Abstract
The origin recognition complex (ORC) binds origins of replication and directs the assembly of a higher order protein complex at these sites. ORC binds and hydrolyzes ATP in vitro. ATP binding to the largest subunit of ORC, Orc1p, stimulates specific binding to origin DNA; however, the function of ATP hydrolysis by ORC is unknown. To address the role of ATP hydrolysis, we have generated mutants within Orc1p that are dominant lethal. At physiological ATP concentrations, these mutants are defective for ATP hydrolysis but not ATP binding in the absence of DNA. These mutants inhibit formation of the prereplicative complex when overexpressed. The dominant lethal phenotype of these mutant ORC complexes is suppressed by simultaneous overexpression of wild-type, but not mutant, Cdc6p. Our findings suggest that these hydrolysis-defective mutants inhibit growth by titrating Cdc6p away from the origin. Based on these observations, we propose that Cdc6p specifically recognizes the ATP-bound state of Orc1p and that ATP hydrolysis is coupled to preRC disassembly.
Initiation of DNA replication requires the precise and timely assembly of protein factors at each origin of replication. Recent work by a number of labs has identified a set of factors that localize to origins during the G1 phase of the cell cycle (1–5). In Saccharomyces cerevisiae, these higher-order complexes are nucleated by the origin recognition complex (ORC), which binds origin DNA in vitro and in vivo (1, 3, 6). ORC appears to be bound to origins throughout the cell cycle (4, 7) and is required for the stepwise recruitment of Cdc6p and a complex of six related proteins, Mcm2–7p, to the origin. This complex containing at least ORC, Cdc6p, and Mcm2–7p is known as the prereplicative complex (preRC; reviewed in ref. 8). Although originally identified in the yeast S. cerevisiae, subsequent studies have identified analogs of ORC and a similar preRC assembly process in multiple other eukaryotic species (reviewed in ref. 8).
Ten of the 14 polypeptides known to be present in the S. cerevisiae preRC contain consensus nucleotide binding motifs within their sequences (9). In prokaryotic replication systems, ATP plays multiple roles in the initiation process (10, 11). Thus, it is likely that understanding the role of nucleotides in the eukaryotic initiation process will be important to determine the molecular details of this critical cellular event. Orc1p, Orc4p, Orc5p, Cdc6p, and Mcm2–7p all are members of a class of ATPases known as the AAA+ family (standing for ATPases associated with a variety of cellular activities; ref. 12). This family contains a region of sequence similarity that extends over 220–250 aa and includes the Walker A and B motifs common to many nucleotide binding proteins (reviewed in ref. 15). AAA+ members carry out diverse functions within the cell, including proteolysis, transcription, DNA replication, and recombination. A common functional theme for the role of ATP in these proteins is the regulation of the formation, rearrangement, and dissociation of macromolecular complexes (12). Typically, ATP binding stimulates the formation of the macromolecular complex and ATP hydrolysis stimulates disassembly (reviewed in ref. 13).
We have previously demonstrated that the largest subunit of ORC, Orc1p, binds and hydrolyzes ATP (14). ATP binding to Orc1p is essential for ORC to specifically recognize and bind origins (6, 14). Furthermore, when ORC is bound to origin DNA, the ATPase activity of Orc1p is inhibited and ATP remains stably bound to Orc1p. We have hypothesized that this bound ATP could be hydrolyzed in a reaction coupled to a downstream step in replication, such as recruitment of other preRC components, initiation of replication, or inactivation of origins after initiation (14). To better understand the role of ATP hydrolysis in the function of ORC, we sought to generate Orc1p mutants that retain the ability to bind ATP, but lack hydrolysis activity.
Mutation of the conserved Walker A motif within Orc1p leads to a loss of both ATP binding and hydrolysis activities. A second region common to many ATPases, the Walker B motif, is hypothesized to coordinate nucleotide hydrolysis activity rather than nucleotide binding. Consistent with this hypothesis, mutations within the B-motif of a number of ATPases have been identified that specifically affect ATP hydrolysis but not ATP binding (for examples, see refs. 16 and 17). Mutations that inhibit ATP hydrolysis but not ATP binding often possess a dominant lethal phenotype in vivo. For example, a mutant of the Escherichia coli initiator protein, DnaA, that can bind but not hydrolyze ATP causes overinitiation and lethality in a dominant manner (18). Similarly, a mutation in the Walker B motif of yeast Cdc6p leads to dominant lethality when overexpressed, whereas overexpression of wild-type Cdc6p or a mutant that is presumed to lack both ATP binding and hydrolysis activities does not cause lethality (19).
Here, we describe the isolation and characterization of mutants within Orc1p that are dominant lethal when overexpressed and have hydrolysis-specific defects in vitro. We demonstrate that these mutants block replication by inhibiting preRC formation. Co-overexpressing Cdc6p suppresses the lethality of mutant ORC overexpression, suggesting that these mutant ORC complexes specifically titrate Cdc6p away from the origin. These findings support a model in which ATP binding to Orc1p stimulates higher order complex assembly and in which ATP hydrolysis by ORC stimulates preRC disassembly.
Materials and Methods
Plasmids and Strains.
