC |
| PTBrev2 | GTCCTGAGCACATGCTACTG |
| PTBrev4 | CCTACTTCCTTATCCAGAACG |
| SSPfor1 | GAAGCTAAGGACGCTATGAAC |
| SSPrev1 | TCATAGCGTCCTTAGCTTCC |
| SSPrev2 | TCGACATAACCCCTTCCTGC |
| 27f | AGAGTTTGATCMTGGCTCAG (8-27b) |
| rP2 | ACGGCTACCTTGTTACGACTT (1491-1511b) |
| 519f | CAGCMGCCGCGGTAATWC (518-535b) |
| 519r | GWATTACCGCGGCKGCTG (518-535b) |
| 926f | AAACTCAAAKGAATTGACGG (906-925b) |
| 926r | CCGTCAATTCMTTTRAGTTT (906-925b) |
H = A, C, or T; I = inosine; K = G or T; M = A or C; N = A, C, G, or T; R = A or G; W = A or T; Y = C or T.
Positions are numbered according to the E. coli numbering system (12).
Degenerate PCR.
Amino acid sequences of the genes ptb and buk from C. acetobutylicum, Clostridium beijerinckii, Clostridium perfringens, Bacillus subtilis, Thermotoga maritima, Listeria monocytogenes (buk only), Listeria innocua (buk only), and Enterococcus faecalis (buk only) were obtained from GenBank (8) and aligned by using ClustalW through the BCM search launcher (49). Degenerate primers were designed from those alignments with CODEHOP (41) and by visual inspection. The primers (Table 2) consisted of 15 to 20 degenerate positions at the 3′ end followed by a clamp of 10 to 15 positions based on C. acetobutylicum sequence (GenBank accession number L14744 [52]). The degeneracy was kept below 100-fold by replacing random positions (N) toward the 5′ end with inosine (I).
Amplification was performed with a ramped annealing approach whereby the annealing temperature is stepwise reduced within every cycle (48). The following conditions were used: initial denaturation (2 min at 94°C), 35 cycles of denaturation (30 s at 94°C), annealing (20 s at 55°C, 5 s at 50°C, 5 s at 45°C, 5 s at 40°C, 15 s at 35°C), and elongation (1.5 min at 72°C), with a final extension step (10 min at 72°C). PCR products were excised from agarose gels if multiple bands were encountered and purified or directly purified, cloned, and sequenced with primers M13F and M13R.
Genome walking.
Genome walking was performed using the universal fast walking approach as described previously by Myrick and Gelbart (33) using Exonuclease I from New England Biolabs with a final MgCl2 concentration of 3 mM. The primers used are listed in Table 2. The initial first-strand extension was performed for either 2 or 3.5 min, and all other incubations were performed according to the methods used by Myrick and Gelbart (33). In some cases, PCR products were purified from agarose gels and reamplified. Primers were designed from the newly obtained sequence, and the whole operon was amplified with primers PFSfor1 and SSPrev2 and sequenced with a range of primers (Table 2).
Phylogenetic analysis.
16S rRNA sequences were amplified with universal primers 27f and rP2 (54) (Table 2). Amplified PCR products were purified and directly sequenced with primers 27f, rP2, 519f, 519r, 926f, and 926r (Table 2). Two independent PCR products were sequenced per strain to avoid errors introduced by the Taq polymerase.
The similarity of the 16S rRNA sequences (minimum, 1,400 bases) from the isolates with other organisms was compared with all sequence data in GenBank by using the BLAST algorithm (1). Phylogenetic analysis was performed with the United Kingdom Human Genome Mapping Project computing services tool (www.hgmp.mrc.ac.uk). Nucleotide or amino acid sequences were aligned with ClustalW by using the MAGI interface, and phylogenetic trees were constructed with DNADIST or PROTDIST by using the PIE interface (distance matrix program neighbor, distance model Kimura, 100 times bootstrap resampling).
Enzymatic activities.
