Abstract
Introduction
The transforming growth factor-β (TGF-β) signaling pathway has a pivotal role in tumor suppression and yet, paradoxically, in tumor promotion. Functional context dependent insights into the TGF-β pathway are crucial in developing TGF-β-based therapeutics for cancer.
Areas covered
This review discusses the molecular mechanism of the TGF-β pathway and describes the different ways of tumor suppression by TGF-β. It is then explained how tumors can evade these effects and how TGF-β contributes to further growing and spreading of some of the tumors. In the last part of the review, the data on targeting TGF-β pathway for cancer treatment is assessed. This review focuses on anti-TGF-β based treatment and other options targeting activated pathways in tumors where the TGF-β tumor suppressor pathway is lost. Pre-clinical as well up to date results of the most recent clinical trials are given.
Expert opinion
Targeting the TGF-β pathway can be a promising direction in cancer treatment. However, several challenges still exist, the most important are differentiating between the carcinogenic effects of TGF-β and its other physiological roles, and delineating the tumor suppressive versus the tumor promoting roles of TGF-β in each specific tumor. Future studies are needed in order to find safer and more effective TGF-β-based drugs.
Keywords: cancer, TGF-β, tumor promotion, tumor suppression
1. Introduction
The Transforming Growth Factor-β (TGF-β) signaling pathway is instrumental in mammalian development as well as in tumor suppression through inhibition of proliferation and induction of apoptosis in multiple cell types. Yet paradoxically, TGF-β has a dual role in tumor development as it can also promote tumor cell invasiveness and metastasis mainly through modulation of the immune system as well as of the tumor microenvironment [1,2]. Key functional insights into this powerful pathway are vital for developing new therapeutics in cancer. Current clinical approaches are now aimed at establishing novel cancer drugs that target activated pathways when the TGF-β tumor suppressor pathway is inactivated, and in some cancers they are aimed at targeting TGF-β signaling itself [3,4].
2. Molecular mechanism of TGF-β signaling
2.1 Ligands, receptors and Smads
TGF-β is one of the members in a large family of growth factors that include TGF-βs, activins, inhibins, bone morphogenetic proteins (BMPs) and others [2,5–7]. In mammals three isoforms of TGF-β exist, TGF-β1, -β2 and -β3. TGF-β1, the most abundant and widely studied isoform is a 390 amino acids polypeptide while TGF-β2 and TGF-β3 contain 412 amino acids. All three forms share high degree of homology. TGF-β is secreted from variety of cells, among them platelets play a major role in humans [8]. TGF-β is secreted from the cell as an inactive latent homodimeric polypeptide bound to other extracellular proteins [9–12]. The mature, bioactive ligand is produced on proteolytic cleavage of the latent complex. TGF-β binding results in the activation of type II (TβRII) and then type I (TβRI) receptors. Activated TβRI then initiates cytoplasmic signaling pathways to produce cellular responses (Figure 1).
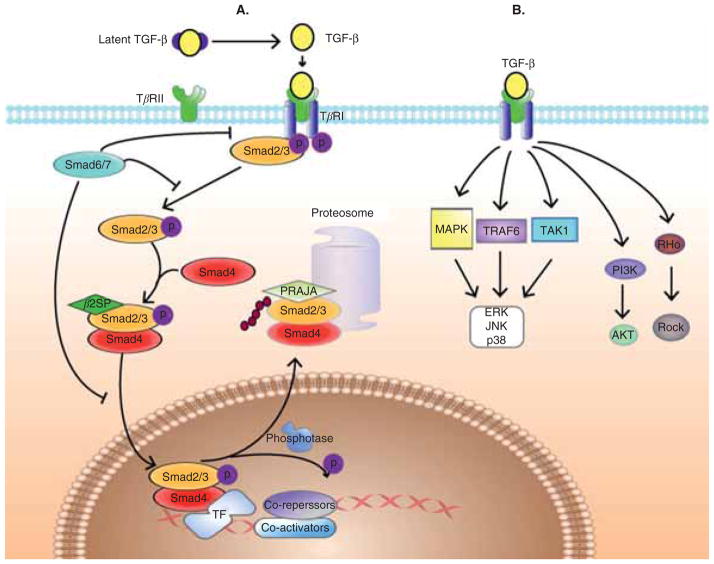
Figure 1. TGF-β signaling pathway.
A. TGF-β ligands signal through distinct receptors and Smads that are modulated by adaptor proteins and ubiquitinators. TGF-β binds to serine threonine kinase receptor complexes that phosphorylate R-Smads as well as adaptor proteins such as β2-spectrin. R-Smads, β2-spectrin and Smad-4 form a heteromeric complex, translocate to the nucleus and regulate target genes expression. At all levels, Smad modulation occurs through adaptor proteins as well as E3 ligases such as PRAJA and Smurfs, generating diverse and complex signals. B. Smad-independent signaling. TGF-β can promote the activity of several signaling pathways other than Smad, including mitogen activated protein kinases (MAPKs), phosphoinositide 3′ kinase (PI3K), TRAF6-TAK1-p38/JNK, Rho-Rock, among others. Such alternative signal transducers often regulate the Smad pathway.
TβRI and TβRII are transmembrane serine/threonine kinases [5]. Seven type I receptors and five type II receptors are encoded in humans [3,13] and paired in different combinations for different ligands (e.g., the combination of ALK5 and TβRII is needed for TGF-β1 signaling in most cells). Two co receptors: endoglin and β-glycan (type III TGF-β receptor) bind soluble ligands and regulate their binding, accessibility and signaling through the signaling receptors (TβRI and TβRII) [14]. β-glycan binds all three isoforms of TGF-β with high affinity and helps the ligands bind more efficiently to the type II receptors [3,13,15]. Endoglin is expressed more specifically in hematopoietic and endothelial cells, does not bind TGF-β2 and interacts with both receptors [14,16].
A heterotetramer of two TβRI and two TβRII molecules comprises the functional receptor [13,17,18]. The association of TβRII with TβRI triggers the cross-phosphorylation of TβRI by TβRII activating its kinase activity and switching this domain into a docking site for substrate Smad proteins [13,19,20].
The current model of ligand-induced response to TGF-β is a canonical signaling pathway from the type II to the type I receptor to Smad activation and target gene transcription (Figure 1) [1,2,21]. Smads are small intracellular effector proteins characterized by homologous regions at their N- and C-termini, known as Mad homology domains MH-1 and MH-2, respectively. An intermediate linker connects the MH-1 and MH-2 domains. This linker recruits ubiquitin ligases and is phosphorylated by other signaling kinases such as MAPKs and cyclin dependent kinases (CDKs) [1]. Three classes of Smads have been defined: the receptor-regulated Smads (R-Smads), which include Smad-1, -2, -3, -5 and -8; the common mediator Smad-4; and the inhibitory Smads, Smad-6 and -7 [5,20,22–28]. R-Smads act as direct substrates of specific type I receptors. Regulation of R-Smads by the receptor kinase confers specificity in this system: Smad-2 and Smad-3 are substrates of TGF-β receptors [29–31], whereas Smad-1, -5 and -8 are targets of BMP receptors [32–35]. Once phosphorylated by TβRI, R-Smads associate with Smad-4 [36] and mediate nuclear translocation of the heterohexameric complex. In the nucleus, Smad complexes activate specific genes, through cooperative interactions with Smad binding elements within the promoter regions of the target genes [13,37], together with other DNA-binding cofactors that increase their affinity and specificity for such target genes. The R-Smad transcription factor complex recruits co-activators and co-repressors to regulate the expression of hundreds of genes [3,9]. The cellular context (i.e., the differential expression of these regulatory cofactors in the cells) determines which specific genes are induced. The antagonistic Smads, Smad-6 and Smad-7, are thought to function by blocking ligand-dependent signaling [25,38]. Smad-6 binds to receptor activated Smad-1, preventing its association with Smad-4. Smad-7 in turn induces Smurf inactivation of TGF-β and BMP receptors.