Genotypes of yeast strains used in this study are listed in Table 1. Strains and plasmids were prepared by using standard laboratory methods (20). Plasmids to overexpress two of the ORC subunits were prepared by first subcloning the GAL1–10 promoter prepared by PCR from yeast genomic DNA into pMDW13 (ORC1,6), pMDW8 (ORC3,4), and pSPB25 (ORC2,5) baculovirus transfer vectors (21). Fragments containing the GAL1–10 promoter-driven ORC genes were then subcloned into the multicloning sites of yeast integrating vectors p404 (to generate p404 Gal1–10 ORC3,4), p405 (to generate p405 Gal1–10 ORC2,5), and p403 (to generate p403 Gal1–10 ORC1,6). The lys2 gene from pRS317 cut with PvuII was subcloned into p405 Gal1–10 ORC2,5 cut with Tth111 I and XhoI to generate pLys2 Gal1–10 ORC2,5. A triple hemagglutinin (HA) tag was introduced at the C terminus of ORC1 in the overexpressing construct to yield p403 Gal1–10 ORC1c-HA,6. To test complementation, mutants were subcloned into p403-ORC1 (with the endogenous ORC1 promoter) and integrated into AIAy20. The plasmid-borne copy of wild-type ORC1 was selected against by plating on media containing 5-fluoroorotic acid (5-FOA). Mutants were also subcloned into pMDW13 for protein expression. CDC6 plasmids pSF320-CDC6 and pSF320-Cdc6K114E contain the CDC6 gene with an N-terminal 10XHis tag and a C-terminal 3XHA tag under the control of the Gal1–10 promoter. The control vector pSF322 expresses only the 3XHA tag.
Table 1.
Strains used in this study
| Strain Name | Genotype |
|---|---|
| AIAy20 | ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 lys2∷HisG Bar1∷HisG orc1∷HisG pSPB162 MATa |
| RKy50 | ade2-1 ura3-1 his3-11,15 can1-100 Bar1∷HisG orc1∷HisG leu2∷ORC1 trp1∷p404-GAL1-10-ORC3,4 lys2∷plys2-GAL1-10-ORC2,5 MATa |
| RKy61 | RKy50 his3∷p403-GAL1-10-ORC1c-HA,6 |
| RKy62 | RKy50 his3∷p403-GAL1-10-Orc1(K485T)c-HA,6 |
| RKy63 | RKy50 his3∷p403-GAL1-10-Orc1-dlc-HA,6 |
| RKy64 | RKy50 his3∷p403-GAL1-10-Orc1-d2c-HA,6 |
| RKy83 | RKy63 ura3∷pSF321-CDC6 |
| RKy84 | RKy64 ura3∷pSF321-CDC6 |
| RKy85 | RKy63 ura3∷pSF323 |
| RKy86 | RKy64 ura3∷pSF323 |
| RKy87 | RKy63 ura3∷pSF321-cdc6K114E |
| RKy88 | RKy64 ura3∷pSF321-cdc6K114E |
| RKy90 | AIAy20 his3∷p403-ORC1 |
| RKy91 | AIAy20 his3∷p403-orc1-K485T |
| RKy92 | AIAy20 his3∷p403-orc1-d1(D569Y) |
| RKy93 | AIAy20 his3∷p403-orc1-d2(D569F) |
| RKy94 | ade2-1 ura3-1 trp1-1 leu2-3,112 can1-100 lys2∷HisG Bar1∷HisG orc1∷HisG his3∷p403-ORC1 MATa |
| RKy95 | ade2-1 ura3-1trp1-1 leu2-3,112 can1-100 lys2∷HisG Bar1∷HisG orc1∷HisG his3∷p403-orc1-d1 MATa |
| RKy96 | ade2-1 ura3-1trp1-1 leu2-3,112 can1-100 lys2∷HisG Bar1∷HisG orc1∷HisG his3∷p403-orc1-d2 MATa |
Screen for Lethal When Overexpressed Mutants.
Four oligonucleotides were used that contain degenerate sequences at the three positions of each of the four codons of the Walker B “DELD” sequence (amino acids 566–569) of Orc1p. The sequences of these oligonucleotides (which hybridize to the sense strand) are as follows: ORC1B1, 5′TTCGTTACCA TGGCATCGAG TTCFENCAAC AAGACTACAA TGGTTTTC-3′; ORC1B2, 5′-TTCGTTACCA TGGCATCGAG FENGTCCAAC AAGACTACAA TGGTTTTC-3′; ORC1B3, 5′-TTCGTTACCA TGGCATCFEN TTCGTCCAAC AAGACTACAA TGGTTTTC-3′; ORC1B4, 5′-TTCGTTACCA TGGCFENGAG TTCGTCCAAC AAGACTACAA TGGTTTTC-3′; where F = G(40%), C(40%), A(20%), and T(0%); where E = G(20%), C(20%), A(30%), and T(30%); and where N = G(25%), C(25%), A(25%), and T(25%). These ratios were chosen to minimize amino acid bias and stop codons, similar to ref. 22. These oligonucleotides were used to PCR amplify the ATP binding domain of ORC1. The mutant PCR products were first cloned into p403-ORC1c-HA (containing a triple HA tag at the C terminus of ORC1). The pool of mutant orc1 genes was then ligated into p403 Gal1–10 ORC1c-HA,6. Plasmids were prepared from individual transformants from the ligation, and were individually tested by integration into RKy50 and streaking on plates containing 2% Galactose.
