Abstract
In our efforts to identify the components participating in electron transport to nitrogenase in Rhodospirillum rubrum, we used mini-Tn5 mutagenesis followed by metronidazole selection. One of the mutants isolated, SNT-1, exhibited a decreased growth rate and about 25% of the in vivo nitrogenase activity compared to the wild-type values. The in vitro nitrogenase activity was essentially wild type, indicating that the mutation affects electron transport to nitrogenase. Sequencing showed that the Tn5 insertion is located in a region with a high level of similarity to fixC, and extended sequencing revealed additional putative fix genes, in the order fixABCX. Complementation of SNT-1 with the whole fix gene cluster in trans restored wild-type nitrogenase activity and growth. Using Western blotting, we demonstrated that expression of fixA and fixB occurs only under conditions under which nitrogenase also is expressed. SNT-1 was further shown to produce larger amounts of both ribulose 1,5-bisphosphate carboxylase/oxgenase and polyhydroxy alkanoates than the wild type, indicating that the redox status is affected in this mutant. Using Western blotting, we found that FixA and FixB are soluble proteins, whereas FixC most likely is a transmembrane protein. We propose that the fixABCX genes encode a membrane protein complex that plays a central role in electron transfer to nitrogenase in R. rubrum. Furthermore, we suggest that FixC is the link between nitrogen fixation and the proton motive force generated in the photosynthetic reactions.
Biological nitrogen fixation catalyzed by nitrogenase is an energy-demanding process, requiring both ATP and low-potential reducing equivalents. The metabolic routes leading to the reduction of nitrogenase have been clearly identified and characterized only in Klebsiella pneumoniae (35, 47). In this bacterium the anaerobic oxidation of pyruvate is linked to nitrogenase activity through the action of a pyruvate-flavodoxin oxidoreductase (the nifJ gene product) and a soluble flavodoxin (the nifF gene product). In Anabaena a similar system has been shown to be active only under iron-limiting conditions (3).
In a number of diazotrophic bacteria the proton motive force (PMF) has been proposed to be involved in generating reductant for nitrogenase (29). In the phototrophic nonsulfur bacterium Rhodobacter capsulatus a set of nitrogen-regulated genes, the rnf genes, as shown by mutational studies, encode proteins involved in electron transfer to nitrogenase (25, 30, 42, 45). The membrane protein complex encoded by the rnf genes shows similarity to sodium-dependent NADH oxidoreductases and is thought to drive the NADH-dependent reduction of ferredoxin I (the fdxN gene product) by utilizing the PMF derived from the photosynthetic activities of the cell. In Rhodospirillum rubrum energization of the chromatophore membrane has been shown to be important for the reduction of nitrogenase (33), and generation of reductant has been proposed to be dependent on the tricarboxylic acid cycle (5).
In some diazotrophic bacteria a set of genes designated fixABCX has been proposed to encode proteins involved in electron transfer to nitrogenase (9, 13, 16, 26). Mutational studies of the fixABCX genes revealed a Nod+ Fix− phenotype in members of the Rhizobiaceae (20). FixAB exhibits similarity to the electron transfer flavoprotein (ETF), and FixCX exhibits similarity to the ETF ubiquinone oxidoreductase (ETF-QO) involved in electron transfer from certain oxidative mitochondrial processes (e.g., β-oxidation) to the Q pool in the inner membrane (2, 50, 53). However, there is no experimental evidence that the proteins encoded by fixABCX are directly involved in electron transfer to nitrogenase. One study with Azorhizobium caulinodans suggested that the fixABCX gene products are involved in maturation of dinitrogenase reductase rather than electron transfer to nitrogenase (26). The complete pathway for electron transfer to nitrogenase in members of the Rhizobiaceae and other diazotrophic bacteria containing fixABCX is not known, but most diazotrophic bacteria containing these genes are dependent on a functional respiratory chain for efficient diazotrophic growth, and it has been proposed that the PMF is involved in driving the reduction of nitrogenase (29). In addition, if FixC, like ETF-QO (11), is an integral membrane protein, it is possible that a protein complex encoded by the fixABCX operon is the long-sought link between the PMF and nitrogen fixation.
In the phototrophic nonsulfur bacterium R. rubrum the pathway for electron transfer to nitrogenase is poorly understood. A nifJ-like gene encoding a pyruvate-ferredoxin oxidoreductase has been identified (6, 32). This pyruvate-ferredoxin oxidoreductase has been shown to support nitrogenase activity in vitro, linking the oxidation of pyruvate to the reduction of nitrogenase by using partially purified ferredoxin fractions (6). Studies of a knockout mutant strain revealed no phenotype with respect to nitrogen fixation, indicating that the nifJ-like gene product is not likely to be part of the major pathway for electron transfer to nitrogenase in R. rubrum (32). Similar results have been reported for the nifJ gene from R. capsulatus (54).
Metronidazole is a specific inhibitor of electron transfer to nitrogenase in a number of diazotrophic organisms (28, 41, 49). This drug is specifically reduced by low-potential electron donors, such as ferredoxins and flavodoxins, and the reduced derivatives are toxic to cells (44). Previous reports have shown that metronidazole enrichment for electron transport mutants can be successful (21, 46).
In this communication we report the use of a random mutagenesis strategy in which a transposable element and then metronidazole enrichment for electron transfer mutants were used. Selection was carried out under diazotrophic conditions in an attempt to identify novel genes encoding proteins involved in electron transfer to nitrogenase in R. rubrum. We describe identification of a fixABCX cluster in R. rubrum and partial characterization of a mini-Tn5 fixC mutant isolated after metronidazole enrichment for mutants affected in nitrogen fixation. Our results show that the fixC gene product most likely is involved in electron transfer to nitrogenase in R. rubrum.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Plasmid-harboring strains and insertion mutants were routinely grown with antibiotics at the following final concentrations: ampicillin, 50 μg/ml for Escherichia coli strains; kanamycin, 50 μg/ml for E. coli strains and 12.5 μg/ml for R. rubrum mutant strains; and tetracycline, 15 μg/ml for E. coli strains and 5 μg/ml for R. rubrum strains.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | Host for pGEM-7z(−) derivatives | 19 |
| One Shot TOP10 | Host for pCR-Blunt II-TOPO derivatives | Invitrogen |
| S17-1 | RP4:2-Tc:Mu:Km Tn7, Tpr Smr | 48 |
| S17-1 (λ-pir) | RP4:2-Tc:Mu:Km Tn7, λ-pir, Tpr Smr | 8 |
| BL21 Star (DE3) | Host for pET101/d-TOPO | Invitrogen |
| R. rubrum strains | ||
| S1 | Wild type | |
| SNT-1 | S1 fixC::mini-Tn5, Kmr | This study |
| Plasmids | ||
| pGEM-7z(−) | Cloning vector, Apr | Promega |
| pUTKm | Mini-Tn5 source, mob, λ-pir oriR, Kmr Apr | 8 |
| pRKD 418 | Broad-host-range plasmid, mob, Tcr Smr | 36 |
| pCR-Blunt II-TOPO | Blunt PCR cloning vector, Kmr | Invitrogen |
| pET101/d-TOPO | Overexpression vector, Apr | Invitrogen |
| pSNF 4.8 | pGEM-7z(−) derivative carrying a 4.8-kb fixC::mini-Tn5 fragment, Apr Kmr | This study |
| pSNF 19.4 | pGEM-7z(−) derivative carrying a 19.4-kb fixC::mini-Tn5 fragment, Apr Kmr | This study |
| pRKDfix | pRKD 418 derivative containing nifW fixABCX, Tcr | This study |
| pFIXop | pCR-Blunt II-TOPO derivative containing nifW fixABCX, Kmr | This study |
| pFIXC | pCR-BluntII-TOPO derivative containing fixC, Kmr | This study |
| pETfixC | pET101/D-TOPO derivative giving recombinant FixC, Apr | This study |
Growth conditions.