2.2 Receptor interacting proteins, adaptors and E3 ligases
Key functional insights into the tight and coordinated regulation of this ubiquitous pathway have been gained from mouse knockout studies. This regulation largely occurs through a multitude of adaptor proteins (β2-Spectrin (β2SP), Filamin, menin among others) [7,20–22,27,39,40], E3 ligases (Smurfs, PRAJA, WWP1 and Nedd4-2) [13,21,22,41–43], as well as interacting proteins at all levels from ligand binding to receptors to Smad signaling. Smad-2/-3 and Smad-4 are thought to be distributed along the microtubule (MT) network such that MT stability is thought to be involved in Smad inactivation [44,45]. Genetic and biochemical studies demonstrate that β2SP, an adaptor protein, is required for Smad-3/-4 activation. Moreover, it is thought that β2SP modulates the recruitment of Smads to the receptor by controlling the subcellular localization of Smad-3 and Smad-4. Some of the proximal signaling events coupling TGF-β receptor activation to biological responses involve proteins such as SARA [46], FKBP12, TβRI associated protein (TRAP) 1 and 2 and others.
Interactions involving ubiquitination are an integral part of the signaling functions of Smads, involving several ubiquitin pathways e.g., Cytoplasmic and nuclear R-Smad (e.g., Smad-1 and -2) ubiquitination and proteasomal degradation mediated by Smurfs [47].
More recently, PRAJA1 has been identified as a RING finger protein that ubiquitinates β2SP and Smad3 in a TGF-β-dependent manner. PRAJA1 is involved in cell proliferation, apoptosis, juxtaposition and architecture [48]. Our studies demonstrate that loss of Smad3/β2SP through ubiquitination by PRAJA1 could play a significant role in the development of liver and gastrointestinal (GI) tumors [22].
Nuclear phosphatases, such as PPM1A, dephosphorylate the C-terminal tails of R-Smads and lead to disassembly of the transcriptionally active R-Smad/Co-Smad, initiating a molecular cascade for termination of the transcriptional Smad signal [39,49].
2.3 Smad-independent signaling
In addition to activation of the Smad pathway, TGF-β promotes the activity of several other signaling pathways, including mitogen activated protein kinases (MAPKs), phosphoinositide 3′ kinase (PI3K), TRAF6-TAK1-p38/JNK, Rho-Rock, among others [50]. Such alternative signal transducers often regulate the Smad pathway and mediate signal transduction by various other effectors. Thus, TGF-β transmits biological signals to cells through Smad-dependent pathway, and also through alternative signaling pathways which offer nodal points for crosstalk with other signal transduction pathways.
3. TGF-β signaling in tumor suppression
3.1 Clinical-genetic data
Genetic studies have identified mutations in genes encoding the components of TGF-β signaling. The most commonly mutated TGF-β associated genes are TβRII, TβRI, Smad-2 and Smad-4. These mutations occur mainly in GI tract cancers. In colorectal cancer (CRC) the TβRII gene mutations occur late during tumorigenesis, at the adenoma to carcinoma transition [51]. These mutations are also abundant in gastric, pancreatic, biliary tract, lung and brain (glioma) tumors [52]. Those inactivating mutations in TβRII occur in most human colorectal and gastric carcinomas with microsatellite instability (MSI), since TβRII is a mutational hotspot due to its 10 base poly-A repeat within its coding sequence [37,53–57]. Mutations in TβRI are less frequent, although they have been described in pancreatic, colorectal, ovarian and head and neck cancers [58–60]. Mutations of TβRII and TβRI are relatively rare in breast and skin cancers [37,52], as well as in hematological malignancies [16]. A number of point mutations have been identified in Smad-2, mainly in ovarian, cervical, liver, CRC and lung cancers [58,61–63].
The most frequently mutated Smad gene in human cancer is Smad-4. It undergoes biallelic loss in one-half of all of pancreatic cancers [64,65], one third of metastatic colon tumors [59,66] and smaller subsets of other carcinomas (hepatocellular, breast, bladder, biliary tract, ovarian, intestinal, colorectal and lung carcinomas as well as tumors of prostate and cervical origin) [52,62,67–70]. In addition, germline mutations in Smad-4 cosegregate in a subgroup of patients with juvenile polyposis syndromes (JPSs), an autosomal dominant disorder characterized by the development of hamartomatous intestinal polyps and increased risk of GI cancers [71].
The genetic evidence from human tumors supports a clear role of the Smad-dependent TGF-β pathway as a tumor suppressor in many types of human cancers, particularly those of the GI tract.
3.2 Mechanisms of TGF-β signaling in tumor suppression (Figure 2)
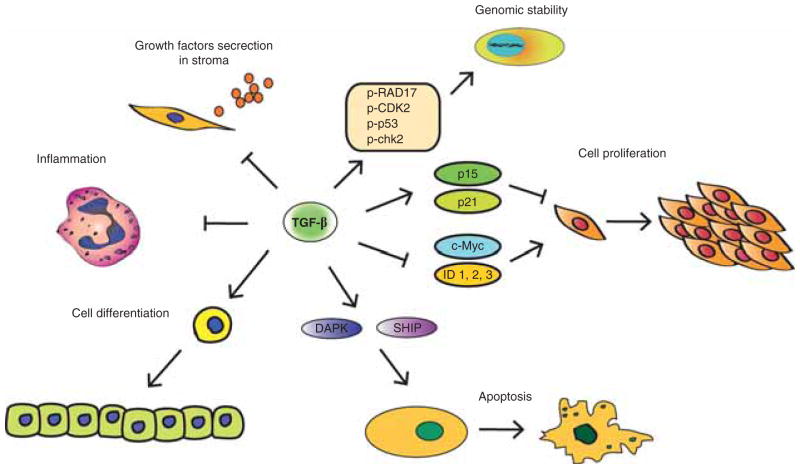
Figure 2. TGF-β signaling in tumor suppression.
TGF-β achieves its tumor suppressive effect by several arms: the most important one is the cytostatic or cell proliferation regulation arm. Here, TGF-β induces expression of cyclin-dependent kinase (CDK) inhibitors p21 and p15 and decrease expression of proliferative drivers such as c-Myc and ID. Other modes of TGF-β action include its effects on apoptosis and cell differentiation, genomic stability and indirect effects on the tumor stroma, such as inhibition of growth factors secretion and anti-inflammatory effects.
TGF-β achieves its tumor suppressive effect by several arms: the most important one is the cytostatic or cell proliferation regulation arm. Other modes of action include its effects on apoptosis and cell differentiation, genomic stability and several indirect effects on the tumor stroma. Very detailed mechanistic description of the TGF-β signaling in tumor suppression and promotion are beyond the scope of this review but can be found in very good other reviews [1,3,4].
3.2.1 Cell proliferation
TGF-β regulates cell proliferation mainly by inhibiting cell cycle progression through G1-arrest. In epithelial cells it does so through a coordinated cytostatic program with dual effects: i) induction of CDK inhibitors p21Cip1 and p15Ink4b to arrest cell proliferation and ii) suppression of proliferative drivers such as c-Myc and ID.
p21Cip1 inhibits the activity of cyclin E/A-cdk2 complex; and p15Ink4b inhibits the interaction between cyclin D and cdk4/6, and also the interaction between cyclin E/A-cdk2 (through mobilization of p27 from cyclin D-cdk4 by p15). The inactivation of the cdk complexes prevents phosphorylation of pRB and the progression from G1 to S phase [4]. In order to induce p21Cip1 and p15Ink4b, Smad-3/-4 form a complex with FoxO and with Sp1 transcription factors [1,4,72–74].