Protein Purification.
ORC mutant complexes were expressed by using baculovirus-infected cells and purified as described (14).
Chromatin Immunoprecipitation (ChIP).
ChIP was performed as described (1) with minor modification. For ORC ChIP, a rabbit polyclonal ORC antibody was used. For MCM ChIP, a monoclonal antibody that recognizes all six MCM subunits was used. Incubation time for this antibody was 6 h, after which protein G beads were added and incubated for an additional hour. PCR was performed for 28 cycles on 1/50 of the immunoprecipitates, and on 1/500 of the input material. Quantification was performed by using the Molecular Dynamics Fluorimager and imagequant software. To assay loading of MCM proteins during orc1 mutant overexpression, cells were grown in 2% raffinose and arrested with 10 μg/ml nocodazole. After 3 h in nocodazole, galactose was added to 2% to induce ORC overexpression or glucose was added to 2% to repress expression. After an additional 90 min, cells were washed three times and resuspended in media containing 50 ng/ml alpha factor and either 2% galactose or 2% glucose. Cells were fixed for ChIP after >95% of cells were in G1.
ATP Hydrolysis Assays and DNase I Protection Assays.
ORC ATP hydrolysis was monitored by using TLC as previously described (14). Hydrolysis reactions contained 1 μg ORC, 50 mM Hepes (pH 7.6), 150 mM KCl, 5 mM MgOAc, 1 mM EDTA, 1 mM EGTA, 0.02% Nonidet P-40, and ATP as indicated. All reactions included 0.5 μCi alpha [32P]ATP. Total reaction volume was 13.3 μl. Aliquots of 1.5 μl were removed and added to 0.38 μl 2% SDS over a time course of 3 h.
DNase I protection assays were performed as described (21). Each reaction contained 50 ng ORC, 50 ng poly(dGdC) competitor DNA, and ≈5 fmol of DNA probe derived from pARS1/WT cut with EcoRI and HindIII (radiolabeled on the T-rich strand of the ARS concensus sequence).
Results
Dominant Lethal Alleles Within ORC1.
To address the role of Orc1p ATP hydrolysis, we sought mutants in the Walker B motif of ORC1 that cause lethality when overexpressed. Our strategy was based on two observations concerning other ATPases. First, studies of other ATPases indicate that mutations that cause lethality when overexpressed frequently are able to bind but not hydrolyze ATP. Second, mutations in the Walker B motif of other ATPases frequently inhibit the ATPase activity but not the ATP binding activity of these proteins. Because Orc1p is stably associated with the other five ORC subunits and has no apparent activity on its own (ref. 23 and data not shown), the screen was performed in a strain that simultaneously overexpressed the mutant Orc1p and wild-type Orc2p-Orc6p after induction with galactose.
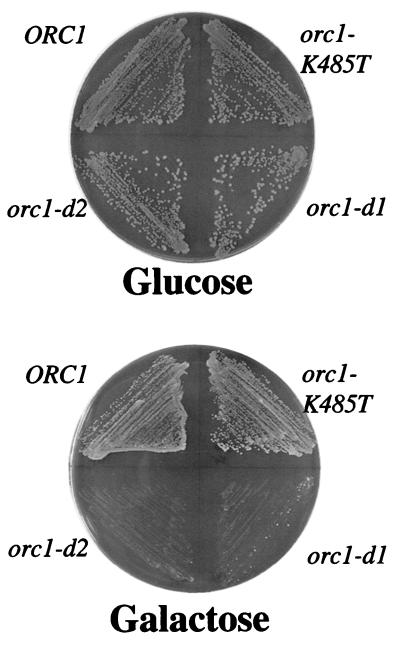
We first tested whether overexpression of wild-type ORC caused lethality, because it was possible that ORC overexpression could titrate critical replication factors away from the origin. There was no growth defect caused by wild-type ORC overexpression (Fig. 1). We also tested whether a previously identified mutant in Orc1p that prevents ATP binding (and hydrolysis) was dominant lethal (14). We found that this mutant, orc1-K485T, also did not exhibit a dominant lethal phenotype when overexpressed (Fig. 1).
Figure 1.
Dominant lethal when overexpressed mutants within ORC1. Strains containing wild-type ORC1 (upper left), a mutant form of ORC1 that cannot bind ATP (orc1-K485T, upper right), orc1-d1 (D569Y, lower right), or orc1-d2 (D569F, lower left) under the control of the GAL1–10 promoter were grown on a plate containing galactose (Lower), which induces overexpression, or glucose (Upper), which does not induce overexpression. All strains also overexpressed wild-type ORC2-ORC6 genes when grown on galactose.
