Abstract
Obesity is associated with induction of endoplasmic reticulum (ER)-stress response signalling and insulin resistance. Protein tyrosine phosphatase (PTP)-1B is a major regulator of adiposity and insulin sensitivity. The aim of this study was to investigate the role of liver-PTP1B in chronically- (high-fat diet) and pharmacologically-induced (tunicamycin, thapsigargin) ER-stress response signalling in vitro and in vivo.
We assessed the effects of ER-stress response induction on hepatic PTP1B expression, and consequences of hepatic-PTP1B deficiency, in cells and mouse liver, on components of ER-stress response signalling.
We found that PTP1B protein and mRNA expression levels were up-regulated in response to acute and/or chronic ER-stress, in vitro and in vivo. Silencing PTP1B in hepatic cell lines or mouse liver (L-PTP1B−/−) protected against induction of pharmacologically- and/or obesity-induced ER-stress. High-fat diet-induced increase in CHOP and BIP mRNA levels were partially inhibited, whereas ATF4, GADD34, GRP94, ERDJ4 mRNAs and ATF6 protein cleavage were completely suppressed in L-PTP1B−/− mice relative to control littermates. L-PTP1B−/− mice also had increased nuclear translocation of spliced XBP-1 via increased p85α binding. We demonstrate that the ER-stress response and liver-PTP1B expression are interlinked in obesity and pharmacologically-induced ER-stress and this may be one of the mechanisms behind improved insulin sensitivity and lower lipid accumulation in L-PTP1B−/− mice.
Keywords: ER stress, PTP1B, insulin resistance, obesity, metabolic syndrome
Introduction
The endoplasmic reticulum (ER) is an eukaryotic intracellular organelle required for synthesis and metabolism of many complex metabolites and biologically active proteins [1]. The ER can also sense and respond to alterations in cellular homeostasis. Conditions interfering with the function of ER are collectively called ER-stress. For example, accumulation of unfolded protein aggregates in the ER lumen leads to the activation of a pathway termed unfolded protein response (UPR) with the aim to return the ER to its normal physiological state. This complex cellular response is mediated initially by three molecules, PKR-like ER kinase (PERK), activated transcription factor 6 (ATF6), and Inositol-requiring enzyme 1 (IRE1) [2, 3]. The ER luminal domain of PERK, IRE1, and ATF6 interacts with the ER chaperone GRP78 (glucose-regulated protein, or BIP).
Activated PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α), thereby reducing the rate of general protein translation, and thus protein load, on the ER [4, 5]. Phosphorylation of eIF2α paradoxically increases translation of activating transcription factor 4 (ATF4) mRNA to produce a transcription factor that activates expression of several UPR target genes [4, 6]. Expression profiling studies have found that PERK, eIF2α and ATF4 are required for expression of genes involved in amino acid biosynthesis and transport, anti-oxidative stress, and apoptosis, e.g. growth arrest and DNA damage 34 (GADD34), and CAAT/enhancer-binding protein homologous protein (CHOP) [7]. Activation of the ER protein kinase IRE1 triggers its endoribonuclease activity to induce cleavage of X box-binding protein 1 (XBP-1) mRNA. XBP-1 mRNA is then ligated by an uncharacterized RNA ligase and translated to produce spliced XBP-1 protein [8]. Spliced XBP-1 protein is a highly active transcription factor and one of the key regulators of ER folding capacity [9]. Some of the genes identified that require activation of the IRE1/XBP-1 pathway are components of the ER-associated degradation (ERAD) machinery, such as ER degradation enhancing α-mannosidade-like protein (EDEM) [10, 11], major ER chaperones such as GRP78/BIP and GRP94, and the endoplasmic reticulum-localized DnaJ homologue (ERDJ4) [9, 10]. Concurrently, ATF6 is released from GRP78 and transits to the Golgi body where it is cleaved to release a transcriptionally active fragment [12]. Cleaved ATF6 acts in concert with spliced XBP-1 protein to induce expression of genes encoding protein chaperones and components of the ERAD machinery [13, 14].
Obesity and type 2 diabetes result in conditions that increase demand on the ER. This is particularly clear in the liver, adipose tissue and pancreas, where changes in tissue architecture, increases in protein synthesis, and perturbations in cellular energy fluxes occur [15]. Recent studies demonstrated that ER-stress is increased in adipose and liver tissues in both dietary and genetic obesity [16-18]. In cellular systems, the induction of ER-stress leads to insulin resistance, at least in part through IRE-1-dependent activation of JNK. Modulation of ER-folding capacity through gain- and loss-of-function studies with XBP-1 showed a close link between ER function and insulin action in vitro and in vivo [17].
PTP1B is the prototype for the superfamily of PTPs and has been implicated in multiple signaling pathways [19]. Gene-targeting studies in mice have established PTP1B as a critical physiological regulator of metabolism and body mass by attenuating insulin and leptin signaling [16, 20-25]. PTP1B-deficient mice exhibit resistance to diabetes and obesity, the two major metabolic diseases in industrialized societies. Not surprisingly, PTP1B is a highly regarded target of the pharmaceutical industry in the treatment of these disorders [26]. Recently, we generated liver-specific PTP1B−/− (L-PTP1B−/−) mice and found that they exhibited improved glucose homeostasis and lipid profiles, independently of changes in adiposity when fed a high-fat diet (HFD). In addition, liver-specific PTP1B deletion also partially attenuated some markers of the hepatic ER-stress response induced by chronic high-fat diet feeding [16]. The aim of the present study was to investigate in detail the contribution of PTP1B in the induction and regulation of ER-stress response in liver using both in vitro and in vivo models of PTP1B deletion. ER-stress response was induced either by chronic feeding of mice with HFD and/or with pharmacological treatment of mice and cells, and subsequently activation of the PERK, IRE1 and ATF6 pathways were examined.
Methods
Animal studies
All animal studies were performed under a project licence approved by the Home Office under the Animals (Scientific Procedures) Act 1986 and the principles of laboratory animal care (NIH publication no. 85–23, revised 1985; http://grants1.nih.gov/grants/olaw/references/phspol.htm) were followed. Mice were maintained on a 12-hour light/dark cycle in a temperature-controlled barrier facility, with free access to water and food. Liver-specific PTP1B knockout (L-PTP1B−/−) and fl/fl control mice were described previously [16, 22]. All mice studied were age-matched littermate males on the mixed 129Sv/C57Bl6 background. Mice were placed either on standard lab chow or HFD (Harlan Teklad, Bicester, UK; 55% fat) at weaning (21 days old), and weights were monitored weekly. The animals were kept on diet for 16 weeks.
For tunicamycin-treatment of animals, mice were given a single 1 μg/g body weight intraperitoneal injection of 0.05 g/l suspension of tunicamycin (Sigma, Gillingham, UK) in saline. The control groups were injected with saline. Three hours later, mice were sacrificed by decapitation, livers removed and frozen in liquid N2.