The repression of the important oncogene c-Myc which stimulates proliferation, but also inhibits the transcriptional activation of p21 and p15 adds another component to the tight regulation of the TGF-β signaling pathway on these two target genes [74–76]. TGF-β inhibits ID1,2,3 – nuclear factors which play a role in cell differentiation and progression from G1 to S phase.
3.2.2 Apoptosis and senescence
TGF-β can both induce and suppress apoptosis [77] depending on cellular and extracellular factors. Unlike the TGF-β cytostatic program, there is not a unique TGF-β-induced apoptotic program. In vitro studies have shown some Smad-dependent and -independent mechanisms, e.g., TGF-β increases the expression of death-associated protein kinase (DAPK) in HCC cell-lines [78], but it induces the expression of SH2-domain-containing inositol-5-phosphate (SHIP) in hematopoietic cell-lines, which in turn inhibits the survival signals from the PI3K-AKT pathway. TGF-β can induce senescence of mammary stem cell population by diminishing their self-renewing capability [37,79]. Other apoptotic related genes affected by TGF-β pathway are DAXX (that normally activates p38MAPK), FAS and BIM (in gastric cancer cell lines) and GADD45b (in hepatocytes) [1,4,38]. The final targets in TGF-β-induced apoptosis are the proapoptotic caspases and several members of the BCL2 family [3].
3.2.3 Genomic stability
Another tumor suppressor function of TGF-β is to maintain the genomic stability. It has been shown that keratinocytes from TGFβ1-null mice exhibit marked genomic instability in vitro and this could accelerate tumor progression [37,80]. TGF-β also functions as an extracellular sensor of DNA damage. Inhibition of TβRI as well as knockout of Tgfb1 impaired phosphorylation of ATM, p53, Chk2 and Rad17, which results in reduced gammaH2AX radiation-induced foci; and increased radiosensitivity compared with TGF-β competent cells [81]. Studies in the Smad-4 conditional knockout mice, that develop head and neck cancers, demonstrate a significant role for Smad-4 in promoting genomic stability through regulation of the Fanconi anemia/BRCA DNA repair pathway [82]. Recently, we have shown that β2SP has a major role in maintaining genomic stability from alcohol-induced DNA damage, also through regulation of the Fanconi Anemia pathway (Shukla V et al., under review).
3.2.4 Tumor microenvironment
Besides its direct effect on epithelial tumor cells, TGF-β further controls tumor development by modulating growth factors produced by the surrounding stroma. A murine model showed that overexpression of dominant negative TβRII in the stroma of the mammary gland increases expression of hepatocyte growth factor (HGF) in the fibroblasts and resulted in increase in the lateral branching of adjacent mammary ducts [1,4,83]. In addition, inactivation of TβRII expression in mouse fibroblasts causes prostatic intraepithelial hyperplasia and squamous cell carcinoma of the forestomach, accompanied by higher expression of HGF and its receptor MET in the TβRII negative fibroblasts and the neighboring epithelial cells, respectively [84].
Another non-cell autonomous role of TGF-β during tumorigenesis is to suppress immune and inflammatory processes. This was demonstrated in several mouse models with deficiency/deletion of TGF-β, TβRII and Smad-3 [1,85]. TGF-β inhibits CD8+ cytotoxic T cells, CD4+ T cells, macrophages, dendritic cells and NK cells and it stimulates the generation of regulatory T cells and Th17 cells [1]. TGF-β disruptive signaling is one of the molecular mechanisms in the pathogenesis of the pre-cancerous inflammatory bowel diseases (IBD) which are characterized by alteration of intestinal mucosal immune tolerance. Indeed, colonic T cells from IBD patients exhibit high level of Smad-7 and decreased responsiveness to TGF-β [86].
4. Escaping the tumor suppressive mechanisms of TGF-β pathway
As previously mentioned, the first way by which tumors evade TGF-β’s tumor suppressive mechanisms, exhibited mainly by GI and head and neck cancers, is through inactivating mutations of the core elements of the TGF-β signaling pathway, the receptors and the Smads genes. However, other types of cancer like breast cancers, melanomas, gliomas, prostate cancers and some hematopoietic cancers retain the core components of the TGF-β pathway, and only inhibit its tumor suppressive arm. In order to lose the tumor suppressive arm, some of the cancer cells alter the Smad-regulated genes that mediate the cytostatic program [3]. This anti-proliferative mechanism is based on dual, and hence redundant, effects: induction of CDK inhibitors to hold up cell proliferation, and suppression of proliferative drivers, so combined alteration is needed for disrupting this cytostatic program. Indeed, TGF-β still inhibit cell proliferation very effectively even in cells that lack its p15 or c-Myc reaction alone [87,88], however, the combined loss of those two responses causes an effective escape from cell proliferation [89]. Studies in breast cancer have shown intact core elements of the TGF-β pathway, but partial to complete loss of TGF-β-induced cell cytostasis due to failure of p15 induction as well as c-Myc suppression in response to TGF-β [3]. The mechanism behind this combined action involves the cofactor C/EBPβ [3,90].
Other mechanisms for the loss of the TGF-β suppressive arm have been described in other types of cancers, e.g., homozygous deletion of p15Ink4b in some gliomas, which prevents TGF-β-mediated induction of this gene in those cancers [91]; over expression of oncogenes like cyclin D1 and c-Myc that reduce the TGF-β effect on CDK inhibitors or expression of Ras signaling which inhibits Smads [1]; and induction of ID1 instead of its suppression in metastatic breast cancer [1,92].
In hematological malignancies, resistance to the suppressive effects of TGF-β takes place mostly through decreased expression of the TGF-β receptors on the cell membrane (e.g., decreased TβRI expression in CLL [93]; TβRII in polycythemia vera [94] and essential thrombocytosis [94]; both receptors in cutaneous T-cell lymphoma [95]; or deficient trafficking of the receptors to the cell surface in multiple myeloma (MM) [96]). Other mechanisms in hematological malignancies include repression of TGF-β signaling by oncoproteins such as Tax and Evi-1 in chronic myeloid leukemia (CML) and adult T-cell acute lymphoblastic leukemia (ALL) [16,97,98], or the t(8;21) in acute myeloid leukemia (AML) M2 that results in the fusion protein AML1 (Runx1)/ETO that binds to Smad-3 as the original AML1 protein, but instead of activating the TGF-β pathway, it represses it [16,99]. More examples for mechanisms of TGF-β pathway disruption in hematological malignancies can be found elsewhere [16].
Mutation of the tumor suppressor p53 can also be responsible for changing the TGF-β response [100,101] through suppression of p63. Other possible mechanisms can be found in more detail elsewhere [100]. The ability of certain tumors to escape the cytostatic program permits the consequent use of the TGF-β pathway for tumor promotion.
5. TGF-β signaling in tumor promotion (Figure 3)
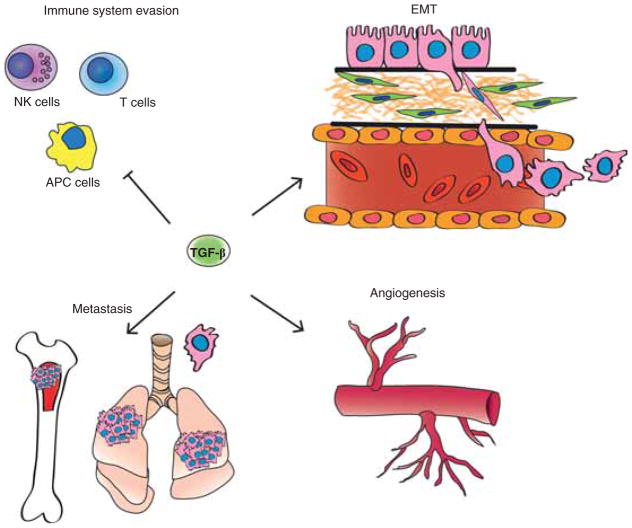
Figure 3. TGF-β signaling in tumor promotion.
