Abstract
The biological and enzymatic function of SIRT4 is largely uncharacterized. We show that the C. elegans SIR-2.2 and SIR-2.3 orthologs of SIRT4 are ubiquitously expressed, also localize to mitochondria and function during oxidative stress. Further, we identified conserved interaction with mitochondrial biotin-dependent carboxylases (PC, PCC, MCCC), key enzymes in anaplerosis and ketone body formation. The carboxylases were found acetylated on multiple lysine residues and detailed analysis of mPC suggested that one of these residues, K748ac, might regulate enzymatic activity. Nevertheless, no changes in mPC acetylation levels and enzymatic activity could be detected upon overexpression or loss of functional SIRT4.
Keywords: sirtuins, SIRT4, C. elegans, biotin-dependent carboxylase, pyruvate carboxylase, protein acetylation
1 Introduction
Sirtuins are highly conserved NAD+-dependent protein deacetylases involved in many cellular processes including genome stability, stress responses, metabolism, and aging (Imai and Guarente, 2010). Three of the seven mammalian sirtuins, SIRT3, SIRT4 and SIRT5, are located in mitochondria (Michishita et al., 2005) and are thought to be important energy sensors and regulators of cellular metabolism (Huang et al., 2010).
The mitochondrion is the major organelle for ATP production, generation of reactive oxygen species (ROS) and apoptotic signaling events (Scheffler, 2001). Dysfunction of mitochondrial metabolism has been associated with many age-related diseases such as diabetes, neurodegenerative disorders, cardiovascular diseases and cancer (Van Houten et al., 2006). Mitochondrial function must therefore be tightly linked to nutrient availability (Huang et al., 2010).
There is growing evidence that reversible lysine acetylation regulates the catalytic activity and/or protein stability of metabolic enzymes and allows cells to respond to changes in nutrient availability (Wang et al., 2010; Zhao et al., 2010). A recent study demonstrated that most enzymes of the major metabolic pathways are acetylated in human liver tissue (Zhao et al., 2010). High glucose levels promote increased protein acetylation thereby inhibiting key enzymes of gluconeogenesis and amino acid catabolism. Reversible lysine acetylation might therefore function as a global mechanism in coordinating energy metabolism (Wang et al., 2010; Yang et al., 2007).
SIRT3 is the major protein deacetylase in mitochondria (Lombard et al., 2007). It has been shown to regulate numerous proteins involved in mitochondrial energy metabolism and stress responses (Lombard et al., 2011; Verdin et al., 2010). SIRT5 regulates urea cycle function by deacetylating carbamoyl phosphate synthetase (CPS1) (Nakagawa et al., 2009). Two recent studies revealed novel desuccinylase and demalonylase activities of SIRT5 (Du et al., 2011; Peng et al., 2011), thereby suggesting that sirtuins might be more general NAD+-dependent deacylases with broader substrate use (Hirschey, 2011).
SIRT4 is the only sirtuin where deacylase activity could so far not be demonstrated (Ahuja et al., 2007; Haigis et al., 2006). However, ADP-ribosylation by SIRT4 down regulates glutamate dehydrogenase (GDH) activity and ATP synthesis through the TCA cycle (Haigis et al., 2006). In addition, the factor has been reported to negatively regulate insulin secretion of pancreatic island cells in mice as well as cultured insulinoma cells (Ahuja et al., 2007; Haigis et al., 2006). SIRT4 interacts with insulin-degrading enzyme (IDE) and adenine nucleotide translocators (ANT2/3), which are both implicated in the pathogenesis of diabetes (Ahuja et al., 2007). SIRT4 knock down in mouse primary hepatocytes and myotubes increases mitochondrial gene expression and fatty acid oxidation (Nasrin et al., 2010). Nevertheless, the exact role of mitochondrial SIRT4 in regulating energy metabolism still needs to be defined in order to manipulate SIRT4 function for therapeutical benefits in diseases such as diabetes.
The nematode C. elegans possesses two genes, sir-2.2 and sir-2.3, which are orthologs of SIRT4, but there are no genes with similarity to the other mitochondrial deacetylases, SIRT3 and SIRT5. Whereas the majority of studies in the worm have focused on the mammalian SIRT1 homologue, SIR-2.1, due to its role in life span determination (Tissenbaum and Guarente, 2001), SIR-2.2 and SIR-2.3 remain by and large uncharacterized. Knock down of sir-2.2 by RNAi was shown to augment huntingtin polyglutamine toxicity (Bates et al., 2006) and sir-2.2 was identified in a genome wide RNAi screen to contribute to genome stability (Pothof et al., 2003). Both sir-2.1 and sir-2.3 are not required for dietary restriction mediated life span extension (Mair et al., 2009). So far, we do not know whether sir-2.2 and sir-2.3 function is conserved from C. elegans to human.
In the present study we characterized the expression profile and subcellular localization of C. elegans SIR-2.2 and SIR-2.3. We further identified mitochondrial biotin-dependent carboxylases, pyruvate carboxylase (PC), propionyl-CoA carboxylase (PCC) and methylcrotonyl-CoA carboxylase (MCCC) as novel interacting factors of these proteins. These interactions are conserved to mammalian SIRT4. We showed that the murine biotin-dependent carboxylases are acetylated at multiple lysine residues and identified one of these conserved sites in mPC to be important for enzymatic activity. Although overexpression or loss of SIRT4 did not result in changes in mPC acetylation levels or activity, our results suggest a possible role as regulator of metabolic adjustments during nutrient deprivation.
2 Materials and methods
2.1 DNA constructs
Cosmid F46G10 was obtained from the Sanger Institute (UK). sir-2.1, sir-2.2, sir-2.3, sir-2.4, pcca-1, mccc-1, pyc-1, acdh-3 cDNAs containing the full-length ORFs were amplified from total C. elegans mRNA after reverse transcription. SIRT4, PCCA, MCCC1 and PC cDNAs were amplified from total mRNA of NIH3T3 cells after reverse transcription. The promoter and genomic sequence of sir-2.2 was cloned into pPD115.62 (Andrew Fire, Stanford University) using PstI and MluI restriction sites thereby removing the myo-3 promoter and introducing a C-terminal Strep-tag and TEV cleavage site before the GFP-tag. For gene knockdown in C. elegans sir-2.2 and sir-2.3 cDNAs were cloned into the L4440 vector (Andrew Fire, Stanford University). For recombinant protein expression using wheat germ extract C. elegans sir-2 cDNAs were cloned into the pEU3-NII-StrepII vector. For transient protein expression in HEK293 cells cDNAs were cloned into pEGFP-N1 (Clontech) or a derivative pcDNA3.1 vector (Invitrogen) generating C-terminal fusion to a 2xFLAG-2xHA epitope tag. For generation of stable cell lines mouse PC cDNA with a C-terminal fusion to 2xFLAG-2xHA epitopes was subcloned into the pGeneV5 vector (Invitrogen). Site-directed mutagenesis was carried out according to the QuickChange protocol (Stratagene). Human SIRT3, SIRT4 and SIRT5 were subcloned into a derivative pcDNA3.1 vector (Invitrogen) generating C-terminal fusion to a MYC-His-tag. Further details on plasmids and cloning procedures are available upon request.
2.2 Western blot analysis
Primary antibodies were used as follows: anti-SIR-2.2 (rabbit), 1:1000; anti-GFP (mouse, Roche, 11814460001), 1:1000; anti-ATP synthase (Complex V) (mouse, Mitosciences, MS507), 1:1000; anti-NUO-2 (Complex I subunit NDUFS3) (mouse, Mitosciences, MS112), 1:1000; anti-cytochrome c (mouse, Mitosciences, MSA06), 1:1000; anti-H3 (rabbit, Abcam, ab1791) 1:10000-30000; anti-FLAG M2 (mouse, Sigma, F3165), 1:1000; anti-MYC (mouse, clone 4A6, Millipore, 05-724), 1:1000; anti-PC (rabbit, (Rohde et al., 1991)), 1:1000; anti-acetylated lysine (rabbit, Cell Signalling, #9441), 1:1000; anti-MnSOD (rabbit, Biomol, #S8060-10b), 1:10000.
2.3 C. elegans maintenance and strains
C. elegans was cultivated on NGM plates seeded with E. coli OP50-1 or in liquid S medium containing E. coli HB101 according to standard procedures (Brenner, 1974; Lewis, 1995; Stiernagle, 2006). N2 was the wild type strain. C. elegans strains with the following genotypes were used: sir-2.2(tm2648)X (National BioResource project (NBRP)) (crossed back to N2 worms six times); sir-2.2(tm2673)X (NBRP) (crossed back to N2 worms five times); sir-2.3(ok444)X (Caenorhabditis Genetics Center (CGC)); sir-2.1(ok434) (CGC), BC14289 (sEX14289[sir-2.2pr::gfp]) and BC15074 (sEX15074[sir-2.2pr::gfp]) (Dupuy et al., 2007; Hunt-Newbury et al., 2007; McKay et al., 2003); SJ4103 (zcls14[myo-3pr::gfp(mit)]) and SJ4143 (zcls17[ges-1pr::gfp(mit)]) (Benedetti et al., 2006).
2.4 Generation of transgenic C. elegans strains
Microinjection of C. elegans was performed as described with some modifications (Mello, 1995; Mello et al., 1991; Stinchcomb et al., 1985). pRF4 plasmid expressing the dominant mutant rol-6(su1006) allele was used as injection marker. To generate sir-2.2::gfp transgenic worms, 25 ng/µl of the plasmid pPD115.62 encoding the promoter and genomic sequence of sir-2.2 fused to a C-terminal Strep-GFP-tag were injected together with 100 ng/µl pRF4 into N2 worms. The reporter gene constructs sir-2.3pr::sir-2.3::ha::gfp and sir-2.3pr::sir-2.3::ha::mcherry were generated by PCR fusion according to a described protocol (Hobert, 2002). The fusion PCR product was microinjected into worms at a final concentration of ~50 ng/µl together with 100 ng/µl pRF4. F1 progeny displaying a roller phenotype and GFP expression were singled to obtain transgenic lines. Chromosomal integration of extrachromosomal arrays of sir-2.2::gfp and sir-2.3::gfp transgenic worms was achieved by treatment with UV radiation using a Stratlinker® UV crosslinker 2400 with an energy setting of 30,000 µJ/cm2 and a wavelength of 254 nm (Evans, 2006). Obtained stable transgenic lines were crossed back three times to wild type N2 worms to eliminate potential background mutations. The following strains were generated in this study: MAJ13 (mpgIs13[sir-2.2pr::sir-2.2::strep::gfp rol-6(su1006)]), MAJ14 (mpgIs14[sir-2.3pr::sir-2.3::ha::gfp rol-6(su1006)]), MAJ16 (mpgIs13[sir-2.2pr::sir-2.2::strep::gfp rol-6(su1006)]; mpgEx16[sir-2.3pr::sir-2.3::ha::mcherry])
2.5 RNA interference in C. elegans
cDNA sequences of sir-2.2 and sir-2.3 were cloned into the vector L4440 (Timmons and Fire, 1998). RNAi was induced by microinjecting dsRNA into the gonad of C. elegans and further enhanced by feeding injected worms with E. coli HT115 (DE3) expressing the specific dsRNA as described (Jedrusik and Schulze, 2004). The sir-2.2 dsRNA was microinjected into sir-2.3(ok444) mutant worms at a concentration of 2 µg/µl. As control M9 buffer was injected into wild type N2 worms. Microinjected worms were singled to sir-2.2 feeding or control plates and incubated at 24.5°C. Every 12 h, 24 h, 36 h and 48 h the hermaphrodites were transferred to fresh feeding plates.
2.6 Oxidative stress assay
Sensitivity towards oxidative stress was analyzed as described with some modifications (Masse et al., 2008). Worm strains were cultured synchronously for two generations on RNAi feeding plates at 24.5°C. N2, sir-2.2::gfp and sir-2.3::gfp transgenic worms were grown on control feeding plates seeded with E. coli HT115(DE3), transformed with the empty L4440 vector, sir-2.3(ok444) and sir-2.2(tm2648) mutant worms were maintained on sir-2.2 RNAi and sir-2.3 RNAi feeding plates, respectively. 200 µl 250 mM paraquat solution (methylviologendichloride hydrat, Sigma, dissolved in dH2O) was spread on top of already seeded RNAi feeding plates. Of each strain five plates with 20 worms (L4 larvae) per plate were prepared and survival of worms was checked every day. Worms were scored as dead when they did not respond to prodding with a pick. The mean life spans and average survival curves were determined from at least three independent experiments. A two-tailed Student’s t-test was used to calculate the significance of differences in mean life span to wild type worms.