Metabolic measurements
Glucose from tail blood was assessed using a glucometer (Accu-Check, Burgess Hill, UK). Serum insulin was determined by ELISA (CrystalChem, Downers Grove, USA) as described previously [16, 22, 23].
Cell culture
The human liver carcinoma cell line (HepG2) was routinely cultivated in Dulbecco’s modified Eagle’s Medium (DMEM) (Gibco, Paisley, UK) supplemented with 10% foetal bovine serum (FBS, Invitrogen, Paisley, UK), 2 mmol/l glutamine and 1% penicillin/streptomycin (Gibco) and maintained at 37°C in a humidified atmosphere with 5% CO2.
Small-interfering RNA (siRNA) duplexes specific for human PTP1B were obtained from Santa Cruz and control non-silencing siRNA from Eurogentec (Southampton, UK). Transient transfection of cells was done using the Lipofectamine™ RNAiMAX transfection reagent (Invitrogen). Then, cells were treated with tunicamycin (5 mg/l) or vehicle (DMSO) for 7 hours, followed by RNA or protein extraction for further analysis.
In another set of experiments, stable knock-down of PTP1B in HepG2 cells was performed using specific short hairpain RNA (shRNA) constructs designed against human PTP1B using Gateway System (Invitrogen). The viruses were transduced into the cells in the presence of 5 g/l of polybrene (Millipore, Chandlers Ford, UK). Then, cells were treated with thapsigargin (300 nM) or vehicle (DMSO) for 7 hours, followed by RNA extraction for further analysis.
mRNA expression analysis
Total RNA was isolated from transfected/transduced HepG2 cells or mouse livers using TRIReagent (Ambion, Warrington, UK) according to the manufacturer’s protocol. First strand cDNA was synthesized from 1 μg of total RNA employing the Bioline Biorscript™ Preamplification System (Bioline, London, UK) and an oligo(dT)12-18 primer as reverse primer. Then, target genes were amplified by real-time PCR using GoTaq qPCR Master Mix (Promega, Southampton, UK) in Roche LightCycler® 480 System (Roche diagnostics, Burgess Hill, UK). Relative gene expression was calculated using the comparative Ct (2–δδCt) method. The primer sequences used for PCR, real time PCR or sequencing are listed in Supplemental table 1.
XBP-1 splicing assay
We analyzed the splicing of XBP-1 from cDNA using Fast Start Taq DNA polymerase. The PCR conditions were as follows: 94 °C for 3 min; 35 cycles of 94 °C for 10 seconds, 65 °C for 30 seconds, and 72 °C for 30 seconds; and 72°C for 10 minutes. We used the following primers: Sens “5’ A AAC AGA GTA GCA GCT CAG ACT GC 3’” and reverse “5’ TC CTT CTG GGT AGA CCT CTG GGA G 3’”. To distinguish the unspliced from the spliced band, we resolved the PCR products on a carefully prepared 2% agarose gel.
Western Blotting
Tissue lysates were prepared by extraction in radioimmunoprecipitation assay (RIPA) as described previously [16, 23]. Immunoblots were performed using antibodies against PTP1B (Millipore), SHP2, CHOP, ERK1, ERK2, human PTP1B (Santa Cruz by Insight Biotechnology, Wembley, UK), lamin A/C, phospho-eIF2-α (Ser 51), BIP (GRP78), IRE1-α,Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Cell Signalling by NEB, Hitchin, UK), XBP-1 (spliced and unspliced forms) (Abcam, Cambridge, UK) and ATF6 (full length and cleaved forms) (IMGENEX, Cambridge, UK), phosphatase and tensin homolog (PTEN) (Abgent, Oxford, UK). Proteins were visualized using enhanced chemiluminescence and quantified either by scanning densitometry (Image J) or Bio1D software on PeqLab Fusion gel imaging system (PeqLab, Fareham, UK).
Nuclear and cytosolic extraction
For nuclear extracts, we cut the liver tissue in small pieces and washed once with ice-cold PBS then lysed in NE-PER extraction reagent (Thermoscientific, Loughborough, UK) according to the manufacturer’s instructions and as described in [22]. Samples were subjected to SDS-PAGE gel electrophoresis, transferred to a nitrocellulose membrane, and immunobots performed as described above.
Immunoprecipitation
For XBP-1–p85-α co-immunoprecipitation, we subjected both nuclear and cytosolic liver extracts from HFD-fed mice to immunoprecipitation using Pierce Classic IP kit (Thermoscientific) according to the manufacturer’s protocol with minor modification as described by Park et al. [27]. Briefly, we added 5 μg of a goat anti-XBP-1 antibody to 400 μg of liver nuclear or cytosolic lysates and incubated overnight at 4 °C with gentle rotation. The next day, we added the antibody/lysate sample to Protein A/G Plus Agarose in spin columns and incubated with gentle end-over-end mixing or shaking for 2 hours (Eppendorf, St Albans, UK). Proteins from the immunoprecipitates were resolved on SDS-PAGE gel, transferred to a nitrocellulose membrane, and immunolots were performed as described above using rabbit anti-XBP-1 (Abcam) and p85-α (Santa Cruz).
Data analysis
Data are expressed as mean ± SEM, and n represents the number of mice or biological replicates. Statistical analyses were performed using ANOVA (2-way or 1-way, as appropriate), and Mann-Whitney U tests. P < 0.05 was considered to be statistically significant.
Results
ER-stress induction leads to increased PTP1B protein and mRNA expression levels
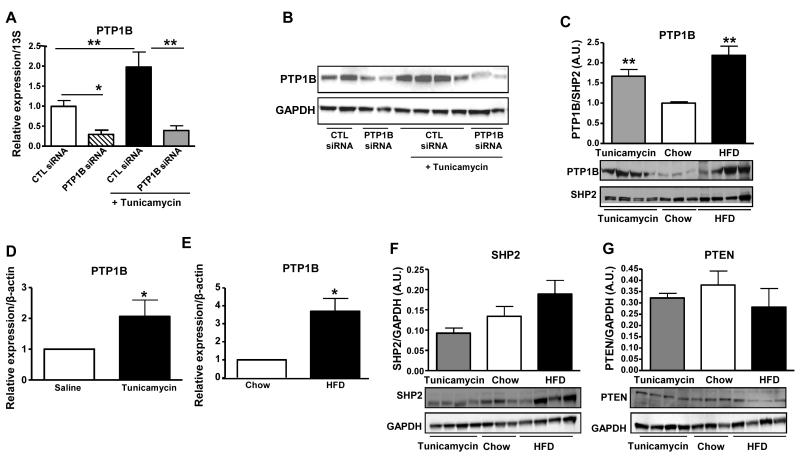
We aimed to investigate the impact of ER-stress induction on hepatic-PTP1B expression in cultured hepatocytes and mice. In HepG2 cells, tunicamycin-treatment led to a ~2 fold-increase in PTP1B mRNA expression and protein levels, but not in those transfected with siRNA against PTP1B (Figure 1A and 1B). Chronic ER-stress [17], caused by prolonged HFD-feeding for 16 weeks, led to an increase in hepatic PTP1B protein levels compared to the mice fed standard chow diet (Figure 1C). Interestingly, acute pharmacological-induction of ER-stress in mice with tunicamycin treatment also resulted in an increase of hepatic-PTP1B protein expression (Figure 1C). The increase in mouse liver-PTP1B protein levels was due to increased mRNA levels with both HFD-feeding and tunicamycin-treatment (Figure 1D and 1E). These data indicate that pharmacological induction of ER-stress by tunicamycin treatment leads to increased PTP1B mRNA and protein levels, both in vitro and in vivo.Importantly, tunicamycin- -induced ER stress did not affect expression of other phosphatases such as SHP2 or PTEN (Figure 1F and 1G); although there was a trend towards an increase in SHP2 protein levels with HFD-feeding in comparison to chow-fed animals (Figure 1F).