TGF-β achieves its tumor promoting effect by several mechanisms: EMT, evasion of the immune system, promotion of cancer cell proliferation by modulation of the tumor microenvironment and effect on the metastatic process.
APC: Antigen presenting cells; EMT: Epithelial-to-mesenchymal transition; NK: Natural killer.
Once the tumor has undergone the genetic changes necessary for escaping TGF-β’s tumor suppressive mechanisms, augmented expression of TGF-β can paradoxically result in tumor progression and metastasis. The most important mechanisms of tumor progression caused by TGF-β are epithelial-to-mesenchymal transition/transdifferentiation (EMT), evasion of the immune system and, promotion of cancer cell proliferation by modulation of the tumor microenvironment [102,103]. These mechanisms result in enhanced tumor cell invasion and migration which lead to tumor progression and metastatic dissemination.
5.1 Epithelial-to-mesenchymal transition
EMT is a well-coordinated process during which cells lose many of their epithelial characteristics and acquire fibroblast-like properties. EMT is a cardinal process during embryogenesis and plays a role in wound healing; however, EMT also takes place in pathological process of fibrogenesis and tumorigenesis.
During EMT, the cells lose their polarity and cell–cell contact by downregulating the expression of E-cadherin and other components of the cell junction [104]. Concomitantly, they upregulate the expression of mesenchymal cell associated transcription factors such as Snail, Slug, Twist and FoxC3 among others, and cytoskeleton associated genes such as Fibronectin, α-smooth muscle actin and Vimentin, [3,105,106], which are essential for enhanced motility and invasiveness.
In vitro evidence has demonstrated that TGF-β is a major regulator of the EMT process. Notably, cells that overexpress Smad-7 or have reduced expression of Smad-3/-4 show significantly decreased EMT in response to TGF-β1 [4,107]. Conversely, overexpression of Smad-3/-4 results in increased EMT [107]. In human carcinomas, cells that have undergone EMT are found in the invading tumor edges which are usually areas rich in TGF-β and other related cytokines.
EMT is a reversible process until the mesenchymal phenotype becomes fixed by other genetic and epigenetic changes. The plasticity and reversibility of this process are TGF-β-dependent and respond to the local TGF-β level [37]. It is important to mention that TGF-β is not the only determinant factor of EMT, and other cytokines such as HGF also regulates EMT, even in the absence of TGF-β [108].
Besides acquiring mesenchymal cell properties during EMT, the epithelial cells also obtain some stem cell characteristics under the regulation of TGF-β [3,4]. In immortalized mammary epithelial cells, induction of EMT by TGF-β, Snail or Twist, stimulates expression of surface markers associated with cancer stem cells. These cells share high homology to bone marrow-derived mesenchymal stem cells [109].
5.2 Immune evasion
Despite of its anti-inflammatory properties which result in tumor suppression, when the immunosuppressive effects of TGF-β become more dominant, the net effect is towards tumor progression [1]. In mouse model with T cell specific dominant negative form of TβRII challenged with melanoma or thymoma cell lines, growth and metastasis formation were repressed [110]. TGF-β suppresses transcription of pro-apoptotic and cytolytic factors in CTLs like granzyme A and B, perforin, interferon-γ and FAS ligand [4,111]. TGF-β can inhibit the function of antigen presenting cells, thereby further decreasing T cell activation [112]. TGF-β acts on both CD4+ and CD8+ T cells as well as on natural killer (NK) cells. The inhibition of the NK cells is caused by transcriptional repression of NKG2D and NKp30 [4,113,114]. Inhibition of TGF-β increases NK cells activity to suppress metastasis formation in breast cancer cell line [112].
TGF-β drives the immune response from type 1 differentiated anti-tumor cells into the more immature type 2. This modulation occurs in the innate immune system (neutrophils and macrophages) as well as in the T cells level. These immature cells release more TGF-β and IL-11 into the tumor environment, which result in a tumorigenic effect [37,115].
5.3 Invasion and angiogenesis
TGF-β promotes the production and secretion of matrix metalloproteases MMP-2 and MMP-9, and it downregulates the expression of the protease inhibitor TIMP [3,66,116]. TGF-β also potently stimulates hyaluronan synthesis through upregulation of hyaluronan synthase 2 in mammary epithelial cells [117].
TGF-β can stimulate angiogenesis by its effects on local angiogenic factors such as vascular endothelial growth factor (VEGF) and connective tissue growth factor (CTGF) [3,118,119]. Impairment of TGF-β signaling in mouse models lacking TβRI or TβRII has revealed defects in angiogenesis leading to death of those mice [120,121], and increased expression of TGF-β either in tumor cells or their environment resulted in amplification of angiogenesis [3,122].
5.4 Metastasis
TGF-β has an impact on the metastatic process; however, this effect is complicated and context-dependent [3,4]. A detailed description of the roles of TGF-β in metastasis can be found elsewhere [3].
Clinical evidence ties TGF-β to the metastatic process. The extent of TβRII expression in estrogen receptor (ER) negative breast cancer is negatively associated with overall survival [123], and higher expression of TGF-β is seen in metastatic breast cancer than in the primary tumors [124]. Pre- and postoperative plasma levels of TGF-β are correlated to the presence of metastases in different types of cancer like breast, prostate, CRC, pancreas and more [1].
Mouse models have shown that radiotherapy and chemotherapy cause increased TGF-β1 levels as well as circulating tumor cells and lung metastases in breast metastasis model [125], while administration of anti-TGF-β neutralizing antibodies prevented the enhanced metastasis [1,4,125].
However, not all studies point to the same direction. While short-term stimulation with TGF-β increases metastasis formation, persistent TGF-β stimulation reduces lung metastases [126]. Furthermore, while expression of activated TβRI enhances lung metastasis in transgenic mouse model of breast cancer, targeted deletion of TβRII results in the same phenomenon [4,127,128]. Approximately 40% of the human breast cancers show a positive TGF-β gene response signature, that is context dependent, and appears more in ER- tumors (as opposed to ER+ tumors) and in lung metastasis (as opposed to bone metastasis) [1]. The mechanism of the TGF-β induced lung metastasis in breast cancer is related to the induction of the angiopoietin-like 4 (ANGPTL4) gene by TGF-β in the primary tumor, enabling the cells which leave the breast to disrupt the lung capillary walls [92]. The fenestrated capillaries of the bone marrow do not have any advantage from the action of ANGPTL4, and that might explain why the impact of TGF-β is directed to lung and not to bone metastasis [1].
Besides its role in priming the tumor cells to distal metastasis, TGF-β affects the growth of the metastases themselves. TGF-β is a major player in the formation of bone metastases. TGF-β is released from bone matrix when metastatic cells activate osteoclasts that degrade the bone matrix. On its discharge, TGF-β stimulates releasing of other osteolytic cytokines from the metastatic cells, such as parathyroid hormone related protein (PTH-rP), IL-11 and CTGF to perpetuate the metastatic process [129]. Smad-3 and -4 are necessary for the metastatic expansion in bone, while positive staining of phospho-Smad-2 is presented in lung, liver and brain metastases of breast cancer [1,4,130].
6. Targeting TGF-β signaling – therapeutic applications
High serum or tissue TGF-β levels are associated with worse prognosis for breast cancer, HCC and gastric cancer [131–133], the rationale for targeting the tumor promoting side of TGF-β is clear [134]. Indeed, powerful anti-TGF-β strategies have been developed and tested in pre-clinical studies and some even in clinical trials.
We will first describe the current knowledge of anti-TGF-β treatments and then suggest other treatment options based on the enhancement of TGF-β’s tumor suppressive properties.