2.7 NAD+ and NADH assays
Adult wild type, sir-2.2 and sir-2.3::gfp transgenic hermaphrodites were transferred from 20°C to 26°C for 3 h. Animals were rinsed three times with M9 buffer, suspended in 100 mM NaCl, 1% NP-40, 1 mM DTT, 10% glycerol, and 100 mM Tris-HCl, pH 8 and homogenized by sonication using a BRANSON Sonifier 150. Protein concentration was determined by the Bradford method. Samples were mixed with 100 µL of NAD+ or NADH extraction buffer and heated at 60°C for five minutes (NAD+/NADH assay kit from Gentaur, Kampenhout, Belgium). 20 µl of assay buffer and 100 µL of NADH (or NAD+) extraction buffer were added. Samples were then vortexed and centrifuged at 10,000 × g for five minutes. The resulting supernatant was used for NAD+ and NADH assays. For the statistical significance Student’s t-test was used.
2.8 Isolation of C. elegans mitochondria and cellular subfractionation
Mitochondria were isolated according to published procedures (Li et al., 2009). Equal protein amounts of post nuclear supernatant (PNS), post mitochondrial supernatant (PMS) and mitochondrial pellet were analyzed by SDS PAGE and Western blotting.
2.9 Electronmicroscopy
For immunoelectron microscopy of ultrathin cryosections worms were cut, fixed with 2% paraformaldehyde in 0.1 M Na-phosphate buffer (pH 7.4) for 24 h at 4°C, and postfixed with 4% paraformaldehyde-0.1% glutaraldehyde for 2 h on ice. Cryosections were prepared as previously described (Liou et al., 1996; Wenzel et al., 2005), labeled for the indicated antigens, and examined with a Philips CM120 electron microscope and a TVIPS charge-coupled device camera system.
2.10 C. elegans extracts
Total worm protein extract was prepared as described with some modifications (Cheeseman et al., 2004). Approximately 5 g of frozen C. elegans were grounded using a pre-chilled mortar and pestle in liquid N2. An equal volume of 2× extraction buffer 1 (50 mM HEPES-KOH, 2 mM EGTA, 2 mM MgCl2, 200 mM KCl, 20% (v/v) glycerol, 0.1% (v/v) NP-40, 2× EDTA-free Complete Protease Inhibitor Cocktail (Roche), pH 7.4) was added while grounding and then thawed on ice. Grounded worms were sonicated (Branson Digital Sonifier) and the extract was clarified by centrifugation (22,000 × g, 10 min, 4°C followed by 105,000 × g, 20 min, 4°C). Protein concentrations were determined using Coomassie Plus (Bradford) Protein Assay (Thermo Scientific).
For mitochondrial protein extracts frozen crude mitochondria (5–10 mg protein/ml) were thawed on ice and then centrifuged at 12000 × g and 4°C for 10 min. The mitochondrial pellet was resuspended in extraction buffer 2 (25 mM HEPES-KOH, 1 mM EGTA, 1 mM MgCl2, 500 mM KCl, 10% (v/v) glycerol, 0.5% (v/v) NP-40, 1× EDTA-free Complete Protease Inhibitor Cocktail (Roche), pH 7.6) and incubated on a rotating platform for 1 h at 4°C. The extract was clarified by centrifugation (16000 × g, 30 min, 4°C) and the supernatant was saved. The pellet was resuspended in lysis buffer and the extraction procedure was repeated. Supernatants were combined.
2.11 Immunoprecipitation of proteins from C. elegans extract
Immunoprecipitation experiments were performed with total worm protein extracts or mitochondrial protein extracts obtained from sir-2.2::gfp (MAJ13), sir-2.3::gfp (MAJ14), BC14289/BC15074 (negative control for total worm extracts) or SJ4103/SJ4143 (negative control for mitochondrial worm extracts) transgenic worms. GFP-Trap®-A beads (Chromotek) or ProteinG-Sepharose beads (Thermo Scientific) bound with anti-GFP (mouse, Roche) antibody or anti-SIR-2.2 antibody were blocked with 0.1% (w/v) BSA (1 h, 4°C, under constant rotation), washed three times with extraction buffer and then incubated with total worm protein extract or mitochondrial protein extract overnight at 4°C under constant rotation. Beads were washed three times with extraction buffer and boiled SDS sample buffer (5 min).
2.12 Isolation of RNA from C. elegans and reverse transcription
Total RNA was extracted from C. elegans using TRIzol® reagent (Invitrogen). Contaminating genomic DNA was removed by digestion with DNAse I (1 U/µg RNA, 30 min, 37°C) (NEB). First strand synthesis of cDNA was performed using Oligo(dT) primers and the Superscript II Kit (Invitrogen).
2.13 Deacetylase activity assay
Histone deacetylase assays were essentially performed as described (Verdin et al., 2004; Wirth et al., 2009) using a chemically acetylated H4 peptide as substrate. For protein expression using wheat germ extract (Spirin, 2008) full-length sir-2.1, sir-2.2, sir-2.3 and sir-2.4 were cloned into pEU3-NII-StrepII.
2.14 Cell culture
HEK293 and HEK293T cells were grown in DMEM medium supplemented with 10% bovine growth serum (BGS), 2 mM L-glutamine, and 1× Penicillin/Streptomycin (Invitrogen) at 37°C and 5% CO2. Cells were transfected with plasmid DNA using the CalPhos™ Mammalian Transfection Kit (Clontech) or the jetPEI™ Transfection Reagent (Polyplus-transfection SA).
2.15 Generation of stable cell lines
50% confluent GeneSwitch HEK293 cells (Invitrogen) seeded in a 6-well plate were transfected with 3 µg pGENE PC-2xFLAG-2xHA plasmid DNA using JetPEI reagent (Polyplus-transfection). One day after transfection, cells were trypsinized and transferred to 15 cm dishes containing medium supplemented with 50 µg/ml hygromycin and 300 µg/ml zeocin (Invitrogen). After separate colonies were visible, cells were washed with PBS and covered with 37°C warm 0.5% low melting agarose in PBS. PC-2xFLAG-2xHA expressing cells were picked using cloning cylinders and transferred into 24 well plates. After induction with 10 nM mifepristone (Invitrogen), expression of PC-2xFLAG-2xHA was analyzed by fluorescence microscopy (Axiovert 40 CFL, Zeiss) and Western blotting of the inducible colonies. Two colonies of moderately overexpressing cell-lines were frozen for long-term use and storage.
To generate an inducible mPC-2xFLAG-2xHA cell line stably expressing SIRT4 or SIRT3 cells were transfected with pcDNA3.1 hSIRT4-MYC-His or pcDNA3.1 hSIRT3-MYC-His as described above. Stable transfected cells were selected in medium supplemented with with 50 µg/ml hygromycin, 300 µg/ml zeocin and 800 µg/ml G-418 (Invitrogen).
2.16 Immunoprecipitation of proteins from mammalian cells
Immunoprecipitation experiments were performed as described with some modifications (Ahuja et al., 2007). 24 to 48 h after transfection HEK293 cells were washed once with ice-cold PBS and then lyzed with buffer A (50 mM Tris-HCl, 500 mM NaCl, 10 mM CaCl2, 0.5% (v/v) NP-40, 0.5 mM EDTA, 10% (v/v) glycerol, 1× EDTA-free Complete Protease inhibitor (Roche), pH 7.5) containing 20 U/ml micrococcal nuclease (MNase, Calbiochem). The chromatin fraction of the lysates was digested for 30 min at 30°C with agitation. An equal volume buffer B (50 mM Tris-HCl, 150 mM NaCl, 0.5% (v/v) NP-40, 0.5 mM EDTA, 10% (v/v) glycerol, 1× Complete Protease inhibitor (EDTA free, Roche), pH 7.5) was added and the lysate was sonicated (30 min, 30 s on, 30 s off, 4°C, Bioruptor (Diogenode)) and then clarified by centrifugation (16000 × g, 30 min, 4°C). Supernatant was added to equilibrated beads that had been blocked with 0.1% (w/v) BSA (1 h, 4°C, under constant rotation). GFP-tagged proteins were immunoprecipitated with GFP-Trap® A agarose beads (Chromotek). Anti-FLAG® M2 agarose beads (Sigma) or magnetic Dynabeads® M-280 Sheep anti-mouse (Invitrogen) and anti-FLAG® M2 antibody (Sigma) were used to isolate FLAG-tagged proteins. Anti-MYC IP experiments were performed with anti-MYC antibody (Millipore) bound to Dynabeads® M-280 Sheep anti-mouse (Invitrogen). After immunoprecipitation (2 h or overnight, 4°C, under constant rotation) beads were washed six times with buffer A and then boiled in SDS sample buffer. Proteins were separated by SDS-PAGE and analyzed by Western blotting.
2.17 SILAC labelling
Stable inducible PC-2xFLAG-2xHA and PC-2xFLAG-2xHA, SIRT4-MYC-His HEK293 cells were grown in lysine- and arginine-deficient DMEM medium supplemented with 10% dialyzed fetal bovine serum (PAA) and 1× Penicillin/Streptomycin (Invitrogen) at 37°C and 5% CO2 (Ong and Mann, 2006). One cell population was supplemented with normal isotope containing l-lysine and l-arginine (Sigma) and another with heavy isotope labeled 13C6-lysine and 13C6 15N4-arginine (Euriso-Top) generating mass shifts of +6 and +10 Da, respectively. Cells were grown for at least 7 days (2–3 passages) in SILAC medium. Expression of PC was induced by 10 nM mifepristione two days before harvesting of cells.
2.18 LC-MS/MS and data analysis
Proteins were separated by gel electrophoresis using NuPAGE 4–12% gradient gels (Invitrogen) and stained with Coomassie Blue. Entire gel lanes were cut into 23 slices of equal size. Proteins within the slices were digested (Shevchenko et al., 1996) and extracted peptides were loaded onto an in-house packed C18 trap column (1.5 cm, 360 µm o.d., 150 µm i.d., Reprosil-Pur 120 Å, 5 µm, C18-AQ, Dr. Maisch GmbH, Germany) at a flow rate of 10 µl/min. Retained peptides were eluted and separated on an analytical C18 capillary column (15 cm, 360 µm o.d., 75 µm i.d., Reprosil-Pur 120 Å, 5 µm, C18-AQ, Dr. Maisch GmbH, Germany) at a flow rate of 300 µl/min with a gradient from 5 to 38% ACN in 0.1% formic acid for 180 min using an Agilent 1100 nano-flow LC system (Agilent Technologies) coupled to a LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo Electron, Bremen, Germany). CID fragment spectra were searched against NCBInr database using MASCOT (taxonomy filter C. elegans) as search engine. The statistical program R was used to analyze output files and substract output files from each through their gi-numbers (NCBI).
To find acetyl-lysine peptides, LTQ- Orbitrap Velos was operated in data-dependent mode where survey scan were acquired with resolution of 30 000 at m/z 400 and a target value of 1× 10e6. Up to fifteen of the most intense ions with charge two or more from the survey scan were sequentially isolated and fragmented by higher collision-induced dissociation (HCD) with normalized collision energy of 45. Dynamic exclusion was set at 90 s to avoid repeated sequencing of peptides.
Raw MS files of SILAC experiments were analyzed by MaxQuant software (version 1.2.2.25) (Cox and Mann, 2008) using the Andromeda search engine (Cox et al., 2011) against the IPI Mouse protein database (version 3.87) supplemented with commonly observed contaminants (e.g. keratins, serum albumin) and concatenated with the reverse sequences of all entries. Andromeda search parameters were used as follows: carbamidomethylation of cysteine was set as a fixed modification whereas oxidation of methionine and N-terminal protein acetylation and lysine acetylation were set as variable modifications; tryptic specificity with no proline restriction and up to four missed cleavages was used. The MS survey scan mass tolerance used was 7 ppm and for MS/MS 20 ppm. Only peptides with minimal length of six amino acids were considered. False discovery rate was set to 1 % both at peptide and protein level. To be confident about acetylation sites, each candidate acetyl-lysine peptide from MaxQuant output was manually checked using the following criteria: 1) pair of modified and acetyl-lysine peptides should show similar ion series; 2) retention time change compared to unmodified peptide since acetylated peptide are more hydrophobic; 3) MS/MS spectrum must have y- or b- ions flanking the acetyl-lysine residue or marker ions (m/z 126.1 and/or 143.1) of acetyl-lysine must be present (Cong et al., 2011; Trelle and Jensen, 2008). Statistical analysis was done with Perseus tools available in the MaxQuant environment. All plotting and graphics were performed using R statistical software.