R. rubrum strains were routinely grown photoheterotrophically at 30°C in the minimal medium described by Ormerod et al. (40) with 40 mM malate and 28 mM NH4Cl as the carbon and nitrogen sources, respectively (N+). Diazotrophic growth was achieved by sparging nitrogen-free medium aseptically with 95% N2-5% CO2 (N−) (31). nif-derepressing conditions were also achieved by growing R. rubrum with 27 mM glutamate as the nitrogen source. E. coli strains were grown aerobically in Luria-Bertani medium at 37°C. Cell growth was monitored spectrophotometrically by measuring the absorbance at 600 nm. Routine photoheterotrophic growth of strains containing pRKDfix was carried out behind a red filter to reduce the phototoxicity of tetracycline (36).
Protein techniques.
Whole-cell nitrogenase activity was measured by transferring 2 ml of nitrogen-fixing R. rubrum into a 24.5-ml anaerobic vial. Two milliliters of acetylene was added, and nitrogenase activity was measured after 15 min of incubation at 30°C in the light by the acetylene reduction assay (33). Protein concentrations were determined by the Lowry protein concentration assay (34). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blot analyses were done as described previously (51). The enhanced chemiluminescence method (Amersham Biosciences) was used to visualize the blots. The antibodies used were raised against R. rubrum ribulose 1,5-biphosphate carboxylase/oxygenase (RubisCO) (a kind gift from R. Tabita), overexpressed A. caulinodans FixA and FixB (kindly provided by E. Ruston and N. Watmoug), E. coli leader peptidase (Lep) (kindly provided by J. W. deGier), and R. rubrum dinitrogenase reductase. In addition, antibodies against the V5 epitope (Invitrogen) were used. To determine the modification state of the nitrogenase reductase, a low-cross-linking SDS-PAGE system was used (27). Whole-cell SDS-PAGE samples were prepared as described by Grundwald et al. (14).
Cell fractionation and in vitro nitrogenase activity measurements.
R. rubrum cell extracts were prepared by washing diazotrophically grown cells once in 100 mM Tris-HCl (pH 7.6)-2 mM dithionite. The cells were resuspended in 300 mM Tris-HCl (pH 7.6)-1 mM dithionite-1 mM 1,4-dithiothreitol and were broken by passage through a Ribi cell fractionator (Sorvall). The lysate was centrifuged for 10 min at 10,000 × g to sediment unbroken cells and cell debris. The soluble and chromatophore fractions were separated by centrifugation for 90 min at 100,000 × g. The chromatophore fraction was then washed in the same buffer containing 0.5 M NaCl. In vitro nitrogenase activities in cell extracts were determined by the acetylene reduction assay with an ATP-regenerating system in the presence of Mn2+ and with dithionite as the electron donor (39).
Inhibition of nitrogenase activity by metronidazole.
Metronidazole inhibition of R. rubrum nitrogenase activity was measured in the same way as whole-cell activity except that metronidazole in dimethyl sulfoxide was added prior to the addition of acetylene. Nitrogenase activity was monitored for 30 min by taking samples every 5 min. After the nitrogenase activity was measured, the modification state of dinitrogenase reductase was assayed by SDS-PAGE and Western blot analysis.
Mini-Tn5 mutagenesis and metronidazole selection of mini-Tn5 mutants with reduced nitrogen fixation.
pUTKm (8) was transferred to R. rubrum S1 by aerobic diparential mating on nitrocellulose membranes by using pUTKm-harboring strain S17-1 (λ-pir) as the donor strain. After 12 h of conjugation at 30°C on SMN plates (N+ medium supplemented with 0.3% Casamino Acids and 0.3% yeast extract), cells were resuspended in minimal N+ medium. A dilution series was prepared, and the different dilutions were plated on minimal N+ plates with and without kanamycin and were grown photoheterotrophically for 5 days in anaerobic containers (Oxoid). The mini-Tn5 insertion frequency was calculated by counting the colonies on plates with and without kanamycin.
Transfer of pUTKm into R. rubrum for metronidazole selection of mutants affected in electron transfer to nitrogenase was carried out as described above. After 12 h of conjugation at 30°C on SMN plates, the nitrocellulose membranes were cut into smaller pieces to enhance the probability of recovering different mutants. The cells were resuspended in anaerobic N+ medium containing kanamycin and allowed to recover for 10 h in the light. After the photosynthetic apparatus had been induced, cells were harvested and resuspended in anaerobic N− medium and then sparged with argon for 6 h to induce nitrogen fixation. After induction of nitrogen fixation, metronidazole in dimethyl sulfoxide was added at final concentrations of 10, 15, and 20 mM; after this the cells were incubated for 6 h. After metronidazole selection, the cells were washed in N+ medium, plated on kanamycin-containing N+ plates, and grown photoheterotrophically in anaerobic containers (Oxoid).
Cloning and sequencing of fixABCX from R. rubrum.
Genomic DNA from a mini-Tn5 mutant affected in nitrogen fixation, designated SNT-1, was isolated by using standard phenol-chloroform isolation techniques (43). Southern blot analysis of genomic DNA from SNT-1 digested with ApaI revealed a 4.8-kb fragment hybridizing with the aminoglycoside 3′-phosphotransferase gene from the mini-Tn5 element. Genomic fragments of the corresponding size resulting from ApaI digestion of the mutant genome were purified by using QIAEX II (Qiagen) and ligated into pGEM-7z(−) (Promega). Positive clones were selected by plating transformed E. coli DH5α on plates containing ampicillin and kanamycin. The resulting plasmid, designated pSNF 4.8, was sequenced by using an ABI PRISM II dye terminator cycle sequencing kit (Applied Biosystems) with primers starting in the plasmid. Another 19.5-kb genomic fragment containing the aminoglycoside 3′-phosphotransferase gene was isolated in the same way by MfeI digestion of SNT-1 genomic DNA and ligated into EcoRI-digested pGEM-7z(−), yielding pSNF19.5. Once the mini-Tn5 sequence was obtained, primers flanking the mini-Tn5 insertion site were designed. The corresponding DNA fragment was amplified from wild-type R. rubrum genomic DNA by high-fidelity PCR by using Pfu DNA polymerase (Stratagene), and the product was cloned into pCR-zero blunt II (Invitrogen), which resulted in pFIXC. The insert was sequenced in order to determine the sequence at the mini-Tn5 insertion. Two other mutants, SNT-2 and SNT-3, were analyzed by the same procedure.