Having found that neither wild-type ORC nor ORC containing an Orc1p ATP-binding mutant showed a dominant negative phenotype, we initiated a screen to identify mutations in the Orc1p Walker B motif that exhibited such a phenotype. Four conserved amino acid residues within the Walker B motif (DELD) were randomly mutagenized (see Materials and Methods). We individually tested 80 random mutants targeted to each of the four amino acids (320 total) and found 18 that resulted in lethality or decreased viability when overexpressed. The seven mutants that caused a complete loss of viability were sequenced. Of these, four corresponded to a change of the second aspartate to tyrosine (D569Y, named Orc1-d1) and three corresponded to a change of the second aspartate to phenylalanine (D569F, named Orc1-d2). Because these alleles were obtained multiple times with different codons encoding the tyrosine or the phenylalanine, we believe that the library of orc1 mutants was saturated. Interestingly, no dominant mutants were identified in the first two amino acid positions, which are typically the most well conserved among ATP binding proteins. The lack of mutants in this region may be due to inhibition of ATP binding, because previous studies of ORC complexes mutated in these amino acids resulted in a loss of ATP binding as well as ATP hydrolysis (data not shown). Consistent with the hypothesis that the lethal phenotype was due to the overexpression of an intact mutant ORC complex, the dominant lethal phenotype of Orc1-d1 and Orc1-d2 required co-overexpression of ORC2–6 (data not shown).
ORC1-d1 Has a Hydrolysis Defect in Vitro.
To understand the molecular defect associated with these dominant lethal alleles of Orc1p, we expressed and purified the mutant orc1-d1 encoded subunit complexed with the remaining five ORC subunits and characterized this mutant ORC complex (for simplicity hereafter we refer to this complex as ORC-d1 and the complex containing Orc1-d2p as ORC-d2) in vitro. The ORC complex used for these in vitro studies contained a second mutation in the Walker A motif of Orc5p to focus the analysis on ATP binding to Orc1p. Orc5p binds ATP in a DNA-independent manner, and does not hydrolyze ATP at detectable levels (14). We are confident that this change in the complex did not affect our studies of Orc1p function because Orc5p mutants that cannot bind ATP support viability, indicating that ATP binding to Orc5p is not required for any essential ORC function.
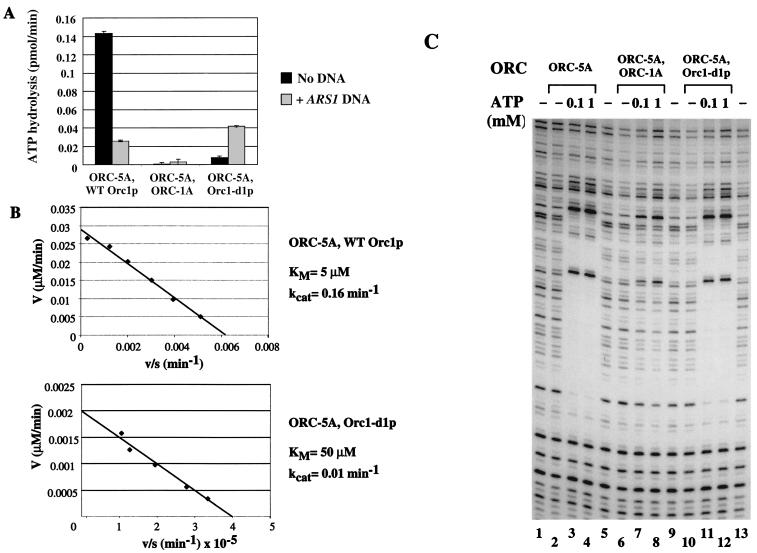
ATP hydrolysis activity was measured for an ORC complex containing wild-type Orc1p, a complex with a point mutation in the Orc1p Walker A motif that eliminates ATP binding (orc1-K485T, ORC-1A), and the ORC-d1 complex (Fig. 2A). As shown previously, ATP hydrolysis by wild-type Orc1p is inhibited by autonomously replicating sequence-1 (ARS1) origin DNA. The ORC-1A complex is defective for ATP hydrolysis both in the presence and absence of ARS1 DNA. The mutant ORC-d1 complex hydrolyzed ATP at a 16-fold reduced rate in the absence of ARS1 DNA. In contrast, in the presence of ARS1 DNA, the ORC1-d1 mutant could hydrolyze ATP at a rate slightly higher than wild-type ORC.
Figure 2.
Biochemical characterization of an Orc1-d1p-containing complex. (A) ORC-d1 is defective in ATP hydrolysis. ATP hydrolysis was measured for ORC complexes containing wild-type Orc1p, an ATP binding-defective mutant Orc1p (ORC-1A) or Orc1-d1p. Hydrolysis was measured in the absence of DNA (filled bars) or in the presence of 7.5 pmol ARS1 DNA (open bars). One microgram of ORC (2.4 pmol) was used in each reaction. Rates are indicated as the amount of ATP hydrolyzed per pmol ORC. Each complex also contained a mutation in Orc5p that prevented ATP binding to this subunit (indicated by ORC-5A). (B) Kinetic analysis of ATP hydrolysis by the ORC complex. ATPase activity was measured for a complex containing wild-type ORC1 (Upper) or ORC-d1 (Lower) with a titration of ATP. Data were plotted as an Eadie-Hofstee plot. Calculated KM and kcat values are stated at the right. As in A, each complex contained the ORC-5A mutation. (C) The ORC-d1 complex can bind ARS1 DNA. Origin binding was assayed by using a DNase I protection assay for complexes containing wild-type Orc1p (lanes 2–4), ORC-1A (lanes 6–8), or Orc1-d1 (lanes 10–12). Lanes 2, 6, and 10 do not contain ATP. Lanes 3, 7, and 11 contain 100 μM ATP. Lanes 4, 8, and 12 contain 1.0 mM ATP. As in A, each complex also contained the ORC-5A mutation.
