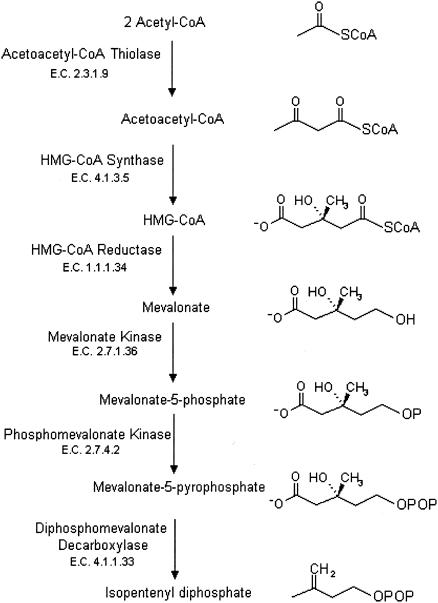
The biosynthesis of isopentenyl diphosphate (isopentenyl pyrophosphate), the precursor of isoprenoids in all forms of life, occurs by two distinct metabolic pathways, the mevalonate pathway (Fig. 1) and the glyceraldehyde 3-phosphate/pyruvate pathway, often termed the nonmevalonate pathway (17). Whereas many gram-negative bacteria employ the nonmevalonate pathway (26), humans, other eukaryotes, archaea, gram-positive cocci, and the spirochete Borrelia burgdorferi utilize the enzymes and intermediates of the mevalonate pathway (15, 16, 20, 21, 26). This review addresses 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, the catalyst for the rate-limiting reaction of the mevalonate pathway for isopentenyl diphosphate biosynthesis.
FIG. 1.
Intermediates and enzymes of the mevalonate pathway of isopentenyl diphosphate biosynthesis.
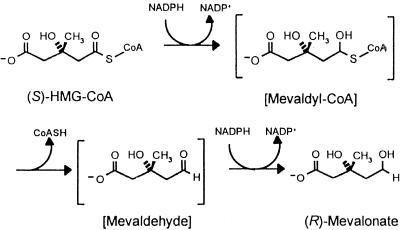
HMG-CoA reductase catalyzes the reductive deacylation of (S)-HMG-CoA to (R)-mevalonate:
The reaction proceeds in three stages, the first and third of which are reductive, and it involves successive formation of enzyme-bound mevaldyl-CoA and mevaldehyde. While mevaldehyde is not released during the course of the reaction,
 |
(1) |
HMG-CoA reductase catalyzes two reactions of free mevaldehyde. Reaction 2 resembles the third stage, and reaction 3 resembles the reverse of stages 1 and 2 of the overall reaction 1.
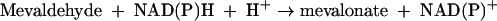 |
(2) |
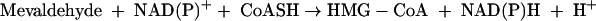 |
(3) |
HMG-CoA reductase also catalyzes the reverse of reaction 1, the oxidative acylation of (R)-mevalonate to (S)-HMG-CoA (reaction 4).
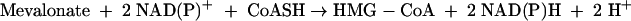 |
(4) |
This enzyme is one of a few four-electron oxidoreductases. Two moles of reduced pyridine nucleotide coenzyme is oxidized during the reduction of 1 mol of the thioester group of HMG-CoA to the primary hydroxyl group of mevalonate.
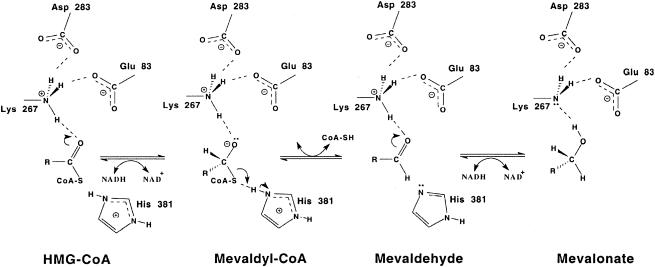
Site-directed mutagenesis of HMG-CoA reductase has implicated a histidine (7, 8), an aspartate (9), and a glutamate (25), residues that are conserved in all forms of the enzyme, as critical for catalysis. An active-site lysine detected in the first crystal structure of this enzyme (18) was confirmed as a fourth critical residue by mutagenesis (4, 5) and by inspection of the crystal structures of ternary complexes (13, 23). The proposed role of the histidine is to protonate the departing CoA thioanion. If retained, this thioanion would attack bound mevaldehyde and block completion of the overall reaction (9). The catalytic lysine, aspartate, and glutamate form a hydrogen bond-linked network that interacts with the carbonyl group of HMG-CoA. The active-site aspartate participates in both reductive stages of the overall reaction, is central to the hydrogen bond network, and may be part of a proton shuttle (9). The active-site glutamate participates in the second reductive stage of the reaction (9, 25), and the active-site lysine appears to stabilize the mevaldyl-CoA intermediate (23) (Fig. 2).