To complement SNT-1 in trans, a fragment starting 811 bp upstream of the fixA start codon and ending downstream of the fixX stop codon was amplified from a wild-type genomic DNA preparation by a high-fidelity PCR. The product was subcloned in pCR-zero blunt II (Invitrogen), which resulted in pFIXop. The whole fragment was purified after partial EcoRI digestion of pFIXop and ligated into EcoRI-digested pRKD 418 (36). The final construct, designated pRKDfix, was transferred into SNT-1 and S1 by diparental mating by using E. coli S17-1 as the donor strain.
Switch-off of mini-Tn5 mutant.
NH4+ switch-off experiments were carried out with 5 ml of cells in anaerobic 50-ml vials containing 10% acetylene; 0.3 mM NH4Cl was added after 20 min, and nitrogenase activity was measured by determining the amount of accumulated ethylene until the activity was recovered. Light switch-off experiments were carried out by using 40 ml of cells in a 50-ml anaerobic vial. Darkness was achieved by wrapping the vial in two layers of aluminum foil. Nitrogenase activity was measured by the acetylene reduction assay by using 2 ml of cells that was withdrawn from the 40-ml culture, transferred to an anaerobic 24.5-ml vial containing 10% acetylene, and incubated in the light at 30°C for 2 min. Aliquots were removed, and the modification state of the dinitrogenase reductase was analyzed by SDS-PAGE and Western blotting.
Cellular localization of FixC.
To investigate the cellular localization of FixC, fixC lacking the stop codon was PCR amplified by using Pfu-Taq DNA polymerase (Stratagene) with pRKDfix as the template and was cloned in pET101/D-TOPO (Invitrogen), which resulted in fixC fused to a C-terminal V5 epitope and a six-histidine tag. Primers were designed according to the suppliers' recommendations. The final construct, designated pETfixC, was transformed into BL21 Star (DE3) cells (Invitrogen), and 500-ml cultures were grown at 30°C. When cultures were induced, they were incubated with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. Cells were harvested by centrifugation for 15 min at 3,000 × g and washed in 25 ml of 50 mM Tris-HCl (pH 7.5), resuspended in 25 ml of 50 mM Tris-HCl (pH 7.5) containing 20 μg of DNase per ml, and broken by passage through a Ribi cell fractionator (Sorvall). The crude extract was centrifuged twice at 2,500 × g for 10 min to sediment unbroken cells and cell debris. The soluble and membrane fractions were prepared from the cell extract by centrifugation at 100,000 × g for 1 h. The membrane fraction was resuspended in 12 ml of 50 mM Tris-HCl (pH 7.5) and subjected to washing with Na2CO3 as described by Molloy et al. (37) by adding 34 ml of ice-cold 0.1 M Na2Co3 (pH 11) to 6 ml of resuspended membranes and stirring the preparation on ice for 1 h. After the carbonate wash the soluble and membrane fractions were separated by centrifugation at 100,000 × g for 1 h. The cellular localization of FixC and E. coli Lep was determined by SDS-PAGE and Western blot analysis.
Northern blot analysis.
Total RNA samples were prepared from R. rubrum by using an RN-easy midi kit (Qiagen). Ten micrograms of total RNA was separated by denaturing formamide agarose gel electrophoresis and transferred to a Hybond XL membrane (Amersham Biosciences) by using standard alkaline transfer techniques (43). The membrane was hybridized in Rapid Hyb buffer (Amersham Biosciences) with a purified Redi-prime II (Amersham Biosciences)-labeled fixABCX probe at 65°C for 2.5 h. The blot was washed twice for 20 min in a high-stringency buffer (0.1× SSC-0.5% SDS [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at 65°C. The blot was developed by exposure overnight on a phosphorimager plate.
RESULTS
Metronidazole inhibition of nitrogenase activity in R. rubrum.
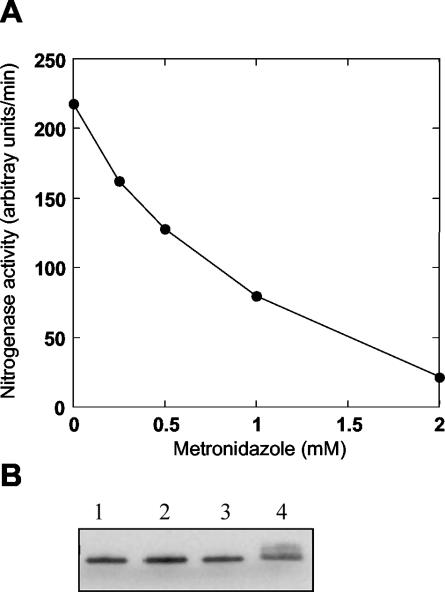
Metronidazole has previously been shown to be specifically reduced by low-potential electron donors, such as ferredoxins and flavodoxins (44), and to inhibit nitrogenase activity in other diazotrophic bacteria (28, 41, 49). The molecular mechanisms underlying this inhibition activity are largely unknown, but metronidazole probably competes for the low-potential reductant required in nitrogen fixation. We therefore investigated the effect of metronidazole on nitrogenase activity in R. rubrum in order to establish if this compound could be used for efficient selection of mutants deficient in electron transfer to nitrogenase. As shown in Fig. 1, metronidazole inhibited in vivo nitrogenase activity in R. rubrum in a concentration-dependent manner, with 50% inhibition observed at a concentration of 0.8 mM. The inhibition was not due to switch-off by ADP ribosylation, as determined by SDS-PAGE and Western blot analysis (Fig. 1B).
FIG. 1.
Metronidazole inhibition of nitrogenase activity in R. rubrum. (A) Inhibition of R. rubrum nitrogenase activity by metronidazole. (B) SDS-PAGE and Western blot analysis, showing the modification state of dinitrogenase reductase after metronidazole addition. Whole-cell samples were taken after 30 min and separated on a low-cross-link SDS-10% PAGE gel. Lane 1, no metronidazole; lane 2, 0.5 mM metronidazole; lane 3, 1 mM metronidazole; lane 4, switch-off control with 0.25 mM NH4Cl added.
Mini-Tn5 mutagenesis and metronidazole enrichment of mini-Tn5 mutants deficient in electron transfer to nitrogenase.
The efficient inhibition of nitrogenase activity in R. rubrum by metronidazole indicated that this compound might be a good selection agent for enrichment of mutants deficient in electron transport to nitrogenase. A random mutagenesis strategy in which a transposable element was used would have several advantages compared to chemical mutagenesis since the element itself would simplify identification of the point of mutation in mutants having the desired characteristics. Tn5 has previously been shown to be inserted into the R. rubrum genome (10), and we used a mini-Tn5 transposable element from pUTKm that has been characterized previously (8). This minitransposon was inserted at a frequency of 2.5 × 10−4 into the R. rubrum genome, which is similar to the frequency previously reported for Tn5 insertion (10). The mini-Tn5 element was inserted into the R. rubrum genome as a single copy, and the insertion was random, as demonstrated by Southern blot analysis of recovered mutants (results not shown).
