Abstract
Analysis of protein interaction networks and protein complexes using affinity purification and mass spectrometry (AP/MS) is among most commonly used and successful applications of proteomics technologies. One of the foremost challenges of AP/MS data is a large number of false positive protein interactions present in unfiltered datasets. Here we review computational and informatics strategies for detecting specific protein interaction partners in AP/MS experiments, with a focus on incomplete (as opposite to genome-wide) interactome mapping studies. These strategies range from standard statistical approaches, to empirical scoring schemes optimized for a particular type of data, to advanced computational frameworks. The common denominator among these methods is the use of label-free quantitative information such as spectral counts or integrated peptide intensities that can be extracted from AP/MS data. We also discuss related issues such as combining multiple biological or technical replicates, and dealing with data generated using different tagging strategies. Computational approaches for benchmarking of scoring methods are discussed, and the need for generation of reference AP/MS datasets is highlighted. Finally, we discuss the possibility of more extended modeling of experimental AP/MS data, including integration with external information such as protein interaction predictions based on functional genomics data.
Keywords: Proteomics, Affinity Purification, Mass Spectrometry, Protein Interactions, Statistical Models, Label-free Quantification, Integrative Analysis
Introduction
The analysis of protein-protein interactions and protein complexes is of great importance in biological research. A combination of affinity purification (AP) and mass spectrometry (MS), AP/MS for short, has become a commonly used method for the analysis of protein complexes and interaction networks both on small and large scale [1–8]. Owing to recent technological advances in MS instrumentation and sample preparation methods, there have been a steady increase in the number of groups utilizing AP/MS approaches. At the same time, the development of computational tools and algorithms for processing of AP/MS data has lagged behind. In particular, one of the foremost computational challenges is the need to deal with a large number of false positive interactions present in unfiltered AP/MS datasets.
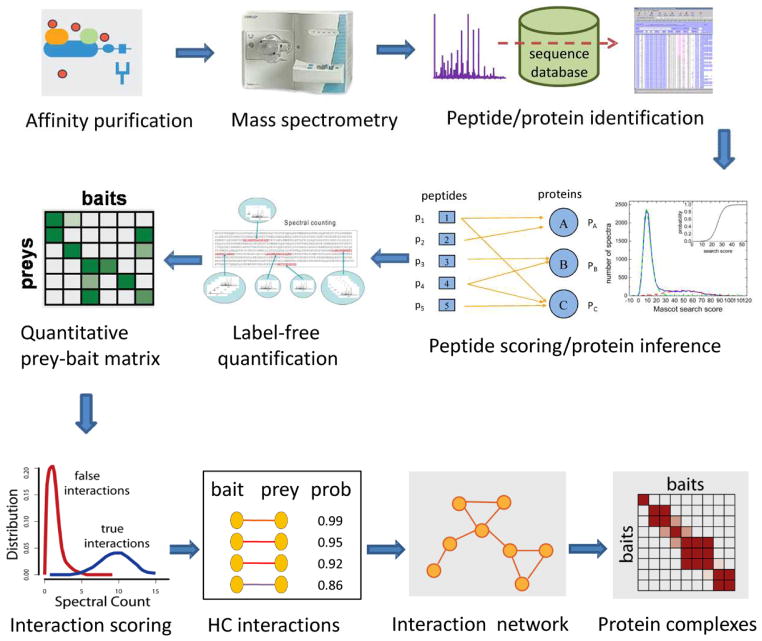
In a typical AP/MS experiment, selected proteins of interest (commonly referred to as “baits”) are purified along with their interactors (“preys”) through one or more AP steps (see Figure 1 for an illustration of the entire AP/MS workflow for mapping protein interactions). Several different tags (e.g. FLAG-tag [9]) or tag combinations (as in tandem affinity purification, TAP [3, 5]) can be used [10–12]. Proteins in the affinity purified sample are digested with trypsin, and peptides are separated using liquid chromatography (LC) coupled online to a mass spectrometer [13, 14]. Eluting peptides are ionized, transferred into a gas phase, and selected peptide ions are fragmented to generate tandem MS (MS/MS) spectra. Database searching of MS/MS spectra is used to identify the peptides and proteins [15, 16]. The initial outcome of such an AP/MS experiment is a protein list containing the bait protein and its co-purifying partners (preys), which in the ideal case can be interpreted as the list of bait-prey protein interaction pairs. In practice, however, such AP/MS datasets contain a large number of false positive interactions.
Figure 1. Mapping protein interaction networks using AP/MS.
Bait proteins and their interaction partners are purified using AP. Resulting protein samples are digested into peptides and peptides are sequenced using tandem mass spectrometry. Peptides are identified from acquired MS/MS spectra via sequence database searching. Computational tools are used to assign confidence scores to peptide identifications, map peptides to proteins, and to summarize the results at the protein level. Label-free quantification (e.g. spectral counting) is used to estimate the abundance of proteins in each experiment. Data from all AP/MS runs in the experiments are summarized in the form of quantitative prey-bait matrix. This matrix is computationally analyzed to compute a confidence score for each bait-prey pair. The interaction network assembled using high confidence (HC) protein interactions is computationally analyzed, e.g. to reconstruct protein complexes or signalling pathways.
There are several sources of false positive interactions in AP/MS data. The first source is incorrect protein identifications resulting from incorrect assignment of peptide sequences to MS/MS spectra. The problem of false positive identifications is common to all MS-based proteomic methods and is discussed in details elsewhere [17]. The second source of false positive interactions is background or non-specifically binding proteins. These include highly abundant cellular protein (e.g. tubulins and ribosomal proteins), proteins that bind to unfolded polypeptides (e.g. heat-shock proteins), and proteins that interact with affinity matrices [12, 18, 19]. In the case of single step AP experiments, the true interaction partners of the bait often represent less than 10% of all identified proteins [19].
Experimentally, strategies employing more stringent washes or multiple purification steps have been able to reduce the co-purification of non-specific binders. However, these approaches may also lead to the loss of true interaction partners. Thus, strategies aiming at identifying true interactors amongst the noise are essential. Conceptually, the simplest approach - and the one used most frequently - is the subtraction of the AP/MS results from a negative control (e.g. cells expressing the epitope tag only, without the bait) from those of the bait protein. However, this strategy has so far been applied without underlying statistical modeling. Coupling AP/MS with stable isotope labeling-based proteomic strategy using ICAT, iTRAQ or SILAC labeling is another strategy for distinguishing specific from non-specific interactions [20–24]. The quantitative ratios observed in these experiments are amenable to computational modeling to detect non-specific binders [21, 25]. An in-depth review of the labeling strategies with application to protein interaction data can be found elsewhere [12]. At the same time, labeling based methods are largely limited to small-scale studies due to increased complexity of the experimental protocols and high cost of reagents.
The need for computational filtering of AP/MS data was identified in early genome-scale studies. In these studies promiscuity was used as a filter to remove proteins which exhibited a lack of specificity for the bait from the data [3, 26–29]. Such simple filters are clearly arbitrary and non-quantitative in nature. Importantly, they have the potential to negatively impact datasets through the elimination of proteins that serve as network hubs, as well as true interaction partners for a given protein that also appear frequently across the dataset [30]. On the opposite end, a number of sophisticated bioinformatics approaches utilizing external data sources such as gene expression, sequence homology, and structural information to assist with the calculation of a confidence score for an interaction have been developed [31–36]. While additional data sources are generally expected to improve the quality of filtered data, these approaches rely on the selection of training data for building a predictive model. Finding appropriate training datasets is challenging in practice. Other computational methods attempt to calculate the confidence measure of a protein interaction from the topology of the network, i.e. using network properties such as connectivity. The scoring in these methods involves such concepts as matrix/spoke models and their extensions that can be collectively called “affinity scores” [2, 37–40], likelihood-based tests [41], or direct application of graph theory based methods [42]. However, these tools were optimized for a very specific type of interaction data, namely global yeast TAP studies [2, 3, 5, 9]. As such they rely on multiple purifications, ideally in situations where each protein identified in the interaction network is used sequentially as bait and the entire network is highly connected. Furthermore, these methods were developed and applied to datasets composed of binary calls (observed, not observed). A common weakness in most of these computational approaches is that they do not fully utilize the quantitative protein abundance information that can be extracted even from label-free AP/MS data.
In human cells, no single group has developed methodologies and throughput to perform AP/MS on the scale of the yeast studies of Gavin et al. [2] and Krogan et al. [5]. Instead, most recent publications focus on individual baits, on small interconnected networks (e.g. [4, 43–47]), or on bigger networks that are largely unconnected ([1, 46]). It is becoming increasingly likely that an AP/MS map of the human interactome will be a decentralized enterprise, with multiple groups contributing a portion of the interactions. While the topology methods may work to some extend for the intermediate size networks, they are not appropriate for unconnected networks regardless of the number of baits used to generate the dataset. Thus, there is a growing need in the research community to design methods specifically for scoring interactions stemming from label-free AP/MS data in other types of projects than global (i.e. genome-wide) studies.