Figure 1. ER stress induction leads to increased PTP1B expression.
(A) Effect of tunicamycin-induced ER stress on PTP1B mRNA expression in HepG2 cells treated with PTP1B- or control-siRNA (*P <0.05, **P<0.01 vs. the indicated group). (B) Effect of tunicamycin-induced ER stress on PTP1B protein levels expression in HepG2 cells treated with PTP1B- or control-siRNA. (C) PTP1B protein levels in livers from control mice fed a chow diet, mice fed a chow diet and injected with tunicamycin and mice fed HFD. Bar graphs represent pooled, normalized data to total amount of SHP2 protein expressed as arbitrary units (n = 3–4 per group). (D) PTP1B mRNA expression in livers from mice injected either with saline or tunicamycin. (E) PTP1B mRNA expression in livers from mice fed chow and HFD (n=3-4 per group) (*P<0.05, **P<0.01 vs. chow or saline). SHP2 (F) and PTEN (G) protein levels in livers from control mice fed a chow diet, mice fed a chow diet and injected with tunicamycin and mice fed HFD. Bar graphs represent pooled, normalized data to total amount of GAPDH protein expressed as arbitrary units (n = 3–4 per group). Data (A-G) were analyzed by t-test or one-way ANOVA, followed by a Tukey’s multiple comparison test.
PTP1B knockdown protects against pharmacologically-induced ER-stress in HepG2 cells
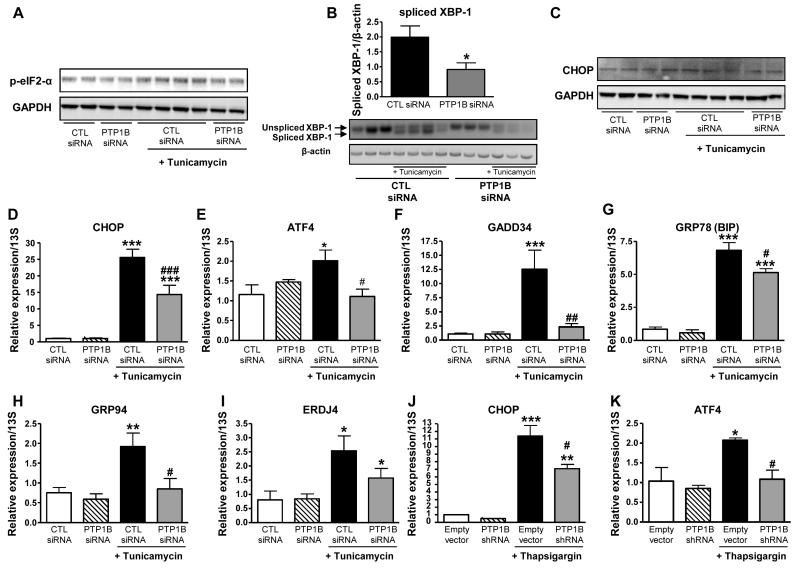
Since PTP1B siRNA decreased PTP1B mRNA expression in cells by ~75 % compared to scrambled control siRNA (Figure 1A and 1B), we used these cells to analyze ER stress activation. Interestingly, as shown in Figure 2A, tunicamycin treatment of HepG2 cells transfected with control siRNA enhanced eIF2-α phosphorylation whilst PTP1B silencing prevented this. In addition, tunicamycin treatment lead to XBP-1 splicing in scrambled siRNA- and PTP1B siRNA-transfected cells (Figure 2B); however, PTP1B silenced cells exhibited lower amount of spliced XBP-1 (Figure 2B). Interestingly, tunicamycin-induced increase in CHOP protein levels was lower in PTP1B silenced cells compared to controls (Figure 2C). Furthermore, we analysed mRNA expression levels of the main markers of ER-stress response by real-time PCR. As expected, we found that treatment of control cells with either tunicamycin or thapsigargin led to enhanced expression of ER-stress response markers regulated through the PERK/eIF2-α pathway (CHOP, ATF4 and GADD34) and through IRE1-α and ATF6 pathways (BIP, GRP94 and ERDJ4) (Figure 2D-2K). Importantly, silencing of PTP1B in these cells partially (CHOP and BIP) or completely (ATF4, GADD34, GRP94 and ERDJ4) protected against tunicamycin, as well as thapsigargin, effects on the main ER-stress response components, suggesting that PTP1B may be required for the full induction of ER-stress response signalling (Figure 2D-2K).
Figure 2. PTP1B knockdown protects against pharmacologically-induced ER stress in HepG2.
Effect of tunicamycin-induced ER stress on eIF2-α phosphorylation (A), XBP-1 splicing (B), and CHOP protein levels (C) in HepG2 cells treated or not with PTP1B siRNA. (D-K) Effect of tunicamycin-induced ER stress on mRNA expression CHOP (D), ATF4 (E), GADD34 (F), BIP (G), GRP94 (H), ERDJ4 (I) in HepG2 cells treated or not with PTP1B siRNA. (J and K) Effect of thapsigargin-induced ER stress on mRNA expression CHOP (J) and ATF4 (K) in HepG2 cells treated either with empty vector or specific PTP1B shRNA (*P<0.05, **P<0.01, ***P<0.001 vs. CTL siRNA or empty vector; #P< 0.05, ## P< 0.01, ### P<0.001 vs. CTL siRNA or empty vector +tunicamycin). Data (B, and D-K) are expressed as mean±SEM and were analyzed two-way ANOVA, followed by a Bonferroni’s multiple comparison test (n =3-7 per group).
In vivo liver PTP1B deficiency and glucose/lipid homeostasis
To better understand the physiological role of hepatic-PTP1B in ER-stress response signalling, we used an in vivo mouse model for the rest of our study. We used the liver-specific PTP1B knockout (L-PTP1B−/−) mice which are previously published [16, 22]. L-PTP1B−/− and control mice were weaned either onto normal chow diet (4.5% fat) or HFD (55% kcal from fat). The absence of PTP1B in hepatocytes had no effect on body weight on either diet (data not shown), and as previously published [16, 22].