In the metronidazole enrichment three different metronidazole concentrations were used, and there was a negative correlation between the number of viable cells and the amount of metronidazole; 44, 34, and 14 colonies were recovered after selection with 10, 15, and 20 mM metronidazole, respectively. Three of the mutants recovered after the selection were clearly affected in nitrogenase activity in vivo. Chromosomal mini-Tn5-containing DNA fragments from the mutants were cloned and sequenced. In one of the mutants the sequence at and around the site of the mini-Tn5 element insertion exhibited a high level of similarity to fixABCX genes found in several diazotrophs. This mutant was designated SNT-1 and characterized further. The localization of insertion in the other two mutants was determined to be sequences in the R. rubrum genome draft (http://genome.ornl.gov/microbial/rrub/) similar to a putative hyfF for SNT-2 and to the sequence for a conserved putative membrane protein for SNT-3.
Sequence analysis of R. rubrum fixABCX.
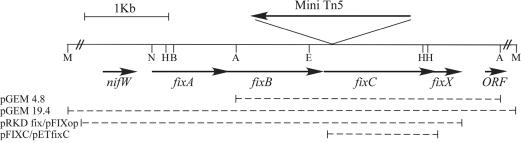
The physical map of fixABCX in R. rubrum is shown in Fig. 2, and the GenBank accession number of the whole sequence is AY362827.
FIG. 2.
Physical map of the fixABCX region of R. rubrum. Coding regions and directions of transcription are indicated by arrows. The flanking MfeI sites are not to scale, as indicated by broken lines. The dashed lines below the coding regions represent fragments cloned in the plasmids indicated to the left. Restriction site abbreviations: A, ApaI; B, BamHI; E, EcoRI; H, HindIII; M, MfeI; N, NsiI.
Sequencing of pSNF4.8 showed that the mini-Tn5 element was inserted in the 5′ region of fixC (Fig. 2). Four potential open reading frames (ORFs) were found to correspond to fixA, fixB, fixC, and fixX, in that order. An ORF exhibiting a high level of identity to nifW was identified 201 bp upstream of the fixA start codon, and an ORF exhibiting identity to the N terminus of a proton-transporting ATPase was found 291 bp downstream of the fixX stop codon. No typical NifA or σ54 binding sites were found within the fixABCX cluster or the nifW-fixA intergenic region.
At the protein level the ORFs exhibited the highest levels of identity with the following proteins: for NifW, 46% identity to Azotobacter vinelandii NifW; for FixA, 72% identity to Bradyrhizobium japonicum FixA; for FixB, 72% identity to A. caulinodans FixB; for FixC, 63% identity to B. japonicum FixC; and for FixX, 61% identity to Gluconoacetobacter diazotrophicus FixX. All ORFs are preceded by putative Shine-Delgarno sequences. The fixX start codon overlaps the fixC stop codon, an organization also seen in the G. diazotrophicus, Azospirillum brasilense, and E. coli fixABCX operons. The region was analyzed for potential stem-loops at the RNA level since overlapping start and stop codons have been shown to be a mechanism for translational coupling in the nifLA operon of K. pneumoniae (12), but no obvious stem-loop structure was detected.
fixA and fixB were predicted to encode soluble proteins that have molecular masses of 30.2 kDa (292 amino acids) and 39.2 kDa (368 amino acids), respectively, and exhibit identity with ETF-β and ETF-α, respectively, including a conserved flavin adenine dinucleotide-binding motif (53). fixC was predicted to encode a 47.8-kDa protein (433 amino acids) that exhibits identity with the N-terminal portion of ETF-QO, including a putative N-terminal dinucleotide-binding motif (11). fixX was predicted to encode a 10.7-kDa soluble protein (100 amino acids) that exhibits identity with the C-terminal portion of ETF-QO, including a conserved ferredoxin-like iron-sulfur cluster-binding cysteine motif. Although the sequence of the N terminus of R. rubrum FixC is similar to that of Rhizobium meliloti FixC, which has been proposed to contain a cleavable N-terminal signal peptide (9), it was not predicted to contain a cleavable signal peptide when SignalP V2.0 (http://www.cbs.dtu.dk/services/SignalP-1.1/) was used. Furthermore, the putative dinucleotide-binding motif located in the N-terminal portion of R. rubrum FixC makes cleavage unlikely. In fact, when different FixC proteins and ETF-QO were compared, this putative dinucleotide-binding site was one of the most conserved regions (results not shown).
When the FixC amino acid sequence was analyzed for possible transmembrane helices by using a number of prediction programs, no well-defined helices were found. Depending on the program and hydrophobicity scale used, zero to nine putative helices were found. Two regions in R. rubrum FixC that could be transmembrane regions were also found in other FixC proteins. It should be noted that human ETF-QO, which has been shown to be an integral membrane protein (11), also showed inconclusive results when three different prediction programs were used (11).
Expression and cellular localization of FixA, FixB, and FixC.
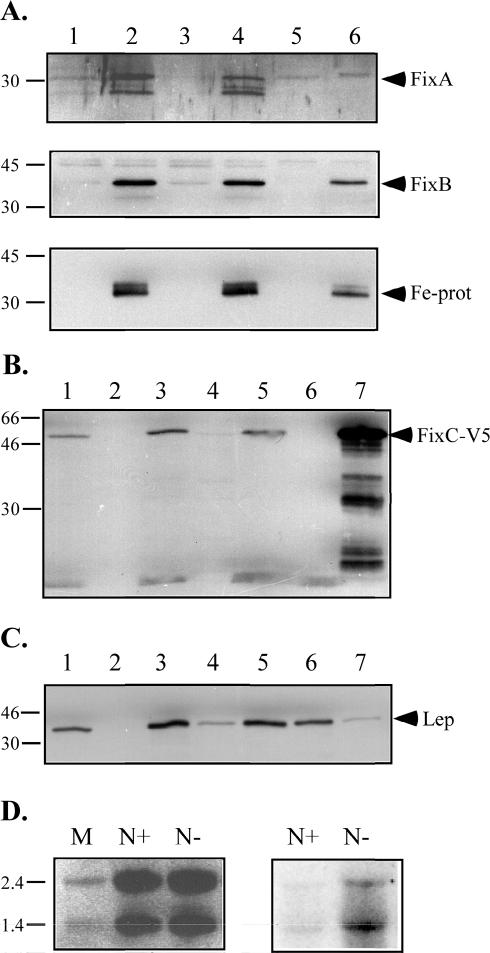
Western blot analyses of cell extracts from R. rubrum grown with NH4+ and diazotrophically are shown in Fig. 3A. FixA and FixB were shown to be present in soluble extracts from nitrogen-fixing cells at the predicted molecular masses. The nif-dependent expression was verified by Northern blot analyses of total RNA from R. rubrum by using radiolabeled fixABCX as the probe. Two transcripts were detected, at ∼2.6 and ∼1.4 kb, respectively, which together corresponded to the entire fixABCX cluster (Fig. 3D). The transcripts were strongly induced during diazotrophic growth of R. rubrum. Both FixA and FixB were found predominantly in the cytosolic fraction of R. rubrum, as expected for soluble proteins.