Diversity of AP/MS datasets
To further elaborate on the important issue raised above, there is a wide range of AP/MS datasets that are currently being generated and published in the scientific literature. First, datasets range in size, from a single bait and up to hundreds of baits in large-scale studies. Second, when AP/MS datasets are generated in a more targeted way using selected baits of interest, the underlying interaction networks display a varying degree of interconnectivity among the participating proteins. Third, each bait protein in the study may be profiled once or in multiple replicates. Fourth, all AP/MS experiments in the datasets may or may not be performed under identical conditions (e.g. different AP conditions may be used for a challenging bait to increase the sensitivity of detecting its interacting partners). Furthermore, all or a subset of baits can be purified using multiple different epitope tags, which is also done to increase the interactome coverage. Finally, in parallel with AP/MS experiments using bait proteins, one may perform a number of negative control experiments using cells expressing the epitope tag only (i.e. without the bait or other types of control). This diversity of AP/MS datasets creates a computational challenge in that a single computational model or a scoring scheme may not be applicable to all datasets. Because global AP/MS dataset are rare and can be reasonably well processed using the existing methods such as socio-affinity scores or their extensions, we focus on the analysis of typical AP/MS datasets that are “incomplete” in nature. Below we summarize several recently developed approaches for these data. While all these methods utilize label-free quantitative information as the basis for scoring the interactions, they differ significantly in what type of AP/MS datasets they were developed and tested for, in the label-free quantification strategy used, and in model assumptions and underlying statistical (or empirical) methods used.
Label-free protein quantitation as the basis for scoring interaction data
As an alternative to using network topology as a primary way to score protein interactions, non-specific binding proteins can be detected with a help of quantitative information regarding the abundance of proteins in the affinity-purified sample. Label-free MS-based protein quantification methods can be divided into two categories depending on the type of quantitative information they use: continuous (MS1-based integrated peptide ion intensities [48–51], MS/MS-based summed fragment ion intensities [52, 53], or fragment ion intensities in selected reaction monitoring, SRM, strategies [54]) and count-based (MS/MS based spectral counting [48, 55–65]).
Spectral count is the number of times a peptide from a certain protein was successfully selected for sequencing by the mass spectrometer (number of acquired MS/MS spectra on that protein). With proper normalization, spectral counts can be used as a quantitative measure of the protein abundance in the sample [58, 66, 67]. This method is conceptually similar to the approach of measuring gene expression using SAGE [68], EST [69] or RNA-Seq [70] count data. Spectral counting can be an accurate and informative approach for detecting differential protein expression in comparative studies [55, 71] and in the analysis of AP/MS data [45, 72]. As a practical advantage, spectral counting does not require modifications to the experimental protocols - all the necessary information is already present in AP/MS data. It is compatible with most commonly used instrumentation, and does not require additional software for accessing raw MS files [73, 74]. A number of tools are available for extracting spectral counts from proteomic data, e.g. Abacus [75] that is compatible with the widely used Trans-Proteomic pipeline (TPP) [17, 76].
The alternative approach is based on using continuous data. Peptide ion intensity-based methods require additional data processing steps and high mass accuracy instrumentation [63]. Peptide intensities can be extracted using one of the several available tools, e.g. msInspect [77], SuperHirn [78], and IDEAL-Q [79] that are compatible with TPP file formats, using the integrated system MaxQuant [80], or commercial tools such as Progenesis (Nonlinear Dynamics). The peak intensities of all peptides in each MS run are often normalized, e.g. to the total peak intensity based on all identified peptides in the analysis. For computing protein intensities, one common approach is to average the normalized peak intensities of the three most intense peptides for each protein. The intensity based methods can quantify a protein identified by a single peptide (although multiple peptides are recommended) and are generally more accurate than spectral counting provided the data is of sufficiently high quality and expertly processed using advanced bioinformatics tools. In particular, the accuracy of intensity-based methods can be affected by such data problems as overlapping peptide intensity signals due to the presence of co-eluting peptides having similar molecular weight.
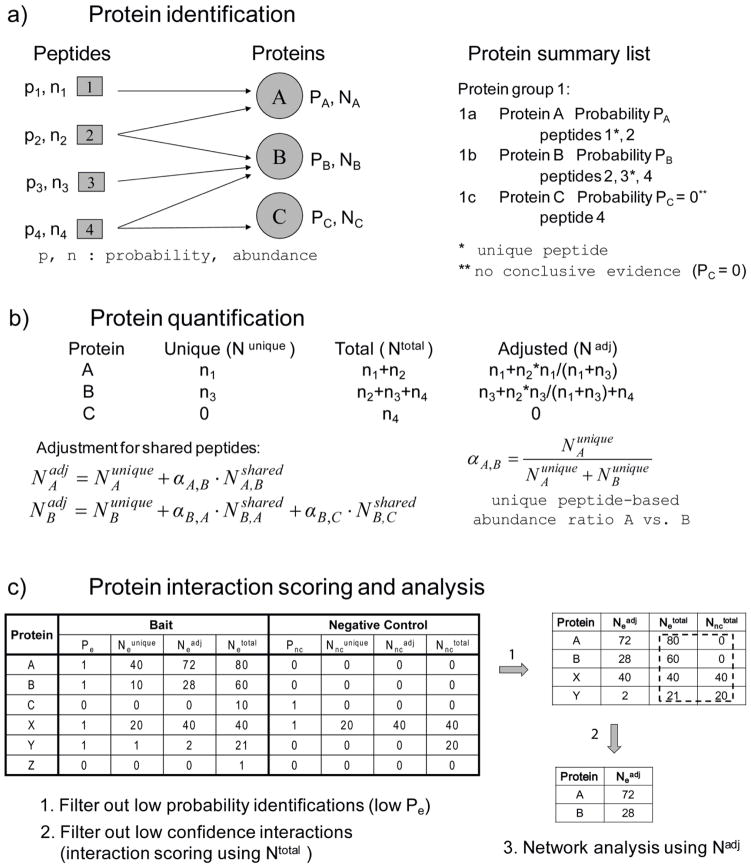
It should also be noted that peptide level quantification is not always an unambiguous measure of protein abundance. In many organisms, including higher eukaryotes, a large fraction of peptides sequences are shared across multiple proteins or protein isoforms [81, 82] (see Figure 2 for an illustration of the protein inference issue and its implications for protein quantification and analysis of AP/MS data). There are several strategies for computing isoform-specific quantification. A simple approach is to consider only unique (isoform specific) peptides. This is a reasonable strategy only when it is possible to restrict the analysis to just a few selected peptides per protein (e.g. when peptides are selected in advance for SRM-based quantification). This is generally not the case with spectral counting, which works most effectively as a protein-level summary statistics because individual peptide spectral counts are usually too low. Intensity-based methods provide more accurate quantitative estimates for individual peptides, however accurate absolute protein quantification still requires multiple peptides (e.g., top three most intense peptides per protein [83], or all peptides [84, 85]). A more accurate approach to count-based quantification is to apportion the peptide count among all its corresponding proteins using weighting factors estimated based on unique peptide counts [75, 86] (see Figure 2b for illustration using spectral counts; similar adjustment procedure should be applicable to continuous quantification measures as well). In some cases, however, is very difficult or even impossible to distinguish between proteins having a high degree of sequence homology. Relying on unique peptides only, and even using adjusted spectral counts may underestimate the protein abundance in such cases. Unfortunately, many of the common contaminants appearing in AP/MS studies are members of large homologous protein families (e.g. ribosomal proteins, tubulins, histones). Thus, it is recommended to be extra cautious and take a conservative approach of using all peptides for quantification when determining abundance levels of these proteins (especially in the negative controls runs, when available; see Figure 2c for illustration). For example, when scoring protein interactions using spectral count data in SAINT (see below), we routinely utilize total spectral counts in order to perform more conservative analysis and elimination of false interactions. After elimination of non-specific binders, however, adjusted spectral counts are more suitable for reconstruction of protein complexes [87] and for comparison of abundances of proteins that are part of the same complex [75, 88].