The loss of hydrolysis activity by the ORC-d1 complex could be due to a defect in binding ATP or in the rate of catalysis. To clarify this issue, we measured the ATPase activity of the wild-type and ORC-d1 complexes at various concentrations of ATP to determine KM and kcat. Our findings are shown as an Eadie-Hofstee plot (Fig. 2B). The complex containing wild-type Orc1p had a KM of 5 μM and a kcat of 0.16 min−1. In contrast, the ORC-d1 complex had a KM of 50 μM and a kcat of 0.01 min−1. These results indicate that the mutant has defects in both the binding component and the catalysis component of the reaction. However, the concentration of ATP in the cell is between 1.5 mM and 4.0 mM (24, 25)—at least 30-fold higher than the KM of this mutant. At this concentration of ATP, the ORC-d1 complex is expected to be >96% saturated with ATP (vs. >99% for wild-type) but have a 16-fold hydrolysis defect. Thus, the primary defect of ORC-d1 at physiological concentrations of ATP is a reduced rate of ATP hydrolysis, not ATP binding.
To detect ATP binding to Orc1p, we assayed the ability of the wild-type and mutant ORC complexes to bind origin DNA by using a DNase I protection assay of ARS1 (origin DNA binding is ATP dependent, Fig. 2C). At concentrations of 100 μM and 1.0 mM, the wild-type and ORC-d1 complexes can efficiently bind DNA, yet no origin binding is seen in the absence of ATP. As shown previously, The ORC-1A mutant is defective in origin binding. Thus, ORC-d1 and wild-type ORC have approximately equivalent DNA and ATP binding activities at physiological ATP levels. When not bound to DNA, our analysis of ATP hydrolysis by ORC strongly suggests that the ORC-d1 complex will be almost exclusively in the ATP bound state yet be strongly defective in ATP hydrolysis. We believe that the loss of ATPase activity when ORC is not bound to DNA is the critical defect, because chromatin precipitation analysis of these mutant yeast strains indicates that the majority of overexpressed protein is found within the soluble fraction (data not shown).
orc1-d1 and orc1-d2 Support DNA Replication in Low Copy.
In theory, the dominant lethal phenotype resulting from overexpressing the mutant ORC1 alleles could be caused by at least two different mechanisms. First, the mutant could compete with wild-type ORC for replication origin binding, but once bound be unable to perform an essential replication step, leading to lethality. Second, if the mutant ORC assumes a conformation that is preferentially recognized by an essential interacting factor, overexpression of the mutant ORC could titrate this factor away from the origin, leading to lethality. The first but not the second mechanism requires that the mutant ORC be unable to support DNA replication when the mutant allele is the only copy of ORC1 present in the cell.
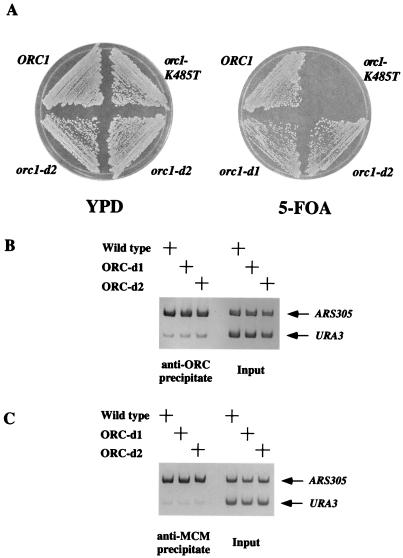
To differentiate between these mechanisms, we tested whether the orc1-d1 and orc1-d2 mutants could complement a deletion of ORC1 when expression of the mutant genes was driven by the ORC1 promoter. Although the strains expressing orc1-d1 or orc1-d2 as the only copy of the ORC1 gene do have a growth defect (the generation time is 10–20% longer than wild-type controls), we found that both alleles complemented a deletion of ORC1 (Fig. 3A). We also used ChIP to test the ability of these mutants to bind origin DNA and assemble the preRC in vivo. ORC complexes containing Orc1-d1p or Orc1-d2p bound the origin efficiently (Fig. 3B). In addition, both mutant complexes assembled the preRC, as measured by MCM recruitment to the origin (Fig. 3C). These findings are consistent with the biochemical studies described above indicating that the ORC-d1 complex can bind ATP and DNA normally. We conclude that these mutants are capable of performing their essential function in replication and that the lethality induced by their overexpression is most likely due to titration of another essential replication factor away from origin bound ORC. Because neither wild-type nor ATP-binding-deficient ORC exhibits the lethal phenotype when overexpressed, we suggest that the inappropriately stabilized binding of non-origin bound ORC1-d1 or ORC1-d2 to ATP is responsible for titrating away the proposed replication factor.