FIG. 3.
Expression and cellular localization of FixA, FixB, and FixC. Numbers on the left are molecular masses in kilodaltons (A through C) and lengths in kilobases (D). (A) SDS-PAGE and Western blot analysis of R. rubrum extracts, showing cellular localization and expression of FixA (top panel), FixB (middle panel), and dinitrogenase reductase (bottom panel). Three micrograms of total protein was loaded in each lane and separated on an SDS—12% PAGE gel for the FixA and FixB blots, while 1 μg of total protein was loaded for the dinitrogenase reductase blot. Lane 1, N+ extract; lane 2, N− extract; lane 3, N+ cytosolic fraction; lane 4, N− cytosolic fraction; lane 5, N+ chromatophore fraction; lane 6, N− chromatophore fraction. Fe-prot, dinitrogen reductase. (B) SDS-PAGE and Western blot analysis of E. coli extracts, showing cellular localization of recombinant FixC. Five micrograms of total protein from different fractions from uninduced BL21 Star (DE3) harboring pETfixC was loaded in each lane. Lane 1, cell extract; lane 2, soluble fraction; lane 3, membrane fraction; lane 4, soluble fraction after Na2CO3 wash; lane 5, membrane fraction after Na2CO3 wash; lane 6, membrane fraction of BL21 Star (DE) without plasmid (corresponding to lane 3); lane 7, membrane fraction of induced cells (0.5 μg). (C) SDS-PAGE and Western blot analysis of E. coli extracts, showing cellular localization of Lep. The contents of the lanes were the same as those in panel B, except that one-half as much protein was used. (D) Northern blot analysis of 10 μg of R. rubrum total RNA separated on a 1.5% denaturing formamide gel. Left panel, loading control showing ethidium bromide staining of the blot; right panel, autoradiography of the blot hybridized with radiolabeled fixABCX.
Since no antibodies against FixC were available, the Invitrogen pET101/D-TOPO system was used in an attempt to determine the cellular localization of FixC in E. coli. This system adds a C-terminal histidine tag preceded by a V5 epitope to the protein of interest, and antibodies against the epitope can be used to detect the fusion protein. Inducing the system with 0.5 mM IPTG for 2 h resulted in strong induction of the 51-kDa fusion protein and a number of smaller products cross-reacting with the V5 epitope antibodies (Fig. 3B, lane 7). Uninduced BL21 Star (DE3) harboring pETfixC contained a detectable amount of the full-length fusion protein. Fractionation of uninduced BL21 Star (DE3) harboring pETfixC followed by SDS-PAGE and Western blot analysis showed that recombinant FixC was recovered in the membrane fraction (Fig. 3B, lane 3). FixC remained in the membrane fraction after the carbonate wash (Fig. 3B, lane 5). There was no cross-reaction with the V5 epitope antibody in the FixC area in either induced or uninduced BL21 Star (DE3) lacking pETfixC (Fig. 3B, lane 6). The cellular localization of E. coli Lep, a well-characterized transmembrane protein, was the same as that of recombinant FixC throughout this process (Fig. 3C).
Growth and nitrogenase activity.
Growth rates and nitrogenase activities of SNT-1 are summarized in Table 2. Growth under nitrogen-surplus conditions was unaffected, while diazotrophic growth was clearly reduced. Under diazotrophic conditions the mutant had 23% of the nitrogenase activity of the wild type in vivo, and the growth of the mutant was approximately three times slower than the growth of the wild type. Interestingly, the nitrogenase protein levels were essentially the same in the mutant and the wild type, as determined by Western blot analysis (Fig. 4B). Furthermore and importantly, the in vitro nitrogenase activity in cell extracts was the wild-type the same as that in extracts. SNT-1 growth on glutamate was not affected even though the in vivo nitrogenase activity remained low, indicating that maximal utilization of the nitrogenase present is not required for growth on glutamate.
TABLE 2.
Growth and nitrogenase activities of SNT-1 and wild-type R. rubrum grown on different nitrogen sources
| Strain (plasmid) | Nitrogen source | Doubling time (h) | Nitrogenase activity (nmol h−1 mg−1)
|
|
|---|---|---|---|---|
| In vivo | In vitro | |||
| S1 | N2 | 10.3 | 2,405 ± 425 | 252 ± 23 |
| SNT-1 | N2 | 29.7 | 515 ± 95 | 358 ± 26 |
| S1 | NH4+ | 8.4 | ||
| SNT-1 | NH4+ | 8.7 | ||
| S1 | Glutamate | 9.4 | 1,759 ± 68 | 324 ± 23 |
| SNT-1 | Glutamate | 9.0 | 806 ± 13 | 208 ± 18 |
| SNT-1 (pSNCfix) | N2 | 13.5 | 2,253 ± 222 | 503 ± 26 |
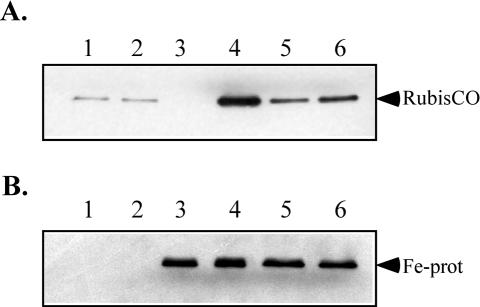
FIG. 4.
SDS-PAGE and Western blot analysis of R. rubrum wild-type strain S1 and SNT-1 whole-cell extracts, showing RubisCO levels (A) and dinitrogenase reductase levels (B) under different growth conditions. Lane 1, NH4+-grown S1; lane 2, NH4+-grown SNT-1; lane 3, diazotrophically grown S1; lane 4 diazotrophically grown SNT-1; lane 5, glutamate-grown S1; lane 6, glutamate-grown SNT-1. One microgram of total protein was loaded in each lane and separated on an SDS—12% PAGE gel.
The correlations between optical density and protein concentration were different for diazotrophically grown SNT-1 and wild-type R. rubrum; the mutant had a lower protein concentration at a given optical density. We suggest that this was due to increased deposition of poly-(β-hydroxyalkanoates) (PHA), which accounted for at least 10% of the dry weight as determined after crude purification by the extraction method described by Brandl et al. (4). The PHA were recovered as a massive white pellet after low-speed centrifugation of cell extracts. Diazotrophically grown SNT-1 was also shown to induce increased amounts of RubisCO, as demonstrated by Western blot analysis (Fig. 4A), suggesting that the redox status of the cell is markedly affected during diazotrophic growth. Reconstitution of SNT-1 with pRKDfix in trans restored the wild-type growth rates and activities (Table 2), showing that the nitrogen-source-dependent phenotype of SNT-1 was due to the mini-Tn5 mutagenesis and not to some other mutation acquired during the metronidazole selection process.
Nitrogenase switch-off experiments.