Figure 2. The protein inference problem and its implication for protein quantification in AP/MS datasets.
a) Peptides are identified and quantified from MS data and mapped to proteins. Protein probabilities P and abundances N are estimated based on the probabilities and abundances of their corresponding peptides. Proteins A, B, and C are identified from four peptides (summarized as protein groups 1). Protein C does not have any unique (non-shared) peptides identified in the dataset, and thus it receives zero probability (Pc=0; to be interpreted as the absence of conclusive evidence for its presence in the sample). b) Protein abundance can be estimated using all peptides equally (Ntotal), using unique peptides only (Nunique), or with apportioning the abundances of shared peptides among their corresponding proteins using weighting factors determined based on unique peptide abundances (Nadj). c) Quantitative prey-bait matrix (one bait, one negative control). A, B, C: a family of homologous proteins that are specific interactors of the bait, identified from four, as in panel a, high probability peptides with spectral counts n1=n2=40 and n3=n4=10. X, Y, Z: a group of homologous non-specific binders, also identified by four peptides (spectral counts n5=n6=20 and n7=n8=1). In the negative control, peptides 7 and 8 were not identified, resulting in PY=PZ =0. Only high probability proteins (high Pe) are considered for subsequent analysis (A, B, X, and Y). Protein interaction confidence is best determined using Ntotal (not to underestimate the abundance of inconsistently identified non-specific binders such as protein Y). For high scoring interactions, subsequent analysis (e.g., network modeling) is best performed using Nadj.
Data filtering using fold change and p-value thresholds
Simple methods such as subtraction of all proteins identified in the control runs, or applying various ad hoc thresholds based on the ratio of spectral counts in the AP/MS experiment with bait protein vs. controls have long been used for filtering the data (see, e.g. [89]). As a more rigorous extension, standard statistical methods such as t-test can be applied to compute significance scores (p-values) for each interaction (bait-prey pair) [90]. This p-value measures whether the observed fold change in the abundance of the prey protein identified in the AP/MS experiments with a given bait, compared to negative controls, is statistically significant given the observed variance of abundance measurements across all replicates. The application of t-test requires that each bait protein is analyzed in at least three biological replicates and a similar number of matching negative controls. In general, the same controls can be used for t-test analysis for all baits as long as all data were generated on the same experimental platform. T-test is more applicable to continuous data such as intensity-based abundance estimates than to spectral count data because of this test’s underlying assumption of the normality of the distribution. Furthermore, regardless of the quantitation method, t-test statistics are sensitive to missing data, especially for proteins that are identified in bait purifications at very low levels and not identified in the controls. This problem (which also applies to other scoring methods described below) can be addressed with the help of missing data imputation procedures.
For improved separation between true and false interactions, statistical scores such as p-value can be combined with the fold change in the abundance values. This is commonly done by making a volcano plot, i.e. plotting fold change against the negative log transformed p-values [91]. Proteins that have both a high magnitude of change (fold change) and high significance (low p-value) are selected as true interactors. The significance line corresponding to a desired False Discovery Rate (FDR) for interactors can be estimated by a permutation-based method. The accuracy of FDR estimations cannot be guaranteed, though. Thus, it is recommended that the FDR threshold is selected in such a way that none or only a few prey proteins are located in the ‘down regulated’ region (i.e. lower abundance compared to that in the negative controls) of the volcano plot which should not contain any preys that are true interactions [92].
Empirical scoring approaches for large datasets
While it is of course ideal that all baits are analyzed in at least triplicates, most published studies do not conform to this guideline. Furthermore, AP/MS datasets are sometimes even generated without performing parallel negative control experiments. In such cases, application of standard statistical methods such as t-test is not possible. Thus, several empirical approaches were developed that measure the significance of a particular prey i – bait j interaction by comparing the observed spectral count of prey i in the AP/MS experiment with bait j with respect to spectral counts observed for prey i in purifications with other baits. In one example of this strategy, CompPASS [46] calculates two different scores, Z and D scores. Z score is computed from the original spectral count by mean centering and scale normalization, using mean and standard deviation of spectral counts of prey i across all AP/MS experiments. As noted by the authors of the original paper, this score is less effective. This is so because the spectral count distribution is typically skewed in AP/MS datasets due to a large fraction of observations having zero counts (i.e. absence of protein identification). The more effective D-score is based on the spectral count adjusted by a scaling factor that reflects the reproducibility over replicate purifications of the same bait and the frequency of appearance of this prey in the dataset, and computed as
where Xij denotes the spectral count of prey i in purification with bait j, k is the total number of baits profiled in the experiment, fi is the number of experiments in which prey i was detected, and pij is the number of replicate experiments of bait j in which prey i was detected. After computing the scores, a threshold DT is selected using simulated data so that 95% of the simulated data falls below the chosen threshold. Note that CompPASS merges replicate data for bait j to produce a unique spectral count Xij for a given pair. In doing so, it considers only nonzero counts and then averages counts over multiple replicates.
Another empirical approach, termed E-filter, was used in [93] for the analysis of a related type of data generated using endogenous immunoprecipitation on a large scale. The distribution of total spectral counts for each protein (including zero counts for experiments where the protein was not identified) was tested by a simple boxplot-type analysis. The extreme upper outliers were considered as likely specific identifications. Based on a manual investigation of protein interaction scores for members of several well-known complexes, a prey i with spectral count Xij was called specific to bait j if Xij > Xi95 +3·(Xi95 − Xi5), where Xi95 and Xi5 are the 95th and 5th percentile values, respectively, in the spectral count distribution for prey i. It should be noted that this E-filter was applied only as a pre-processing method in a rather complicated ad hoc filtering scheme which included application of a “complex enrichment” (CE) filter.
The method presented by Sardiu et al. [45, 94, 95] takes a two-step approach to filtering non-specific binders. The method utilizes spectral counts normalized to protein length and the total spectral count in each experiment (normalized spectral abundance factors, NSAF). The first filtering step involves removing most obvious non-specific binders. In this step, the NSAF values of proteins in each of the individual purifications are compared with the average NSAF value observed for that protein in the negative controls. If the ratio of NSAF values is less than a certain empirically selected threshold, the protein is considered to be a non-specific binder for that particular bait. The NSAF value in this case is replaced by 0, otherwise it remains unchanged (the original manuscript utilized a similar vector length ratio approach [45] – a less practical method requiring an equal number of bait purifications and negative controls). After removing the proteins that are non-specific in all purifications in the dataset, the remaining bait-prey NSAF matrix is subjected to further filtering via singular value decomposition (SVD). SVD is used to find a group of proteins in the dataset that contributes most to the NSAF matrix by using a ranking estimation method. The subsequent analysis is restricted to the first left singular vector (LSV), which represents a weighted average of the overall protein NSAF values in the dataset. Only those proteins are retained that have the corresponding LSV coefficient large than a certain cutoff value. This threshold is determined manually for each dataset by analyzing the LSV scores of proteins that are known to be a part of the protein complexes under investigation. At the end, NSAF values for protein interactions passing the filter are converted to posterior probabilities. These probabilities, however, are not measures of confidence in the protein interaction and are not used for any additional filtering. Instead, they are used as scores reflecting the preference between a prey and a particular bait relative to all other baits. To summarize, this method is best-suited for processing datasets centered on reasonably well characterized protein complexes, and when the main goal is not as much the identification of novel interactions but in-depth analysis of the relationships between the components of these complexes.
Probabilistic modeling
In parallel with empirical scoring methods described above, an advanced probability approach, SAINT, for scoring AP/MS data has been described [30, 96, 97] (see Figure 3 for illustration). The first version of this method (SAINT 1) and the corresponding software was designed in the course of the analysis of the yeast kinase and phosphatase interactome [98]. Similar to CompPASS, SAINT 1 was designed to detect non-specific binders based on the analysis of spectral counts of prey proteins across a large number of purifications with different baits, and with only a minimal use of negative controls. The approach was then significantly extended in subsequent work (SAINT 2) [97]. Furthermore, while SAINT was originally developed to work with spectral count data, it has been recently extended to model continuous intensity-based data [99] (for reasons of clarity, SAINT will be reviewed and discussed below in the context of spectral count data).