6.1 Treatments targeted against the TGF-β pathway (Figure 4)
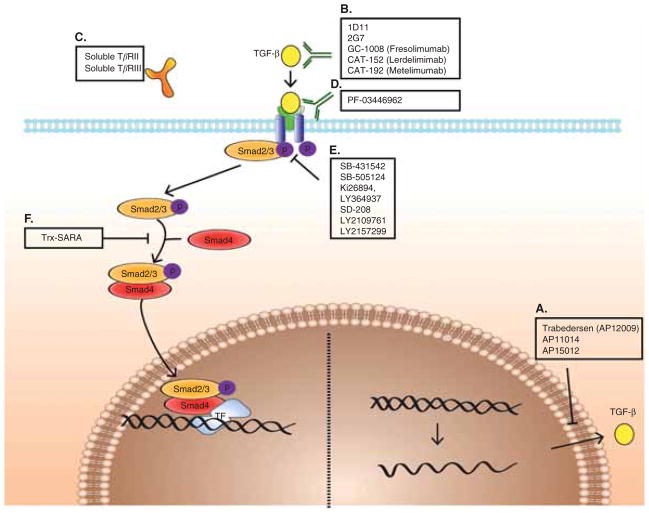
Figure 4. Treatments targeted against the TGF-β pathway.
(A) antisense molecules prevent TGF-β synthesis (B – D) monoclonal antibodies, soluble receptors and anti-receptor monoclonal antibodies prevent ligand–receptor interaction (E,F) receptor kinase inhibitors and peptide aptamers prevent signal transduction.
Therapeutic strategies against TGF-β can be divided into three levels:
Ligand level: prevention of TGF-β synthesis by using antisense molecules.
Ligand–receptor level: prevention of ligand–receptor interaction by ligand traps (monoclonal antibodies and soluble receptors) and anti-receptor monoclonal antibodies.
Intracellular level: prevention of signal transduction by receptor kinase inhibitors and peptide aptamers.
6.1.1 Antisense molecules
Antisense molecules (oligonucleotides) are single stranded oligonucleotide molecules containing 13 – 25 nucleotides that bind complementary sequences on specific mRNA, thereby preventing its translation and accelerating its degradation [37,135]. Since TGF-β production is usually increased during tumor progression, blocking its synthesis has the potential to reduce excess TGF-β levels within the tumor microenvironment. AP12009 (Trabedersen, Antisense, Pharma) is an anti-sense molecule against TGF-β2, whose expression is correlated with poor prognosis in glioblastoma and pancreatic cancer. In vitro studies in glioma cells and in a mouse model of pancreatic cancer have shown the efficacy of this drug in decreasing proliferation, migration, tumor growth and metastasis [136,137]. AP11014 and AP15012 are other two antisense molecules in pre-clinical trials for treatment of non-small cell lung cancer, prostate carcinoma and CRC; and MM, respectively [138].
Trabedersen (AP12009) was successfully tested in Phase I/II study in patients with refractory high grade glioma that showed significant increase in median survival compared with chemotherapy [136]. The results of an open-label, Phase I/II study of Trabedersen, in patients with advanced tumors known to overproduce TGF-β2 (pancreatic cancer, multiple melanoma and CRC – all of them in stage III/IV) were presented in the ASCO meeting of 2012. Trabedersen was safe and well-tolerated. The only expected adverse reaction identified was thrombocytopenia. Survival analysis of pancreatic cancer patients revealed a median overall survival (mOS) of 13.4 months (n = 9). One patient had a complete response of liver metastases and was still alive after 75 months. Promising efficacy data were also seen in MM patients with a current mOS of 9.3 months (n = 14) [139].
6.1.2 Ligand traps
The excess of TGF-β production in the tumor microenvironment can be controlled by using ligand traps. Ligand traps include monoclonal neutralizing antibodies against TGF-β; soluble TGF-β receptors and TGF-β receptor antibodies.
6.1.2.1 Neutralizing antibodies
Treatment of metastatic breast cancer mouse model with 1D11 (Genzyme Corp., Sanofi), a monoclonal antibody (mAB) that binds TGF-β1, 2 and 3, resulted in suppression of lung metastasis, mainly by significant increase in the anti-tumor response of CD8+ T-cells [140]. It also decreased bone loss by reduced expression of PTHrP and its regulator Gli2 [37]. Another mAB in pre-clinical trials is 2G7, which has shown efficacy in inhibiting breast cancer metastasis, increasing NK cells activity and preventing radiation induced acceleration of metastases [112,125,141].
Three fully humanized mAB were developed by Genzyme and tried in clinical trials: GC-1008 (Fresolimumab), CAT-152 (Lerdelimimab) and CAT-192 (Metelimumab). GC-1008 was tested in Phase I/II clinical trial on patients with advanced renal cell carcinoma (RCC) (n = 1) and MM (n = 22). The results of this trial were presented at the 2008 ASCO meeting [142]. Overall, no dose limiting toxicities were reported; however, several adverse events were reported such as dose-dependent skin rash (mainly non-malignant keratoacanthomas), fatigue and gingival bleeding. Five patients achieved stable disease and continued with treatment. Two current trials of GC-1008 are in recruitment phase: Fresolimumab and radiotherapy in metastatic breast cancer (NCT01401062) and safety and imaging study of GC1008 in glioma (NCT01472731). The other two mABs have not been tried yet on cancer patients.
6.1.2.2 Soluble TGF-β receptors
Another way to block TGF-β before it interacts with its receptor is by adding soluble TGF-β receptors. Soluble TβRII and TβRIII (betaglycan) have been tested in pre-clinical studies [4]. Expression of soluble TβRII reduced breast cancer and pancreatic cancer metastasis [143–145], and soluble TβRIII inhibited pulmonary metastasis when administered intraperitoneally to athymic nude mice [146]. No clinical trials have been undertaken with those soluble receptors.
6.1.2.3 Monoclonal antibodies against the receptors
PF-03446962 is an anti-TβRI mAB which competes highly efficiently with the binding of the TβRI ligands BMP9 and TGF-β to TβRI. This antibody inhibits endothelial cell sprouting and can serve as an anti-angiogenesis agent [147]. This is the first compound of this family to be tested in a Phase I clinical trial, reported in the 2012 ASCO meeting to be a promising anti-angiogenic agent [148]. A Phase II clinical trial of PF-03446962 in patients with advanced malignant pleural mesothelioma is recruiting patients now (NCT01486368).
6.1.3 Signal transduction blockade
Two therapeutic strategies exist in order to block signal transduction after binding of the ligand and its receptors. The first is the use of receptor kinase inhibitors, and the second is targeting the intracellular TGF-β signaling pathway molecules, such as the Smads, with peptide aptamers [37].
6.1.3.1 Receptor kinase inhibitors
Targeting receptor kinases has been extensively investigated in cancer treatment during the last years, mainly because of the ease of drug production and the ability to administer it through the oral route [37]. Blocking of the downstream signaling is more efficient by receptor kinase inhibitors than by ligand traps or antisense molecules, however, it is less specific. Most of the receptor kinase inhibitors act by inhibition of the catalytic ATP-binding site of TβRI.
SB-431542 (GlaxoSmithKline) is a small molecule inhibitor of TβRI, preventing phosphorylation of Smad-2 and Smad-3. SB-431542 inhibits TGF-β-induced proliferation of osteosarcoma cell lines as well as proliferation, motility and angiogenesis of glioma cells, and transcription of collagen and fibronectin in renal carcinoma cells [149,150]. This compound also induces dendritic cells maturation, CD8+ T cell activity and releases stromal cells from MM-induced differentiation arrest [37,151,152]. Another inhibitor SB-505124 has found to be 3–5 times more potent [135,153]. However, these two inhibitors are unstable and non-specific. This lack of specificity can lead to unpredictable results and side effects. Ki26894, LY364937 and SD-208 are other TβRI inhibitors which have shown promising results in in vitro and in vivo experiments using breast and gastric cell line [154,155], xenografts [154,156] and mouse models of glioma [157] and metastatic MM [158].