Since the fixC mutant was severely affected in nitrogen fixation, the switch-off response of the mutant was studied. Halbleib and coworkers have previously shown that oxidized dinitrogenase reductase is the preferred substrate for dinitrogenase reductase ADP ribosyltransferase (DRAT), whereas the reduced form is the preferred substrate for dinitrogenase reductase-activating glycohydrolase (DRAG) (17, 18). Glutamate-grown SNT-1 showed a normal switch-off response to ammonia and light, while the response of diazotrophically grown SNT-1 was variable; sometimes diazotrophically grown SNT-1 showed a normal switch-off response, while at other times little or no switch-off was observed. In general, it is difficult to assess the significance of these observations since the mutant had much lower in vivo nitrogenase activity, which can be assumed to lead to a situation in which cells experienced nitrogen starvation. It has previously been shown that the switch-off effect is lost under nitrogen starvation conditions (38), and these growth conditions have in fact been used to isolate unmodified, active dinitrogenase reductase.
DISCUSSION
Metronidazole inhibition of R. rubrum nitrogenase activity in vivo is similar to the inhibition reported previously for R. capsulatus and A. vinelandii (28, 41) and is not due to switch-off catalyzed by DRAT and DRAG. The only other effector reported to date that inhibits nitrogenase activity in R. rubrum without involving the DRAT-DRAG system is methylviologen (27). Like metronidazole, methylviologen can act as a low-potential electron acceptor, opening the possibility that the inhibitory effect is an effect on electron transfer to nitrogenase per se. Metronidazole has been shown to be specific for reduced flavodoxins and ferredoxins (44), supporting the hypothesis that they have a role as electron donors to nitrogenase in R. rubrum. In this study we showed that random mini-Tn5 mutagenesis followed by metronidazole selection for electron-transfer-deficient mutants is a potent tool to study low-potential redox reactions in R. rubrum.This method provides positive selection for all potential mutations, leading to less reduced metronidazole under the conditions studied. Since reduced metronidazole is mutagenic, caution should be used when mutants are characterized after metronidazole selection and workers should make sure that the phenotype is not derived from a mutation acquired through the selection process.
The complete sequence of the R. rubrum fixABCX cluster is available in the recently presented R. rubrum genome draft (http://genome.ornl.gov/microbial/rrub/). The sequence in the draft is identical to the sequence which we report here; in fact, we did not continue our sequencing efforts when the draft became available. In this study we found that expression of the fixABCX cluster in R. rubrum is regulated according to the nitrogen status of the cell, as determined by Northern and Western blot analyses, although no typical nif regulatory elements directly upstream of the fixABCX gene cluster were identified. All fixABCX operons identified in other diazotrophic bacteria to date are preceded by their own nif promoters. In the R. rubrum draft sequence, the fix genes are in a nif cluster whose organization is similar to that in G. diazotrophicus (i.e., orf6 nifU nifV nifw fixABCX) (31). When this region was analyzed for nif promoter-like sequences, a potential σ54 binding sequence preceded by a NifA binding site was found upstream of the conserved ORF, from which nitrogen-dependent transcription of fixABCX of R. rubrum could start. A fixABCX transcript starting here should be ∼6 kb, while a transcript starting in the 201-bp nifW-fixA intergenic region should be ∼3.6 kb. The fixABCX transcripts detected were ∼2.6 and ∼1.4 kb. One possible explanation for the two transcripts could be processing of a large, complete fixABCX transcript. The transcription and expression of the fixABCX genes in B. japonicum were shown to have peculiar characteristics (15, 16), and mRNA processing of a fixABCX transcript has previously been proposed for A. caulinodans (1). Furthermore, mRNA processing of a glnBA transcript has previously been reported to occur in R. rubrum (22).
SNT-1 is severely affected in diazotrophic growth and in vivo nitrogenase activity compared to the wild type. The deposition of PHA seen in diazotrophically grown SNT-1 indicates that the redox status of the cells is affected, as PHA have been shown to be deposited in R. rubrum under unbalanced growth conditions (4). The high level of induction of RubisCO seen in diazotrophically grown SNT-1 also indicates that the redox status of the cells is altered, as RubisCO is known to act as an electron sink during photoheterotrophic growth (23, 24, 52). Nitrogenase normally plays this role during diazotrophic growth, explaining the low RubisCO levels detected in diazotrophically grown wild-type R. rubrum (Fig. 4A). Expression of nif genes has been reported when an R. rubrum RubisCO mutant is grown under normal photoheterotrophic growth conditions in the presence of NH4+ (23); this has been proposed to be due to the RegA/RegB global regulatory system, which up-regulates nitrogenase as an alternative electron sink (23, 52). It is possible that the high levels of RubisCO in diazotrophically grown SNT-1 are due to a similar system. Interestingly, the growth rate of SNT-1 was not affected when the organism was grown with glutamate as a nitrogen source. No alterations in the RubisCO levels were detected, indicating that the level of nitrogenase activity detected (30% of the wild-type level) was sufficient to support glutamate growth (i.e., to relieve the reductant pressure). The normal growth rate on glutamate also indicates that diazotrophically grown SNT-1 is nitrogen limited, since decreased nitrogenase activity did not affect the growth rate with glutamate as the nitrogen source.
FixABCX can be assumed to be part of a pathway for electron transfer to nitrogenase in R. rubrum, as shown by the low in vivo nitrogenase activities compared to the in vitro nitrogenase activities, which were the same as in the wild type. An electron transfer role for FixABCX has previously been proposed, but there is no experimental evidence for this proposal. Rather, the few reports to date point to involvement of the FixABCX gene products in maturation of dinitrogenase reductase in A. caulinodans (25). The possibility that the FixABCX gene products play this role in R. rubrum can be excluded, since the in vitro nitrogenase activity was not reduced in SNT-1.
The FixABCX proteins have also been proposed to be involved in utilization of alternative carbon sources to meet the increased demand for ATP during nitrogen fixation (53). Again, this is not likely to be the case for R. rubrum since ATP is not limiting in photoheterotrophically grown R. rubrum, in which the photosynthetic activities of the cell meet the ATP demand of nitrogen fixation. The basis for the metronidazole selection used to identify SNT-1 also supports the hypothesis that the FixABCX proteins are involved in electron transfer to nitrogenase in R. rubrum.
SNT-1 has approximately 25% of the nitrogenase activity of the wild type during diazotrophic growth, indicating that FixC is not absolutely required for electron transfer to nitrogenase. The remaining activity is probably due to other metabolic processes that generate reduced low-potential ferredoxins as electron donors to nitrogenase (e.g., the NifJ-like protein [6]), which most likely are identical to the pyruvate-feredoxin oxidoreductase described previously (7) or the α-ketoglutarate-ferredoxin oxidoreductase present in R. rubrum (7).