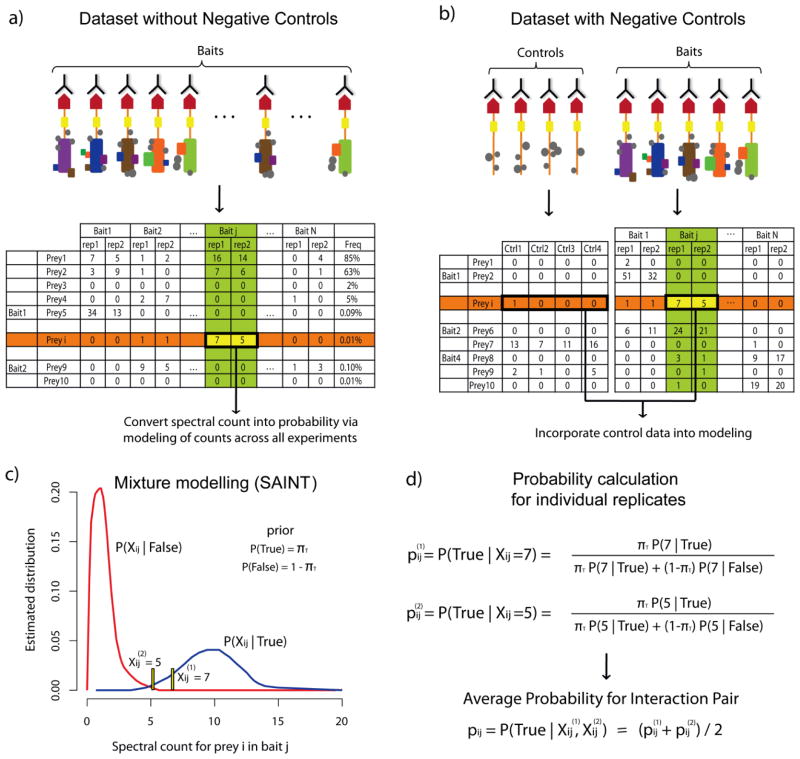
Figure 3. Scoring protein interactions using SAINT.
a–b) AP/MS protein interaction data in the absence (a) or presence (b) of negative control purifications. Schematic of the experimental AP/MS procedure and the resulting quantitative prey-bait matrix (illustrated using spectral counts). Each bait protein is profiled in two biological replicates. c) Modeling quantitative distributions for true and false interactions. For the interaction between prey i and bait j, SAINT uses all relevant data for the two proteins shown in panels a and b, i.e. abundance of prey i across all N baits (the data in the row of the prey, highlighted in orange) and abundance of other preys identified in the purification with bait j (the data in the column of the bait, highlighted in green). In the presence of negative control data, as in panel b, prey abundance across the negative controls is also incorporated in the modeling. d. Probability is calculated for each replicate using Bayes rule. Probabilities from the independent biological replicates, p(1) and p(2), are averaged to compute a summary probability for the interaction pair (i,j).
The underlying assumption in the mixture-model based approach of SAINT is that interactions identified in the experiment can be categorized into one of the two categories, true or false. The first category includes preys that are background contaminant proteins appearing with consistent spectral counts with a large fraction of baits (and also in the negative controls, when available). It also includes preys detected with low spectral counts, some of which could be false protein identifications. Preys that are true interactors appear with high spectral counts in a few baits but generally not across many experiments. SAINT compares normalized spectral count distributions for each of the preys in the dataset and across all purifications, and models these distributions to calculate the posterior probability of interaction between prey i and bait j using Bayes rule:
where Xij is the spectral count of prey i in purification experiment with bait j. P(x|λ) denotes the Poisson distribution for count data x with bait-prey specific mean parameters, λtrue and λfalse, for true and false interactions, respectively. The mixing proportion πT denotes the proportion of true interactions in the data. The distributional parameters (and the mixing proportion) are estimated from the data for each dataset. In the absence of multiple replicates for each bait, these parameters cannot be estimated only from the observed data for each bait-prey pair. Instead, they are estimated using a hierarchical Bayes technique that is an example of a commonly used statistical strategy called data pooling. In this strategy, the prey i - bait j specific parameters and are estimated by considering the spectral count distributions for prey i across all purifications and also considering spectral counts for other prey proteins identified in purification with bait j. Once these parameters are estimated and the posterior probabilities are computed for all interaction pairs, interactions can be sorted in a decreasing order of probabilities. The associated FDR for the filtered interactions can then be approximated at any threshold probability by averaging the complement of probability (1−P) for all selected interactions.
SAINT 1 was designed for the analysis of datasets containing a substantial number of different bait purifications (ideally more than 30 baits) that were acquired on a set of baits not significantly interconnected. In the case of highly interconnected baits, many valid interactions may appear across all or most purifications, thus looking like contaminant proteins. This is also true for empirical scoring methods, e.g. it necessitated the introduction of a weighted D score (WD-score) in a more recent application of CompPASS [100]. Probabilistic modeling of all false interactions (regardless of abundance level) using a single model specification represented another challenge and required addition of an empirical frequency-based filtering step. These challenges were addressed in SAINT 2. The statistical model was strengthened by directly incorporating the data from the negative controls (Figure 3b). This allowed a more uniform treatment of all false interactors regardless of their level abundance. It also allowed more accurate estimation of the false distribution because the mean λfalse parameter can then be estimated solely from the negative control data via semi-supervised mixture modeling. In addition, the model specifications were changed to use the generalized Poisson distribution with a variance component. As a result, SAINT 2 is able to more effectively process challenging datasets, e.g. datasets cont aining a large number of true interactions involving prey proteins that also frequently appear in AP/MS datasets as contaminants or non-specific binders [30].
In addition to SAINT, another probabilistic scoring approach, Decontaminator, was recently presented [101]. Decontaminator builds a model (null model) of contaminants using a small number of representative negative control experiments. As a proxy for protein abundance, the method uses Mascot protein identification scores and not MS intensities or spectral counts. The null model is then used to determine whether the Mascot score of a prey is significantly larger than what is observed for that prey in the negative controls and computes a p-value. The p-value distribution is then used to estimate FDR. The author demonstrated that their model was effective in reducing the number of non-specifically binding proteins in TAP experiments. As with SAINT, the advantage of the model is that it does not rely on the topological data and should not be significantly affected by the size of the dataset or the type of the network analyzed. At this time it is difficult to access the applicability of Decontaminator to single step affinity purification experiments in which the observed number of contaminant proteins is significantly higher than in TAP experiments.
Combining data from multiple biological replicates and experimental platforms
Another important problem that requires careful consideration is how to combine data from multiple replicates. Several computational approaches have to be considered depending on the nature of these replicates. Under the assumption of independence, multiple biological replicates can be naturally used (and are required) to compute p-values, and can be built in the probabilistic model as well. In the case of SAINT, the model by default reports the average of the probabilities of interaction from all replicates of the same bait (Figure 3d). Alternatively, the geometric mean can be used when a conservative approach is desired effectively penalizing protein interactions that were not identified consistently in all replicates. Selecting an appropriate way of combining the information from multiple biological replicates is necessary for empirical scoring schemes as well. In CompPASS, spectral counts from biological replicates are averaged prior to computing the D score. A relevant issue is how to treat technical replicates, i.e. repeated LC-MS/MS analyses of the same affinity purified sample. Technical replicates capture only non-biological, post-AP sources of variability such as MS and liquid chromatography which are easier to control and to minimise. Thus, it is best to combine data from multiple technical replicates within the same biological replicate. This can be done, e.g., by averaging the quantitative information for all technical replicates prior to scoring.
In certain biological applications, however, the replicate experiments for selected baits are performed under different AP conditions. This is commonly done for “challenging” baits (i.e., baits that, for various technical reasons and due to the properties of the bait protein itself [102], are not easily purified using AP/MS) to maximize the likelihood of capturing low stoichiometry or transient interactions that are sensitive to the experimental conditions. In such cases, it may be advantageous to select the maximum probability score from each replicate rather than the average. To eliminate spurious interactions, it is still advisable to generate multiple biological replicates for each AP condition, average the probabilities across the replicates generated under the same condition, and then select the maximum probability across all AP conditions for each bait-prey pair.
Furthermore, an increasing number of studies are utilizing the strategy of using multiple affinity purification tags (e.g., FLAG, HA, and TAP tags in [98]). Data generated using different epitope tags is best analysed separately at first, with appropriate controls when available, and then merged only after combining data from multiple biological replicates generated using the same epitope tag. In doing so, the less conservative approach of taking the union of interactions identified with different affinity tags should be justifiable.
Assessment of scoring methods and the need for reference datasets
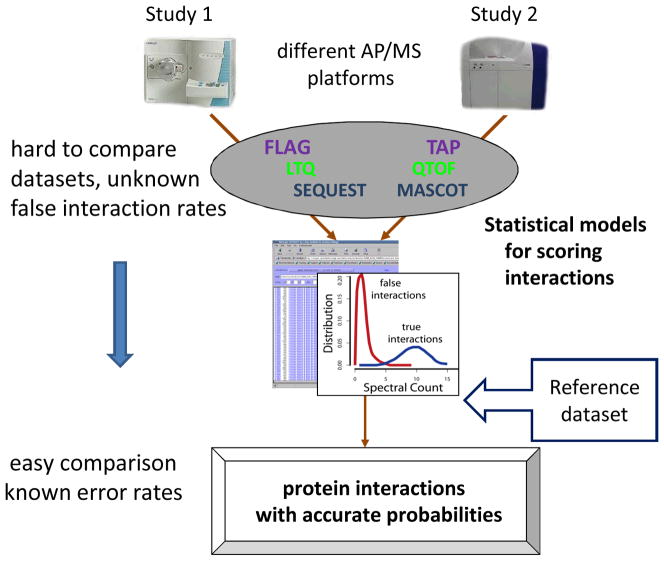
The purpose of developing more advanced computational methods to score protein interactions is two-fold. The first task is to achieve improved separation between true and false interactions, allowing more efficient filtering of AP/MS data. The second goal is to provide an accurate estimation of probabilities of individual interactions, as well as global error rates (FDR) in filtered datasets (Figure 4). Error rate estimation is particularly important in the case of large datasets where subsequent biological validation of identified protein interactions is not feasible or not intended, or when submitting data to public repositories. Of great importance here is the ability to perform accurate estimation of the error rates.