Figure 3.
orc1-d1 and orc1-d2 Support Replication in Low Copy. (A) orc1-d1 and orc1-d2 supported cell viability. A yeast strain with the chromosomal copy of ORC1 deleted and containing the ORC1 gene on a URA3 plasmid was transformed with an integrating plasmid containing either wild-type ORC1 (RKy90), orc1-K485T (RKy91), orc1-d1 (RKy92), or orc1-d2 (RKy93) under the control of the ORC1 promoter. These strains were then struck on a plate containing 5-fluoroorotic acid to select for loss of the URA3 plasmid (Right) or on a nonselective YPD plate (Left). (B) ORC-d1 and ORC-d2 bound origins when present in low copy. Strains containing wild-type ORC1 (RKy94), orc1-d1 (RKy95), or orc1-d2 (RKy96) under control of the ORC1 promoter as the only copy of ORC1 in the cell were assayed for ORC binding to ARS1305 by using ChIP with antibodies directed against ORC using asynchronous cultures. (C) ORC-d1 and ORC-d2 facilitated preRC formation when present in low copy. The same strains in B were tested for MCM association with ARS305 DNA. For MCM loading, cells were first arrested in G1 with alpha factor. Samples of the immunoprecipitated DNA and the input DNA were subjected to PCR.
Overexpression of Cdc6p Suppresses Orc1-d1 Lethality.
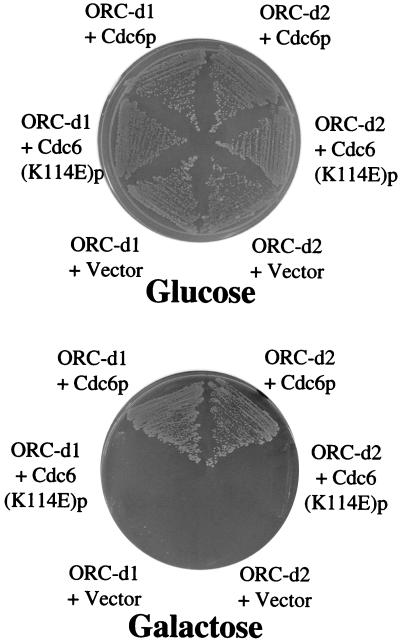
We reasoned that, if a replication factor were being specifically titrated by the overexpressed mutants, it would be possible to suppress the lethality by simultaneously overexpressing this factor. Cdc6p is a good candidate for such a factor, because it has been demonstrated to interact directly with ORC (26, 27). Indeed, co-overexpression of Cdc6p with ORC-d1 or ORC-d2 suppressed the dominant lethality of both mutant complexes (Fig. 4). This suppression is not simply a consequence of overexpressing preRC components, because co-overexpression of Mcm2p or Mcm4p did not lead to suppression (data not shown). Previous studies suggested that the ATP binding motif of Cdc6p is important for association with Orc1p (27). Therefore, we tested whether a nonviable mutant of Cdc6p that contains a point mutation within the Walker A motif (K114E) could suppress the ORC dominant negative mutants. Overexpression of Cdc6-K114E did not suppress the defect in these mutants (Fig. 4), consistent with evidence that this mutant is defective in vivo (2, 19).
Figure 4.
Overexpression of wild-type, but not mutant Cdc6p suppresses ORC-d1 and ORC-d2 lethality. Strains overexpressing ORC-d1 or ORC-d2 (when grown on galactose) were transformed with plasmids containing either wild-type Cdc6p or mutant Cdc6-K114Ep under the control of the GAL1–10 promoter as indicated. Vector overexpressing only a 3xHA tag was used as a control. Strains were grown on plates containing galactose (Lower) or glucose (Upper).
Overexpression of ORC-d1 and ORC-d2 Inhibits preRC Formation.
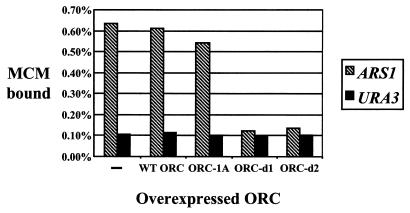
If Cdc6p were titrated away from the origin by the dominant negative ORC alleles, then overexpression of these alleles would alter the efficiency of preRC formation. To test this possibility, we measured the amount of MCM recruited to origins after overexpression of wild-type or mutant ORC was induced. Cells were first synchronized in G2/M with nocodazole. Galactose (or glucose as a control) was added for 90 min, and then cells were released from the nocodazole block into media containing galactose (or glucose) and α-factor to arrest the cells in G1. MCM localized to ARS1 was measured by using ChIP (Fig. 5). Overexpressing the ORC-d1 and ORC-d2 complexes decreases the amount of MCM loaded to levels equivalent to the non-origin background, indicating that preRC formation was disrupted by ORC-d1 or ORC-d2 overexpression. In contrast, overexpression of wild-type ORC or the mutant ORC-1A complex had no effect on preRC formation.
Figure 5.