2.19 PC activity assay
PC activity assays were performed as described (Janke et al., 2010). Cells were resuspended in PC extraction buffer (50 mM HEPES-KOH, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton, 17.4% (v/v) glycerol, (10 µM TSA, 20 mM NAM where indicated), 1 mM PMSF, 1× EDTA-free Protease inhibitor (Roche), pH 7.9) and sonicated for 30 min (30 s on, 30 s off, Bioruptor). After centrifugation (16,000 × g, 4°C, 30 min) supernatants were directly used for the microplate based PC activity assay..
For glucose starvation mPC-FLAG expression was induced for two days using 5 nM mifepristone. One day prior harvesting cells were washed once with PBS and then incubated with glucose free medium (DMEM (Gibco, #11966), w/o glucose, w/o pyruvate, supplemented with 10% heat inactivated and dialysed FBS (PAA), 2 mM L-glutamine and 1× Penicillin/Steptomycin (Invitrogen)) containing 5 nM mifepristone overnight (14–16 h). Cells were lysed as described above and cell lysates were directly used for PC activity assays.
Livers were isolated from 12–16 weeks old male wild type and SIRT4 knock out mice (Haigis et al., 2006) fed or fasted for 24 h, snap frozen in liquid nitrogen and kept at −80°C until use. Frozen livers were homogenized in five volumes PC extraction buffer (containing 10 µM TSA and 20 mM NAM) using a dounce homogenizer and sonicated for 30 min (30 s on, 30 s off, Bioruptor). Lysates were cleared by centrifugation at 16,000 × g and 4°C for 30 min. The supernatant was diluted 1:32 or 1:64 with PC extraction buffer and then directly used for the activity assay.
Enzyme activities were calculated from the maximal reaction velocities (ΔA(Absorbance)/min) per relative amounts of PC. PC protein levels were determined by Western blot analysis. Western Blots were developed using anti-PC antibodies and the Odyssey® imaging system (Licor), and quantified using ImageJ.
2.20 Analysis of PC protein stability
Stable inducible PC-2xFLAG-2xHA, PC-2xFLAG-2xHA, SIRT4-MYC-His and PC-2xFLAG-2xHA, SIRT3-MYC-His HEK293 cells were split into 6 well plates. The next day cells (30–40 % confluent) were treated with 1 µM TSA and 10 mM NAM overnight. On day 3 cells (60–80% confluent) were treated with 20 µM MG132, 20 µg/ml cycloheximide, 1 µM TSA, and 20 mM NAM as indicated for 6 h. Cells were washed of plates with ice-cold PBS and lysed in SDS sample buffer.
3 Results
3.1 C. elegans SIR-2.2 and SIR-2.3 localize to mitochondria
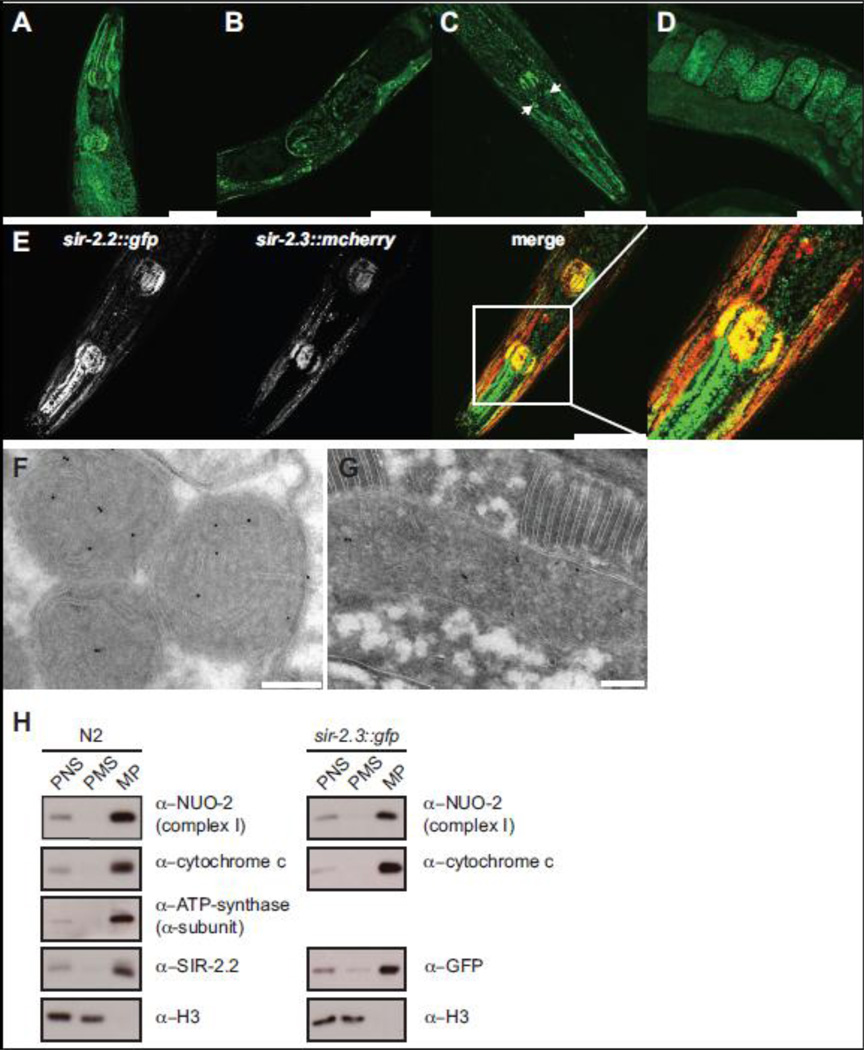
The genome of C. elegans contains four sir-2 gene orthologs, sir-2.1 (R11A8.4), sir-2.2 (F46G10.7), sir-2.3 (F46G10.3) and sir-2.4 (C06A5.11) (Frye, 2000). Of these, SIR-2.2 and SIR-2.3 show highest sequence similarity to mammalian SIRT4 (Supplementary Fig. 1). In order to analyze the expression and localization of SIR-2.2 and SIR-2.3 we generated stable transgenic worm strains expressing C-terminally GFP-tagged SIR-2.2 and SIR-2.3 under control of the endogenous promoters. Both SIR-2.2 and SIR-2.3 were expressed ubiquitously in C. elegans and exhibited a particularly high expression in the pharynx and bodywall muscles (Fig. 1A, C). While the expression patterns were overall very similar, there were also differences. Strong SIR-2.3::GFP expression was detected in a subset of cells in the head, which did not costain with the neuronal marker DiI (Hedgecock et al., 1985) (Fig. 1C, arrows). Further, the fusion protein was seen in somatic cells of the gonad (not shown). Expression of SIR-2.3::GFP started early in embryogenesis (approximately at the 100 cell stage, Fig. 1D). In contrast, SIR-2.2::GFP expression was first detected in the three-fold stage (as it also has been reported for sir-2.1 (Wang and Tissenbaum, 2006), Fig. 1B).
Fig. 1. C. elegans SIR-2.2 and SIR-2.3 are mitochondrial proteins.
(A) and (B) representative fluorescent confocal images of sir-2.2::gfp transgenic worms showing SIR-2.2::GFP expression in the head region and gonad of adult C. elegans. (C) and (D) SIR-2.3::GFP expression and localization in the head and gonad of adult worms. Arrows point at unidentified cells in the head. (E) Expression and localization of SIR-2.2::GFP and SIR-2.3::mCherry in the head of double transgenic sir-2.2::gfp; sir-2.3::mcherry worms. Scale bars represent a magnification of 50 µm. (F) Immunogold labelling of wild type N2 worm using the anti-SIR-2.2 antibody. (G). Representative cryo section of sir-2.3::gfp transgenic worm stained with anti-GFP antibody. Immunocomplexes were visualized with protein A-conjugated gold beads (10 nm). High-density black dots indicate the localization of SIR-2.2 and SIR-2.3 to mitochondria. Scale bars represent a magnification of 200 nm. (H) Wild type N2 worms and sir-2.3::gfp transgenic worms were homogenized and subjected to subcellular fractionation by differential centrifugation. Equal protein amounts of the post nuclear supernatant (PNS), mitochondrial pellet (MP) and post mitochondrial supernatant (PMS) were analyzed by SDS-PAGE and Western blotting, using the indicated antibodies.
Analysis of sir-2.2::gfp; sir-2.3::mcherry double transgenic worms verified that SIR-2.2 and SIR-2.3 localized to the same filamentous subcellular structures (Fig. 1E). Since we could only obtain a SIR-2.2-specific antibody (Supplementary Fig. 2) the subcellular localization of SIR-2.3 was further analyzed in sir-2.3::gfp transgenic worms using an antibody against GFP. Electron microscopy analysis using these reagents identified both proteins in the mitochondria (Fig. 1F, G). Further, biochemical subcellular fractionation showed that SIR-2.2 and SIR-2.3 distributed with the mitochondrial NUO-2, cytochrome c and ATP-synthetase proteins, but not the nuclear histone H3 (Fig. 1H). The results demonstrate that SIR-2.2. and SIR-2.3 are bona fide orthologs of mammalian SIRT4.
3.2 SIR-2.2 and SIR-2.3 function during oxidative stress
We next analyzed the biological function of SIR-2.2 and SIR-2.3 using the sir-2.2(tm2648), sir-2.2(tm2673) and sir-2.3(ok444) mutant worm strains (Supplementary Fig. 2A). RT-PCR and Western blot analysis showed that the deletion mutations of the tm2648 and tm2673 alleles resulted in deletion of exons 3 and 4. The resulting mRNAs splices exon 2 to exon 5 of the sir-2.2 gene thereby giving raise to a truncated SIR-2.2 protein lacking 75 aa of the conserved sirtuin domain (Supplementary Fig. 2B, D, F). Sequencing of the sir-2.3(ok444) RT-PCR product revealed that the ok444 allele encodes a truncated protein where a frame shift mutation in exon 4 results in replacement of the 154 aa of the C-terminus corresponding to exons 4 to 7 by eight unrelated aa (Supplementary Fig. 2C, D).
All three mutant worm strains did not exhibit any obvious phenotype under standard growth conditions. SIR-2.2 and SIR-2.3 share 75.3% sequence identity (Supplementary Fig. 1). Since the sir-2.2 and sir-2.3 genes are located directly next to each other on chromosome X it was not possible to generate a double mutant worm strain. To test whether there is functional redundancy we knocked down sir-2.2 in sir-2.3(ok444) worms by RNAi. Depletion of sir-2.2 in sir-2.3(ok444) mutant worms caused a weak phenotype with 4.5% of worms exhibiting growth and egg-laying defects (Table 1) compared to 1.9% in the wild type control worms.
Table 1.
Analysis of knock down of sir-2.2 in sir-2.3(ok444) mutant worms.
| genotype | % Dpy and Egl ± S.E.M | n (F1) |
|---|---|---|
| wild type (N2) | 1.9 ± 0.3 | 5 (518) |
| sir-2.3(ok444); sir-2.2(RNAi) | 4.5 ± 1.3 | 7 (1012) |
sir-2.2 dsRNA was microinjected into sir-2.3(ok444) mutant hermaphrodites. Injected hermaphrodites were subsequently singled onto sir-2.2 RNAi feeding plates. For control, wild type (N2) hermaphrodites were microinjected with M9 buffer and transferred to feeding plates seeded with E. coli Ht115(DE3) containing an empty L4440 vector. The F1 generation was analysed for dumpy (Dpy) and egg-laying defective (Egl) phenotypes. S.E.M: standard error of mean; n: number of microinjected hermaphrodites, F1: number of analysed F1 generation progeny; a p-value > 0.05 (0.05003) was calculated using the standard Student t-test.