Bacterial strains were grown in 10 ml of M2GSC medium (31) for approximately 20 h, and the bacterial cell pellets were harvested by centrifugation (10,000 × g for 10 min) at 4°C and disrupted by sonication as described previously by Duncan et al. (18). Acetate kinase and butyrate kinase activities were determined by using the colorimetric method of Rose (40). Butyryl-CoA:acetate CoA-transferase activities were determined by the method described previously by Barker et al. (6) and adapted for use in a microtiter assay. Protein measurements of each extract were determined with a bicinchoninic acid protein assay reagent kit (Pierce). All activities were determined from a minimum of three independent cultures of each bacterial strain.
Fermentation product analysis.
Acidic fermentation end products were determined by capillary gas chromatography analysis (37) after 22 to 24 h of growth on M2GSC medium.
Nucleotide sequence accession numbers.
DNA sequences of 16S rDNA genes are deposited under GenBank accession numbers AY305305 to 305322 and AY350746 (see Fig. 2). The accession number for the butyrate kinase operon and adjacent genes from strain L2-50 is AY357288, and the sequences of the butyrate kinases of other strains are deposited under numbers AY357289 to AY357292 and AY487175 to 487176 (see Fig. 3).
FIG. 2.
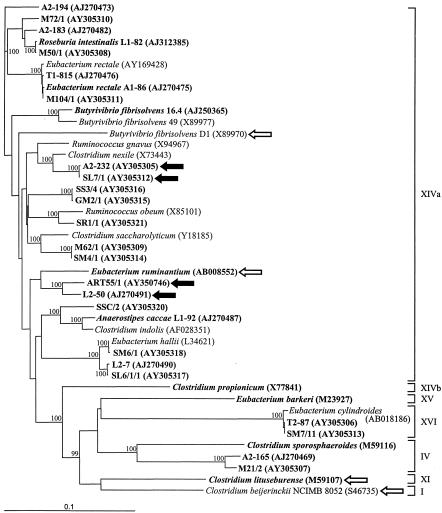
Phylogenetic tree of low mol% G+C-content gram-positive bacteria based on 16S rRNA sequence corresponding to positions 100 to 1447 of the E. coli numbering system (12). Strains examined in this study are shown in boldface, while the remaining strains served as reference sequences. Accession numbers for the sequences used are given in brackets. Bootstrap values greater than 95 (per 100 replications) are shown at branch points. Arrows indicate strains that carry the butyrate kinase pathway for butyrate formation (solid arrows, human fecal strains; open arrows, strains from other environments). Clostridial clusters (14) are indicated by roman numbers. The scale bar represents genetic distance (10 substitutions per 100 nucleotides).
FIG. 3.
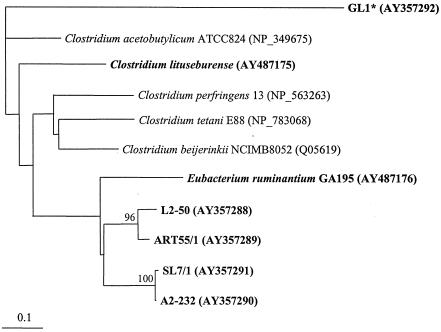
Phylogenetic tree of part of the deduced protein sequence (amino acid positions 22 to 127 of strain L2-50) of the butyrate kinase gene (buk) from bacteria examined in this study (shown in boldface) and several clostridial sequences (13, 34, 35, 47). Accession numbers for the sequences used are given in brackets. Bootstrap values greater than 95 (per 100 replications) are shown at branch points. The scale bar represents genetic distance (10 substitutions per 100 nucleotides). *, no corresponding isolate available.
RESULTS
Identification of butyrate kinase pathway genes from strain L2-50.