LY2109761 is a small molecule inhibiting the kinase activity of both TβRI and TβRII. This compound inhibits matastasis formation in mouse models of breast cancer, CRC and pancreatic cancer [159–161]. However, long-term use of this drug in a skin cancer mouse model resulted in resistance and cancer progression [162], suggesting that more than one drug may be needed for long-term inhibition of one signaling pathway [37].
Another approach to target the kinase is by blocking the substrate-binding site of the TβRI kinase by peptides that mimic Smad-2 as was shown in Mv1Lu cells [135,163].
LY2157299 (Eli-Lilly & Co) is a TβRI kinase inhibitor that reduces growth of lung and breast cell lines [164]. This is the only TGF-β receptor kinase inhibitor currently in clinical trials. During the 2011 ASCO meeting the results of the first human dose study were reported. Twenty-eight patients with Grade IV glioma were treated, and LY2157299 was well tolerated at all doses. There were two drug-related dose limiting toxicities, a pulmonary embolism and thrombocytopenia. Three patients taking the lowest dose, 160 mg/day, were treated for > 20 cycles. Totally there were three confirmed and two unconfirmed partially responses [165]. At present the drug is tested in four clinical trials, all of them are still recruiting patients: Phase Ib/II in stage II – IV pancreatic cancer of LY2157299 combined with gemcitabine versus gemcitabine plus placebo (NCT01373164); Phase II in HCC patients who have had disease progression on Sorafenib or are not eligible to receive sorafenib (NCT01246986); Phase Ib/IIa study combining LY2157299 with standard Temozolomide-based radiochemotherapy in patients with newly diagnosed malignant glioma (NCT01220271); and Phase II Study of LY2157299 monotherapy or LY2157299 plus Lomustine therapy compared to Lomustine monotherapy in patients with recurrent glioblastoma (NCT01582269).
6.1.3.2 Peptide aptamers
Peptide aptamers are small peptide molecules containing a target binding site and a scaffolding domain that impedes the function of the target. They are designed against specific targets, such as the Smad proteins and other targets downstream the TGF-β signaling pathway. Trx-SARA is an example of a peptide aptamer which reduces the levels of TGF-β-induced Smad-2/-3 in complex with Smad-4 [37], and inhibit EMT after TGF-β stimulation in breast cancer epithelial cells [166]. No clinical trials have been undertaken with peptide aptamers.
6.2 Therapeutic targets that arise from enhancing TGF-β’s tumor suppressive properties
In several tumor cell type, activation of cell cycle proteins such as CDK4, c-Myc, β-catenin and h-TERT occurs when TGF-β signaling is inactivated. Thus, those molecules could represent new functional targets for therapeutics of lethal cancers that evade TGF-β [167–169]. Most human cancers appear to have lost their growth-inhibitory response to TGF-β. However, only about 10% of the tumors (mainly GI and head and neck tumors) appear to exhibit loss of expression of TGF-β receptors or Smads, suggesting that other mechanisms such as amplification and overexpression of cell cycle regulatory proteins such as Cyclin D1 and/or CDKs, or loss of expression of scaffolding proteins, such as β2SP, may account for the loss of TGF-β signaling in human tumors. Foregut cancers with inactivation of TGF-β signaling express high levels of cyclin D1 and CDK4 levels. Small molecule inhibitors that specifically inhibit CDK4 but do not exhibit cross-reactivity with other known CDKs could be very useful in cancer therapy. The results of a Phase I clinical trial of P1446A-05, a CDK4 inhibitor, were presented at the 2012 ASCO meeting. A total of 29 patients were dosed. Six SAEs including one death related to study drug were reported. Stable disease for 4 – 6 cycles was reported in five patients, however, no objective responses were observed in this group of heavily pre-treated patients [170]. Other clinical trials with this drug and other CDK4 inhibitors are currently ongoing (Table 1).
Table 1.
Drugs targeting signaling pathways which are activated with loss of TGF-β signaling.
| Agent name | Type | Target | Indications |
|---|---|---|---|
| Targeting Wnt signaling | |||
| Sulindac and derivatives | NSAID | β-catenin | Hereditary forms of colon cancer |
| Retinoids | Vitamin A | β-catenin | Colon cancer |
| 1α,25-Dihydroxyvitamin D3 and synthetic derivatives | Vitamin D | β-catenin | Colon, breast and prostate cancers |
| Targeting CDKs | |||
| P1446A-05 | Small molecule inhibitors | CDK4 | Phase I, advanced refractory malignancies |
| PD-0332991 | Small molecule inhibitors | CDK4, CDK6 | Phase I, advanced cancer |
| Targeting telomerase | |||
| GV1001 | Peptide vaccine | TERT epitopes | Phase III trial for advanced pancreatic patients |
| Telomelysin® | Adenovirus | Containing the hTERT promoter | Phase I solid tumor clinical trials |
| Targeting Stat3 signaling | |||
| PY*LKTK, Y*LPQTV | Peptide | STAT3 SH2 | Preclinical |
| NSC 74859 | Small molecule inhibitors | STAT3 SH2 | Activated STAT3 in HCCs with increased cancer stem cells4, 5 |
| Targeting TGF-β signaling | |||
| Belagenpumatucel-L | Anti-TGF-β2 vaccine | TGF-β2 | Nonsmall-cell lung cancer |
| AP 12009 | Antisense oligonucleotide | TGF-β2 | Glioma, pancreatic carcinoma, melanoma |
Pathways that control stem-cell proliferation are other options for cancer treatment. The canonical Wnt signaling maintains the growth of stem cells. In the intestine, the presence of TGF-β-signaling and the absence of Wnt signaling in the villus compartment result in rapid cell cycle arrest and differentiation. Thus, Tcf4 (affected by Wnt signaling) and Smad-4 constitute a dominant switch between the proliferative progenitor and the transitional progenitor of differentiated epithelial cell. At all stages of CRC this switch is permanently reversed because TGF-β signaling is inactivated while Tcf4 is constitutively activated by mutations in the Wnt cascade, leading to aberrant crypt foci and the long lived adenomatous polyps. These observations make the Wnt signaling pathway a useful target in GI cancers. A vitamin D3 analog, Seocalcitol, has been known to be able to inactivate β-catenin [171,172], the key protein in the wnt signaling. Our preliminary data demonstrate the promising effects of vitamin D in treatment of colon and liver cancers. Other drugs targeting Wnt signaling are listed in the Table 1.
Cross-talk between TGF-β/Smad and JAK/STAT signaling pathways has been reported. TGF-β can downregulate IL-6-induced phosphorylation of STAT3 [173]. Our data shows that STAT3 level is remarkably increased in HCC tissues in β2SP knockout mice model, in which TGF-β signaling is disturbed [174]. Moreover, NSC 74859, a STAT3-specific inhibitor, markedly inhibits STAT3 phosphorylation in HCCs with inactivation of the TGF-β/β2SP pathway, indicating that IL6/STAT3, can provide a novel approach to the treatment of specific HCCs [175]. Aberrant activation of STAT3 occurs in many human tumors. Up to now, many STAT3 inhibitors have been developed. The strategies of targeting STAT3 are shown in Table 1.
Carcinoembryonic antigen (CEA) is a cellular glycoprotein [176] which has been widely used as a marker for metastatic CRC. Increased CEA expression in metastatic CRC may enhance metastasis to liver [177,178]. We recently identified a key mechanism by which CEA plays a role in CRC metastasis [179]. We observed that CEA inhibits downstream TGF-β tumor suppressor signaling by interacting directly with TβR1. Targeting CEA with either an anti-CEA specific antibody or siRNA-mediated CEA silencing restores the tumor suppressive properties of TGF-β signaling. Future studies are needed to evaluate the therapeutic potential of small molecule inhibitors, blocking peptides or antibodies which block the interaction between CEA and TβR1 and thereby restores the tumor suppressive function of TGF-β signaling.