Overexpression of ORC-d1 and ORC-d2 prevents assembly of the preRC. Strains capable of overexpressing wild-type ORC (RKy61), orc1-K485T (RKy62), ORC-d1 (RKy63), or ORC-d2 (RKy64) were grown in 2% raffinose and then arrested in the G2/M phase of the cell cycle with nocodazole for 3 h. Two percent galactose was added to induce overexpression of ORC, or, as a control, 2% glucose was added to one culture of RKy61 to repress ORC expression (first two bars marked with dash). Cells were allowed to grow for an additional 90 min and were then washed to release from the nocodazole block. Cells were resuspended in galactose (or glucose for the control) containing media including alpha factor to arrest in G1. Cells were subjected to ChIP using a monoclonal antibody that recognizes all six MCM proteins. Samples of the immunoprecipitated DNA and input DNA were subjected to PCR by using primers specific for origin DNA (ARS1) or non-origin DNA (URA3). Quantified immunoprecipitate as a percentage of total input DNA is shown.
Discussion
We report the isolation, in vivo characterization, and in vitro characterization of a class of ORC mutant that caused lethality when overexpressed. In solution, the primary biochemical defect of these mutant complexes at physiological ATP concentration was a strong reduction of ATP hydrolysis activity. When overexpressed in vivo, these mutants inhibited the formation of the preRC. Because these mutant orc1 genes could support growth when expressed at physiological levels, we suggest that these mutants act by titrating a limiting essential replication factor away from origin bound ORC. In addition, our studies support the hypothesis that this factor recognized the ATP bound state of ORC, because an Orc1p mutant that cannot bind ATP (ORC-1A) or wild-type ORC did not cause lethality when overexpressed. This lethality was suppressed by co-overexpression of Cdc6p, suggesting that Cdc6p is the titrated factor and that Cdc6p preferentially interacts with ATP bound ORC.
ATP Bound ORC and preRC Formation.
We have previously demonstrated that binding of ORC to origin DNA stabilizes the ATP bound state of Orc1p and inhibits ATP hydrolysis. Free ORC or ORC that is bound nonspecifically to DNA converts between ATP and ADP bound forms frequently, because hydrolysis is inhibited only when ORC binds specific origin sequences. The data presented here suggest that Cdc6p preferentially interacts with ATP-bound Orc1p. We have sought to directly assay the in vivo association of Cdc6p with ORC in both wild-type and mutant backgrounds by co-immunoprecipitation; however, we were unable to detect any stable interaction. We suspect that this lack of detection is due to ATP hydrolysis by ORC during the isolation of the complexes leading to dissociation of Cdc6p. This hypothesis is consistent with recent data demonstrating that in vitro Cdc6p association with ORC requires the presence of origin DNA and that this interaction enhances ORC DNA binding (26). Thus, ATP binding to Orc1p may direct an additional level of origin specificity (beyond increasing the intrinsic ORC-origin DNA affinity) by enhancing the affinity of ORC for Cdc6p when it is bound at the origin. Such a mechanism could be used in vivo to ensure that preRCs are assembled only on ORC complexes bound to origins and not to ORC free in solution or bound to non-origin sequences.
Another possible explanation for the suppression of lethality by co-overexpressing Cdc6p is that Cdc6p activates the ATPase of ORC-d1 and ORC-d2 when not bound to origins and thus rescues the lethality. We have tested the ability of Cdc6p to stimulate the ATPase activity of ORC and have found no effect of Cdc6p in the absence or presence of origin DNA (R. Austin and S.P.B., unpublished results). If this were the mechanism of suppression by Cdc6p, however, this model would suggest that a different factor is titrated by ORC-d1 and ORC-d2 causing lethality.
We have found that overexpression of Cdc6-K114E does not suppress the Orc1-d1- and Orc1-d2-induced lethality. This mutant is expected to be defective for ATP binding because mutation of a similar residue in human Cdc6p eliminates the ability to bind and hydrolyze ATP (28). This Cdc6p mutant cannot interact with Orc1p in vitro (27), exhibits decreased efficiency of chromatin association, and is lethal in vivo (2). Cdc6-K114E also cannot support a partial in vivo footprint that depends on wild-type Cdc6p, consistent with decreased origin association (19). Furthermore, CDC6-K114E is not dominant lethal when overexpressed in vivo. One might expect that, if this mutant were able to bind ORC but be unable to support replication, it would induce lethality when overexpressed. However, it has recently been observed that this mutant Cdc6p can interact with the ORC complex by using an in vitro pull-down assay (26). We suggest that the efficient interaction between the Orc1p subunit and Cdc6p requires ATP binding to Cdc6p, but other interactions between the ORC complex and Cdc6p may not require Cdc6p ATP binding. This hypothesis is consistent with recent results suggesting that ATP is required for the association of Cdc6p and the MCM complex with ORC at origins of replication (29). Alternatively, the origin dependence of the in vitro pull-down assay may change the requirement for ATP binding by Cdc6p. We propose that Cdc6p may be regulated in a manner similar to Orc1p: ATP binding is required for origin localization, and this association stabilizes the ATP bound state.