FIG. 2.
Reaction mechanism proposed for P. mevalonii HMG-CoA reductase. The three stages of the reaction and the roles proposed for the catalytic residues Lys267, Asp283, Glu83, and His381 (23) are shown.
Inspection of primary-structure alignments of representative HMG-CoA reductases from eukaryotes, archaea, and bacteria led Bochar et al. (3) to distinguish two distinct classes of HMG-CoA reductases. The distinction between classes rested initially on the observation that the number of conserved residues and sequences was significantly higher within a single class of HMG-CoA reductases than across the two classes and on the different location of the active-site lysine (Fig. 3). Subsequent comparison of the crystal structures of Pseudomonas mevalonii and human HMG-CoA reductases established that this lysine, which is conserved only within each class of the enzyme, was present on a different structural element in the two classes (13).
FIG. 3.
Partial sequence alignments of four class I and four class II HMG-CoA reductases. Sequences were aligned by using T-COFFEE (www.expasy.ch). The regions selected include the catalytic glutamate, lysine, aspartate and histidine residues, which are indicated by a green background. Amino acids with a black background are identical for both classes of enzymes. Amino acids identical in the class I enzymes and in the class II enzymes are indicated by blue and red backgrounds, respectively. Abbreviations: S. sol, Sulfolobus solfataricus; S. cer, Saccharomyces cerevisiae (yeast); M. aur, Mesocricetus auratus (Syrian hamster); H. sap, Homo sapiens (human); S. aur, Staphylococcus aureus; E. fae, Enterococcus faecalis; A. ful, Archaeoglobus fulgidus; P. mev, Pseudomonas mevalonii.
The primary sequence differences between the two classes parallel the evolutionary diversity of the organisms that harbor enzymes belonging to each class (26). Class I includes the enzymes from eukaryotes and most archaea, and class II includes the HMG-CoA reductases of certain prokaryotes and archaea. In addition to the divergence in the sequences, the enzymes of the two classes also differ with respect to inhibition by statin drugs. The inhibition constant values for the class I enzymes are nanomolar, whereas the class II reductases are over 4 orders of magnitude less sensitive to inhibition by statins (Table 1) (1, 12, 27).
TABLE 1.
Kinetic parameters of characterized class II HMG-CoA reductases
The best-studied class I HMG-CoA reductases are those from mammals, plants, yeast, and certain archaea. Eukaryotic HMG-CoA reductases consist of a highly conserved C-terminal catalytic domain and a poorly conserved N-terminal domain that comprises from two to eight transmembrane helices (2). The crystal structure of the catalytic domain of the human enzyme (13) revealed a tetramer with active sites lying at subunit interfaces. The activity of human and other eukaryotic HMG-CoA reductases is regulated by reversible phosphorylation (19), but no eubacterial or archaeal HMG-CoA reductase appears to be regulated by phosphorylation in vivo.
The interest in class II HMG-CoA reductases arises from their presence in certain bacterial pathogens and the discovery that a functional mevalonate pathway is essential for survival of these pathogens (26). The differences in structure, regulation, and sensitivity to statin drugs between the two classes of enzymes suggested, furthermore, that it may be possible to design inhibitors for use as antibiotics that target a class II HMG-CoA reductase. These considerations led to characterization of the class II reductases from Staphylococcus aureus and Enterococcus faecalis in order to supplement the information for the previously characterized class II enzymes from P. mevalonii and Archaeoglobus fulgidus. Inspection of gene assignments by The Institute for Genome Research (www.tigr.org) also suggested that the following bacteria and archaea also encode a class II HMG-CoA reductase: B. burgdorferi, Lactococcus lactis, Listeria innocua, Listeria monocytogenes, Oceanobacillus iheyensis, Streptococcus agalactiae, Streptococcus mutans, Streptococcus pneumoniae, Streptococcus pyogenes, Thermoplasma acidophilum, and Thermoplasma volcanium. This review summarizes the properties of the class II HMG-CoA reductases that have been characterized.