The two genes (ptb and buk) of the butyrate kinase pathway enzymes, phosphotransbutyrylase and butyrate kinase, form an operon in clostridia. Degenerate primers for both genes were designed by using ClustalW alignments of deduced amino acid sequences of the respective genes from database entries. Genomic DNA prepared from the human colonic strain L2-50, which is known to harbor enzyme activity for butyrate kinase (18), was used as a template for PCR with the degenerate primers. A strong product of the expected size with primers PTBfor2 and BUKrev1 was obtained by using a ramped annealing amplification cycle. The PCR product was cloned and sequenced. A BLASTX search of related sequences revealed high similarity to the ptb-buk operon from Clostridium species. The whole operon and adjacent genes were obtained from strain L2-50 by using the genome-walking method. The deduced amino acid sequences of the genes from strain L2-50 showed high similarity to their first match in a BLASTP search (Fig. 1). An alignment of the deduced protein sequences encoded by the ptb and buk genes from strain L2-50 with the phosphotransbutyrylase and phosphotransacetylase, as well as butyrate kinase and acetate kinase sequences, respectively, of C. perfringens strain 13 and C. acetobutylicum ATCC 824 confirmed that the L2-50 sequences were more closely related to the butyrate pathway genes (data not shown).
FIG. 1.
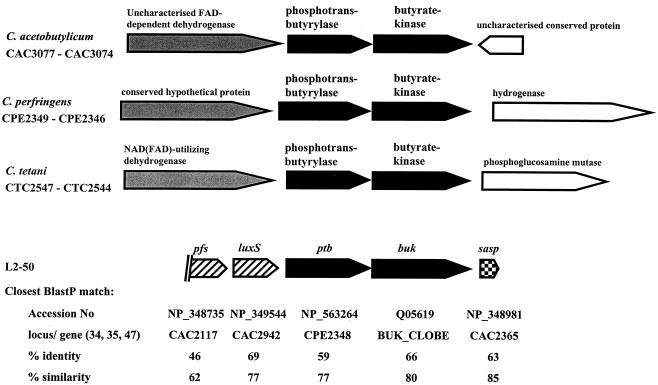
Arrangement of the genes for phosphotransbutyrylase and butyrate kinase (ptb and buk) and neighboring genes in C. acetobutylicum ATCC 924 (34), C. perfringens 13 (47), Clostridium tetani E88 (13), and strain L2-50. For the genes of strain L2-50, the closest relatives and their identity and similarity (shown as percentages) according to a BLASTP search are also shown. FAD, flavin adenine dinucleotide.
The gene arrangement upstream of ptb is conserved in the three clostridia whose genomes have been sequenced but is not conserved in strain L2-50, in which a homologue of the gene luxS is present instead of a dehydrogenase gene (Fig. 1). The next open reading frame upstream of luxS showed similarity to a S-adenosylhomocysteine nucleosidase (Fig. 1). These two gene products are known to be involved in methyl group metabolism in bacteria by converting S-adenosylhomocysteine, the toxic product of the methylation reaction (with S-adenosylmethionine as methyl donor), to homocysteine (55). LuxS has also been proposed to have a role in bacterial cell-to-cell communication (quorum sensing), as one of the reaction products is the precursor of an autoinducer molecule (AI-2) (43). The luxS gene is widespread among various gram-positive and gram-negative bacteria (30, 55). Downstream of buk in strain L2-50 lies a short open reading frame with high homology to a small acid-soluble spore protein (45). Since spores have not been observed in this organism, the function of this gene is not clear.
Screening of butyrate-producing isolates for presence of the butyrate kinase pathway.
In order to establish how widespread the butyrate kinase pathway is among the human colonic microflora, a wide range of butyrate-producing human gut bacterial isolates (Table 1) were screened for the presence of genes ptb and buk. The study included 38 strains, of which 15 had been isolated previously from four donors (4, 5, 42), in addition to 23 new isolates from fecal samples from four further donors in order to ensure good coverage of the phylogenetic diversity within this group of bacteria. The 16S rRNA genes were sequenced, and a phylogenetic tree was constructed to show the phylogenetic relationship of the different isolates (Fig. 2). All strains were related to the low mol% G+C-content gram-positive anaerobic bacteria and fell into clusters IV, XIVa, and XVI (14). The isolates were subjected to degenerate PCR targeted against the butyrate kinase operon. The primer set PTBfor2 and BUKrev1 gave bands of various intensity in the expected size range for several bacteria (data not shown). Bands from representatives of the different phylogenetic groups were cloned and sequenced. Only the PCR products from E. ruminantium-like strains (L2-50 and ART55/1) and the Clostridium nexile-like strains (A2-232 and SL7/1) were confirmed to possess the ptb and buk genes.