7. Conclusions
The TGF-β signaling pathway has a pivotal role in tumor suppression through inhibition of proliferation and induction of apoptosis in multiple cell types, as well as effect on tumor microenvironment. Yet, TGF-β has a paradoxical role in tumorigenesis by which it can also promote tumor development by stimulating EMT, tumor cell invasiveness and metastasis [1,2,10]. Functional context dependent insights into the TGF-β pathway are crucial in developing new therapeutics in cancer. Effective anti-TGF-β compounds have been developed and tested in pre-clinical studies, and Phase I and II clinical trials. These drugs are working in three different levels: the ligand level, the ligand–receptor interaction level and the intracellular one. Other therapeutic approaches are aimed at targeting activated pathways in tumors where the TGF-β tumor suppressor pathway is lost. In spite of the concerns from severe adverse events due to the multifunctional role of TGF-β in normal physiology, the results from these trials are encouraging and call for further research and drugs development.
8. Expert opinion
Although the name of TGF-β was given to this cytokine in recognition to its ability to transform fibroblasts [180,181], it is known today to be one of the most important growth inhibitors of normal epithelial and hematopoietic cells as well as of transformed cells. TGF-β has many roles in physiological processes from embryogenesis to wound healing and from cell proliferation to apoptosis. However, it is also tightly related to pathological processes such as fibrosis and carcinogenesis.
More than 58,000 articles were published through the last 30 years on TGF-β, > 13,000 of them dealing with its role in cancer. Numerous studies on the TGF-β signaling pathway and its context-dependent properties have helped us better understand the paradox of this cytokine that can be both tumor suppressor and tumor promoter. We know that timing is a critical factor because during early phases of the cancer process TGF-β serves as a tumor suppressor, while later on it becomes tumor promoter. We also understand now that the type of tumor determines if this cytokine will act as a tumor suppressor or a tumor promoter.
Through its Smad-dependent and -independent branches the TGF-β pathway cross-talks with other signal transduction pathways and together with other cofactors, coactivators and corepressors it is responsible for the activation or inhibition of numerous genes. These actions are context specific and explain the diversity of influences of this one molecule.
Several challenges exist in the development of TGF-β pathway-related drugs: first, delineating the tumor suppressive versus the tumor promoting roles of TGF-β in each specific tumor, and even more important, differentiating between the carcinogenic effects of TGF-β and its other physiological roles. In order to prevent systemic side effects, we should be able to target only the tumor promoting arm of TGF-β pathway. This can be done by considering factors such as patient selection and timing before starting the treatment. This personalized treatment can take place by using future tools such as genetic screens and biomarkers [4]. Future research must focus on this issue.
Another caveat in anti-TGF-β based therapy is the lack of impressive success in clinical trials, especially in primary tumors. Future research should focus on combination treatments containing anti-TGF-β drugs + ionized irradiation/ chemotherapy. TGF-β inhibitors can sensitize the tumor to radiation treatment and some chemotherapeutic drugs. Concomitant targeting of several targets (e.g., EGFR, TGF-β and src) may be more effective than targeting each of them alone, due to its impact on deleterious cross-talks between those pathways [182].
In summary, inhibition of the TGF-β pathway and targeting activated pathways in tumors which the TGF-β tumor suppressor pathway is lost are two attractive options for cancer treatment. Future studies are needed in order to find more safe and effective drugs, based on a better understanding of all the diverse functions of TGF-β and their molecular mechanisms.
Article highlights.
The TGF-β signaling pathway is instrumental in tumor suppression, yet paradoxically, it can also promote tumor cell invasiveness and metastasis.
Through its Smad-dependent and -independent branches TGF-β is responsible for the activation or inhibition of numerous genes. These actions are context specific and explain the diversity of influences of this one molecule.
TGF-β achieves its tumor suppressive effect by cytostatic or cell proliferation regulation, effects on apoptosis and cell differentiation, genomic stability and indirect effects on the tumor stroma.
Tumors can evade TGF-β’s suppressive mechanisms by inactivating mutations of the core elements of the TGF-β signaling pathway or by escaping the cytostatic program, thus, permiting the consequent use of the TGF-β pathway for tumor promotion.
The most important mechanisms of tumor progression caused by TGF-β are EMT, evasion of the immune system and, promotion of cancer cell proliferation by modulation of the tumor microenvironment.
Therapeutic strategies against TGF-β are based on prevention of TGF-β synthesis by using antisense molecules; prevention of ligand–receptor interaction by ligand traps and anti-receptor monoclonal antibodies; and prevention of signal transduction by receptor kinase inhibitors and peptide aptamers.
Targeting activated pathways in tumors which the TGF-β tumor suppressor pathway is lost is another promising therapeutic strategy.
Future research should focus on finding safer and more effective drugs, based on a better understanding of all the diverse functions of TGF-β and their molecular mechanisms.
This box summarizes key points contained in the article.
Footnotes
Declaration of interest
Grant Support: NIH grants RO1 CA106614 (L Mishra), RO1 CA042857 (L Mishra), PO1 CA130821 (L Mishra), RC2 AA019392 (H Tsukamoto, L Mishra), Multidisciplinary Research Program (MRP) Proposal (L Mishra), Science & Technology Acquisition and Retention Funding (STARs) (L Mishra), P30 CA016672 (R DePinho), P30 DK56338 (M Estes). The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1••.Massague J. TGFbeta in cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. A comprehensive review on TGFβ signaling and its role in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2••.Mishra L, Derynck R, Mishra B. Transforming growth factor-beta signaling in stem cells and cancer. Science. 2005;310(5745):68–71. doi: 10.1126/science.1118389. A comprehensive review on TGFβ signaling – focus on stem cells and cancer. [DOI] [PubMed] [Google Scholar]
- 3••.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. doi: 10.1038/cr.2008.316. A comprehensive review on TGFβ signaling in the metastatic process, emphasizing its role in EMT. [DOI] [PubMed] [Google Scholar]
- 4••.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. J Pathol. 2011;223(2):205–18. doi: 10.1002/path.2785. A comprehensive review on TGFβ signaling dealing with its dual role in cancer. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 6.Souchelnytskyi S, Moustakas A, Heldin CH. TGF-beta signaling from a three-dimensional perspective: insight into selection of partners. Trends Cell Biol. 2002;12(7):304–7. doi: 10.1016/s0962-8924(02)02300-0. [DOI] [PubMed] [Google Scholar]
- 7.Katuri V, Tang Y, Li C, et al. Critical interactions between TGF-beta signaling/ ELF, and E-cadherin/beta-catenin mediated tumor suppression. Oncogene. 2006;25(13):1871–86. doi: 10.1038/sj.onc.1209211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childs CB, Proper JA, Tucker RF, Moses HL. Serum contains a platelet-derived transforming growth factor. Proc Natl Acad Sci USA. 1982;79(17):5312–16. doi: 10.1073/pnas.79.17.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zu X, Zhang Q, Cao R, et al. Transforming growth factor-beta signaling in tumor initiation, progression and therapy in breast cancer: an update. Cell Tissue Res. 2012;347(1):73–84. doi: 10.1007/s00441-011-1225-3. [DOI] [PubMed] [Google Scholar]