P. MEVALONII
The eubacterium P. mevalonii was isolated from soil based on its ability to grow on mevalonate as the sole source of carbon (10). The HMG-CoA reductase from this organism, which was the first class II HMG-CoA reductase characterized, plays a biodegradative role in vivo, converting (R)-mevalonate to (S)-HMG-CoA, and it utilizes NAD(H) exclusively as the oxidoreductant (14).
The three-dimensional crystal structure of the P. mevalonii apoenzyme (18) revealed that the active site is a large open cavity located at the subunit interface of this dimeric enzyme (Fig. 4). The active site contains three distinct subsites that bind cofactors and substrates. The large domain of one monomer binds the CoA portion of HMG-CoA. The small domain of the second monomer forms the NAD(H) binding site. Located at the interface of the two monomers is a smaller, deeper pocket that binds the HMG portion of HMG-CoA. Subsequently determined structures of two nonproductive ternary complexes (23) provided further insights into the mode of substrate binding and the mechanism of catalysis. The HMG-CoA/NAD+ ternary structure revealed that HMG-CoA is bound in an extended conformation, with the HMG moiety present in the central pocket (Fig. 5A). NAD+ also binds in an extended conformation, with its nicotinamide ring approaching the thioester group of HMG-CoA in an anti/pro-S relationship. In the mevalonate/NADH structure (Fig. 5B), NADH binds in the same conformation as NAD+. Mevalonate sits in the central pocket in an open linear conformation that overlies the HMG portion of HMG-CoA. The structures of these ternary complexes revealed that binding of substrates and cofactors induces folding of the last 50 residues in the C-terminal region of the monomer. These 50 residues, which are disordered in the apoenzyme structure (18), form a flap domain that closes over the active site. This conformational change isolates the reaction from the solvent and positions the catalytic histidine proximal to the sulfur atom of HMG-CoA, which is consistent with its proposed role in protonating the departing CoAS− anion (9). The ternary structures also revealed that Lys267 participates in catalysis and substrate binding, which was subsequently confirmed by mutagenesis and kinetic studies (4, 5).
FIG. 4.
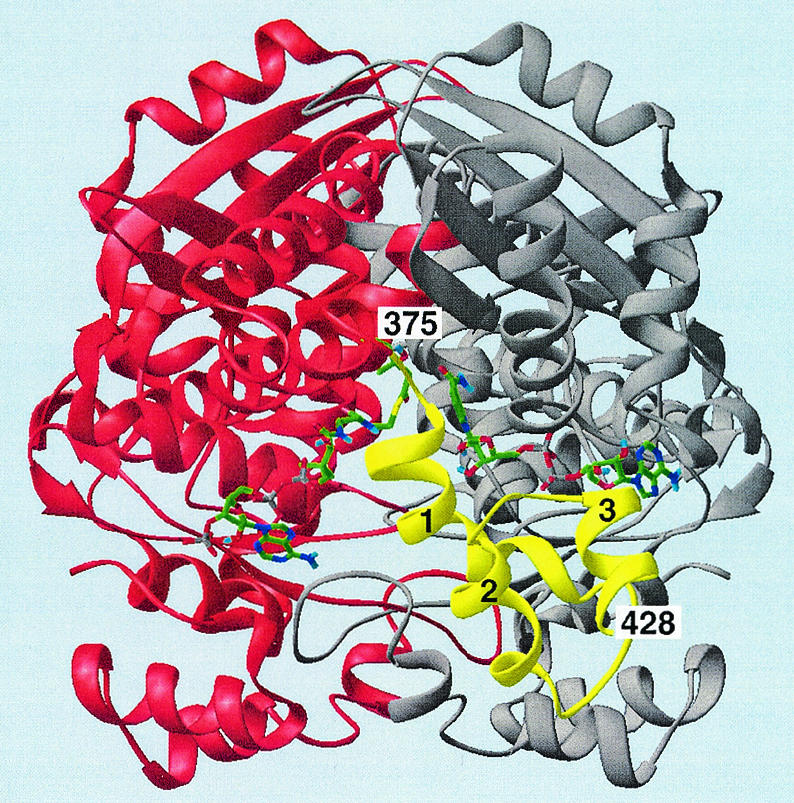
Ribbon diagram of the HMG-CoA reductase homodimer with HMG-CoA and NAD+ bound. This view looks directly into the active-site cavity located at the interface between the two monomers, colored red and gray. The third or flap domain of the red monomer, colored yellow, closes over the active site when substrates bind. The flap domain has three helical segments (designated 1 to 3) that extend from residue 375 to the C-terminal residue 428. The flap domain of the gray monomer lies behind and thus is not visible. HMG-CoA and NAD+ are shown as stick models (green = C; blue = N; red = O; gray = P; yellow = S). Both the substrate and the cofactor bind in an extended conformation, HMG-CoA to the large domain of the red monomer and NAD+ to the small domain of the gray monomer. Reproduced by permission from reference 23 (copyright 1999, National Academy of Sciences).