Degenerate PCRs performed with primers BUKfor1 and BUKrev1 on all 38 isolates gave a band of the correct size only in the E. ruminantium-like strains (L2-50 and ART55/1) and the C. nexile-like strains (A2-232 and SL7/1), a finding which is in accordance with the results we obtained with the primer set PTBfor2 and BUKrev1 (data not shown).
Several bacteria isolated from other environments that belong to different clostridial clusters (14) and are reported to produce butyrate were also screened for the presence of the butyrate kinase pathway with both degenerate primer pairs. No PCR product was obtained for C. propionicum (cluster XIVb), C. sporosphaeroides (cluster IV), and E. barkeri (cluster XV). C. lituseburense (cluster XI) and E. ruminantium (cluster XIVa), however, gave bands of the expected size. The sequences obtained from those PCR products corresponded to the butyrate kinase/phosphotransbutyrylase genes (data not shown).
The deduced protein sequences of the amplified region of the buk gene were aligned with the respective sequences of four known clostridial sequences, and a phylogenetic tree was constructed (Fig. 3). The sequences from the human colonic isolates group into two pairs (as in the phylogenetic tree based on their 16S rRNA sequences) and cluster together with the ruminal isolate E. ruminantium, the closest relative of strains L2-50 and ART55/1 based on 16S rRNA sequence. A fifth human colonic sequence related to ptb and buk was obtained from isolate GL1, but this strain could not be maintained in culture. The recovery of this divergent buk gene does, however, demonstrate that the degenerate primer combinations were able to amplify a wide range of ptb- and buk-related genes. The function of the GL1 gene could not be established, of course, and it should be noted that certain kinases involved in the metabolism of branched-chain fatty acids in various bacteria have sequences that are closely related to butyrate kinases (16, 53).
Screening for enzymatic activities.
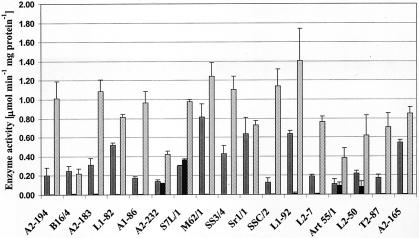
Seventeen human butyrate-producing strains representing different phylogenetic groups (according to the phylogenetic tree based on 16S rRNA sequences [Fig. 2]) were also tested for their butyrate kinase and acetate kinase activities. All strains displayed acetate kinase activity, whereas only E. ruminantium-like strains (L2-50 and ART55/1) and C. nexile-like strains (A2-232 and SL7/1) displayed significant levels of butyrate kinase activity (Fig. 4). This finding therefore corresponds with the distribution of the butyrate kinase gene as detected by the degenerate PCR approach. Butyryl-CoA:acetate CoA-transferase activity was detected in all isolates examined, including the ones displaying butyrate kinase activity (Fig. 4). The butyryl-CoA:acetate CoA-transferase activity measured in this study was higher than that observed previously (18), a finding reflecting better optimization of the assay. In the present study, all four strains that produced butyrate kinase were found to be net acetate producers. Of the strains that possessed butyryl-CoA:acetate CoA-transferase, but not butyrate kinase activity, approximately two-thirds (23 out of 34) were net acetate utilizers, while the remaining third were net acetate producers, under the growth conditions examined herein (Table 1).
FIG. 4.
Enzyme activities for acetate kinase (solid gray bars), butyrate kinase (black bars), and butyryl-CoA:acetate CoA-transferase (light gray bars) from human colon bacteria. The butyryl-CoA:acetate CoA-transferase values are divided by 1,000.