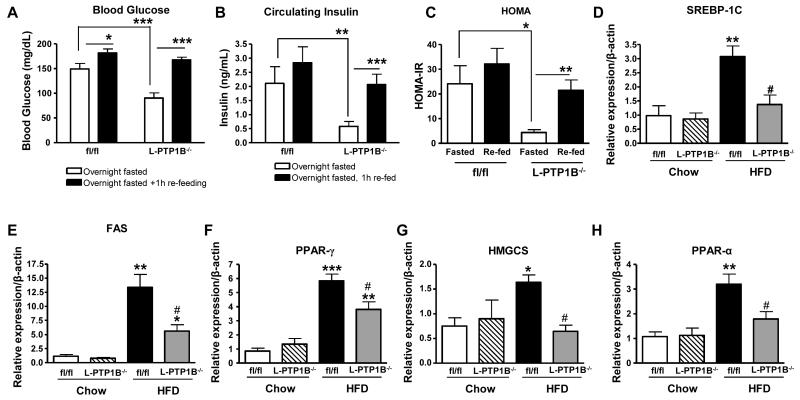
To analyze this mouse model further, we investigated if liver-PTP1B played a role in fasting/re-feeding control of glucose homeostasis. We fasted the mice overnight (at 8 weeks HFD), and allowed them to freely access food for one hour afterwards. After an overnight fast, L-PTP1B−/− exhibited markedly lower blood glucose levels (Figure 3A), circulating insulin levels (Figure 3B) and homeostatic model assessment (HOMA)-insulin resistance (IR) index (Figure 3C), consistent with some of our previous observations [16]. PTP1B however does not seem to play an important role in the fasting/re-feeding response of glucose maintenance, as glucose levels increased to the same level in both groups of mice (Figure 3A), and while L- PTP1B−/− had a moderately lower increase in re-feeding circulating insulin levels (Figure 3B) and HOMA-IR index (Figure 3C) in comparison to controls, this was not statistically significant.
Figure 3. Liver PTP1B deletion improves glucose homeostasis.
(A) Blood glucose from fl/fl (n =11) and L-PTP1B−/− (n = 7) either fasted overnight or fasted overnight then re-fed for 1h (*P<0.05, ***P<0.001 vs. the indicated group). (B) Circulating insulin from fl/fl and L-PTP1B−/− either fasted overnight or fasted overnight then re-fed for 1h (**P<0.01, ***P<0.001 vs. the indicated group). (C) Homeostatic model assessment (HOMA) calculated for fl/fl and L-PTP1B−/− either fasted overnight or fasted overnight then re-fed for 1h (*P<0.05, **P<0.01 vs. the indicated group). (D-H) Relative mRNA expression, measured by quantitative real-time PCR normalized against β-actin mRNA, of SREBP-1C (D), FAS (E), PPAR-γ (F), HMGCS (G) and PPAR-α (H) in livers of male fl/fl and L-PTP1B−/− mice fed chow or HFD (n=6-13) (*P<0.05, **P<0.01, ***P<0.001 vs. chow fl/fl; #P<0.05 vs. HFD fl/fl). Data (A-J) are expressed as mean±SEM and were analyzed by two-way ANOVA, followed by a Bonferroni’s multiple comparison test.
We also showed previously that liver cholesterol and triglyceride levels were significantly lower in L-PTP1B−/− mice after 5 weeks of HFD-feeding [16]. Since ER-stress response is known to play a major role in lipid regulation [28], we wanted to investigate expression of genes involved in synthesis of fatty acids. De novo synthesis of fatty acids requires sterol regulatory element-binding protein (SREBP)-1, and peroxisome proliferator-activated receptor (PPAR)-γ [29]. Analysis of SREBP-1c expression levels, as well as its target gene fatty acid synthase (FAS), and PPAR- γ, revealed that HFD-fed L-PTP1B−/− mice had lower expression levels of these genes relative to control littermates (Figure 3D, 3E and 3F). Expression of genes involved in cholesterol synthesis, such as 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (HMGCS1) was also significantly decreased in HFD-fed L-PTP1B−/− mice (Figure 3G), as expected from our published phenotype of these mice. Interestingly, however, fatty acids are catabolized to acetyl-CoA for use in energy production mainly by β-oxidization in mitochondria and peroxisomes and PPARα is a known master transcriptional regulator of genes involved in β-oxidization in mitochondria and peroxisomes [29]. We found that HFD-fed L-PTP1B−/− mice also had significantly lower mRNA expression levels of PPARα when compared to control mice (Figure 3H). Collectively, these data indicate that, in our hands, L-PTP1B−/− exhibit improved glucose and lipid homeostasis compared to their control littermates and that these mice are a useful tool to dissect out the in vivo role of PTP1B in ER-stress response signaling.
Liver PTP1B-deficiency decreases ER-stress response regulated through PERK/eIF2-α and ATF6 pathways
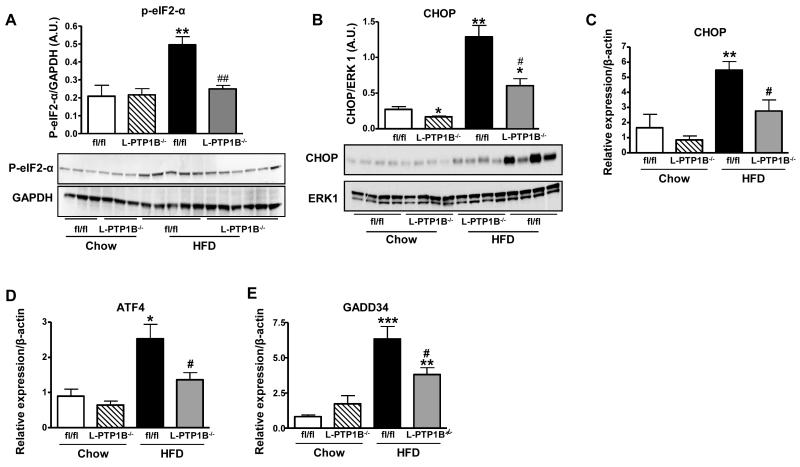
In light of the present findings in cells that hepatic-PTP1B may be required for full induction of ER-stress response; we used L-PTP1B−/− mice to delineate the in vivo role of hepatic-PTP1B in ER-stress response regulated through different branches of the pathway. Mice fed chow or HFD for 16 weeks were sacrificed and livers harvested for analysis of ER-stress response components. First, we analyzed the involvement of liver-PTP1B in regulation of ER-stress response via the PERK/eIF2-α pathway. Whilst HFD-feeding led to an increase in phosphorylation of eIF2-α at Ser 51 in control mice in comparison to chow-fed mice (Figure 4A), liver-PTP1B deletion protected against this. Since phosphorylation of eIF2-α is regulated through PERK activation, this suggests that there is a lower activity of the PERK/eIF2-α pathway in L-PTP1B−/− mice (Figure 4A). In addition, HFD-feeding, as expected, enhanced mRNA expression levels of several components of ER-stress response under the control of PERK/eIF2-α pathway: CHOP, ATF4, and GADD34 (Figure 4C-4E). Importantly, liver-PTP1B deletion partially or completely prevented the HFD-induced increase in protein expression of CHOP (Figure 4B) and mRNA expression of CHOP, ATF4 and GADD34 (Figure 4C-4E).