FIG. 5.
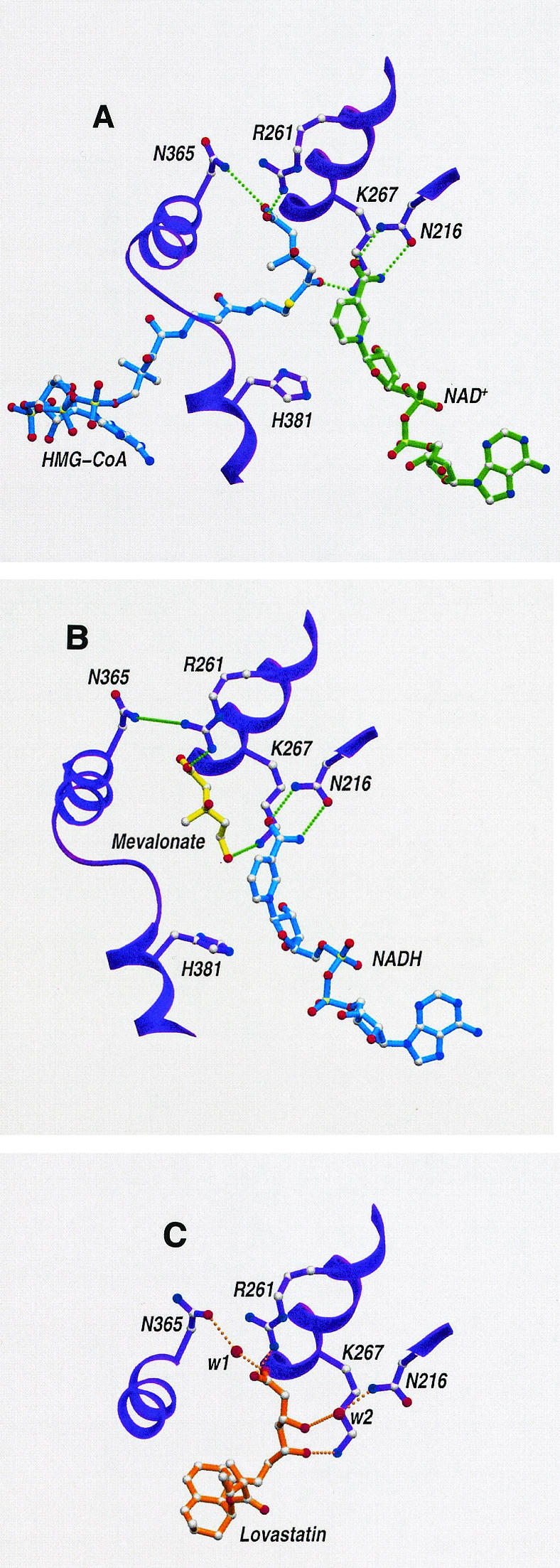
Ribbon diagrams of the active site for complexes of the class II HMG-CoA reductase of P. mevalonii with NAD+ and HMG-CoA (A), NADH and mevalonate (B), and lovastatin (C). Both substrates and coenzymes bind in an extended conformation between the two monomers. HMG-CoA and mevalonate bind to the large domain of one monomer, and NAD(H) binds to the small domain of the other monomer. Mevalonate binds in an extended conformation and occupies the same position as the HMG moiety of HMG-CoA. The nicotinamide ring of a coenzyme approaches the substrate in an orientation that facilitates hydride transfer. The interactions of lovastatin (C) mimic substrate binding in the central portion of the active-site cavity. Two water molecules (w1 and w2) mediate interactions with active-site residues. Hydrogen bond interactions are represented by dotted lines. The data for this figure appear in references 23 and 24.