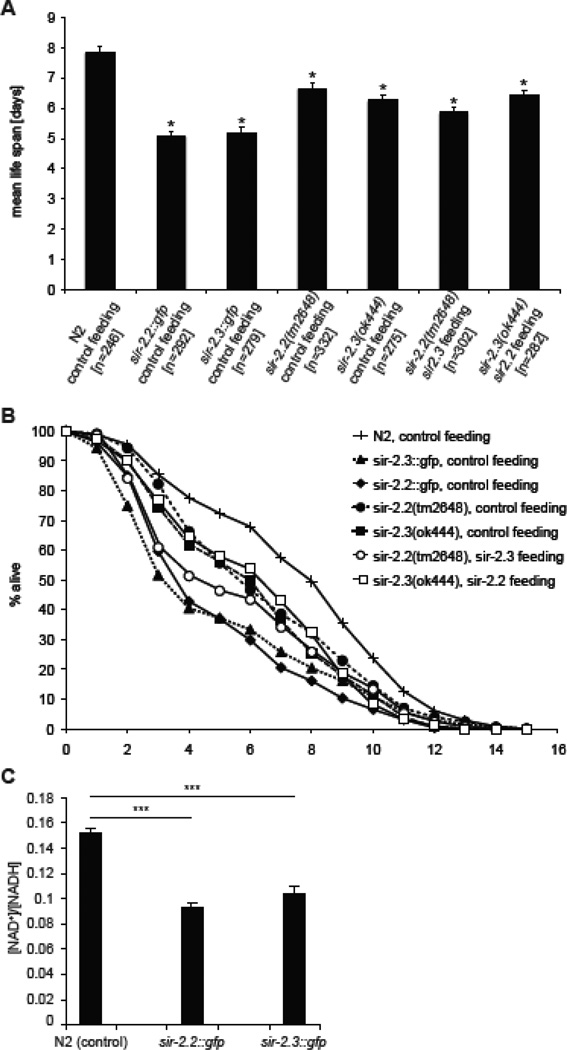
Knockout mice of SIRT3, SIRT4 and SIRT5 grow and develop normally under physiological conditions (Haigis et al., 2006; Lombard et al., 2007; Nakagawa et al., 2009), but display defects in response to different stresses (Haigis and Sinclair, 2010). Thus, we analyzed tolerance of sir-2.2(tm2648) and sir-2.3(ok444) mutant worms as well as sir-2.2::gfp and sir-2.3::gfp overexpression strains to oxidative stress (see Supplementary Fig. 3A and B for analysis of protein levels). Survival of worms in the presence of paraquat that generates superoxide anions was scored daily (Masse et al., 2008). The highest sensitivity towards oxidative stress was observed in sir-2.2::gfp and sir-2.3::gfp transgenic worms having a mean survival of 5.07 ± 0.16 (S.E.M.) and 5.16 ± 0.21 (S.E.M.) days, respectively, compared to wild type N2 worms that had a mean survival of 7.86 ± 0.21 (S.E.M.) days (Fig. 2A and B). sir-2.2(tm2648) (6.66 ± 0.16 (S.E.M.)) and sir-2.3(ok444) (6.26 ± 0.18 (S.E.M.)) mutant worms also showed significantly reduced survival in the presence of paraquat. Whereas knockdown of sir-2.3 in sir-2.2(tm2648) mutant worms led to a slight increase in sensitivity (5.86 ± 0.15 (S.E.M.)) compared to the single mutant worm strains, no significant difference was observed when knocking down sir-2.2 in the sir-2.3(ok444) mutant background (6.42 ± 0.18 (S.E.M.)).
Fig. 2. C. elegans overexpressing or deficient in SIR-2.2 and/or SIR-2.3 show increased sensitivity to oxidative stress.
(A) Calculated mean life span of different worms grown at 24.5°C on RNAi feeding plates containing paraquat (200 µl of 250 mM paraquat solution added onto plate). Wild type (N2), worms overexpressing SIR-2.2 (sir-2.2::gfp) or SIR-2.3 (sir-2.3::gfp), worms mutant in sir-2.2 (sir-2.2(tm2648)) or sir-2.3 (sir-2.3(ok444) were fed on E. coli HT115(DE3) containing an empty L4440 vector (control feeding). Worms mutant in sir-2.2 (sir-2.2(tm2648)) were fed on E. coli HT115(DE3) containing a plasmid encoding for sir-2.3 (sir-2.3 feeding) and worms mutant in sir-2.2 (sir-2.2(tm2648)) were fed on E. coli HT115(DE3) containing a plasmid encoding for sir-2.2 (sir-2.2 feeding). At least 3 trials were conducted per strain. Error bars show standard error of mean (S.E.M.); asterisks indicate a significant difference (p-value < 0.001) to average life span of wild type (N2) worms; p-values were calculated using standard Student t-test; n: number of analyzed worms. (B) Average survival curves of the worm strains analyzed in (A). (C) Analysis of [NAD+]/[NADH] levels in whole worm lysates of wild type N2, sir-2.2::gfp and sir-2.3::gfp overexpressing worms. Error bars represent the standard error of mean; *** P<0.01 as compared with control animals (wild type N2 worms) using Student’s t-test.
To obtain insights into the mechanism of sensitivity to oxidative stress, we first analyzed the levels of MnSOD, a key antioxidant mitochondrial enzyme, which has been shown to be regulated by mammalian SIRT3 (Tao et al., 2010). However, Western blot analysis of extracts from mutant worms did not reveal any differences in the expression of this factor (Supplementary Fig. 3C). We then measured the NAD+/NADH ratio. As Fig.2 C shows this parameter was significantly lower in sir-2.2::gfp and sir-2.3::gfp transgenic worms compared to wild type animals. The results indicated that the fusion proteins are functional and that both factors work enzymatically in consuming NAD+. From the other experiments, we further concluded that sir-2.2 and sir-2.3 are not functionally redundant. We then asked which targets are regulated by SIR-2.2 and SIR-2.3.
3.3 SIR-2.2 and SIR-2.3 interact with mitochondrial biotin dependent carboxylases
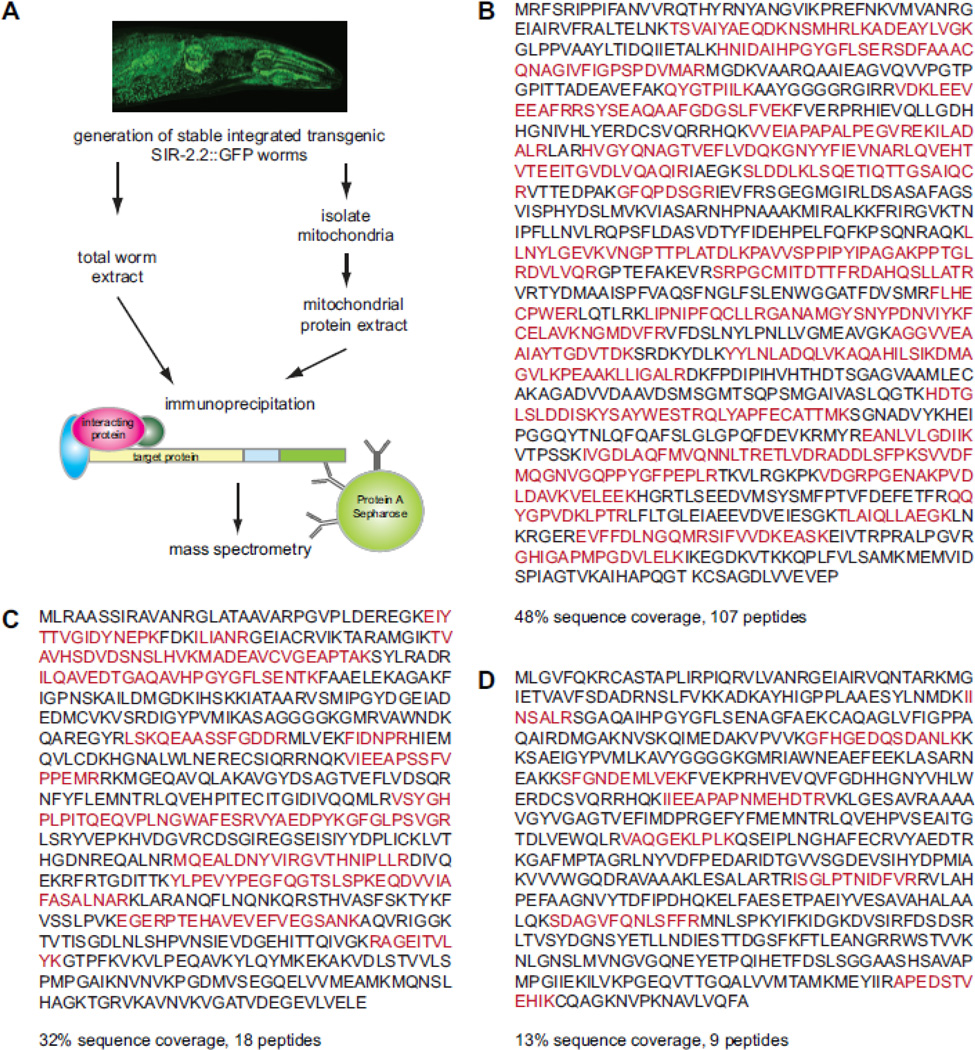
To identify factors interacting with SIR-2.2 we immunoaffinity purified GFP-tagged SIR-2.2 from total or mitochondrial protein lysates of stable sir-2.2::gfp worms using SIR-2.2- or GFP-specific antibodies (Fig. 3A). Control immunoprecipitation experiments were performed using total worm lysates prepared from the transgenic strains BC15074 and BC14289, which express GFP under control of the endogenous sir-2.2 promoter, or with mitochondrial lysates of the transgenic strains SJ4104 and SJ4143, which express GFP with a mitochondrial targeting sequence (Benedetti et al., 2006). Bound proteins were analyzed by mass spectrometry and proteins present only in the SIR-2.2::GFP bound fraction (identified with at least two peptides or a protein score of at least 80) were extracted from the data (Supplementary Table 1). Among the identified factors D2023.2 (pyc-1, pyruvate carboxylase), F27D9.5 (pcca-1, propionyl-coenzyme A (-CoA) carboxylase alpha subunit), and F32B6.2 (ortholog to human alpha methylcrotonoyl-coenzyme A (-CoA) carboxylase 1 (alpha subunit), in this study referred to as mccc-1) were particularly interesting, since these represent the mitochondrial members of the biotin-dependent carboxylase protein family. Sequence coverage obtained for these proteins were 48% with 107 identified peptides for PYC-1 (Fig. 3B), 32% with 18 peptides for PCCA-1 (Fig. 3C) and 13% with 9 peptides for MCCC-1 (Fig. 3D).
Fig. 3. Identification of mitochondrial biotin carboxylases as SIR-2.2 interacting factors.
(A) Schematic overview of the strategy used to identify interaction partners of SIR-2.2. A stable integrated transgenic worm strain expressing SIR-2.2 with a C-terminal Strep-GFP-tag under control of the endogenous promoter was generated and used for extract preparation. GFP-tagged SIR-2.2 was immunoprecipitated with anti-GFP-specific antibodies or the anti-SIR-2.2-specific antibodies from total or mitochondrial worm extracts. Isolated proteins were subjected to SDS-PAGE and analyzed by mass spectrometry. (B) Protein sequence of C.elegans D2023.2 (PYC-1). (C) Protein sequence of C. elegans F27D9.5 (PCCA-1). (D) Protein sequence of F32B6.2 (MCCC-1). Peptides identified by mass spectrometric analyses are highlighted on the protein sequence. Total sequence coverage is indicated.
Pyruvate carboxylase (PC) has an important anaplerotic function for the tricarboxylic acid (TCA) cycle. The enzyme catalyzes the carboxylation of pyruvate to oxaloacetate, an important TCA cycle intermediate (Jitrapakdee et al., 2006) (Supplementary Fig. 4). Propionyl-CoA carboxylase (PCC) is essential for the catabolism of branched chain amino acids (Thr, Val, Ile, and Met), odd-chain length fatty acids and cholesterol. Since it carboxylates propionyl-CoA to methylmalony-CoA, which feeds into the TCA cycle after conversion to succinyl-CoA, it is also important for anaplerosis (Gravel et al., 1980; Jitrapakdee and Wallace, 2003). Methylcrotonyl-CoA carboxylase (MCCC) catalyzes the carboxylation of 3-methylcrotonyl-CoA to 3-methylglutaconyl-CoA in catabolism of leucine, which as an exclusively ketogenic amino acid is degraded to acetoacetate and acetyl-CoA. Therefore, MCCC does not function in anaplerosis but formation of ketone bodies (Jitrapakdee and Wallace, 2003; Lau et al., 1980). Gluconeogenesis, amino acid catabolism, β-oxidation and formation of ketone bodies are essential pathways for maintaining energy homeostasis supporting the hypothesis that sirtuins regulate adjustments in energy metabolism during stress such as nutrient deprivation (Haigis and Sinclair, 2010; Imai and Guarente, 2010).