Figure 4. Toward transparent analysis of AP/MS protein interaction data.
Statistical models for computing confidence in protein interactions should enable transparent analysis and comparison of AP/MS data generated in different groups on different platform (tag types, MS instruments, database search tools used to identify proteins). Establishing the accuracy of scoring methods requires generation of reference AP/MS datasets.
In the absence of appropriate benchmark datasets, objective validation and comparison of the performance of a computational method and the accuracy of its error rate estimation procedure is a challenging task. One approach is to create a “gold standard” set of protein interactions using well characterized stable protein complexes such as those annotated in MIPS database [103]. The interactions between proteins in the same complex can be considered as true positives, and between proteins from different complexes as false positives. This strategy, however, can only be applied in the context of genome-wide AP/MS studies. In addition, interactions involving members of stable protein complexes are not representative of all true interactions, especially transient interactions. Thus, one cannot extrapolate the performance of a particular scoring scheme on a set of “gold standard” interactions to the entire dataset.
One can also consider the overlap of the entire set of detected protein interactions with the literature. For a particular dataset, true positive interactions can be compiled with a help of protein interaction databases such as BioGRID [104], HPRD [105], IntAct [106], DIP [107], MINT [108], or using iRefWeb [109] and PSICQUIC [110] which combine data from multiple sources. One caveat is that these databases contain false interactions, though this problem may be decreased when considering only those interactions that are detected multiple times, preferably by different approaches [111]. At the same time, the existing databases are incomplete, especially with respect to weak or transient interactions and less frequently studies proteins. A complementary approach is to assess the co-annotation rate of interaction partners to common Gene Ontology (GO) terms. This approach is also limited in utility because many proteins are annotated to multiple GO categories, and some do not have any annotation. As a result, using known interactions or GO co-annotation is more useful for comparative analysis of the specificity of different scoring methods rather than for evaluation of the absolute performance of one particular tool.
The need for more objective assessment of various scoring methods calls for generation of more appropriate reference datasets. Such datasets can be experimentally generated using a set of selected baits that are comprehensively analysed using multiple AP/MS strategies and possibly using complementary approaches as was done to benchmark binary interaction approaches [112]. This would require a substantial and organized effort, and the resulting reference datasets would still be limited in their utility given a large number of available experimental AP/MS platforms. Nevertheless, it would represent an important step toward the development of common data analysis standards and quality control procedures. One can envision that such experimentally generated reference datasets could be used to establish the guidelines for the application of various empirical scoring strategies, e.g. selection of score cut-offs. In the case of more advanced statistical methods, they can be helpful for establishing the accuracy of computed protein interaction scores, or for calibrating computed probabilities so they would correspond to the actual rate of true AP/MS interactions.
Limitations of scoring methods
The scoring methods for AP/MS data, including probabilistic methods, continue to evolve. With the SAINT model being increasingly applied to a wider range of AP/MS experiments, additional model specifications are being added to ensure accurate modeling of different types of datasets. For example, in SAINT, the probability of a particular bait–prey interaction is affected by the abundance of the prey in purification with other baits (the estimate of λtrue is influenced by the highest count values observed for that prey). As a result, if the prey is observed in purifications with several baits with high counts, and with several other baits with much lower counts (a commonly observed feature in datasets with high degree of interconnectivity), the probability of the interaction in the lower count experiments may be reduced. In essence, the model penalizes secondary interactions (some of which can be indirect interactions mediated by interaction of the prey and bait protein with a third protein), and may not be a desired feature in all cases. A number of limitations in the model assumptions in Decontaminator regarding the distribution of Mascot scores have been acknowledged as well [101]. The results of scoring interactions, probabilistically or empirically, are also dependent on the details of data pre-processing, e.g. ways to count spectra when computing spectral counts, and on details of missing data imputation (especially for intensity-based abundance measures). It is unreasonable to expect that a single statistical model or an empirical scoring scheme could equally well analyze drastically different datasets. Ultimately, the optimal analysis may involve multi-step strategies that combine the elements of probabilistic and empirical scoring, and also use contaminant repositories and integration with external data, as discussed below.
Repositories of non-specific proteins
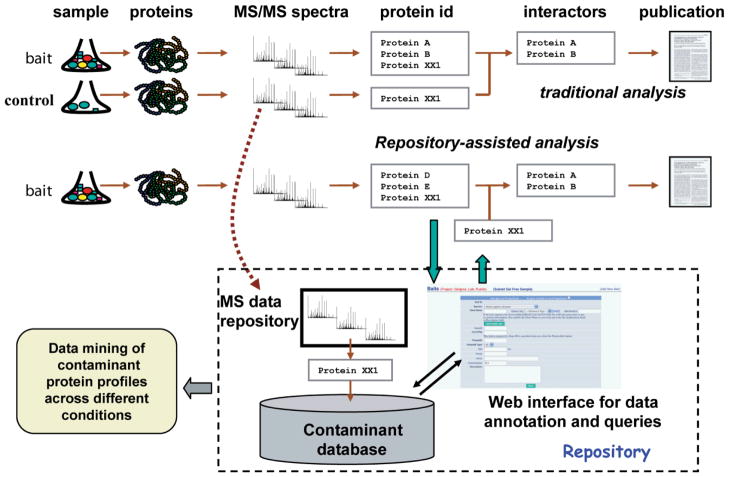
AP/MS experiments should ideally include a sufficient number of matching negative controls, allowing statistical modelling as outlined above. However, in some experiments, an insufficient number of high quality control runs may be collected due to cost/time considerations or technical reasons. The detection of non-specific binders can be assisted by referencing the experiment-specific protein lists against the databases of known contaminants identified in control runs from a wide variety of AP/MS experiments (Figure 5).
Figure 5. Building and using repositories of non-specific binders.
In the conventional analysis, negative controls are generated in parallel with experimental purifications and used to eliminate non-specifically binding proteins. Negative control data, from different studies and generated under different conditions, can be assembled to create a common repository of non-specific binders and background contaminants. In new experiments, this repository can be queried to extract negative control data generated under similar condition, which can then be used for filtering (either on its own as shown, or ideally in combination with negative control data generated as a part of the new experiment). The repository can also be used for additional computational analysis and data mining.
Many laboratories have independently assembled their own background contaminant lists based on the analyses of in-house generated control data [1, 44, 45, 113, 114]. However, at the moment there is no publicly available repository that would allow easy access to these datasets. We are involved in an on-going collaborative effort to create such a repository for AP/MS with intent to make it a growing resource (D. Mellacheruvu et al., in preparation). The repository is populated with negative control data obtained through the scientific community. Experiments are annotated to keep key information regarding the experimental conditions such as experimental system (cell type, expression system), purification (affinity matrix employed), and LC-MS/MS (e.g. gel based or gel-free). The database implements various search functions and allows selection of control datasets that most closely match the experimental conditions of the experiment under consideration.
Such a repository may serve multiple purposes. It can be used to perform various statistical and data mining analyses, e.g. to determine the propensity of various non-specific binding proteins to associate with different epitope tags or affinity matrices under different conditions. It can be used to gain a better understanding of the nature of non-specific background, and what experimental parameters have an effect on this background. Importantly, it can be used as a part of statistical or empirical scoring schemes such as those discussed above. Alternatively, it can be used as aid in manual data validation and for quality control.