DISCUSSION
The strains studied here were isolated essentially nonselectively from fecal samples provided by eight different individuals over a period of 10 years. The samples represent a broad range of bacterial diversity within the clostridial clusters IV, XIVa, and XVI, including at least four as yet undescribed species (strains A2-194, SS3/4 and GM2/1, L2-50 and ART55/1, and SSC/2 and SS2/1, the sequence identity of which to known species is less than 95%) from this ecosystem. The results indicate that the butyrate kinase operon is not at all widespread among human butyrate-producing bacteria recovered from human feces. Significant enzymatic activity and amplifiable gene sequences corresponding to butyrate kinase could be detected only in 4 out of 38 butyrate-producing strains tested. Interestingly, these four strains fall into only two branches in the phylogenetic tree defined by 16S rRNA sequences, a finding which strongly supports the view that butyrate kinase has a limited distribution in the microflora of the human colon. In C. acetobutylicum ATCC 824, a second butyrate kinase isoenzyme has been described (26), and the question of whether our screening approach could have missed such a gene arises. The fact that we obtained a sequence that was only weakly related to the ptb-buk operon (Fig. 3, GL1) supports the assumption that our degenerate primers do amplify diverse members of this gene family. Furthermore, our enzymatic data fully support the results obtained with the degenerate PCR approach. We therefore conclude that it is unlikely that strains that were negative in our screen possess the butyrate kinase pathway.
A phylogenetic tree of the butyrate kinase sequences shows a similar branching structure to the one based on 16S rRNA sequence. Sequences of the new human colonic species diverge from those known for other clostridia (clostridial clusters I and XI) but cluster together with the more closely related ruminal strain, E. ruminantium (cluster XIVa). This arrangement appears more consistent with a progressive loss of butyrate kinase from some clostridial lineages rather than with its acquisition by a few species from other bacteria. The significance of the butyrate kinase pathway in butyrate-producing bacteria from other environments remains to be established. Of the five strains examined within this study, only two seemed to carry the butyrate kinase gene, one originating from the rumen (E. ruminantium) and one isolated from soil (C. lituseburense). Some bacteria isolated from soil, however, might have their main habitat within the mammalian gut and merely survive in soil—for example as spores. The different fermentation pathways present in different butyrate producers (for example, the presence or absence of solventogenesis) might also play a role.
Activity for the enzyme butyryl-CoA:acetate CoA-transferase was detected in all the bacteria tested, including those possessing the butyrate kinase pathway. This finding is in contrast to a study on rumen B. fibrisolvens strains, which exhibited enzyme activities for either butyrate kinase or butyryl-CoA:acetate CoA-transferase but not both (17). The CoA-transferase pathway appears to be most important for butyrate formation in the human colonic ecosystem. Besides butyryl-CoA, the enzyme needs acetate as a substrate. Acetate is usually present in the colonic environment at high concentrations (30 to 80 mM), as it is formed as a fermentation product by many different bacteria. The pH in the colon, especially the proximal region where the fermentation activity is presumably highest due to the availability of nutrients entering from the ileum, has been reported to be slightly acidic (10, 15). At a pH of 6, about 5% of the acetate pool, which amounts to a several-millimolar concentration, is protonated and is therefore expected to enter the cell readily by diffusion across the membrane. This mechanism may help to account for the prevalence of the CoA-transferase route for butyrate synthesis in human colonic bacteria. If the internal pH of these bacteria is in the neutral range, a supposition which remains to be shown at this stage, acetate would be expected to accumulate within the cells to much higher concentrations than those present externally, as observed in Escherichia coli (39). The usage of the CoA-transferase route under those circumstances could even be seen as a mechanism to detoxify excess acetate. However, further studies are necessary to investigate this aspect of the butyrate metabolism in more detail.