A mutation within the CDC6 Walker B motif has been isolated with similar genetic properties to ORC-d1. Overexpression of Cdc6-E224Gp causes lethality when overexpressed and inhibits recruitment of the MCMs (19). Like Orc1-d1 and Orc1-d2, this mutant supports viability when expressed in low copy (2). In light of this fact, it is possible that Cdc6-E224Gp causes lethality in a similar manner to ORC-d1 and ORC-d2. This mutant may be inappropriately stabilizing the ATP bound state and titrating an essential replication factor. Because ATP binding to Cdc6p appears to be important for interaction with Orc1p, we suggest that the ATP bound state is the form present when bound to the origin. Isolating suppressors of this mutation could identify replication factor(s) titrated by Cdc6-E224Gp.
The Role of ORC ATP Hydrolysis.
Previously, we have posited that ATP hydrolysis by Orc1p could be coupled to a step in replication downstream of the initial origin-binding event, such as preRC formation, initiation, or inactivation of replication complexes. The mutants we have generated can hydrolyze ATP at low but near normal rates when bound to origin DNA. Therefore, these mutants cannot be used directly to discern the role of hydrolysis on DNA. Overexpression of wild-type ORC does not cause lethality, nor does it inhibit preRC formation assayed by MCM recruitment. When wild-type ORC is not bound to origins, it will be almost exclusively in the ATP bound state, but will frequently turn over bound ATP. We expect that the overexpressed wild-type ORC complex in solution will bind ATP, and therefore be able to interact with Cdc6p. Because wild-type ORC overexpression does not appear to titrate Cdc6p away from the origin, this interaction must be transient. Based on these results, we suggest that hydrolysis of Orc1p-bound ATP may be a mechanism to release Cdc6p. Recently, we have shown that ORC can interact with single-stranded DNA and that the rate of Orc1p ATP hydrolysis is stimulated when bound to ssDNA (30). We have proposed that the association of ORC with the unwound origin could act as a trigger for Orc1p ATP hydrolysis. If altering the ATP bound state of ORC alters its association with other replication factors, triggering ATP hydrolysis could lead to disassembly of complexes bound at the origin and contribute to a transition between initiating and elongating replication complexes. Such a mechanism would be similar to that observed during promoter clearance when the C-terminal domain of RNA polymerase II is phosphorylated to stimulate its transition to an elongating state (31). Similarly, ORC ATP-hydrolysis-dependent replication complex disassembly could also contribute to the block to reinitiation during a single S-phase by ensuring the dismantling of preRCs after replication initiation.
Acknowledgments
The authors thank R. Austin for CDC6 plasmids and ORC antibody, and A. Schwacha for MCM antibody. This work was supported by National Institutes of Health Grant GM52339 and the Howard Hughes Medical Institute. R.D.K. was supported by a training grant from the National Institutes of Health.
Abbreviations
- ORC
origin recognition complex
- MCM
mini chromosome maintenance
- ARS
autonomously replicating sequence
- preRC
prereplicative complex
- HA
hemagglutinin
- ChIP
chromatin immunoprecipitation
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Weinreich M, Liang C, Stillman B. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 4.Donovan S, Harwood J, Drury L S, Diffley J F. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye B K. Genes Dev. 2000;14:913–926. [PMC free article] [PubMed] [Google Scholar]
- 6.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 7.Diffley J F, Cocker J H, Dowell S J, Rowley A. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 8.Kelly T J, Brown G W. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 9.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 10.Sekimizu K, Bramhill D, Kornberg A. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 11.Wahle E, Lasken R S, Kornberg A. J Biol Chem. 1989;264:2463–2468. [PubMed] [Google Scholar]
- 12.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 13.Vale R D. J Cell Biol. 2000;150:F13–F20. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 15.Patel S, Latterich M. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- 16.Pause A, Sonenberg N. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brosh R M, Jr, Matson S W. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima T, Nishida S, Kurokawa K, Katayama T, Miki T, Sekimizu K. EMBO J. 1997;16:3724–3730. doi: 10.1093/emboj/16.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins G, Diffley J F. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 21.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 22.Jellis C L, Cradick T J, Rennert P, Salinas P, Boyd J, Amirault T, Gray G S. Gene. 1993;137:63–68. doi: 10.1016/0378-1119(93)90252-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee D G, Bell S P. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Hollander J A, Ugurbil K, Brown T R, Shulman R G. Biochemistry. 1981;20:5871–5880. doi: 10.1021/bi00523a034. [DOI] [PubMed] [Google Scholar]
- 25.Coste H, Brevet A, Plateau P, Blanquet S. J Biol Chem. 1987;262:12096–12103. [PubMed] [Google Scholar]
- 26.Mizushima T, Takahashi N, Stillman B. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B, Feng L, Hu Y, Huang S H, Reynolds C P, Wu L, Jong A Y. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- 28.Herbig U, Marlar C A, Fanning E. Mol Biol Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seki T, Diffley J F. Proc Natl Acad Sci USA. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D G, Makhov A M, Klemm R D, Griffith J D, Bell S P. EMBO J. 2000;19:4774–4782. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conaway J W, Shilatifard A, Dvir A, Conaway R C. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]