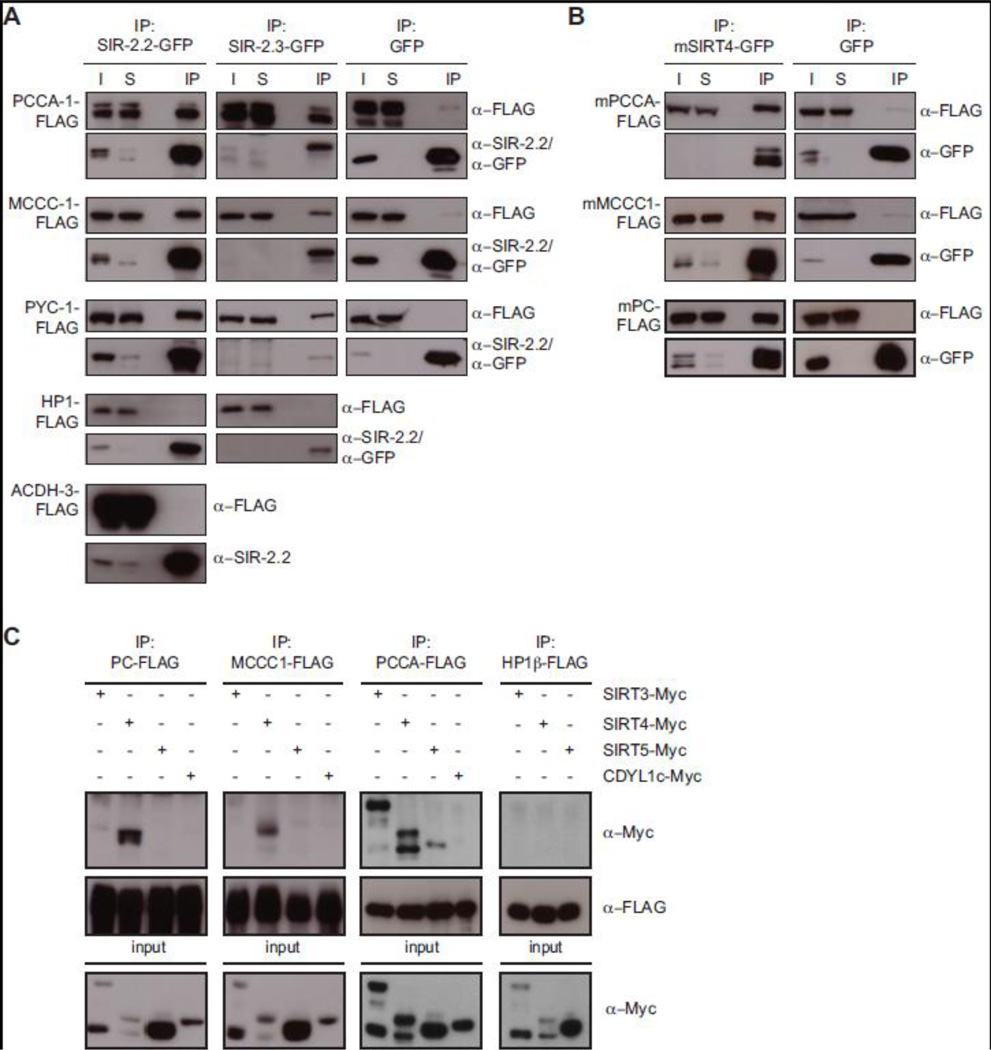
To independently verify interaction between SIR-2.2 and the biotin-dependent carboxylases we expressed the proteins in HEK293 cells and performed immunoprecipitation experiments. As Fig. 4A shows, PYC-1, PCCA-1 and MCCC-1 co-immunoprecipitated specifically with GFP-tagged SIR-2.2, but not with GFP alone. Importantly, the mitochondrial protein ACDH-3 (acyl-CoA dehydrogenase-3) and the nuclear protein HP1β did not associate with SIR-2.2::GFP under the same experimental conditions. In similar assays SIR-2.3 also specifically interacted with all three proteins (Fig. 4A). Indeed, when analyzing proteins co-immunoprecipitated with SIR-2.3-GFP isolated from mitochondrial extracts of stable sir-2.3::gfp transgenic worms using mass spectrometry we found peptides of all three mitochondrial biotin-dependent carboxylases (Supplementary Fig. 5 and Supplementary Table 2). The results indicated that both SIRT4 orthologous factors of C. elegans interact with PYC-1, PCCA-1 and MCCC-1 enzymes.
Fig. 4. Mammalian SIRT4 specifically interacts with mitochondrial biotin-dependent carboxylases.
(A) Expression vectors encoding SIR-2.2-GFP, SIR-2.3-GFP or GFP alone were cotransfected in HEK293 cells with expression vectors for either FLAG-tagged C. elegans PCCA-1, MCCC-1 or PYC-1 or mouse ACDH-3 or HP1β. GFP-tagged proteins were immunoprecipitated with GFP-TrapA and analyzed by SDS-PAGE and Western blotting using anti-FLAG, anti-SIR-2.2 and anti-GFP antibodies. I: input (2%); S: supernatant after immunoprecipitation (2%); IP: immunoprecipitated material. (B) Similar analysis as in (A) but using vectors encoding mouse SIRT4-GFP, PCCA-FLAG, MCCC1-FLAG and PC-FLAG as well as GFP. (C) Expression vectors for human SIRT3, SIRT4, SIRT5 and CDYL1c (MYC-tagged) were cotransfected with expression vectors encoding either murine PC, MCCC1, PCCA of HP1β (FLAG-tagged) in HEK293 cells. Proteins were immunoprecipitated with anti-FLAG antibodies and analyzed by Western blotting with anti-MYC and anti-FLAG antibodies (top). Cell extracts (input) were directly analyzed in Western blotting with anti-MYC antibodies to determine expression levels of tagged protein in each experiment (bottom).
3.4 The biotin carboxylase domain mediates evolutionarily conserved interaction of mammalian SIRT4 with mitochondrial biotin-dependent carboxylases
To test whether the observed interaction is conserved between C. elegans and mammals we performed co-immunoprecipitation experiments. We transiently expressed mouse SIRT4-GFP or GFP alone together with FLAG-tagged mouse propionyl-CoA carboxylase α-subunit (mPCCA), mouse methylcrotonyl-CoA carboxylase α-subunit (mMCCC1), or mouse pyruvate carboxylase (mPC) in HEK293 cells. Immunoprecipitation of total cell extracts using anti-GFP antibodies specifically recovered all three biotin-dependent carboxylases with SIRT4-GFP but not GFP (Fig. 4B). In similar experiments the C. elegans PYC-1, PCCA-1 and MCCC-1 proteins also specifically bound to SIRT4-GFP (data not shown).
Next, we asked whether binding to biotin-dependent carboxylases is specific to SIRT4 compared to the other mitochondrial mammalian sirtuins, SIRT3 and SIRT5 (Fig. 4C). After transient expression in HEK293 cells FLAG-tagged mPC and mMCCC1 specifically co-immunoprecipitated only with MYC-tagged SIRT4 but not with MYC-tagged SIRT3 or SIRT5. FLAG-tagged mPCCA bound strongly to SIRT3-MYC and SIRT4-MYC and much weaker to SIRT5-MYC. MYC-tagged SIRT3 and SIRT4, but not SIRT5 showed multiple bands in Western blot analysis. We think these are due to incomplete mitochondrial import of the overexpressed proteins, which excludes normal N-terminal processing (Haigis et al., 2006; Schwer et al., 2002). For unknown reasons MYC-tagged SIRT3 and SIRT4 reached higher expression levels when in presence of mPCCA-FLAG as compared to being coexpressed with mPC-FLAG or mMCCC1-FLAG. In control immunoprecipitation experiments, none of the biotin-dependent carboxylases did bind to CDYL1c (Chromodomain Y-like protein 1c), nor did we find interaction of the sirtuins with FLAG-tagged HP1β (Heterochromatin protein 1β).
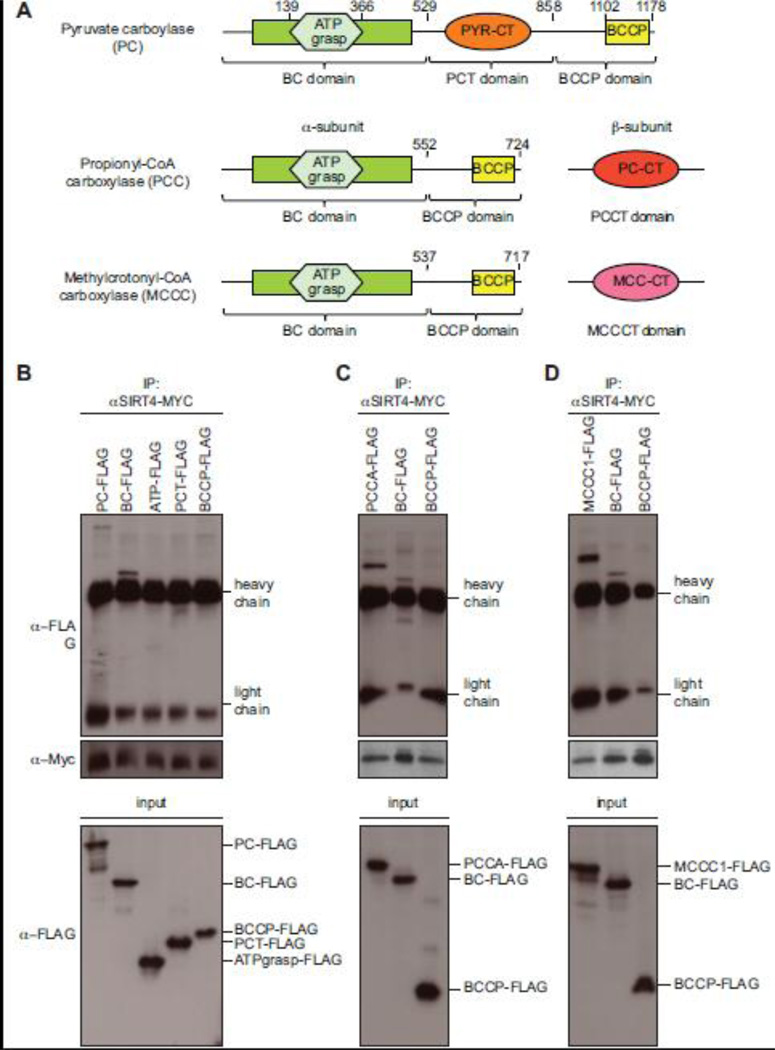
PC, PCCA and MCCC1 catalyze metabolically important carboxyl group transfer reactions and possess highly conserved functional domains (Fig. 5A). PC consists of four identical subunits (α4-form) of 1178 aa, which contain a N-terminal biotin carboxylase domain (BC) featuring a conserved ATP grasp domain (ATP), a substrate-specific carboxyltransferase domain (PCT), and a C-terminal biotin carboxyl carrier protein (BCCP) domain (Jitrapakdee et al., 2008; Jitrapakdee and Wallace, 2003). To determine, which domain mediates interaction with SIRT4, four different deletion constructs of mPC representing these different regions of the protein were generated with a C-terminal FLAG-tag and transiently expressed in HEK293 cells together with MYC-tagged SIRT4. Since expression levels and consequently recovery rates of SIRT4 were not equal when co-expressed with the different mPC constructs, we normalized material immunoprecipitated with anti-MYC antibodies to the amounts of SIRT4-MYC for Western blot analysis (Fig. 5B). Next to full-length mPC only the biotin carboxylase domain associated with SIRT4. Since the conserved ATP grasp domain was not sufficient for binding, we concluded that residues outside this domain must mediate binding of the N-terminal BC domain to SIRT4.
Fig. 5. The biotin carboxylase domain of PC, PCCA and MCCC1 specifically binds to SIRT4.
(A) Schematic representation of the domain organization of mitochondrial biotin carboxylases. All three proteins have a highly conserved N-terminal biotin carboxylase domain (BC) and a C-terminal biotin carboxyl carrier protein domain (BCCP). Brackets indicate the deletion constructs generated to map the region interacting with SIRT4. Domain boundaries of the mutant proteins are indicated by the amino acid positions. Analysis of interaction with PC (B), with PCCA (C) and MCCC1 (D). For co-immunoprecipitation experiments, the indicated FLAG-tagged deletion constructs of the biotin-dependent carboxylases were transiently expressed together with human SIRT4-MYC in HEK293 cells. Equal amounts of immunoprecipitated MYC-tagged SIRT4 were loaded on SDS-PAGE gels and analyzed by Western blotting using anti-FLAG and anti-MYC antibodies (top). Cell extracts (input) were directly analyzed in Western blotting with anti-FLAG antibodies to determine expression levels of tagged protein in each experiment (bottom). Running positions of FLAG-tagged proteins and of the antibody’s heavy and light chains are indicated on the right.