The existence of some human colonic species that possess both mechanisms for butyrate synthesis, however, also requires explanation. So far, all isolates we have found that are capable of net uptake of acetate during growth lack the butyrate kinase gene, but further investigations of metabolic regulation, energy formation, and hydrogen disposal in these little-studied organisms are called for before their fermentative metabolism can be properly interpreted. It seems likely that these physiologically distinct types of butyrate-producing bacteria will be found to occupy different ecological niches within the human gut.
Acknowledgments
This work was supported by the Scottish Executive Environment and Rural Affairs Department.
M. Jackson thanks CSIRO Division of Health Sciences and Nutrition for supporting her on a visit to RRI as part of the Chief Study Award Program. We thank Pauline Young, Donna Henderson, and Gill Campbell for automated DNA sequencing, and we thank John Wallace and Nest McCain for providing strain Eubacterium ruminantium GA195.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1997. In L. C. Parks (ed.), Handbook of microbiological media, p. 1245. CRC Press, Cleveland, Ohio.
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1998. Current protocols in molecular biology. John Wiley & Sons Inc., New York, N.Y.
- 4.Barcenilla, A. 1999. Ph.D. thesis. Robert Gordon University, Aberdeen, United Kingdom.
- 5.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, H. A., E. R. Stadtman, and A. Kronberg. 1955. Coenzyme A transferase from Clostridium kluyveri. Methods Enzymol. 1:599-600. [Google Scholar]
- 7.Bennett, G. N., and F. B. Rudolph. 1995. The central metabolic pathway from acetyl-CoA to butyryl-CoA in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:241-249. [Google Scholar]
- 8.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2003. GenBank. Nucleic Acids Res. 31:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaut, M., M. D. Collins, G. W. Welling, J. Doré, J. van Loo, and W. de Vos. 2002. Molecular biological methods for studying the gut microbiota: the EU human gut flora project. Br. J. Nutr. 87:S203-S211. [DOI] [PubMed] [Google Scholar]
- 10.Bown, R. L., J. A. Gibson, G. E. Sladen, B. Hicks, and A. M. Dawson. 1974. Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut 15:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boynton, Z. L., G. N. Bennett, and F. B. Rudolph. 1996. Cloning, sequencing, and expression of clustered genes encoding β-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J. Bacteriol. 178:3015-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 15.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. E. Naylor, and G. T. Macfarlane. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debarbouille, M., R. Gardan, M. Arnaud, and G. Rapoport. 1999. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diez-Gonzalez, F., D. R. Bond, E. Jennings, and J. B. Russell. 1999. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch. Microbiol. 171:324-330. [DOI] [PubMed] [Google Scholar]
- 18.Duncan, S. H., A. Barcenilla, C. S. Stewart, S. E. Pryde, and H. J. Flint. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan, S. H., G. L. Hold, A. Barcenilla, C. S. Stewart, and H. J. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. E vol. Microbiol. 52:1615-1620. [DOI] [PubMed] [Google Scholar]
- 20.Duncan, S. H., G. L. Hold, H. J. M. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium gen. nov., comb. nov. Int. J. Syst. E vol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 21.Frank, D. N., and N. R. Pace. 2001. Molecular-phylogenetic analyses of human gastrointestinal microbiota. Curr. Opin. Gastroenterol. 17:52-57. [DOI] [PubMed] [Google Scholar]
- 22.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 24.Hobson, P. N. 1969. Rumen bacteria. Methods Microbiol. 3B:133-149. [Google Scholar]
- 25.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 26.Huang, K.-X., S. Huang, F. B. Rudolph, and G. N. Bennett. 2000. Identification and characterization of a second butyrate kinase from Clostridium acetobutylicum ATCC 824. J. Mol. Biotechnol. 2:33-38. [PubMed] [Google Scholar]
- 27.Macfarlane, G. T., and G. R. Gibson. 1997. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine, p. 269-318. In R. I. Mackie and B. A. White (ed.), Gastrointestinal microbiology, vol. 1. Chapman & Hall, New York, N.Y. [Google Scholar]