The crystal structure of P. mevalonii HMG-CoA reductase has been solved by using a complex with the inhibitor lovastatin (24). Lovastatin binds in the central HMG pocket, accommodating itself in the same position as mevalonate and the HMG portion of HMG-CoA (Fig. 5C). The β-hydroxy-δ-acid moiety of lovastatin sits in the HMG pocket and forms hydrogen bond interactions with active-site residues. The decalin ring and the α-methylbutyrate ester moiety twist inward and face the hydrophobic side wall of the pocket (Fig. 5C). Lovastatin completely invades the HMG-binding site, partially occupies the site for the CoA portion of HMG-CoA, and blocks access of the substrate to critical catalytic residues. In addition, its decalin ring protrudes from the active site and prevents closure of the flap domain, a conformational change essential for catalysis.
A. FULGIDUS
The first example of an archaeal class II HMG-CoA reductase was that of A. fulgidus. The gene encoding the A. fulgidus enzyme was cloned and expressed in Escherichia coli, and the gene product was purified to apparent homogeneity. Its kinetic parameters, pH profile, and statin inhibition profile resemble those of the P. mevalonii enzyme. However, A. fulgidus HMG-CoA reductase, unlike the P. mevalonii enzyme, can use NAD(H) or NADP(H) equally well as the oxidoreductant [the kcat(NADPH)/kcat(NADH) ratio is 0.5]. In addition, NADPH behaves like a competitive inhibitor for NADH, suggesting that both coenzymes bind to the same site. Another unusual feature of the A. fulgidus reductase is its temperature profile, which shows that the activity is optimal at 85°C (16).
S. AUREUS
The first HMG-CoA reductase from a pathogen to be investigated was that of S. aureus (27). Genetic disruption experiments showed that the mvaA gene that encodes S. aureus HMG-CoA reductase was essential for growth of this organism, and mvaA null mutants were severely attenuated in a mouse pylonephritis model (26). The mvaA gene was cloned as a His-tagged construct, and its protein product was expressed, purified, and characterized. The kinetic parameters, statin inhibition values, and pH profiles were similar to the corresponding parameters for P. mevalonii HMG-CoA reductase. The enzyme can use either NAD(H) or NADP(H) as a cofactor, but it exhibits a preference for NADP(H) [the kcat(NADPH)/kcat(NADH) ratio is 25]. The presence in S. aureus of other mevalonate pathway enzymes suggests that S. aureus HMG-CoA reductase is a true biosynthetic enzyme (27).
FUSION PROTEIN OF E. FAECALIS
A unique feature of the class II HMG-CoA reductases from enterococci is their location on the same polypeptide as acetoacetyl-CoA thiolase, the first enzyme of isopentenyl diphosphate biosynthesis (26). The single open reading frame mvaE encodes the protein. The recombinant, N-terminally His-tagged mvaE product has been expressed and purified (12). While the Vmax for the E. faecalis enzyme was significantly lower than those of other class II reductases studied, the Km values, pH profiles, and statin Ki values were comparable to those of other class II HMG-CoA reductases. The E. faecalis enzyme is presently the only known class II HMG-CoA reductase that uses NADP(H) exclusively.
The portion of the mvaE gene that encodes HMG-CoA reductase was subcloned, and the encoded protein was expressed separately as an N-terminally His-tagged construct and purified (11). Although the Vmax was higher than that of the fused enzyme, the kinetic parameters of this unfused enzyme were similar to those of the HMG-CoA reductase portion of the fusion protein. Analytical ultracentrifugation and size exclusion chromatography showed that while the fusion protein exists in several oligomeric states, the unfused HMG-CoA reductase forms dimers in solution (22). Conceivably, the fusion protein may act as a scaffold for assembly of a multienzyme complex for isopentenyl diphosphate biosynthesis.
As noted by Wilding et al. (26), the enterococci are uniquely predicted to synthesize a fusion protein containing the activities of both HMG-CoA reductase and acetoacetyl-CoA thiolase (Fig. 1). While direct evidence for the activities exists only for E. faecalis, this dual-function protein indeed appears to occur in other enterococci. Western blot analysis of cell lysates of E. faecalis, Enterococcus faecium, and Enterococcus hirae performed with antibodies raised against the purified dual-function enzyme of E. faecalis revealed the presence of a similar-size mvaE gene product in all three organisms (12). The enterococcal mvaE gene product thus does not appear to undergo posttranslational cleavage in vivo. However, since genetic evidence does not suggest that this fusion protein occurs in organisms other than the enterococci, the phenomenon appears to be clade specific.