Both PCC and MCCC are heteropolymeric enzymes with six heterodimers of one α- and one β-subunit arranged in a (αβ)6 configuration (Gravel et al., 1980; Jitrapakdee and Wallace, 2003; Lau et al., 1980). The α-subunits contain the N-terminal biotin carboxylase (BC) domain and the C-terminal biotin carboxyl carrier protein (BCCP) domain (Fig. 5A). The substrate-specific carboxyltransferase (CT) activities are located on separated β-chains (Jitrapakdee and Wallace, 2003), which were not detected as binding partners of C. elegans SIR-2.2 and SIR-2.3 in the proteomic analyses (Supplementary Table 1 and 2). Indeed, in coimmunoprecipitation experiments after transient overexpression the BC domains of PCCA and MCCC1 were sufficient to mediate binding to SIRT4 while the BCCP domain showed no interaction (Fig. 5C, D). From these experiments we deduced that – like the C. elegans SIR-2.2 and SIR-2.3 proteins the mammalian SIRT4 binds specifically to all three mitochondrial biotin-dependent carboxylases and that the biotin carboxylase domain of the enzymes mediates this interaction.
3.5 Mitochondrial biotin-dependent carboxylases are acetylated on multiple lysine residues
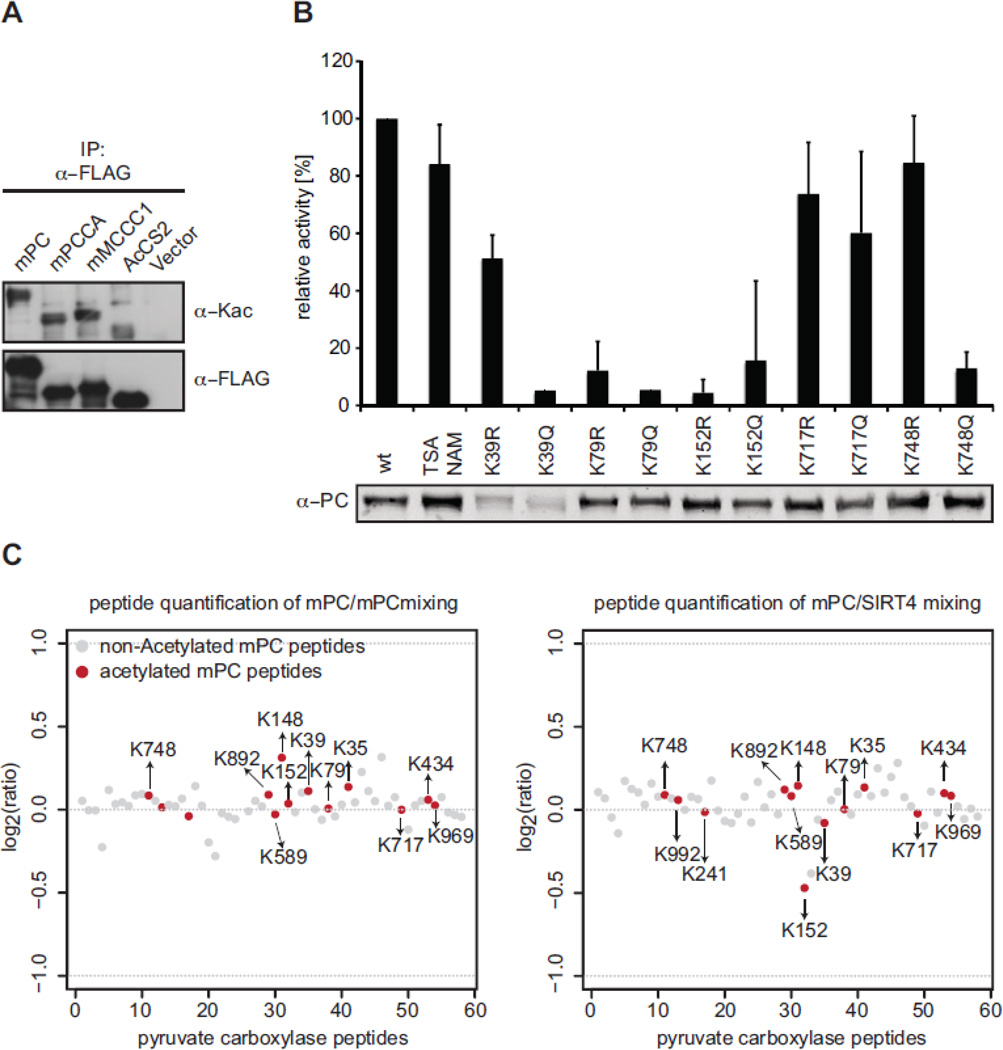
We hypothesized that the mitochondrial biotin-dependent carboxylases might be substrates of SIRT4 enzymatic activity. No ADP-ribosylation has been reported for PC, PCCA and MCCC1, but several global proteomic screens have identified putative lysine acetylation in PC (mapping to K237, K316, K992 and K1090 in mouse PC, Supplementary Fig. 7) (Choudhary et al., 2009; Fritz et al., 2012; Kim et al., 2006; Zhao et al., 2010) and in PCCA (mapping to K61 and R549 in mouse PCCA) (Fritz et al., 2012; Zhao et al., 2010). In a mass spectrometry based proteomic approach using mouse liver we further identified K128, K150, K223, K403, K460, K492 and K509 in mouse PCCA as well as K180 and K717 in mouse MCCC1 as putative sites of acetylation (data not shown). To verify that the mitochondrial biotin-dependent carboxylases are indeed acetylated proteins, we immunoprecipitated FLAG-tagged mPC, mPCCA and mMCCC1 from HEK293 cells after transient transfection. In Western blot analysis a pan anti-acetyl lysine antibody indeed recognized these factors, as well as the mitochondrial AceCS2 control protein (Hallows et al., 2006; Schwer et al., 2006) (Fig. 6A). Since pyruvate carboxylase is a major enzyme in anaplerosis, since it is the only monomeric mitochondrial biotin carboxylase and as a robust enzymatic assay for this enzyme has been described (Janke et al., 2010; Scrutton and White, 1974), we decided to further focus our analysis on this factor.
Fig. 6. Analysis of PC acetylation in dependence of SIRT4.
(A) Expression vectors encoding FLAG-tagged mPC, mPCCA and mMCCC1 or the empty vector (vector) were transfected into HEK293 cells. Proteins were immunoprecipitated from total cell extracts with anti-FLAG antibodies. Western blot analysis of the recovered material using anti-acetyllysine-specific antibodies (AcK) and anti-FLAG antibodies is shown. (B) Enzymatic activities of different mPC-FLAG mutants overexpressed in HEK293T cells. Pyruvate carboxylase activity of total cell lysates was measured using a citrate synthase-coupled assay. The carboxylation of pyruvate by PC is coupled to the conversion of oxaloacetate to citrate by citrate synthase. Reduction of DTNB by generated CoASH was recorded by measuring an increase in absorbance at 405 nm. Activities were calculated from the maximal reaction velocities (ΔA/min) per relative amount of mPC protein (quantified from Western blot analyses). Relative activities (normalized to wt mPC) are represented as means of at least three independent experiments. Error bars indicate standard deviation. (C) HEK293 cells expressing mPC-FLAG under inducible control alone or together with SIRT4-MYC were metabolically labeled using SILAC. After anti-FLAG immunoprecipitation the recovered material from mPC-FLAG (labeled in light medium) and mPC-FLAG, SIRT4-MYC (labeled in heavy medium) cells (left panel) or mPC-FLAG (labeled in light medium) and mPC-FLAG (labeled in heavy medium) cells (right panel) were mixed, run on SDS-PAGE and analyzed by quantitative mass spectrometry. Normalized ratios of identified acetylated (red) and non-acetylated (grey) peptides derived from mPC of one cellular labeling experiment are blotted.
First, we generated an inducible stable transfected mPC-FLAG cell line to verify and further map lysine acetylation sites in mPC. FLAG-tagged mPC was immunoprecipitated from total cell extract, run on SDS-PAGE and digested with trypsin. In contrast to the proteomic screening experiments, we did not use pan anti-acetyl-lysine antibodies for enrichment of acetylated peptides before mass spectrometry analysis. According to our observations these antibodies have certain sequence bias and do not detect all Kac sites. Also, the algorithms used for assigning lysine acetylation in such experiments assign false positive hits with a certain frequency (our own observations).
Of the total mPC-derived peptides we could assign 13 acetylation sites with a mass increase of 42 Da compared to the unmodified state. These correspond to the following residues in mPC:: K35, K39, K79, K148, K152, K241, K434, K589, K717, K748, K892, K969 and K992. While only K992 of these matched the previously described sites (see above), we verified several Kac sites by the presence of marker ions in the fragment spectra (MSMS) of sequenced peptides (immonium ion (143.11 m/z) and/or an ammonia derivative of the immonium ion (129.09 m/z) of acetylated lysine, Supplementary Fig. 6) (Trelle and Jensen, 2008). While this manuscript was in preparation independent work on mouse liver indeed verified many of these sites (Lundby et al., 2012). Five of our newly identified acetylation sites K39, K79, K152, K717 and K748, map to lysine residues that are conserved from C. elegans to human (see Supplementary Fig. 7 for annotation of the acetylation sites identified by different studies). Further annotation of the mass spectrometry results did not reveal any indication of succinylation, malonylation or ADP-ribosylation of mPC. We therefore focused our further studies on a putative role of SIRT4 in regulating mPC acetylation levels.
3.6 Acetylation of K748 might regulate mPC activity
There has been increasing evidence that reversible lysine acetylation is an important posttranslational modification regulating the activity of metabolic enzymes (Zhao et al., 2010). To test whether acetylation of any of the five highly conserved lysine residues in mPC might play a role in regulating the enzymatic activity we mutated these sites either to arginine (mimicking the non-acetylated state) or to glutamine (mimicking acetylated lysine). The FLAG-tagged mutant mPC proteins were transiently expressed in HEK293 cells and total cell lysates were analyzed for their pyruvate carboxylase activity using a citrate synthase coupled enzymatic assay (Janke et al., 2010; Scrutton and White, 1974). Enzymatic activity was afterwards normalized to mPC protein levels determined by Western blotting. As Fig. 6B shows, mutation of K39 resulted in reduced protein stability, with the Q mutation displaying reduced activity compared to the R mutation. In contrast, all other mutations had no apparent affect on mPC protein expression levels. Mutation of K79 and K152 to R or Q generally abrogated mPC activity, while mutation of K717 did not significantly affect mPC activity. The most interesting effect was observed on K748. Here mutation to R did not change the enzymatic activity, but the K748Q mutation almost completely abolished the enzymatic activity. The results indicate that the activity of mPC might be regulated by reversible acetylation of K748.
3.7 SIRT4 does not deacetylate mPC
Multiple sequence alignment of the catalytic core domain of the enzymatically active protein deacetylases mammalian SIRT1 and SIRT6 as well as S. cerevisiae Hst2 and SIR2 with SIRT4 and C. elegans SIR-2.2 and SIR-2.3 proteins showed a high degree of sequence conservation (Supplementary Fig. 1). Residues important for the enzymatic activity such as the active site histidine, the majority of residues involved in NAD+ and acetyl-lysine peptide binding and the Zn2+ binding motif are also present in SIRT4 and C. elegans SIR-2.2 and SIR-2.3.
No NAD+-dependent deacetylase activity could be demonstrated for human SIRT4 using a standard histone deacetylase (HDAC) activity assay (Ahuja et al., 2007). Apart from SIR-2.1 (Wood et al., 2004), the enzymatic activities of C. elegans SIR-2.2 and SIR-2.3 have not been analyzed. We performed in vitro deacetylase activity assays using a per-3H-acetylated histone H4 tail peptide as substrate to analyze the enzymatic activity of C. elegans sirtuins. Consistently with previous reports (Wood et al., 2004), SIR-2.1 exhibited robust activity on this substrate. However, no activity was detected for the other two variants using either in vitro translated protein or GFP-tagged SIR-2.2 or SIR-2.3 isolated from the transgenic sir-2::gfp strains (Supplementary Fig. 8).
An acetylated H4 peptide is not a natural target of the mitochondrial sirtuins and the lack of enzymatic activity might be due to very strict substrate specificity. To determine whether SIRT4 can possibly deacetylate mPC we wanted to analyze the acetylation status of the identified Kac sites under conditions of overexpression of SIRT4. For this purpose, we stably introduced SIRT4-MYC into the cell line containing inducible mPC-FLAG. To quantify differences in mPC acetylation of specific sites we performed stable isotope labeling with amino acids in cell culture (SILAC) (Ong and Mann, 2006). After adopting the cells to the growth conditions, mPC-FLAG expression was briefly induced and the fusion protein was purified by anti-FLAG immunoprecipitation. Mass spectrometric comparison of the relative abundance of acetylated and non-acetylated peptides derived from mPC-FLAG in absence (Fig. 6C, right panel) and presence of SIRT4-MYC (Fig. 6C, left panel) revealed only minor differences. Of the 13 peptides suggested to contain acetylation only the one containing K152ac showed slightly changed levels in the samples, which were larger than the variation observed in direct SILAC-MS comparison of mPC-FLAG grown in light and heavy medium in absence of SIRT4-MYC. However, the differences in repetitive experiments were smaller than one fold. Since the variability in quantification at peptide level is also much higher than at the protein level, we concluded that increased SIRT4 levels did not affect the acetylation status of the analyzed sites of mPC.