- 28.Miller, T. L., and S. E. Jenesel. 1979. Enzymology of butyrate formation by Butyrivibrio fibrisolvens. J. Bacteriol. 138:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, T. L., and M. J. Wolin. 1979. Fermentations by saccharolytic intestinal bacteria. Am. J. Clin. Nutr. 32:164-172. [DOI] [PubMed] [Google Scholar]
- 30.Mitsumori, M., L. Xu, H. Kajikawa, M. Kurihara, K. Tajima, J. Hai, and A. Takenaka. 2003. Possible quorum sensing in the rumen microbial community: detection of quorum-sensing signal molecules from rumen bacteria. FEMS Microbiol. Lett. 219:47-52. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. bovis) B14. Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 32.Mullany, P., C. L. Clayton, M. J. Pallen, S. Slone, and A. al-Saleh. 1994. Genes encoding homologues of three consecutive enzymes in the butyrate/butanol-producing pathway of Clostridium acetobutylicum are clustered on the Clostridium difficile chromosome. FEMS Microbiol. Lett. 124:61-68. [DOI] [PubMed] [Google Scholar]
- 33.Myrick, K. V., and W. M. Gelbart. 2002. Universal fast walking for direct and versatile determination of flanking sequence. Gene 284:125-131. [DOI] [PubMed] [Google Scholar]
- 34.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Markarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oultram, J. D., I. D. Burr, M. J. Elmore, and N. P. Minton. 1993. Cloning and sequence analysis of the genes encoding phosphotransbutyrylase and butyrate kinase from Clostridium acetobutylicum NCIMB 8052. Gene 131:107-112. [DOI] [PubMed] [Google Scholar]
- 36.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 37.Richardson, A. J., G. C. Calder, C. S. Stewart, and A. Smith. 1989. Simultaneous determination of volatile and non-volatile fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 9:5-8. [Google Scholar]
- 38.Rigottier-Gois, L., A. G. Le Bourhis, G. Gramet, V. Rochet, and J. Doré. 2003. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 43:237-245. [DOI] [PubMed] [Google Scholar]
- 39.Roe, A. J., D. McLaggan, I. Davidson, C. O'Byrne, and I. R. Booth. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, I. A. 1955. Acetate kinase of bacteria. Methods Enzymol. 1:591-595. [Google Scholar]
- 41.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rumney, C. J., S. H. Duncan, C. Henderson, and C. S. Stewart. 1995. Isolation and characteristics of a wheat bran-degrading Butyrivibrio from human faeces. Lett. Appl. Microbiol. 20:232-236. [DOI] [PubMed] [Google Scholar]
- 43.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 44.Schwiertz, A., G. L. Hold, S. H. Duncan, B. Gruhl, M. D. Collins, P. A. Lawson, H. J. Flint, and M. Blaut. 2002. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 25:46-51. [DOI] [PubMed] [Google Scholar]
- 45.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 46.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Doré. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skantar, A. M., and L. K. Carta. 2000. Amplification of Hsp90 homologs from plant-parasitic nematodes using degenerate primers and ramped annealing PCR. BioTechniques 29:1182-1185. [DOI] [PubMed] [Google Scholar]
- 49.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davison, and K. C. Worley. 1996. BCM search launcher—an integrated interface to molecular biology data base search and analysis services available on the world wide web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 50.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wächtershäuser, A., and J. Stein. 2000. Rationale for the luminal provision of butyrate in intestinal diseases. Eur. J. Nutr. 39:164-171. [DOI] [PubMed] [Google Scholar]
- 52.Walter, K. A., R. V. Nair, J. W. Cary, and G. N. Bennett. 1993. Sequence and arrangement of two genes of the butyrate-synthesis pathway of Clostridium acetobutylicum ATCC 824. Gene 134:107-111. [DOI] [PubMed] [Google Scholar]
- 53.Ward, D. E., P. Ross, C. C. van der Weijden, J. L. Snoep, and A. Claiborne. 1999. Catabolism of branched-chain α-keto acids in Enterococcus faecalis: the bkd gene cluster, enzymes, and metabolic route. J. Bacteriol. 181:5433-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. G. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]