In conclusion, we propose that the FixABCX proteins constitute an electron transfer pathway dedicated to the generation of reductant for nitrogenase in R. rubrum. In this context the cellular localization of FixC is of central importance. Our results show that R. rubrum FixC expressed in E. coli was found exclusively in the membrane. Although not conclusive, these results suggest that FixC, by analogy with the mammalian ETF-QO protein, is a membrane protein. Previous studies have shown that the PMF is important for electron transport to nitrogenase in R. rubrum and the FixABCX system might be the link between nitrogen fixation and the photosynthetic activities. In this model the nature of FixC is crucial, and further studies are needed to determine the cellular localization and topology of FixC in R. rubrum. It is tempting to propose a PMF-driven process in which the Fix complex catalyzes the reduction of a soluble low-potential electron donor (e.g., ferredoxin) with electrons from reduced quinone in the chromatophores or from a soluble FixAB complex. Such a model implies that the Fix system should be coupled, i.e., translocate protons to drive the reduction of a soluble electron donor. The identity of such a soluble electron donor is not known in R. rubrum, but a low-potential ferredoxin (ferredoxin I) has previously been shown to donate electrons to nitrogenase at high rates in a reconstituted system with illuminated thylakoid membranes (55).
This is the first demonstration of fixABCX genes and of the involvement of the gene products in nitrogen fixation in a phototrophic bacterium. The R. rubrum system may well be an excellent model to determine in detail the role(s) of the FixABCX proteins in nitrogenase activity and nitrogen fixation.
Acknowledgments
We are grateful to J. Janson for providing the mini-Tn5 system. Antibodies were kindly given to us by R. F. Tabita (antibody against RubisCO from R. rubrum), J. W. deGier (antibody against E. coli Lep), and by N. Watmough (antibody against FixA and FixB from A. caulinodans).
This study was supported by grants to S.N. from the Swedish Research Council.
REFERENCES
- 1.Arigoni, F., P. A. Kaminski, J. Celli, and C. Elmerich. 1992. Transcriptional analysis of the fixABCXORF1 region of Azorhizobium caulinodans suggests post-transcriptional processing of the fixABCXORF1 mRNA. Mol. Gen. Genet. 235:422-431. [DOI] [PubMed] [Google Scholar]
- 2.Arigoni, F., P. A. Kaminski, H. Hennecke, and C. Elmerich. 1991. Nucleotide sequence of the fixABC region of Azorhizobium caulinodans ORS571: similarity of the fixB product with eukaryotic flavoproteins, characterization of fixX, and identification of nifW. Mol. Gen. Genet. 225:514-520. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, C. C., L. Scappino, and R. Haselkorn. 1993. Growth of the cyanobacterium Anabaena on molecular nitrogen: nifJ is required when iron is limited. Proc. Natl. Acad. Sci. 90:8812-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl, H., E. J. Knee, Jr., R. C. Fuller, R. A. Gross, and R. W. Lenz. 1989. Ability of the phototrophic bacterium Rhodospirillum rubrum to produce various poly(beta-hydroxyalkanoates): potential sources for biodegradable polyesters. Int. J. Biol. Macromol. 11:49-55. [DOI] [PubMed] [Google Scholar]
- 5.Brostedt, E., A. Lindblad, J. Jansson, and S. Nordlund. 1997. Electron transport to nitrogenase in Rhodospirillum rubrum: the role of NAD(P)H as electron donor and the effect of fluoroacetate on nitrogenase activity. FEMS Microbiol. Lett. 150:263-267. [Google Scholar]
- 6.Brostedt, E., and S. Nordlund. 1991. Purification and partial characterization of a pyruvate oxidoreductase from the photosynthetic bacterium Rhodospirillum rubrum grown under nitrogen-fixing conditions. Biochem. J. 279:155-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan, B. B., M. C. Evans, and D. I. Arnon. 1967. Ferredoxin-dependent carbon assimilation in Rhodospirillum rubrum. Arch. Mikrobiol. 59:32-40. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earl, C. D., C. W. Ronson, and F. M. Ausubel. 1987. Genetic and structural analysis of the Rhizobium meliloti fixA, fixB, fixC, and fixX genes. J. Bacteriol. 169:1127-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, R., D. J. Elder, R. Saegesser, D. J. Kelly, and R. Bachofen. 1994. An improved procedure and new vectors for transposon Tn5 mutagenesis of the phototrophic bacterium Rhodospirillum rubrum. Gene 150:97-100. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, S. I., K. M. Axtell, L. A. Bindoff, S. E. Beard, R. E. Gill, and F. E. Frerman. 1994. Molecular cloning and expression of a cDNA encoding human electron transfer flavoprotein-ubiquinone oxidoreductase. Eur. J. Biochem. 219:277-286. [DOI] [PubMed] [Google Scholar]
- 12.Govantes, F., E. Andujar, and E. Santero. 1998. Mechanism of translational coupling in the nifLA operon of Klebsiella pneumoniae. EMBO J. 17:2368-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gronger, P., S. S. Manian, H. Reilander, M. O'Connell, U. B. Priefer, and A. Puhler. 1987. Organization and partial sequence of a DNA region of the Rhizobium leguminosarum symbiotic plasmid pRL6JI containing the genes fixABC, nifA, nifB and a novel open reading frame. Nucleic Acids Res. 15:31-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunwald, S. K., D. P. Lies, G. P. Roberts, and P. W. Ludden. 1995. Posttranslational regulation of nitrogenase in Rhodospirillum rubrum strains overexpressing the regulatory enzymes dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase activating glycohydrolase. J. Bacteriol. 177:628-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gubler, M., and H. Hennecke. 1988. Regulation of the fixA gene and the fixBC operon in Bradyrhizobium japonicum. J. Bacteriol. 170:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubler, M., T. Zurcher, and H. Hennecke. 1989. The Bradyrhizobium japonicum fixBCX operon: identification of fixX and of a 5′ mRNA region affecting the level of the fixBCX transcript. Mol. Microbiol. 3:141-148. [DOI] [PubMed] [Google Scholar]
- 17.Halbleib, C. M., Y. Zhang, and P. W. Ludden. 2000. Regulation of dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase by a redox-dependent conformational change of nitrogenase Fe protein. J. Biol. Chem. 275:3493-3500. [DOI] [PubMed] [Google Scholar]
- 18.Halbleib, C. M., Y. Zhang, G. P. Roberts, and P. W. Ludden. 2000. Effects of perturbations of the nitrogenase electron transfer chain on reversible ADP-ribosylation of nitrogenase Fe-protein in Klebsiella pneumoniae strains bearing the Rhodospirillum rubrum dra operon. J. Bacteriol. 182:3681-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In M. Clover (ed.), DNA cloning, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 20.Hirsch, A. M., and C. A. Smith. 1987. Effects of Rhizobium meliloti nif and fix mutants on alfalfa root nodule development. J. Bacteriol. 169:1137-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, C. Z., and D. C. Yoch. 1990. Complementation of a pleiotropic Nif-Gln regulatory mutant of Rhodospirillum rubrum by a previously unrecognized Azotobacter vinelandii regulatory locus. Arch. Microbiol. 154:528-535. [DOI] [PubMed] [Google Scholar]