Figure 4. Liver PTP1B-deficiency decreases ER stress response regulated through PERK/eIF2-α pathway.
(A) eIF2-α phosphorylation on S51 in livers of male fl/fl and L-PTP1B−/− mice on chow or HFD (n=3-6 per group). (B) CHOP protein expression in livers from male fl/fl and L-PTP1B−/− mice fed chow or HFD. Bar graphs represent pooled, normalized data to total amount of ERK1 protein expressed as arbitrary units (n =4 per group) (*P < 0.05, **P < 0.01 vs. chow fl/fl; #P < 0.05 vs. HFD fl/fl). (C-E) mRNA expression of CHOP (C), ATF4 (D) and GADD34 (E) in livers of male fl/fl and L-PTP1B−/− mice fed chow or HFD (n=6-13 per group) (*P<0.05, **P<0.01 vs. chow fl/fl; #P<0.05 vs. HFD fl/fl). Data (B-E) are expressed as mean±SEM and were analyzed two-way ANOVA, followed by a Bonferroni’s multiple comparison test.
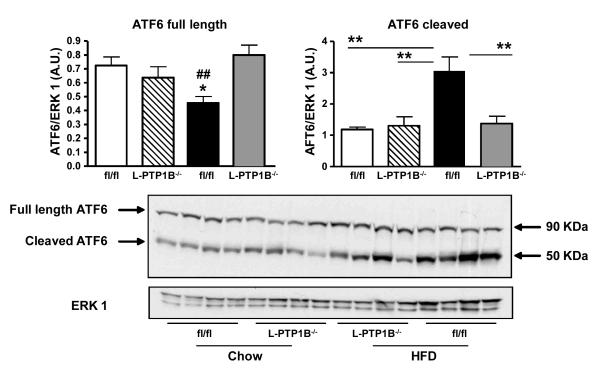
The second arm of the ER-stress response pathway is mediated by ATF6. To investigate ATF6 activation, we analyzed the expression of the full length (90 KDa) and the active, cleaved (50 KDa) forms of ATF6, by western blotting. HFD-feeding enhanced cleavage of ATF6 and reduced its full length form expression in control animals (Figure 5). Importantly, liver-PTP1B deletion protected against HFD-induced ATF6 cleavage (Figure 5) suggesting that liver-PTP1B is also required for maximal cleavage of ATF6 with HFD-feeding.
Figure 5. Liver PTP1B-deficiency decreases ER stress response regulated through ATF6 pathway.
ATF6 protein expression in livers from male fl/fl and L-PTP1B−/− mice fed chow or HFD. Bar graphs represent pooled, normalized data to total amount of ERK1 protein from expressed as arbitrary units (n=4 per group) of full length and cleaved forms of ATF6 (*P<0.05 vs. chow fl/fl; **P<0.01 vs. the indicated group; ##P<0.01 vs. HFD L-PTP1B−/−). Data are expressed as mean±SEM and were analyzed two-way ANOVA, followed by a Bonferroni’s multiple comparison test.
Liver PTP1B-deficiency decreases ER-stress response regulated through IRE1-α sensor
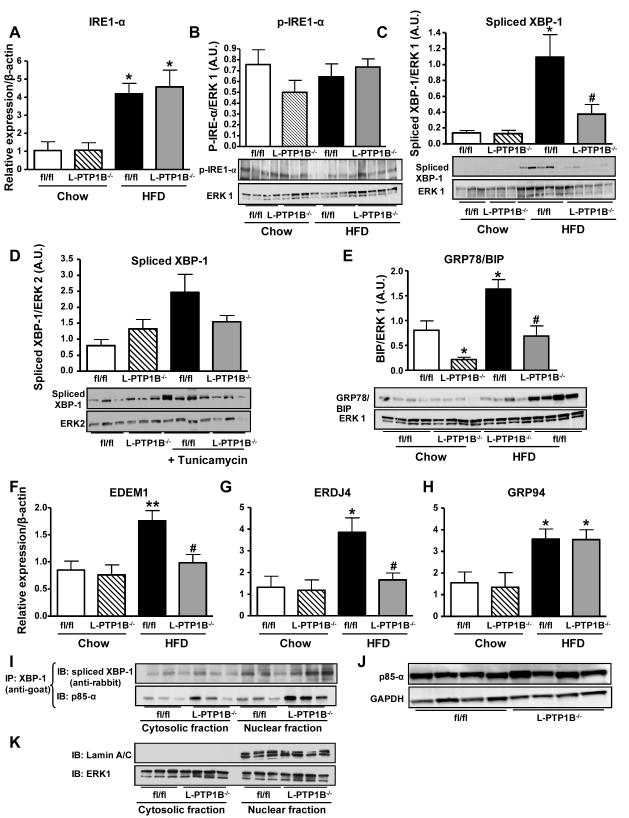
Next we analyzed the involvement of hepatic-PTP1B in the IRE1-α pathway. HFD-feeding enhanced the expression of IRE1-α mRNA in livers from both L-PTP1B−/− and control mice compared to chow animals (Figure 6A). Its phosphorylation was also unaffected by liver-PTP1B deficiency (Figure 6B). However, we observed that chronic (Figure 6C) induction of ER-stress by HFD-feeding, increased the protein expression of spliced form of XBP-1 (XBP-1s) in control fl/fl mice, whilst L-PTP1B−/− mice were protected (Figure 6C). In addition, tunicamycin-injected fl/fl mice also had increased protein expression of XBP-1s, while L-PTP1B−/− mice were completely protected against this (Figure 6D).
Figure 6. Liver PTP1B-deficiency decreases ER stress response regulated through IRE1-α sensor and increases XBP-1 nuclear translocation through p85-α interaction.