Integrative modeling of AP/MS data
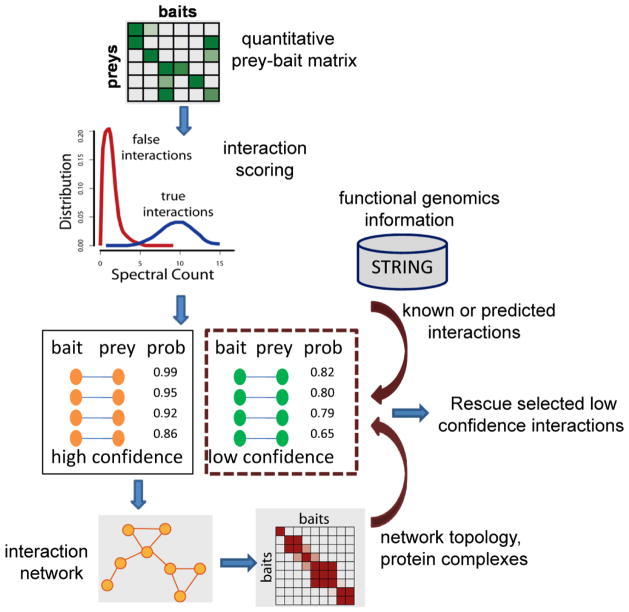
AP/MS, or any method for that matter, has limited sensitivity of detection depending on the selection of baits, experimental conditions, and the technology used. Even when a true interactor of the bait is identified by MS, the bait-prey interaction may receive a low confidence score due to low abundance (spectral count) of the prey. However, in many studies the list of protein interactions generated by AP/MS represents an entry point for subsequent higher level analysis. This includes network visualization and clustering to identify protein complexes and signaling modules, detection of cross-talk among different protein complexes and pathways, and additional experiments aiming to get a better understanding of the functional significance of the results. Some of the low scoring interactions can thus potentially be ‘rescued’ if they are corroborated by higher level or external evidence, e.g. in agreement with the prior biological knowledge, the higher level structure of the local network, or computational predictions based on functional genomics data (illustrated in Figure 6).
Figure 6. Integrative strategies for scoring protein interactions.
Starting with quantitative prey-bait matrix, protein interactions are scored using empirical or statistical models (e.g. using SAINT). High confidence interactions are used to assemble the interaction network, followed by clustering to reconstruct protein complexes. The higher level representation of the network (e.g. membership of a protein in a certain protein complex), as well as external information such as functional genomics data (e.g. prediction of the interaction in the STRING database), can be used to rescue low confidence interactions. These interactions can then be added to the network that was first reconstructed using high confidence interactions only.
To illustrate the concept of using higher level data, consider a low scoring interaction involving a prey protein identified with low spectral count. This prey protein, however, may also be identified as a high probability interactor of several other baits. If all these bait proteins are assigned to the same cluster (protein complex) based on a subsequent clustering analysis [87, 115], it increases the confidence in the interaction in question. A related strategy based on defining a complex enrichment score (CE filter) in addition to the spectral count-based E-filter (described earlier in this manuscript) was recently explored in [93].
An example of external data is prior biological knowledge. This includes previously reported protein interactions, and prior data regarding the composition of the protein complexes of interest, their cellular localization, or function. There are several examples of methods that attempted using such prior information [4, 5]. More generally, one can utilize many sources of functional genomics data, commonly used to assign function or place observations in a biological context, in the analysis of protein interactions or complexes identified from protein interaction screens [116]. External functional genomics data can be used to predict protein-protein interactions or gene functional associations, e.g. using Bayesian integration of multiple sources of biological information [117–122]. A number of computational systems are available that make such predictions, e.g. STRING [123] and GeneMania [124]. The confidence in the experimentally detected interaction should generally be higher if the interaction is predicted based on functional genomics data (illustrated in Figure 6).
One such strategy of combining experimental and predicted protein interaction data was recently used in [125]. For every bait-prey pair observed in the experimental TAP AP/MS data, the probability score was calculated as P = 1− (1−PEXP)(1−PSTRING), where PEXP and PSTRING are probabilities of interaction based on the experimental evidence (computed using a modified socio-affinity score taking into account spectral count information) and predicted by STRING, respectively. Thus, a low scoring TAP AP/MS interaction was accepted for subsequent analysis (clustering) if it had a sufficiently high score in STRING. However, such an approach assumes that both PEXP and PSTRING are accurate probability measures. This may be the case for PEXP, which was obtained by scaling the modified socio-affinity scores to fit the expected rates of true interactions based on a benchmark dataset created using known protein complexes [125]. The accuracy of STRING computed probabilities, however, has yet to be assessed. There is also a possibility of introducing a bias toward certain types of protein interactions or functional categories [126].
Thus, what represents the best way to combine the experimental evidence for a particular protein interaction with external or higher level information remains an open question. Computational validation of any rescoring method that utilizes external information is very challenging since the benchmark dataset cannot be created based on the same information that is used in making the predictions. The safest way is to restrict the use of external information to simple visualization. For example, one can visualize the interaction data filtered stringently based on the experimental AP/MS evidence, and then add to the network additional weaker interactions corroborated by external data. This way, the burden of utilizing the additional information would be shifted to the biologist interrogating such data and interested in a specific protein or protein complex.
Computational tools and informatics platforms
The development of computational methods for scoring label-free AP/MS protein interaction data is an active area of research. However, with respect to the availability of computational tools the choices at the moment are limited (not considering topology-based tools developed for genome-wide AP/MS data). MaxQuant (and the downstream data analysis system Perseus) is a powerful option for the analysis of AP/MS datasets generated using high mass accuracy instrumentation and with at least three biological replicates per bait (necessary for computing p-values in Perseus) [92]. To our knowledge, the CompPASS software is not available as a download; however its well-designed scoring scheme can be easily re-implemented in any in-house computational pipeline. The SAINT software is available as an open source tool (http://saint-apms.sourceforge.net), and is capable of processing both spectral count and intensity-based quantitative data.
Among the most dedicated efforts to develop a complete informatics solution in support of AP/MS experiments is ProHits data management system (http://www.prohitsms.com) [127]. The full version of ProHits provides secure storage of MS data, integration with search engines and MS analytical tools (including the open source X! Tandem search tool and the TPP pipeline), and allows web-based queries of the results. Alternatively, MS data can be processed outside of ProHits and uploaded in the database in, e.g., pepXML, protXML file formats (TPP analysis), or as unprocessed database search results files (e.g. Mascot output files). The Analyst module of ProHits allows easy visualization of data, comparison of multiple experiments, and permits export to third-party software, including the network visualization system Cytoscape [128]. ProHits is also fully integrated with SAINT, and allows export of filtered protein interaction data in the common PSI-MI file format [129] supported by most protein interaction data repositories. A smaller version of ProHits is now available as ProHits Lite virtual machine, providing an easy to install yet powerful integrated interaction proteomics solution for desktop computers.
Concluding remarks
Analysis of protein interactions and protein complexes using AP/MS is one of the most widely used and successful applications of MS-based proteomics in biological research. However, the development of practical computational tools for AP/MS data has lagged behind. A number of approaches have being developed for topology-based analysis of genome-wide interaction networks, but these methods are not applicable to AP/MS data that are generated in most studies. The reliability of results from on-going proteomics studies is of general concern, and there is a need for the development of methods for statistical assessment of protein interaction data in particular.
The importance of accurate bioinformatics analysis of proteomic data has been recognized by leading journals. However, so far most efforts in this area have focused on the problem of peptide and protein identification [15, 130–133], and in the context of the interactome work, on the reliability assessment of Y2H screens [112, 134, 135]. Fortunately, an increasing number of groups are now working to develop new methods and tools for the analysis of AP/MS data. The common denominator among these efforts is the use of label-free quantitative protein information such as spectral counts or integrated peptide intensities that can be extracted from AP/MS data. The methods used for scoring range from standard statistical approaches, to empirical scoring schemes optimized for a particular type of data, to advanced computational frameworks. While only a few computational software solutions are available at the moment, it is hoped that more tools will emerge in the future. Importantly, an increasing number of AP/MS experiments are now being deposited to public repositories, including both processed data and raw MS files. This should assist computational scientists in the development of novel bioinformatics methods and tools for AP/MS data.
It is hoped that eventually a number of competitive publicly available computational tools will emerge, and their performance will be tested using experimental AP/MS reference datasets. Generation of such reference datasest thus should be one of the priorities for the field of interaction proteomics. It is unrealistic to expect that different research groups involved in generation and analysis of their own data will all agree on using the same statistical approaches or computational tools. Still, it should be required by the scientific community that researchers conform to certain data analysis guidelines and also carefully document all data analysis steps. This could be facilitated by extending journal data publication guidelines [136, 137] to the domain of AP/MS protein interaction data.
Finally, proteomics technology is still evolving. For example, targeted proteomics approaches such as those based on selected reaction monitoring have emerged as a powerful addition to conventional shotgun proteomics [138]. These new strategies will be increasingly applied to the analysis of protein interactions and protein complexes, including monitoring their dynamic behavior in a quantitative manner [139]. This will undoubtedly require new computational and software developments in support of these new workflows. Thus, the development of new methods and tools for AP/MS-based analysis of protein interaction data should continue to be an active area of research for a foreseeable future.