CONCLUSIONS
The recent identification and characterization of several class II HMG-CoA reductases allowed assessment of specific properties of this class of enzymes. While all class II reductases exhibit high levels of primary structure identity and have similar Ki values for inhibition by statin drugs (1, 3, 6, 12, 27), notable differences characterize individual class II enzymes. Unlike the class I enzymes, which utilize NADPH exclusively, the class II HMG-CoA reductases vary in the ability to discriminate between NADPH and NADH. The P. mevalonii enzyme is the only known HMG-CoA reductase whose biologic function is biodegradative (14). The E. faecalis HMG-CoA reductase/acetoacetyl-CoA thiolase fusion protein is the only characterized example of a catalytically active HMG-CoA reductase fused to acetoacetyl-CoA thiolase (12). However, both genetic comparisons and Western blot analyses suggest that this phenomenon occurs in other enterococci, and genetic comparisons indicate that it probably is unique to the enterococci. Since many gram-positive pathogens require a class II HMG-CoA reductase for survival (26), the differences between the two classes of enzymes and the individual properties of specific class II enzymes might be exploited to design antibiotics that could be used to treat infections caused by these pathogens.
Acknowledgments
We acknowledge American Heart Association grant 0150503N (M.H. and V.W.R.) and NIH grant HL 52115 (L.T. and C.V.S.) for research support. Support for crystallographic instrumentation was provided by the Purdue Cancer Center (grant NCI CA 82673) and the Markey Center for Structural Biology.
Footnotes
Journal paper 17112 from the Purdue University Agricultural Experiment Station.
REFERENCES
- 1.Bischoff, K. M., and V. W. Rodwell. 1996. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Haloferax volcanii: purification, characterization, and expression in Escherichia coli. J. Bacteriol. 178:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochar, D. A., J. A. Friesen, C. V. Stauffacher, and V. W. Rodwell. 1999. Biosynthesis of mevalonic acid from acetyl-CoA, p. 15-44. In D. Cane (ed.), Comprehensive natural products chemistry, vol. 2. Isoprenoids, including carotenoids and steroids. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 3.Bochar, D. A., C. V. Stauffacher, and V. W. Rodwell. 1999. Sequence comparisons reveal two classes of 3-hydroxy-3-methyl coenzyme A reductase. Mol. Genet. Metab. 66:122-127. [DOI] [PubMed] [Google Scholar]
- 4.Bochar, D. A., C. V. Stauffacher, and V. W. Rodwell. 1999. Investigation of the conserved lysines of Syrian hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochemistry 38:15848-15852. [DOI] [PubMed] [Google Scholar]
- 5.Bochar, D. A., L. Tabernero, C. V. Stauffacher, and V. W. Rodwell. 1999. Aminoethylcysteine can replace the active site lysine of Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochemistry 38:8879-8883. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, Y., H. Huber, S. L'Haridon, K. O. Stetter, and W. F. Doolittle. 2001. Bacterial origin for the isoprenoid biosynthesis enzyme HMG-CoA reductase of the archaeal orders Thermoplasmatales and Archaeoglobales. Mol. Biol. E vol. 18:1378-1388. [DOI] [PubMed] [Google Scholar]
- 7.Darnay, B. G., and V. W. Rodwell. 1993. His865 is the catalytically important histidyl residue of Syrian hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 268:8429-8435. [PubMed] [Google Scholar]
- 8.Darnay, B. G., Y. Wang, and V. W. Rodwell. 1992. Identification of the catalytically important histidine of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 267:15064-15070. [PubMed] [Google Scholar]
- 9.Frimpong, K., and V. W. Rodwell. 1994. Catalysis by Syrian hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proposed roles of histidine 865, glutamate 558, and aspartate 766. J. Biol. Chem. 269:11478-11483. [PubMed] [Google Scholar]
- 10.Gill, J. F., M. J. Beach, and V. W. Rodwell. 1985. Mevalonate utilization in Pseudomonas sp. M. Purification and characterization of an inducible 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 260:9393-9398. [PubMed] [Google Scholar]
- 11.Hedl, M. 2003. Isopentenyl pyrophosphate biosynthesis in bacteria: genes and enzymes of the mevalonate pathway, p. 36-38. Ph.D. thesis. Purdue University, West Lafayette, Ind.