(A) IRE1-α mRNA expression in livers from male fl/fl and L-PTP1B−/− mice fed chow or HFD (n =6-13 per group) (*P<0.05 vs. chow fl/fl). (B) IRE1-α phosphorylation on S724 in livers of male fl/fl and L-PTP1B−/− mice on HFD (n=4 per group). (C) Spliced XBP-1 protein expression in livers from male fl/fl and L-PTP1B−/− mice fed chow or HFD (n =3-5 per group) (*P<0.05 vs. chow fl/fl). (D) Spliced XBP-1 protein expression in livers from male fl/fl and L-PTP1B−/− mice injected either with saline or tunicamycin (n =3-4 per group) (*P < 0.05 vs. chow fl/fl; #P<0.05 vs. HFD fl/fl). (E) BIP protein expression in livers from male fl/fl and L-PTP1B−/− mice fed chow or HFD (n =4 per group) (*P<0.05 vs. chow fl/fl; #P<0.05 vs. HFD fl/fl). BIP and CHOP (Figure 2B) were blotted on the same gel; hence they share the same ERK1 control blot. Bar graphs represent pooled, normalized data to total amount of ERK1 or ERK2 protein expressed as arbitrary units. (F-H) mRNA expression of EDEM1 (F), ERDJ4 (G) and GRP94 (H) in livers of male fl/fl and L-PTP1B−/− mice fed chow or HFD (n =6-13 per group) (*P<0.05, **P<0.01 vs. chow fl/fl; #P<0.05 vs. HFD fl/fl). Data (A-H) were analyzed by two-way ANOVA, followed by a Bonferroni’s multiple comparison test. (I) XBP-1 was immunoprecipitated from nuclear and cytosolic liver lysates (fl/fl and L-PTP1B−/− mice on HFD) and immunoblotted for the p85-α regulatory subunit of phosphatidylinositol 3-kinase and spliced XBP-1 (n =3 per group). (J) Total p85-α protein levels in livers from fl/fl and L-PTP1B−/− mice fed HFD (n =4 per group). (K) To assess the purity of nuclear and cytosolic fractions we blotted for a nuclear marker, lamin A/C, and for ERK1 as total.
HFD-feeding also increased protein levels of BIP and mRNA expression of EDEM1, ERDJ4 and GRP94 (Figure 6E-6H) in fl/fl mice compared to their respective chow groups; however, L-PTP1B−/− mice had dramatically lower protein expression of BIP and mRNA levels of EDEM1 and ERDJ4, but not GRP94, in comparison to their controls (Figure 6E-6H).
Overall, these data suggest that liver-PTP1B also plays a role in the HFD-feeding and pharmacologically-induced ER-stress response pathway downstream of IRE1-α sensor but may not be required for its full induction.
Liver-PTP1B deficiency increases XBP-1 nuclear translocation through p85-α interaction
Recently, phosphatidyl inositol 3-kinase (PI3-K) regulatory subunit, p85, was identified as an interacting partner with XBP-1, and a main modulator of its translocation to the nucleus to resolve ER-stress [27, 30]. Liver-PTP1B deficiency is associated with increased IRS1 and IRS2 association with p85 [16]; thus, we hypothesized that PTP1B may play a role in the regulation of p85α-XBP1 interaction. We immunoprecipitated XBP-1 (using a goat anti-XBP-1 antibody) from cytosolic and nuclear fractions, followed by blotting for p85α. Interestingly, in liver lysates from L-PTP1B−/− mice, XBP-1s nuclear translocation was enhanced (Figure 6I) and p85-α co-immunoprecipated with XBP-1 both in the nuclear and cytosolic fractions. However, the amount of p85-α interacting with XBP-1 was greater in liver lysates from L-PTP1B−/− mice (Figure 6I) in comparison to controls. It is important to note here, that in order to capture this interaction, we used a rabbit anti-XBP-1 antibody for immunoblotting, in order to avoid interference with the goat heavy chain Immunoglobulin which runs closely to the molecular weight of spliced XBP-1 (~54KDa) (Supplemental Figure 1). Importantly, the total levels of p85-α were unchanged between genotypes (Figure 6J), in concurrence with our previously published data [16]. To assess the purity of nuclear and cytosolic fractions we blotted for a nuclear marker, lamin A/C, and for ERK1 as total loading control (Figure 6K).
Discussion
The pathogenesis of obesity and associated insulin resistance is thought to involve over-activation of the ER-stress response signaling [17, 18, 31, 32]. Whole-body and tissue-specific knockout studies in mice have revealed that PTP1B is a major regulator of insulin sensitivity and adiposity via regulation of insulin and leptin signaling pathways in muscle, liver, fat and hypothalamus [16, 20-22]. In this paper we demonstrate that ER-stress response and PTP1B expression are interlinked and that directly down-regulating PTP1B expression in liver can relieve over-activation of the ER-stress response associated with HFD-feeding, obesity and insulin resistance.
Previous studies have shown that PTP1B is over-expressed in multiple insulin- and leptin-responsive tissues in mice with diet-induced or genetic obesity or in cells with pro-inflammatory cytokine or free-fatty acid treatment [33-35]. We now show that in addition to chronic HFD-feeding, acute pharmacological induction of ER-stress response leads to an elevation of PTP1B mRNA and protein levels in mouse livers and hepatic cell lines. The mechanism(s) for this elevation may involve ATF6, since over-expression of ATF6 in hepatic cells reportedly increases mRNA expression of PTP1B [36] and we show here that the active (cleaved) form of ATF6 is increased in HFD-fed mice. These findings suggest that over-activation of the ER-stress response with diet-induced obesity promotes insulin resistance via elevation of PTP1B. However, PTP1B also appears to contribute to the ER-stress response since liver-specific deficiency in mice or siRNA knockdown in hepatic cells blunts the full activation of all three ER-stress pathways, namely IRE1-α/XBP-1, PERK/eIF2-α and ATF6 ([16, 37] and this paper). Collectively, these studies suggest there may be a positive feedback loop between PTP1B expression and full activation of the ER-stress response.
We have shown previously that liver-specific deficiency of PTP1B enhanced insulin-stimulated IRS1- and IRS2- associated p85α protein, without changes in p85α expression levels [16]. Here, we report that liver-PTP1B deletion increases the interaction between p85α and XBP-1 in both cytoplasmic and nuclear fractions from liver, and increases the nuclear translocation of spliced XBP-1 in association with increased interaction with p85-α. This is despite finding that mRNA levels of spliced XBP-1 are decreased in L-PTP1B−/− mice under HFD- and tunicamycin-treated conditions (this paper and [16]). These findings are consistent with recent studies that have revealed a novel link between the IRE1-α pathway and insulin signaling [27, 30]. These studies identified PI3-K, regulatory subunit, p85, as an interacting partner with XBP-1 and a major modulator of its translocation to the nucleus to resolve ER-stress [27, 30]. Thus, it appears that PTP1B may play a direct role in inhibiting insulin-stimulated XBP-1 nuclear translocation via p85, to promote ER-stress. However, further work is required to elucidate if this is the case or secondary to its role in regulating glucose and lipid homeostasis.