3.8 Overexpression of SIRT4 does not affect mPC protein stability
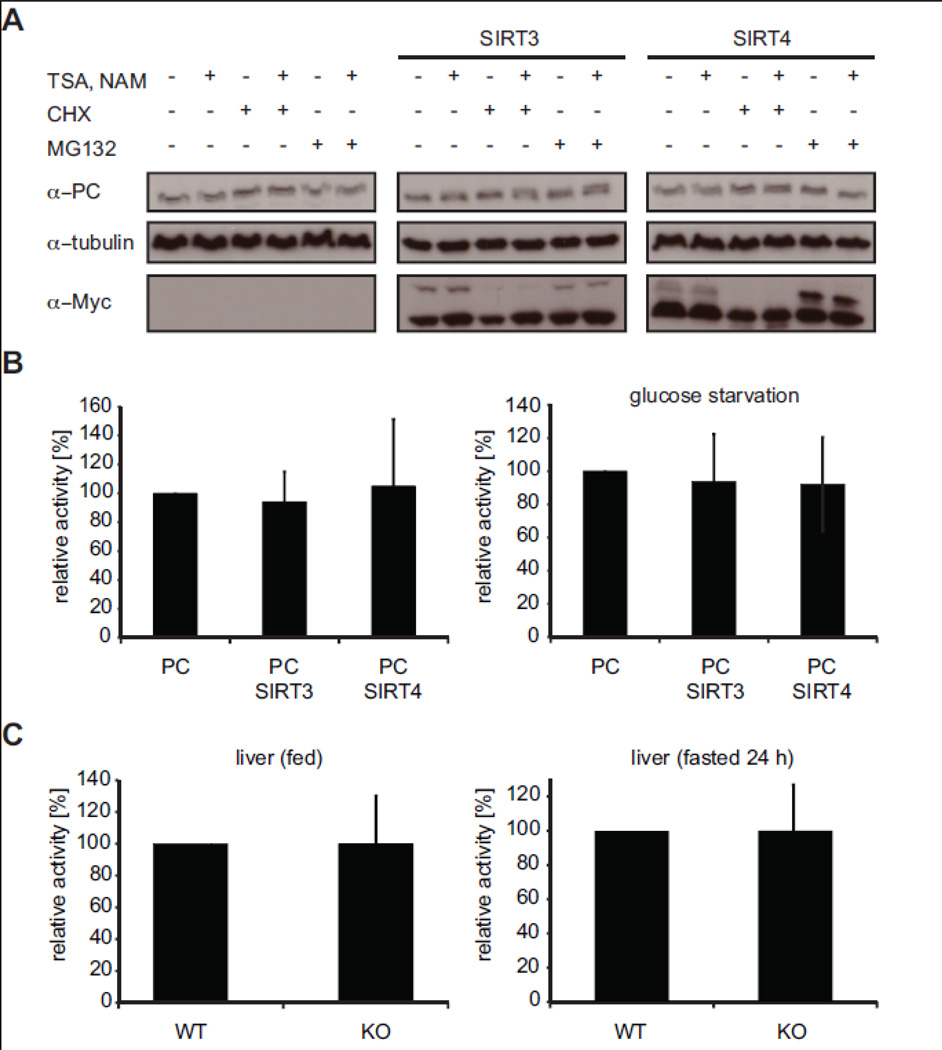
Reversible lysine acetylation is an important posttranslational modification regulating stability or activity of metabolic enzymes (Zhao et al., 2010). Since we failed to detect an effect of SIRT4 on mPC acetylation, we tested whether interaction between these factors might regulate mPC stability. Posttranslational modification different from acetylation or mechanisms independent of chemical conversion might play a role here. We analyzed mPC protein levels in the HEK293 cell line containing inducible mPC-FLAG in dependence of stable expression of MYC-tagged SIRT3 or SIRT4. After short induction of mPC-FLAG expression, cycloheximide or MG132 was added to the culture media to block protein translation and protein degradation, respectively. TSA/NAM were used for inhibition of protein deacetylation. mPC-FLAG levels were analyzed by Western blotting after 6 h of drug treatment. As Fig. 7A shows, neither presence of overexpressed SIRT3 or SIRT4 had any effect on mPC stability. In addition, general inhibition of deacetylation also did not result in changes of mPC levels compared to the controls.
Fig. 7. SIRT4 does not affect mPC stability or enzymatic activity.
(A) Analysis of mPC protein stability. Stable inducible mPC-FLAG HEK293 cells alone or also stably overexpressing human SIRT3-MYC or SIRT4-MYC were treated with 1 µM TSA and 10 mM NAM overnight. Protein synthesis and proteasomal protein degradation was inhibited with cycloheximide (20 µg/ml) and MG132 (20 µM), respectively, for 6 h. Cell lysates were analyzed by Western blot using the indicated antibodies. (B) Pyruvate carboxylase activity in stable inducible mPCFLAG HEK293 cells alone or also stably overexpressing human SIRT3-MYC or SIRT4-MYC. For glucose starvation (right graph) cells were cultured in glucose free medium overnight. Relative activities (normalized to mPC-FLAG in absence of SIRT3 or SIRT4) are represented as means of at least three experiments. Error bars represent standard deviation. (C) Enzymatic activities of mPC in liver lysates of wild type and SIRT4−/− mice fed (left) or fasted for 24 h (right). Relative activities (normalized to wild type) are represented as means of at least three experiments using two (fed) to three (fasted) mice per condition. Error bars represent standard deviation.
3.9 Overexpression or absence of SIRT4 does not affect mPC activity
Finally, we asked whether SIRT4 might affect mPC enzymatic activity. To this end, mPC activity was assayed from total cell lysates of the stable inducible mPC-FLAG HEK293 cell line in dependence of stable overexpression of SIRT3-MYC or SIRT4-MYC. Enzymatic activity was determined as before in a citrate synthase-coupled assay and normalized to mPC protein levels after Western blotting (Janke et al., 2010; Scrutton and White, 1974). Neither overexpression of SIRT4-MYC nor SIRT3-MYC in these cell lines had any significant effect on the enzymatic activity of mPC-FLAG (Fig. 7B, left panel). Since pyruvate carboxylase is a central enzyme in anaplerotic pathways we reasoned that its activity might be differentially regulated in cells under metabolic stress. We therefore repeated the experiment with mPC-FLAG cell extract prepared from cells that were starved overnight for glucose. Also under these conditions no differences in mPC enzymatic activity in presence of overexpressed SIRT4 or SIRT3 were detected (Fig. 7B, right panel).
C-terminal tagging of SIRT4 (MYC-tag) might interfere with the biological and/or enzymatic activity of the factor. Also, overexpression of the protein might not necessarily yield in increased SIRT4 cellular activity, since essential cofactors might be limiting. To further analyze putative effects of SIRT4 onto mPC activity in an in vivo system we turned to knock out mice where SIRT4 gene expression has been ablated (Haigis et al., 2006). Livers were isolated from young males of wild type and knock out mice held under the same conditions of feeding ad libido or after 24 h of starvation. Liver extracts were directly assayed for pyruvate carboxylase enzymatic activity. As Fig. 7C shows absence of SIRT4 protein had no significant effect on mPC enzymatic activity under normal conditions or after inducing metabolic stress by starvation.
4 Discussion
4.1 The biology of C. elegans SIR-2.2 and SIR2.3 is conserved with mammalian SIRT4
SIR-2 proteins constitute a highly conserved protein family (Frye, 2000) and the nematode C. elegans possesses two SIR-2 variants, SIR-2.2 and SIR-2.3, with high sequence conservation to mammalian SIRT4 (49% and 42% identity, respectively). In this study we show that the localization of C. elegans SIR-2.2 and SIR-2.3 to mitochondria is conserved to mammalian SIRT4. All three factors interact with biotin-dependent carboxylases, pyruvate carboxylase, propionyl-CoA carboxylase and methylcrotonoyl-CoA carboxylase, also across species.
SIR-2.2 and SIR-2.3 share high sequence similarity (75.3% identity) and are located next to each other on chromosome X, suggesting that one gene might have evolved from the other by sequence duplication. Our expression analysis indicates that neither sir-2.2 nor sir-2.3 became non-functionalized during evolution. Several lines of evidence indicate that SIR-2.2 and SIR2.3 are not functionally redundant. SIR-2.3 is earlier present than SIR-2.2 in embryogenesis and shows a distinct, prominent expression in a subset of cells in the head as well as in the somatic gonad. While sir-2.2(tm2648), sir-2.2(tm2673) and sir-2.3(ok444) C. elegans do not show any obvious phenotype under standard growth conditions, the single mutant worms are more sensitive to oxidative stress compared to wild type worms. Also, knock down of sir-2.2 or sir-2.3 by RNAi in sir-2.3(ok444) and sir-2.2(tm2648) mutant worms, respectively, does not result in more severe phenotypes indicating that both genes are independently needed for oxidative stress resistance.
It is surprising that expression of SIR-2.2 and SIR-2.3 from additional gene copies in a transgenic context results in even higher sensitivity to oxidative stress and does not increase stress resistance as it might be expected. Our results indicate this might be due to depletion of NAD+ in mitochondria. Mitochondrial NAD+ levels were shown to be essential for cell survival during genotoxic stress and nutrient deprivation (Yang et al., 2007). Worms mutant for the nicotinamidase pnc-1, which catalyzes the first rate-limiting step of NAD+ synthesis from NAM, are more sensitive to oxidative stress. Moreover, increased stress resistance of pnc-1 overexpressing worms depends on sir-2.1 (van der Horst et al., 2007). In mammals Nampt promotes increased mitochondrial NAD+ levels and survival during stress in a SIRT3 and SIRT4 dependent manner (Yang et al., 2007).
4.2 Are SIRT4-type sirtuins active as deacetylases?
As previously shown for SIRT4 (Ahuja et al., 2007), we could not detect broad deacetylase activity of SIR-2.2 and SIR-2.3 on a generic histone H4 tail substrate. Currently, SIRT4-type factors are the only sirtuins for which no protein deacetylase activity could be demonstrated. Glutamate dehydrogenase (GDH) is the only known SIRT4 substrate. Its ADP-ribosylation that inhibits enzymatic activity is the only sirtuin-catalyzed ADP-ribosylation with physiological relevance (Haigis et al., 2006). Due to the low efficiency of sirtuins compared to bacterial ADP-ribosyltransferases and given that most sirtuins have NAD+-dependent deacetylase activity, the physiological relevance of sirtuin mediated ADP-ribosylation has been questioned (Du et al., 2009). Indeed, even SIRT6, which was reported to exhibit strong auto-ADP-ribosyltransferase activity (Liszt et al., 2005), was recently shown to deacetylate a particular site in histone H3, H3K9ac, with very high specificity (Michishita et al., 2008). Others and we have hypothesized that the right substrates for SIRT4 deacetylation have just not been identified.
Mass spectrometric screens showed that all three mitochondrial biotin-dependent carboxylases that we identified as novel SIRT4 interacting factors are heavily acetylated proteins (Choudhary et al., 2009; Fritz et al., 2012; Kim et al., 2006; Lundby et al., 2012; Zhao et al., 2010). We mapped 13 novel sites of acetylation in PC. Of these, one site, K748ac, might indeed regulate enzymatic activity. Nevertheless, using different experimental approaches we failed so far to demonstrate deacetylation of mPC by SIRT4. Cellular overexpression of C-terminally tagged SIRT4 did neither change mPC enzymatic activity nor affect the acetylation status. We have no means to verify whether the overexpressed protein is functional and/or catalytically active. The tag might have a negative effect, also a cellular cofactor might be limiting.
We also were not able to detect any change in PC activity in liver extracts of SIRT4−/− mice. If SIRT4 deacetylates PC in this system and under the applied conditions, this has no consequence on enzymatic activity. Alternatively, there might be functional redundancy with other factors. Knock down of SIRT4 in primary murine hepatocytes increases SIRT3 gene expression (Nasrin et al., 2010) and it is possible that mitochondrial SIRT3 but also SIRT5 mask a possible phenotype. The regulation of PC by SIRT4 might be also tissue or cell type specific. Indeed, GDH is inhibited by SIRT4 in pancreatic β-cells (Haigis et al., 2006), whereas in liver deacetylation of GDH by SIRT3 seems to increase its catalytic activity and promote anaplerosis (Lombard et al., 2007; Schlicker et al., 2008).