Acknowledgments
I would like to thank Anne-Claude Gingras and Hyungwon Choi for numerous discussions and collaborations that resulted in this work, and also Anne-Claude Gingras for reading the manuscript. I also would like to thank Ruedi Aebersold, Gary Bader, Ashton Breitkreutz, Ileana Cristea, Rob Ewing, Damian Fermin, Timo Glatter, Matthias Gstaiger, Brett Larsen, Anna Malovannaya, Dattatreya Mellacheruvu, Jeff Ranish, Brian Raught, Michaela Sardiu, Mike Tyers, Michael Washburn, and Anastasia Yoccum for useful discussions, collaborations, or sharing data at various stages of this work. This work has been funded in part with NIH grants R01-CA-126239 and R01-GM-094231.
References
- 1.Ewing RM, Chu P, Elisma F, Li H, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Molecular Systems Biology. 2007;3 doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavin AC, Aloy P, Grandi P, Krause R, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 3.Gavin AC, Bosche M, Krause R, Grandi P, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Jeronimo C, Forget D, Bouchard A, Li Q, et al. Systematic Analysis of the Protein Interaction Network for the Human Transcription Machinery Reveals the Identity of the 7SK Capping Enzyme. Molecular Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krogan NJ, Cagney G, Yu H, Zhong G, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 6.Link AJ, Eng J, Schieltz DM, Carmack E, et al. Direct analysis of protein complexes using mass spectrometry. Nature Biotechnology. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 7.Rigaut G, Shevchenko A, Rutz B, Wilm M, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnology. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 8.Rinner O, Mueller LN, Hubalek M, Muller M, et al. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
- 9.Ho Y, Gruhler A, Heilbut A, Bader GD, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 10.Bauer A, Kuster B. Affinity purification-mass spectrometry: Powerful tools for the characterization of protein complexes. Eur J Biochem. 2003;270:570–578. doi: 10.1046/j.1432-1033.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- 11.Figeys D. Mapping the human protein interactome. Cell Research. 2008;18:716–724. doi: 10.1038/cr.2008.72. [DOI] [PubMed] [Google Scholar]
- 12.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 13.Wu CC, MacCoss MJ. Shotgun proteomics: Tools for the analysis of complex biological systems. Current Opinion in Molecular Therapeutics. 2002;4:242–250. [PubMed] [Google Scholar]
- 14.Kislinger T, Emili A. Multidimensional protein identification technology: current status and future prospects. Expert Review of Proteomics. 2005;2:27–39. doi: 10.1586/14789450.2.1.27. [DOI] [PubMed] [Google Scholar]
- 15.Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nature Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 16.Sadygov RG, Cociorva D, Yates JR. 3rd, Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat Methods. 2004;1:195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen GI, Gingras A-C. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 19.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Biol Chem. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 21.Kim B, Nesvizhskii AI, Rani PG, Hahn S, et al. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranish JA, Yi EC, Leslie DM, Purvine SO, et al. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 23.Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. Journal of Proteome Research. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 24.Vermeulen M, Hubner NC, Mann M. High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Current Opinion in Biotechnology. 2008;19:331–337. doi: 10.1016/j.copbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Margolin AA, Ong S-E, Schenone M, Gould R, et al. Empirical Bayes Analysis of Quantitative Proteomics Experiments. PLoS ONE. 2009;4:e7454. doi: 10.1371/journal.pone.0007454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Chiba T, Ozawa R, Yoshida M, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steen H, Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 29.Uetz P, Giot L, Cagney G, Mansfield TA, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 30.Skarra DV, Goudreault M, Choi H, Mullin M, et al. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011;11:1508–1516. doi: 10.1002/pmic.201000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nat Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 32.Cloutier P, Al-Khoury R, Lavallee-Adam M, Faubert D, et al. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods. 2009;48:381–386. doi: 10.1016/j.ymeth.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Deng M, Sun F, Chen T. Assessment of the reliability of protein-protein interactions and protein function prediction. Pac Symp Biocomput. 2003:140–151. [PubMed] [Google Scholar]
- 35.Qi Y, Klein-Seetharaman J, Bar-Joseph Z. Random forest similarity for protein-protein interaction prediction from multiple sources. Pac Symp Biocomput. 2005:531–542. [PubMed] [Google Scholar]
- 36.Sharan R, Suthram S, Kelley RM, Kuhn T, et al. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 38.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Hart GT, Lee I, Marcotte ER. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics. 2007;8:12. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholtens D, Gentleman R. Making sense of high-throughput protein-protein interaction data. Stat Appl Genet Mol Biol. 2004;3:Article 39. doi: 10.2202/1544-6115.1107. [DOI] [PubMed] [Google Scholar]
- 41.Sharp JL, Anderson KK, Hurst GB, Daly DS, et al. Statistically inferring protein-protein associations with affinity isolation LC-MS/MS assays. J Proteome Res. 2007;6:3788–3795. doi: 10.1021/pr0701106. [DOI] [PubMed] [Google Scholar]
- 42.Zhang B, Park BH, Karpinets T, Samatova NF. From pull-down data to protein interaction networks and complexes with biological relevance. Bioinformatics. 2008;24:979–986. doi: 10.1093/bioinformatics/btn036. [DOI] [PubMed] [Google Scholar]
- 43.Gingras AC, Caballero M, Zarske M, Sanchez A, et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Molecular & Cellular Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sardiu ME, Cai Y, Jin J, Swanson SK, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glatter T, Schittenhelm RB, Rinner O, Roguska K, et al. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol Syst Biol. 2011;7:547. doi: 10.1038/msb.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. Journal of Proteome Research. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 49.Jaffe JD, Mani DR, Leptos KC, Church GM, et al. PEPPeR, a platform for experimental proteomic pattern recognition. Molecular & Cellular Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li XJ, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Molecular & Cellular Proteomics. 2005;4:1328–1340. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ. 1st Annual Symposium on Proteome Fractionation; Cambridge, MA. 2004. pp. 144–156. [Google Scholar]
- 52.Trudgian DC, Ridlova G, Fischer R, Mackeen MM, et al. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics. 2011;11:2790–2797. doi: 10.1002/pmic.201000800. [DOI] [PubMed] [Google Scholar]
- 53.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 54.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular Systems Biology. 2008;4 doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi H, Fermin D, Nesvizhskii AI. Significance Analysis of Spectral Count Data in Label-free Shotgun Proteomics. Molecular & Cellular Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gramolini AO, Kislinger T, Alikhani-Koopaei R, Fong V, et al. Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Molecular & Cellular Proteomics. 2008;7:519–533. doi: 10.1074/mcp.M700245-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Fu X, Gharib SA, Green PS, Aitken ML, et al. Spectral index for assessment of differential protein expression in shotgun proteomics. Journal of Proteome Research. 2008;7:845–854. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- 58.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nature Biotechnology. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 59.Xia QW, Wang TS, Park Y, Lamont RJ, Hackett M. Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: Non-label methods comparison, q-values and LOWESS curve fitting. International Journal of Mass Spectrometry. 2007;259:105–116. doi: 10.1016/j.ijms.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blondeau F, Ritter B, Allaire PD, Wasiak S, et al. Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3833–3838. doi: 10.1073/pnas.0308186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishihama Y, Oda Y, Tabata T, Sato T, et al. Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.McAfee KJ, Duncan DT, Assink M, Link AJ. Analyzing proteomes and protein function using graphical comparative analysis of tandem mass spectrometry results. Molecular & Cellular Proteomics. 2006;5:1497–1513. doi: 10.1074/mcp.T500027-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, et al. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of Relative Abundance Ratios Derived from Peptide Ion Chromatograms and Spectrum Counting for Quantitative Proteomic Analysis Using Stable Isotope Labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 65.Usaite R, Wohlschlegel J, Venable JD, Park SK, et al. Characterization of global yeast quantitative proteome data generated from the wild-type and glucose repression Saccharomyces cerevisiae strains: The comparison of two quantitative methods. Journal of Proteome Research. 2008;7:266–275. doi: 10.1021/pr700580m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.States DJ, Omenn GS, Blackwell TW, Fermin D, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 67.Tang HX, Arnold RJ, Alves P, Xun ZY, et al. 14th Conference on Intelligent Systems for Molecular Biology; Fortaleza, BRAZIL. 2006. pp. E481–E488. [Google Scholar]
- 68.Cai L, Huang HY, Blackshaw S, Liu JS, et al. Clustering analysis of SAGE data using a Poisson approach. Genome Biol. 2004;5:9. doi: 10.1186/gb-2004-5-7-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang JPZ, Lindsay BG, Cui LY, Wall PK, et al. Gene capture prediction and overlap estimation in EST sequencing from one or multiple libraries. BMC Bioinformatics. 2005;6:11. doi: 10.1186/1471-2105-6-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wold B, Myers RM. Sequence census methods for functional genomics. Nat Meth. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 71.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. PNAS. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong JWH, Sullivan MJ, Cagney G. Computational methods for the comparative quantification of proteins in label-free LCn-MS experiments. Brief Bioinform. 2008;9:156–165. doi: 10.1093/bib/bbm046. [DOI] [PubMed] [Google Scholar]