The activation of IRE1-α/XBP-1 and PERK/eIF2-α pathways in obese and insulin resistant states have been well described [16-18, 38]. Moreover, Gu et al. [37] showed that PTP1B plays an essential role in potentiating IRE-1-mediated ER-stress signaling pathways in vitro. Consistent with these and our previous findings, we show here that liver-PTP1B deletion decreases eIF2α phosphorylation in HFD-fed mice, indicating a reduced activation of the PERK/eIF2α axis, further confirmed by reduced mRNA levels of the target genes ATF4, CHOP and GADD34. Moreover, we now also show that HFD-feeding results in increased processing of ATF6 into its cleaved active form and that this is reduced to chow levels in L-PTP1B−/− HFD-fed mice. Downstream, GRP78/BIP is coordinately up-regulated in control fl/fl HFD-fed mice, but completely reduced to chow diet levels in L-PTP1B−/− HFD-fed mice. This is consistent with ATF6α being solely responsible for transcriptional induction of ER chaperones e.g. GRP78/BIP [14]. Kammoun and co-workers also found increased levels of cleaved ATF6 in livers of ob/ob mice [39]. However in other studies, cleaved ATF6 levels were found to be reduced in ob/ob, db/db and diet-induced obese mice and p85α-deficient cell lines with reduced nuclear XBP-1 [30, 40]. Thus, further work is required to elucidate the exact role of ATF6 in insulin resistant states.
PTP1B over-expression has been reported in other insulin-sensitive tissues following HFD-feeding or in cells after treatment with ER stress inducers. It has been shown that treatment of C2C12 muscle cells with palmitate, a known inducer of insulin resistance and ER stress, enhanced both mRNA expression and protein levels of PTP1B [41]. Moreover, PTP1B over-expression was observed in liver and white adipose tissue in ob/ob mice and in muscle and white adipose tissue in zucker fatty rats (fa/fa) as well as in livers and white adipose tissue in mice fed HFD [34]. In addition, Zabolotny et al [34] showed that treatment of cultured hepatocytes and 3T3-L1 adipocytes with TNF-α, reported to induce ER stress [42], enhanced PTP1B expression. Also, Bettaieb and co-workers [43] suggested that PTP1B is involved in palmitate-induced ER stress in MIN6 insulinoma β cells. Altogether, these findings suggest a close relationship between chronic or acute ER stress induction and PTP1B over-expression in other insulin-sensitive tissues; however, the involved mechanisms are not yet elucidated.
Further to our previous findings [16], we demonstrate here that PTP1B deletion in liver decreases hepatic SREBP1c and SREBP1a gene expression levels, which is counterintuitive to what would be expected from the liver-insulin receptor knock out phenotype and the enhanced insulin sensitivity observed in L-PTP1B−/− mice. It has been shown that silencing PTP1B in ob/ob mice resulted in a similar decrease in lipogenic gene expression, including SREBP1 [44, 45]. Moreover, studies using high-fructose diet treatment in rats showed that this led to development of insulin resistance with concomitant increase in PTP1B and SREBP1 gene expression in the liver. Further investigation revealed that PTP1B may regulate SREBP1a and SREBP1c mRNA expression via protein phosphatase 2A (PP2A) activity [46]. PTP1B tethers to the ER via its C-terminal tail [47], and changes in the intracellular localization of PTP1B induced by truncation of its C-terminal tail did not affect its negative regulation of insulin signalling [48]. Importantly, Shi et al [49] observed that over-expression of C-terminal truncated PTP1B in rat Fao cells did not induce SREBP-1 gene expression. Furthermore, truncated PTP1B failed to bind PP2A, resulting in impaired PP2A activation. Therefore it seems that PTP1B may affect SREBP1 gene expression via a pathway distinct from the insulin signalling where its location within the ER membrane appears critical.
Altogether, our findings demonstrate that PTP1B plays a critical role in hepatic glucose and lipid homeostasis and in the positive regulation of ER-stress response during diet-induced obesity. Therapeutically, inhibiting the activity of PTP1B in the liver as demonstrated previously with anti-sense oligonucleotide administration should result in improved glucose and lipid homeostasis. The mechanism of this improvement appears not only to involve improved insulin sensitivity through the effects on the insulin receptor signalling, but also alleviation of ER-stress. Thus, developing liver-selective PTP1B inhibitors continues to be an attractive option for treatment of metabolic and cardiovascular diseases such as insulin resistance, type diabetes and dyslipidemia.
Supplementary Material
Acknowledgments
This work was supported by a Diabetes UK project grant to Dr. M. Delibegović (BDARD08/0003597). Dr. Delibegovic is also funded by an RCUK Fellowship, British Heart Foundation, Tenovus Scotland and the Royal Society. Dr. N. Mody is funded by a career development fellowship from the British Heart Foundation. Mr. Carl Owen is funded by a BBSRC postdoctoral training studentship. Dr. Kendra K. Bence is funded by the United States Department of Agriculture (USDA) and NIH grant RO1DK082417.
Abbreviations
- ATF
activated transcription factor
- CHOP
CAAT/enhancer-binding protein homologous protein
- EDEM
enhancing α-mannosidade-like protein
- eIF2α
eukaryotic translation initiation factor 2α
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERDJ4
endoplasmic reticulum-localized DnaJ homologue
- ERK
Extracellular signal-regulated kinases
- GADD34
growth arrest and DNA damage 34
- GRP
glucose-regulated protein
- GTT
glucose tolerance test
- HFD
high-fat diet
- HMGCS
3-hydroxy-3-methylglutaryl-coenzyme A synthase
- IRE1
Inositol-requiring enzyme 1
- JNK
c-Jun N-terminal kinase
- PERK
PKR-like ER kinase
- PI3-K
phosphatidyl inositol 3 kinase
- PPAR
peroxisome proliferator-activated receptor
- PTP
protein tyrosine phosphatase
- shRNA
short hairpain RNA
- siRNA
short interfering RNA
- SREBP
sterol regulatory element-binding protein
- UPR
unfolded protein response
- XBP-1
X box-binding protein 1
Footnotes
Duality of interest: None to declare. Designed experiments: M.D.; Executed experiments: A.A., N.M., C.O., A.C., D.Z., M.B.A, K.K.B; Data analysis: A.A., M.D.; Wrote the paper: A.A., M.D., N.M., K.K.B.