- 12.Hedl, M., A. L. Sutherlin, E. I. Wilding, M. Mazzulla, D. McDevitt, P. Lane, J. W. Burgner 2nd, K. R. Lehnbeuter, C. V. Stauffacher, M. N. Gwynn, and V. W. Rodwell. 2002. Enterococcus faecalis acetoacetyl-coenzyme A thiolase/3-hydroxy-3-methylglutaryl-coenzyme A reductase, a dual-function protein of isopentenyl diphosphate biosynthesis. J. Bacteriol. 184:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Istvan, E. S., M. Palnitkar, S. K. Buchanan, and J. Deisenhofer. 2000. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 19:819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan-Starck, T. C., and V. W. Rodwell. 1989. Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl-CoA reductase characterization and chemical modification. J. Biol. Chem. 264:17913-17918. [PubMed] [Google Scholar]
- 15.Kim, D.-Y., D. A. Bochar, C. V. Stauffacher, and V. W. Rodwell. 1999. Expression and characterization of the class II HMG-CoA reductase of the thermophilic archaeon Sulfolobus solfataricus. Protein Expr. Purif. 17:435-442. [DOI] [PubMed] [Google Scholar]
- 16.Kim, D.-Y., C. V. Stauffacher, and V. W. Rodwell. 2000. Dual coenzyme specificity of Archaeoglobus fulgidus HMG-CoA reductase. Protein Sci. 9:1226-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange, B. M., T. Rujan, W. Martin, and R. Croteau. 2000. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. 97:13172-13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, C. M., V. W. Rodwell, and C. V. Stauffacher. 1995. Crystal structure of Pseudomonas mevalonii HMG-CoA reductase at 3.0 angstrom resolution. Science 268:1758-1762. [DOI] [PubMed] [Google Scholar]
- 19.Omkumar, R. V., and V. W. Rodwell. 1994. Phosphorylation of Ser871 impairs the function of His365 of Syrian hamster 3-hydroxy-3-methyl-glutaryl-CoA reductase. J. Biol. Chem. 269:16862-16866. [PubMed] [Google Scholar]
- 20.Rodwell, V. W., M. J. Beach, K. M. Bischoff, D. A. Bochar, B. G. Darnay, J. A. Friesen, J. F. Gill, M. Hedl, T. Jordan-Starck, P. J. Kennelly, D.-Y. Kim, and Y. Wang. 2000. 3-Hydroxy-3-methylglutaryl-CoA reductase. Methods Enzymol. 324:259-280. [DOI] [PubMed] [Google Scholar]
- 21.Rohmer, M. 1999. A mevalonate-independent root to isopentenyl diphosphate, p. 45-67. In D. Cane (ed.), Comprehensive natural products chemistry, vol. 2. Isoprenoids, including carotenoids and steroids. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 22.Sutherlin, A. L. 2003. The mevalonate pathway of isopentenyl pyrophosphate biosynthesis in Enterococcus faecalis: a potential target for antimicrobial agents, p. 37-41. Ph.D. thesis. Purdue University, West Lafayette, Ind.
- 23.Tabernero, L., D. A. Bochar, V. W. Rodwell, and C. V. Stauffacher. 1999. Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase. Proc. Natl. Acad. Sci. 96:7167-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabernero, L., V. W. Rodwell, and C. V. Stauffacher. 2003. Crystal structure of a statin bound to a class II HMG-CoA reductase. J. Biol. Chem. 278:19933-19938. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Y., B. G. Darnay, and V. W. Rodwell. 1990. Identification of the principal catalytically important acidic residue of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 265:21634-21641. [PubMed] [Google Scholar]
- 26.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilding, E. I., D.-Y. Kim, A. P. Bryant, M. N. Gwynn, R. D. Lunsford, D. McDevitt, J. E. Myers, Jr., M. Rosenberg, D. Sylvester, C. V. Stauffacher, and V. W. Rodwell. 2000. Essentiality, expression, and characterization of the class II 3-hydroxy-3-methylglutaryl coenzyme Areductase of Staphylococcus aureus. J. Bacteriol. 182:5147-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]