SIRT4 is highly expressed specifically in β-cells and seems to play an important role in regulating insulin-secretion in the pancreas, which functions as fuel sensor (Haigis et al., 2006). Interestingly, PC was also shown to be necessary for glucose-induced insulin secretion (Jitrapakdee et al., 2006). Since a recent proteomic study has shown that there are tissue and cell type specific differences in lysine acetylation (Lundby et al., 2012), future studies need to address whether SIRT4 modulates the activities of PC, and possibly PCC and MCCC in a tissue and cell-type specific manner.
Despite the negative findings, we take the facts that PC is directly interacting with SIRT4, that PC is heavily acetylated and that K748ac might be a key regulatory event as indication that the right conditions for SIRT4 deacetylation have just not been found.
Two recent reports describe desuccinylation and demalonylation as novel major enzymatic activities for mammalian SIRT5 (Du et al., 2011; Peng et al., 2011). Proteomic surveys have identified diverse lysine acyl-modifications (Tan et al.; Zhang et al., 2009; Zhang et al., 2011), such as succinylation, propionylation, butyrylation or crotonylation. Although, we failed so far to detect acyl-modification of mPC, it remains a possibility that SIRT4 does not act as NAD+-dependent deacetylase but removes specific acyl-groups from lysine residues of mitochondrial proteins.
5 Conclusions
Most sirtuins were initially characterized as ADP-ribosyltransferases. However, consecutive work has established deacylation and in particular deacetylation of nuclear histone proteins and/or key metabolic enzymes with high substrate and sequence specificity as the major physiological role of all sirtuins but SIRT4. Since SIRT4 as well as its orthologues in C. elegans, SIR-2.2. and SIR-2.3, contain all the amino acids implied in sirtuin catalysis, we favour the view that SIRT4-type factors are indeed NAD+-dependent deacylases, but that the particular substrates and type of enzymatic reactions (i.e. deacetylation or removal of other amid bond-linked protein lysine posttranslational modification) of these factors have just not been identified. We demonstrate that SIRT4, SIR-2.2 and SIR-2.3 interact with mitochondrial biotin-dependent carboxylases, pyruvate carboxylase, propionyl-CoA carboxylase and methylcrotonoyl-CoA carboxylase. Since these proteins are all highly acetylated and play major roles in anaplerosis and energy homeostasis, they might be biological targets of SIRT4-type factors. Indeed, we show that PC activity might be negatively regulated by acetylation of the K748 residue. Although we failed so far to demonstrate direct deacetylation or regulation of enzymatic activity of this factor by SIRT4, a possible physiological link between SIRT4, PC and the two other biotin-dependent carboxylases in regulating insulin secretion makes us speculate that the exact conditions of SIRT4 enzymatic conversion of these metabolic enzymes have just not yet been found.
Supplementary Material
Highlights.
C. elegans SIRT4 orthologs function during oxidative stress in mitochondria.
Conserved interaction of SIRT4 with mitochondrial biotin-dependent carboxylases.
All three carboxylases are acetylated on multiple lysine residues.
Acetylation of K748 changed the enzymatic activity of pyruvate carboxylase (PC).
No deacetylation or regulation of PC by SIRT4 in liver and HEK293 cells was found.
Acknowledgements
We thank Dr. Robert Janke (Max Planck Institute for Dynamics of Complex Technical Systems, Bioprocess Engineering group, Magdeburg) for help in setting up plate reader based pyruvate carboxylase activity assays and Jennifer Seefeldt for help in subcellular fractionations. The anti-PC antibodies were a gift from Drs. Sarawut Jitrapakdee (Mahidol University) and John C. Wallace (University of Adelaide). We acknowledge further the CGC and NBRP for providing C. elegans strains. We are thankful to Drs. Dmitry Agafonov and Timur Samatov for wheat germ extract protein expression, Dr. Nora Koester-Eiserfunke for support in C. elegans work and Drs. Henriette Franz, Kerstin Mosch and Szabolcs Sörös for help with the statistical program R, cell culture and biochemical techniques. This work was supported by the Max Planck Society (WF), the DFG (MJB; JE 505/1-2), an NIA training grant (T32-AG000114, DT), the NIH (1R01GM101171, DBL), the Ellison Medical Foundation (AG-NS-0583-09, DBL), the NIDDK (DK085610, EV) and the Gladstone Institutes (EV).
List of abbreviations
- aa
amino acids
- ac
acetylation
- BC
biotin carboxylation
- BCCP
biotin carboxyl carrier protein
- HP1β
heterochromatin protein 1β
- MCCC
methylcrotonyl-coenzyme A carboxylase
- MCCC1
methylcrotonyl-coenzyme A carboxylase α-subunit
- MCCC-1
C. elegans methylcrotonyl-coenzyme A carboxylase α-subunit
- MCCCT
methylcrotonyl-coenzyme A carboxyltransferase
- NAD+
nicotinamide adenine dinucleotide
- NAM
nicotin amide
- PC
pyruvate carboxylase
- PCC
propionyl-coenzyme A carboxylase
- PCCA
propionyl-coenzyme A carboxylase α-subunit
- PCCA-1
C. elegans propionyl-coenzyme A carboxylase α-subunit
- PCT
pyruvate carboyxltransferase
- PCCT
propionyl-coenzyme A carboxyltransferase
- PYC-1
C. elegans pyruvate carboxylase
- ROS
reactive oxygen species
- SIR-2
silent information regulator-2
- SIRT
sirtuin
- TCA
tricarboxylic acid
- wt
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cong X, Held JM, DeGiacomo F, Bonner A, Chen JM, Schilling B, Czerwieniec GA, Gibson BW, Ellerby LM. Mass spectrometric identification of novel lysine acetylation sites in huntingtin. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M111.009829. M111 009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of proteome research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis AR, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabasi AL, Baillie D, Vidal M. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nature biotechnology. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- Evans TC. Transformation and microinjection WormBook, The C. elegans Research Community. 2006 [Google Scholar]
- Fahien LA, MacDonald MJ. The succinate mechanism of insulin release. Diabetes. 2002;51:2669–2676. doi: 10.2337/diabetes.51.9.2669. [DOI] [PubMed] [Google Scholar]
- Fahien LA, MacDonald MJ, Kmiotek EH, Mertz RJ, Fahien CM. Regulation of insulin release by factors that also modify glutamate dehydrogenase. J Biol Chem. 1988;263:13610–13614. [PubMed] [Google Scholar]
- Farfari S, Schulz V, Corkey B, Prentki M. Glucose-regulated anaplerosis and cataplerosis in pancreatic beta-cells: possible implication of a pyruvate/citrate shuttle in insulin secretion. Diabetes. 2000;49:718–726. doi: 10.2337/diabetes.49.5.718. [DOI] [PubMed] [Google Scholar]
- Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. Journal of proteome research. 2012;11:1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Gravel RA, Lam KF, Mahuran D, Kronis A. Purification of human liver propionyl-CoA carboxylase by carbon tetrachloride extraction and monomeric avidin affinity chromatography. Arch Biochem Biophys. 1980;201:669–673. doi: 10.1016/0003-9861(80)90557-3. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Hirschey MD. Old enzymes, new tricks: sirtuins are NAD(+)-dependent deacylases. Cell Metab. 2011;14:718–719. doi: 10.1016/j.cmet.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. 2010;1804:1645–1651. doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke R, Genzel Y, Wahl A, Reichl U. Measurement of key metabolic enzyme activities in mammalian cells using rapid and sensitive microplate-based assays. Biotechnol Bioeng. 2010;107:566–581. doi: 10.1002/bit.22817. [DOI] [PubMed] [Google Scholar]
- Jedrusik MA, Schulze E. Analysis of germline chromatin silencing by double-stranded RNA-mediated interference (RNAi) in Caenorhabditis elegans. Methods Mol Biol. 2004;254:35–48. doi: 10.1385/1-59259-741-6:035. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S, Vidal-Puig A, Wallace JC. Anaplerotic roles of pyruvate carboxylase in mammalian tissues. Cell Mol Life Sci. 2006;63:843–854. doi: 10.1007/s00018-005-5410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S, Wallace JC. The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr Protein Pept Sci. 2003;4:217–229. doi: 10.2174/1389203033487199. [DOI] [PubMed] [Google Scholar]
- Kibbey RG, Pongratz RL, Romanelli AJ, Wollheim CB, Cline GW, Shulman GI. Mitochondrial GTP regulates glucose-stimulated insulin secretion. Cell Metab. 2007;5:253–264. doi: 10.1016/j.cmet.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Lau EP, Cochran BC, Fall RR. Isolation of 3-methylcrotonyl-coenzyme A carboxylase from bovine kidney. Arch Biochem Biophys. 1980;205:352–359. doi: 10.1016/0003-9861(80)90117-4. [DOI] [PubMed] [Google Scholar]
- Lewis JAaFJF. EHFaSDC, editor. Basic culture methods. Methods in Cell Biology, Caenorhabditis elegans, Modern Biological Analysis of an Organism. 1995 [Google Scholar]
- Li J, Cai T, Wu P, Cui Z, Chen X, Hou J, Xie Z, Xue P, Shi L, Liu P, Yates JR, 3rd, Yang F. Proteomic analysis of mitochondria from Caenorhabditis elegans. Proteomics. 2009;9:4539–4553. doi: 10.1002/pmic.200900101. [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Tishkoff DX, Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb Exp Pharmacol. 2011;206:163–188. doi: 10.1007/978-3-642-21631-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, Olsen JV. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald MJ. Feasibility of a mitochondrial pyruvate malate shuttle in pancreatic islets. Further implication of cytosolic NADPH in insulin secretion. 1995;270:20051–20058. [PubMed] [Google Scholar]
- MacDonald MJ, Fahien LA, Brown LJ, Hasan NM, Buss JD, Kendrick MA. Perspective: emerging evidence for signaling roles of mitochondrial anaplerotic products in insulin secretion. Am J Physiol Endocrinol Metab. 2005;288:E1–E15. doi: 10.1152/ajpendo.00218.2004. [DOI] [PubMed] [Google Scholar]
- Mair W, Panowski SH, Shaw RJ, Dillin A. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS One. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse I, Molin L, Mouchiroud L, Vanhems P, Palladino F, Billaud M, Solari F. A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PLoS One. 2008;3:e3354. doi: 10.1371/journal.pone.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJ, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Mello CaFA. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M111.012658. M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M, Lim F, Wallace JC. Electron microscopic localization of pyruvate carboxylase in rat liver and Saccharomyces cerevisiae by immunogold procedures. Arch Biochem Biophys. 1991;290:197–201. doi: 10.1016/0003-9861(91)90608-l. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Scheffler IE. A century of mitochondrial research: achievements and perspectives. Mitochondrion. 2001;1:3–31. doi: 10.1016/s1567-7249(00)00002-7. [DOI] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton MC, White MD. Purification and properties of human liver pyruvate carboxylase. Biochem Med. 1974;9:217–292. doi: 10.1016/0006-2944(74)90062-3. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Spirin AS, Swartz JR, editors. Cell-free protein synthesis: methods and protocols. Weinheim, Germany: Wiley-VCH Verlag GmbH; 2008. [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Trelle MB, Jensen ON. Utility of immonium ions for assignment of epsilon-N-acetyllysine-containing peptides by tandem mass spectrometry. Anal Chem. 2008;80:3422–3430. doi: 10.1021/ac800005n. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Schavemaker JM, Pellis-van Berkel W, Burgering BM. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128:346–349. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2006;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Fischle W, Frye R, Marshall B, North B. Measurement of mammalian histone deacetylase activity. Methods Enzymol. 2004;377:180–196. doi: 10.1016/S0076-6879(03)77010-4. [DOI] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wenzel D, Schauermann G, von Lupke A, Hinz G. The cargo in vacuolar storage protein transport vesicles is stratified. Traffic. 2005;6:45–55. doi: 10.1111/j.1600-0854.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- Wirth M, Paap F, Fischle W, Wenzel D, Agafonov DE, Samatov TR, Wisniewski JR, Jedrusik-Bode M. HIS-24 linker histone and SIR-2.1 deacetylase induce H3K27me3 in the Caenorhabditis elegans germ line. Mol Cell Biol. 2009;29:3700–3709. doi: 10.1128/MCB.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chen Y, Zhang Z, Zhao Y. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. Journal of proteome research. 2009;8:900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.