References
- 1.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Gregor MF, Hotamisligil GS. Thematic review series: Adipocyte Biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J. Lipid Res. 2007;48:1905–1914. doi: 10.1194/jlr.R700007-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 5.Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- 6.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 7.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 8.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 9.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J. Biochem. 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 12.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 16.Delibegovic M, Zimmer D, Kauffman C, Rak K, Hong EG, Cho YR, Kim JK, Kahn BB, Neel BG, Bence KK. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes. 2009;58:590–599. doi: 10.2337/db08-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 18.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip SC, Saha S, Chernoff J. PTP1B: a double agent in metabolism and oncogenesis. Trends Biochem. Sci. 2010;35:442–449. doi: 10.1016/j.tibs.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramachandran C, Gresser MJ, Tremblay ML, Kennedy BP. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 21.Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim YB, Sharpe AH, Stricker-Krongrad A, Shulman GI, Neel BG, Kahn BB. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 2000;20:5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bence KK, Delibegovic M, Xue B, Gorgun CZ, Hotamisligil GS, Neel BG, Kahn BB. Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat. Med. 2006;12:917–924. doi: 10.1038/nm1435. [DOI] [PubMed] [Google Scholar]
- 23.Delibegovic M, Bence KK, Mody N, Hong EG, Ko HJ, Kim JK, Kahn BB, Neel BG. Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol. Cell. Biol. 2007;27:7727–7734. doi: 10.1128/MCB.00959-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabolotny JM, Bence-Hanulec KK, Stricker-Krongrad A, Haj F, Wang Y, Minokoshi Y, Kim YB, Elmquist JK, Tartaglia LA, Kahn BB, Neel BG. PTP1B regulates leptin signal transduction in vivo. Dev. Cell. 2002;2:489–495. doi: 10.1016/s1534-5807(02)00148-x. [DOI] [PubMed] [Google Scholar]
- 25.Cheng A, Uetani N, Simoncic PD, Chaubey VP, Lee-Loy A, McGlade CJ, Kennedy BP, Tremblay ML. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev. Cell. 2002;2:497–503. doi: 10.1016/s1534-5807(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 26.Johnson TO, Ermolieff J, Jirousek MR. Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat. Rev. Drug Discov. 2002;1:696–709. doi: 10.1038/nrd895. [DOI] [PubMed] [Google Scholar]
- 27.Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K, Ozcan U. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat. Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basseri S, Austin RC. ER stress and lipogenesis: a slippery slope toward hepatic steatosis. Dev. Cell. 2008;15:795–796. doi: 10.1016/j.devcel.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G852–858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 30.Winnay JN, Boucher J, Mori MA, Ueki K, Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat. Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailly-Maitre B, Belgardt BF, Jordan SD, Coornaert B, von Freyend MJ, Kleinridders A, Mauer J, Cuddy M, Kress CL, Willmes D, Essig M, Hampel B, Protzer U, Reed JC, Bruning JC. Hepatic Bax inhibitor-1 inhibits IRE1alpha and protects from obesity-associated insulin resistance and glucose intolerance. J. Biol. Chem. 2010;285:6198–6207. doi: 10.1074/jbc.M109.056648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, Lee AS. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dube N, Tremblay ML. Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochim. Biophys. Acta. 2005;1754:108–117. doi: 10.1016/j.bbapap.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 34.Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J. Biol. Chem. 2008;283:14230–14241. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parvaneh L, Meshkani R, Bakhtiyari S, Mohammadtaghvaie N, Gorganifiruzjaee S, Taheripak G, Golestani A, Foruzandeh M, Larijani B, Taghikhani M. Palmitate and inflammatory state additively induce the expression of PTP1B in muscle cells. Biochem. Biophys. Res. Commun. 2010;396:467–471. doi: 10.1016/j.bbrc.2010.04.118. [DOI] [PubMed] [Google Scholar]
- 36.Wang N, Zhang D, Mao X, Zou F, Jin H, Ouyang J. Astragalus polysaccharides decreased the expression of PTP1B through relieving ER stress induced activation of ATF6 in a rat model of type 2 diabetes. Mol. Cell. Endocrinol. 2009;307:89–98. doi: 10.1016/j.mce.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, Chevet E. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J. Biol. Chem. 2004;279:49689–49693. doi: 10.1074/jbc.C400261200. [DOI] [PubMed] [Google Scholar]
- 38.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–537. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parvaneh L, Meshkani R, Bakhtiyari S, Mohammadtaghvaie N, Gorganifiruzjaee S, Taheripak G, Golestani A, Foruzandeh M, Larijani B, Taghikhani M. Palmitate and inflammatory state additively induce the expression of PTP1B in muscle cells. Biochem. Biophys. Res. Commun. 396:467–471. doi: 10.1016/j.bbrc.2010.04.118. [DOI] [PubMed] [Google Scholar]
- 42.Xue X, Piao JH, Nakajima A, Sakon-Komazawa S, Kojima Y, Mori K, Yagita H, Okumura K, Harding H, Nakano H. Tumor necrosis factor alpha (TNFalpha) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNFalpha. J. Biol. Chem. 2005;280:33917–33925. doi: 10.1074/jbc.M505818200. [DOI] [PubMed] [Google Scholar]
- 43.Bettaieb A, Liu S, Xi Y, Nagata N, Matsuo K, Matsuo I, Chahed S, Bakke J, Keilhack H, Tiganis T, Haj FG. Differential regulation of endoplasmic reticulum stress by protein tyrosine phosphatase 1B and T cell protein tyrosine phosphatase. J. Biol. Chem. 2011;286:9225–9235. doi: 10.1074/jbc.M110.186148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waring JF, Ciurlionis R, Clampit JE, Morgan S, Gum RJ, Jolly RA, Kroeger P, Frost L, Trevillyan J, Zinker BA, Jirousek M, Ulrich RG, Rondinone CM. PTP1B antisense-treated mice show regulation of genes involved in lipogenesis in liver and fat. Mol. Cell. Endocrinol. 2003;203:155–168. doi: 10.1016/s0303-7207(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 45.Bence KK. Hepatic PTP1B Deficiency: The Promise of a Treatment for Metabolic Syndrome? J. Clin. Metab. Diabetes. 2011;1:27–33. [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu S, Ugi S, Maegawa H, Egawa K, Nishio Y, Yoshizaki T, Shi K, Nagai Y, Morino K, Nemoto K, Nakamura T, Bryer-Ash M, Kashiwagi A. Protein-tyrosine phosphatase 1B as new activator for hepatic lipogenesis via sterol regulatory element-binding protein-1 gene expression. J. Biol. Chem. 2003;278:43095–43101. doi: 10.1074/jbc.M306880200. [DOI] [PubMed] [Google Scholar]
- 47.Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG. The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 48.Shi K, Egawa K, Maegawa H, Nakamura T, Ugi S, Nishio Y, Kashiwagi A. Protein-tyrosine phosphatase 1B associates with insulin receptor and negatively regulates insulin signaling without receptor internalization. J. Biochem. 2004;136:89–96. doi: 10.1093/jb/mvh094. [DOI] [PubMed] [Google Scholar]
- 49.Shi K, Ugi S, Shimizu S, Sekine O, Ikeda K, Egawa K, Yoshizaki T, Nagai Y, Nishio Y, Takada T, Torii R, Kimura H, Kashiwagi A, Maegawa H. Membrane localization of protein-tyrosine phosphatase 1B is essential for its activation of sterol regulatory element-binding protein-1 gene expression. Biochem. Biophys. Res. Commun. 2007;363:626–632. doi: 10.1016/j.bbrc.2007.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.