- 22.Johansson, M., and S. Nordlund. 1996. Transcription of the glnB and glnA genes in the photosynthetic bacterium Rhodospirillum rubrum. Microbiology 142:1265-1272. [DOI] [PubMed] [Google Scholar]
- 23.Joshi, H. M., and F. R. Tabita. 1996. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc. Natl. Acad. Sci. 93:14515-14520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi, H. M., and F. R. Tabita. 2000. Induction of carbon monoxide dehydrogenase to facilitate redox balancing in a ribulose bisphosphate carboxylase/oxygenase-deficient mutant strain of Rhodospirillum rubrum. Arch. Microbiol. 173:193-199. [DOI] [PubMed] [Google Scholar]
- 25.Jouanneau, Y., H. S. Jeong, N. Hugo, C. Meyer, and J. C. Willison. 1998. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus—characterization of two membrane-bound iron-sulfur proteins. Eur. J. Biochem. 251:54-64. [DOI] [PubMed] [Google Scholar]
- 26.Kaminski, P. A., F. Norel, N. Desnoues, A. Kush, G. Salzano, and C. Elmerich. 1988. Characterization of the fixABC region of Azorhizobium caulinodans ORS571 and identification of a new nitrogen fixation gene. Mol. Gen. Genet. 214:496-502. [DOI] [PubMed] [Google Scholar]
- 27.Kanemoto, R. H., and P. W. Ludden. 1984. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J. Bacteriol. 158:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley, B. C., and D. J. Nicholas. 1981. Inhibition of nitrogenase activity by metronidazole in Rhodopseudomonas capsulata. Arch. Microbiol. 129:344-348. [DOI] [PubMed] [Google Scholar]
- 29.Klugkist, J., H. Haaker, and C. Veeger. 1986. Studies on the mechanism of electron transport to nitrogenase in Azotobacter vinelandii. Eur. J. Biochem. 155:41-46. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai, H., T. Fujiwara, H. Matsubara, and K. Saeki. 1997. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry 36:5509-5521. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S., A. Reth, D. Meletzus, M. Sevilla, and C. Kennedy. 2000. Characterization of a major cluster of nif, fix, and associated genes in a sugarcane endophyte, Acetobacter diazotrophicus. J. Bacteriol. 182:7088-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindblad, A., J. Jansson, E. Brostedt, M. Johansson, U. Hellman, and S. Nordlund. 1996. Identification and sequence of a nifJ-like gene in Rhodospirillum rubrum: partial characterization of a mutant unaffected in nitrogen fixation. Mol. Microbiol. 20:559-568. [DOI] [PubMed] [Google Scholar]
- 33.Lindblad, A., and S. Nordlund. 1997. Electron transport to nitrogenase in Rhodospirillum rubrum: role of energization of the chromatophore membrane. Photosynth. Res. 53:23-28. [Google Scholar]
- 34.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 35.Ludden, P. W. 1991. Energetics and sources of energy for biological nitrogen fixation. Curr. Top. Bioenerg. 16:369-390. [Google Scholar]
- 36.Mather, M. W., L. M. McReynolds, and C. A. Yu. 1995. An enhanced broad-host-range vector for gram-negative bacteria: avoiding tetracycline phototoxicity during the growth of photosynthetic bacteria. Gene 156:85-88. [DOI] [PubMed] [Google Scholar]
- 37.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 38.Nordlund, S. 2000. Regulation of nitrogenase activity in phototrophic bacteria by reversible covalent modification, p. 149-163. In E. W. Triplett (ed.), Prokaryotic nitrogen fixation: a model system for analysis of a biological process. Horizon Scientific Press, Wymondham, United Kingdom.
- 39.Nordlund, S., and A. Noren. 1984. Dependence on divalent cations of the activation of inactive Fe-protein of nitrogenase from Rhodospirillum rubrum. Biochim. Biophys. Acta 791:21-27. [Google Scholar]
- 40.Ormerod, J. G., K. S. Ormerod, and H. Gest. 1961. Light-dependent utilization of organic compounds and photoproduction of molecular hydrogen by photosynthetic bacteria; relationships with nitrogen metabolism. Arch. Biochem. Biophys. 94:449-463. [DOI] [PubMed] [Google Scholar]
- 41.Peterson, J. B. 1988. Metronidazole inhibition of nitrogenase activity in Azotobacter vinelandii. Biochim. Biophys. Acta 964:354-360. [Google Scholar]
- 42.Saeki, K., K. Tokuda, T. Fujiwara, and H. Matsubara. 1993. Nucleotide sequence and genetic analysis of the region essential for functional expression of the gene for ferredoxin I, fdxN, in Rhodobacter capsulatus: sharing of one upstream activator sequence in opposite directions by two operons related to nitrogen fixation. Plant Cell Physiol. 34:185-199. [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Samuelson, J. 1999. Why metronidazole is active against both bacteria and parasites. Antimicrob. Agents Chemother. 43:1533-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmehl, M., A. Jahn, A. Meyer zu Vilsendorf, S. Hennecke, B. Masepohl, M. Schuppler, M. Marxer, J. Oelze, and W. Klipp. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241:602-615. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, G. W., K. S. Matlin, and N. H. Chua. 1977. A rapid procedure for selective enrichment of photosynthetic electron transport mutants. Proc. Natl. Acad. Sci. 74:610-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah, V. K., G. Stacey, and W. J. Brill. 1983. Electron transport to nitrogenase. Purification and characterization of pyruvate:flavodoxin oxidoreductase. The nifJ gene product. J. Biol. Chem. 258:12064-12068. [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 49.Tetley, R. M., and N. I. Bishop. 1979. The differential action of metronidazole on nitrogen fixation, hydrogen metabolism, photosynthesis and respiration in Anabaena and Scenedesmus. Biochim. Biophys. Acta 546:43-53. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, M. H., and M. H. Saier, Jr. 1995. Phylogenetic characterization of the ubiquitous electron transfer flavoprotein families ETF-alpha and ETF-beta. Res. Microbiol. 146:397-404. [DOI] [PubMed] [Google Scholar]
- 51.Ureta, A., and S. Nordlund. 2002. Evidence for conformational protection of nitrogenase against in Gluconacetobacter diazotrophicus by a putative FeSII protein. J. Bacteriol. 184:5805-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, X., H. V. Modak, and F. R. Tabita. 1993. Photolithoautotrophic growth and control of CO2 fixation in Rhodobacter sphaeroides and Rhodospirillum rubrum in the absence of ribulose bisphosphate carboxylase-oxygenase. J. Bacteriol. 175:7109-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weidenhaupt, M., P. Rossi, C. Beck, H. M. Fischer, and H. Hennecke. 1996. Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch. Microbiol. 165:169-178. [DOI] [PubMed] [Google Scholar]
- 54.Yakunin, A. F., and P. C. Hallenbeck. 1998. Purification and characterization of pyruvate oxidoreductase from the photosynthetic bacterium Rhodobacter capsulatus. Biochim. Biophys. Acta 1409:39-49. [DOI] [PubMed] [Google Scholar]
- 55.Yoch, D. C., and D. I. Arnon. 1975. Comparison of two ferredoxins from Rhodospirillum rubrum as electron carriers for the native nitrogenase. J. Bacteriol. 121:743-745. [DOI] [PMC free article] [PubMed] [Google Scholar]