- 74.Isserlin R, Emili A. Interpretation of large-scale quantitative shotgun proteomic profiles for biomarker discovery. Curr Opin Mol Ther. 2008;10:231–242. [PubMed] [Google Scholar]
- 75.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. Abacus: A computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11:1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.May D, Fitzgibbon M, Liu Y, Holzman T, et al. A platform for accurate mass and time analyses of mass spectrometry data. J Proteome Res. 2007;6:2685–2694. doi: 10.1021/pr070146y. [DOI] [PubMed] [Google Scholar]
- 78.Mueller LN, Rinner O, Schmidt A, Letarte S, et al. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. PROTEOMICS. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 79.Tsou CC, Tsai CF, Tsui YH, Sudhir PR, et al. IDEAL-Q, an Automated Tool for Label-free Quantitation Analysis Using an Efficient Peptide Alignment Approach and Spectral Data Validation. Molecular & Cellular Proteomics. 2010;9:131–144. doi: 10.1074/mcp.M900177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 81.Rappsilber J, Mann M. What does it mean to identify a protein in proteomics? Trends in Biochemical Sciences. 2002;27:74–78. doi: 10.1016/s0968-0004(01)02021-7. [DOI] [PubMed] [Google Scholar]
- 82.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data - The protein inference problem. Molecular & Cellular Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 83.Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE - A virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Nagaraj N, Wisniewski JR, Geiger T, Cox J, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwanhausser B, Busse D, Li N, Dittmar G, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to Label Free Proteome Quantitation: How to Deal with Peptides Shared by Multiple Proteins. Analytical Chemistry. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 87.Choi H, Kim S, Gingras AC, Nesvizhskii AI. Analysis of protein complexes through model-based biclustering of label-free quantitative AP-MS data. Mol Syst Biol. 2010;6:11. doi: 10.1038/msb.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ho L, Ronan JL, Wu J, Staahl BT, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandez E, Collins MO, Uren RT, Kopanitsa MV, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hubner NC, Bird AW, Cox J, Splettstoesser B, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. Journal of Cell Biology. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cui X, Churchill G. Statistical tests for differential expression in cDNA microarray experiments. Genome Biology. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 93.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, et al. Analysis of the Human Endogenous Coregulator Complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mosley AL, Sardiu ME, Pattenden SG, Workman JL, et al. Highly Reproducible Label Free Quantitative Proteomic Analysis of RNA Polymerase Complexes. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sardiu ME, Gilmore JM, Carrozza MJ, Li B, et al. Determining Protein Complex Connectivity Using a Probabilistic Deletion Network Derived from Quantitative Proteomics. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Breitkreutz A, Choi H, Sharom J, Boucher L, et al. Global Architecture of the Yeast Protein Kinase and Phosphatase Interaction Network. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi H, Larsen B, Lin ZY, Breitkreutz A, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature Methods. 2011;8:70–U100. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Breitkreutz A, Choi H, Sharom JR, Boucher L, et al. A Global Protein Kinase and Phosphatase Interaction Network in Yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi H, Glatter T, Gstaiger M, Nesvizhskii AI. SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification – mass spectrometry experiments. Journal of Proteome Research. 2012 doi: 10.1021/pr201185r. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–U84. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lavallee-Adam M, Cloutier P, Coulombe B, Blanchette M. Modeling Contaminants in AP-MS/MS Experiments. Journal of Proteome Research. 2011;10:886–895. doi: 10.1021/pr100795z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saha S, Kaur P, Ewing RM. The Bait Compatibility Index: Computational Bait Selection for Interaction Proteomics Experiments. Journal of Proteome Research. 2010;9:4972–4981. doi: 10.1021/pr100267t. [DOI] [PubMed] [Google Scholar]
- 103.Pagel P, Kovac S, Oesterheld M, Brauner B, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 104.Breitkreutz BJ, Stark C, Reguly T, Boucher L, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Research. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Research. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, et al. IntAct - open source resource for molecular interaction data. Nucleic Acids Research. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Salwinski L, Miller CS, Smith AJ, Pettit FK, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Research. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ceol A, Aryamontri AC, Licata L, Peluso D, et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Research. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Malovannaya A, Li YH, Bulynko Y, Jung SY, et al. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aranda B, Blankenburg H, Kerrien S, Brinkman FSL, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nature Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cusick ME, Yu HY, Smolyar A, Venkatesan K, et al. Literature-curated protein interaction datasets. Nature Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Braun P, Tasan M, Dreze M, Barrios-Rodiles M, et al. An experimentally derived confidence score for binary protein-protein interactions. Nature Methods. 2009;6:91–98. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen GI, Gingras A-C. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 114.Boulon S, Ahmad Y, Trinkle-Mulcahy L, Verheggen C, et al. Establishment of a Protein Frequency Library and Its Application in the Reliable Identification of Specific Protein Interaction Partners. Molecular & Cellular Proteomics. 2010;9:861–879. doi: 10.1074/mcp.M900517-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sardiu ME, Florens L, Washburn MP. Evaluation of Clustering Algorithms for Protein Complex and Protein Interaction Network Assembly. Journal of Proteome Research. 2009;8:2944–2952. doi: 10.1021/pr900073d. [DOI] [PubMed] [Google Scholar]
- 116.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nature Biotechnology. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 117.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 118.Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, et al. Probabilistic model of the human protein-protein interaction network. Nature Biotechnology. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 119.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions Part II. Computational methods to predict protein and domain interaction partners. Plos Computational Biology. 2007;3:595–601. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qiu J, Noble WS. Predicting co-complexed protein pairs from heterogeneous data. Plos Computational Biology. 2008;4 doi: 10.1371/journal.pcbi.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Myers CL, Troyanskaya OG. Context-sensitive data integration and prediction of biological networks. Bioinformatics. 2007;23:2322–2330. doi: 10.1093/bioinformatics/btm332. [DOI] [PubMed] [Google Scholar]
- 122.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat Rev Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jensen LJ, Kuhn M, Stark M, Chaffron S, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucl Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuhner S, van Noort V, Betts MJ, Leo-Macias A, et al. Proteome Organization in a Genome-Reduced Bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 126.Lu LJ, Xia Y, Paccanaro A, Yu HY, Gerstein M. Assessing the limits of genomic data integration for predicting protein networks. Genome Research. 2005;15:945–953. doi: 10.1101/gr.3610305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu G, Zhang J, Larsen B, Stark C, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotech. 2010;28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cline MS, Smoot M, Cerami E, Kuchinsky A, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, et al. The HUPO PSI’s Molecular Interaction format[mdash]a community standard for the representation of protein interaction data. Nat Biotech. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 130.Carr S, Aebersold R, Baldwin M, Burlingame A, et al. The need for guidelines in publication of peptide and protein identification data - Working group on publication guidelines for peptide and protein identification data. Molecular & Cellular Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 131.Fenyo D, Beavis RC. Informatics development: Challenges and solutions for MALDI mass spectrometry. Mass Spectrom Rev. 2008;27:1–19. doi: 10.1002/mas.20152. [DOI] [PubMed] [Google Scholar]
- 132.Martens L, Hermjakob H. Proteomics data validation: why all must provide data. Mol Biosyst. 2007;3:518–522. doi: 10.1039/b705178f. [DOI] [PubMed] [Google Scholar]
- 133.Stead DA, Paton N, Missier P, Embury SM, et al. Information quality in proteomics. Brief Bioinform. 2008;9:174–188. doi: 10.1093/bib/bbn004. [DOI] [PubMed] [Google Scholar]
- 134.Venkatesan K, Rual JF, Vazquez A, Stelzl U, et al. An empirical framework for binary interactome mapping. Nature Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chiang T, Scholtens D, Sarkar D, Gentleman R, Huber W. Coverage and error models of protein-protein interaction data by directed graph analysis. Genome Biol. 2007;8:14. doi: 10.1186/gb-2007-8-9-r186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Carr S, Aebersold R, Baldwin M, Burlingame A, et al. The Need for Guidelines in Publication of Peptide and Protein Identification Data: Working Group On Publication Guidelines For Peptide And Protein Identification Data. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 137.Wilkins MR, Appel RD, Van Eyk JE, Chung MCM, et al. Guidelines for the next 10 years of proteomics. Proteomics. 2006;6:4–8. doi: 10.1002/pmic.200500856. [DOI] [PubMed] [Google Scholar]
- 138.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular Systems Biology. 2008;4:14. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bisson N, James DA, Ivosev G, Tate SA, et al. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011;29:653–U138. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]