- 10.Mishra L, Shetty K, Tang Y, et al. The role of TGF-beta and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene. 2005;24(37):5775–89. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- 11.Keski-Oja J, Koli K, von Melchner H. TGF-beta activation by traction? Trends Cell Biol. 2004;14(12):657–9. doi: 10.1016/j.tcb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280(9):7409–12. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 14.Todorovic-Rakovic N, Milovanovic J, Nikolic-Vukosavljevic D. TGF-beta and its coreceptors in cancerogenesis: an overview. Biomarkers Med. 2011;5(6):855–63. doi: 10.2217/bmm.11.59. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell. 1993;73(7):1435–44. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 16••.Dong M, Blobe GC. Role of transforming growth factor-beta in hematologic malignancies. Blood. 2006;107(12):4589–96. doi: 10.1182/blood-2005-10-4169. A comprehensive review on TGFβ signaling in hematological malignancies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Wrana JL, Attisano L, Wieser R, et al. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370(6488):341–7. doi: 10.1038/370341a0. An important paper about activation of TGF-β receptors. [DOI] [PubMed] [Google Scholar]
- 18.Wrana JL, Tran H, Attisano L, et al. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol Cell Biol. 1994;14(2):944–50. doi: 10.1128/mcb.14.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groppe J, Hinck CS, Samavarchi-Tehrani P, et al. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29(2):157–68. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Katuri V, Dillner A, et al. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299(5606):574–7. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 21.Mishra L, Katuri V, Evans S. The role of PRAJA and ELF in TGF-beta signaling and gastric cancer. Cancer Biol Ther. 2005;4(7):694–9. doi: 10.4161/cbt.4.7.2015. [DOI] [PubMed] [Google Scholar]
- 22.Mishra L, Marshall B. Adaptor proteins and ubiquinators in TGF-beta signaling. Cytokine Growth Factor Rev. 2006;17(1–2):75–87. doi: 10.1016/j.cytogfr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Tang B, de Castro K, Barnes HE, et al. Loss of responsiveness to transforming growth factor beta induces malignant transformation of nontumorigenic rat prostate epithelial cells. Cancer Res. 1999;59(19):4834–42. [PubMed] [Google Scholar]
- 24.Attisano L, Wrana JL. Mads and Smads in TGF beta signalling. Curr Opin Cell Biol. 1998;10(2):188–94. doi: 10.1016/s0955-0674(98)80141-5. [DOI] [PubMed] [Google Scholar]
- 25.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390(6659):465–71. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 26.Kretzschmar M, Massague J. SMADs: mediators and regulators of TGF-beta signaling. Curr Opin Genet Dev. 1998;8(1):103–11. doi: 10.1016/s0959-437x(98)80069-5. [DOI] [PubMed] [Google Scholar]
- 27.Tang Y, Katuri V, Srinivasan R, et al. Transforming growth factor-beta suppresses nonmetastatic colon cancer through Smad4 and adaptor protein ELF at an early stage of tumorigenesis. Cancer Res. 2005;65(10):4228–37. doi: 10.1158/0008-5472.CAN-04-4585. [DOI] [PubMed] [Google Scholar]
- 28.Xu L. Regulation of Smad activities. Biochim Biophys Acta. 2006;1759(11–12):503–13. doi: 10.1016/j.bbaexp.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGFbeta-inducible transcriptional complexes. Genes Dev. 1997;11(23):3157–67. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macias-Silva M, Abdollah S, Hoodless PA, et al. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87(7):1215–24. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 31.Souchelnytskyi S, Tamaki K, Engstrom U, et al. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem. 1997;272(44):28107–15. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Lebrun JJ, Vale W. Regulation of transforming growth factor beta- and activin-induced transcription by mammalian Mad proteins. Proc Natl Acad Sci USA. 1996;93(23):12992–7. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoodless PA, Haerry T, Abdollah S, et al. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85(4):489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 34.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389(6651):618–22. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 35.Nishihara A, Hanai JI, Okamoto N, et al. Role of p300, a transcriptional coactivator, in signalling of TGF-beta. Genes Cells. 1998;3(9):613–23. doi: 10.1046/j.1365-2443.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7(4):270–6. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 37••.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-beta targeted cancer therapy. Int J Biol Sci. 2012;8(7):964–78. doi: 10.7150/ijbs.4564. A good review on TGF-β target cancer therapy with uptodate examples of new drugs and studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Derynck R. Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol. 1999;9(7):274–9. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]
- 39.Lin X, Duan X, Liang YY, et al. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125(5):915–28. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and prometastatic factor in human cancer. Biochim Biophys Acta. 2007;1775(1):21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Monga SP, Tang Y, Candotti F, et al. Expansion of hepatic and hematopoietic stem cells utilizing mouse embryonic liver explants. Cell Transplant. 2001;10(1):81–9. [PubMed] [Google Scholar]
- 42.Saha T, Vardhini D, Tang Y, et al. RING finger-dependent ubiquitination by PRAJA is dependent on TGF-beta and potentially defines the functional status of the tumor suppressor ELF. Oncogene. 2006;25(5):693–705. doi: 10.1038/sj.onc.1209123. [DOI] [PubMed] [Google Scholar]
- 43.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99(11):2107–12. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong C, Li Z, Alvarez R, Jr, et al. Microtubule binding to Smads may regulate TGF beta activity. Mol Cell. 2000;5(1):27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 45.Wrana JL. Crossing Smads. Sci STKE. 2000;2000(23):RE. doi: 10.1126/stke.2000.23.re1. [DOI] [PubMed] [Google Scholar]
- 46.Tsukazaki T, Chiang TA, Davison AF, et al. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95(6):779–91. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Chang C, Gehling DJ, et al. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98(3):974–9. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra L, Tully RE, Monga SP, et al. Praja1, a novel gene encoding a RING-H2 motif in mouse development. Oncogene. 1997;15(19):2361–8. doi: 10.1038/sj.onc.1201405. [DOI] [PubMed] [Google Scholar]
- 49.Chen HB, Shen J, Ip YT, Xu L. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes Dev. 2006;20(6):648–53. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19(1):128–39. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grady WM, Rajput A, Myeroff L, et al. Mutation of the type II transforming growth factor-beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58(14):3101–4. [PubMed] [Google Scholar]
- 52.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Akiyama Y, Iwanaga R, Saitoh K, et al. Transforming growth factor beta type II receptor gene mutations in adenomas from hereditary nonpolyposis colorectal cancer. Gastroenterology. 1997;112(1):33–9. doi: 10.1016/s0016-5085(97)70216-6. [DOI] [PubMed] [Google Scholar]
- 54.Grady WM, Myeroff LL, Swinler SE, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999;59(2):320–4. [PubMed] [Google Scholar]
- 55.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 56.Takenoshita S, Tani M, Nagashima M, et al. Mutation analysis of coding sequences of the entire transforming growth factor beta type II receptor gene in sporadic human colon cancer using genomic DNA and intron primers. Oncogene. 1997;14(10):1255–8. doi: 10.1038/sj.onc.1200938. [DOI] [PubMed] [Google Scholar]
- 57.Parsons R, Myeroff LL, Liu B, et al. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55(23):5548–50. [PubMed] [Google Scholar]
- 58.Wang D, Kanuma T, Mizunuma H, et al. Analysis of specific gene mutations in the transforming growth factor-beta signal transduction pathway in human ovarian cancer. Cancer Res. 2000;60(16):4507–12. [PubMed] [Google Scholar]
- 59.Goggins M, Shekher M, Turnacioglu K, et al. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58(23):5329–32. [PubMed] [Google Scholar]
- 60.Chen T, Yan W, Wells RG, et al. Novel inactivating mutations of transforming growth factor-beta type I receptor gene in head-and-neck cancer metastases. Int J Cancer. 2001;93(5):653–61. doi: 10.1002/ijc.1381. [DOI] [PubMed] [Google Scholar]
- 61.Eppert K, Scherer SW, Ozcelik H, et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86(4):543–52. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 62.Yakicier MC, Irmak MB, Romano A, et al. Smad2 and Smad4 gene mutations in hepatocellular carcinoma. Oncogene. 1999;18(34):4879–83. doi: 10.1038/sj.onc.1202866. [DOI] [PubMed] [Google Scholar]
- 63.Maliekal TT, Antony ML, Nair A, et al. Loss of expression, and mutations of Smad 2 and Smad 4 in human cervical cancer. Oncogene. 2003;22(31):4889–97. doi: 10.1038/sj.onc.1206806. [DOI] [PubMed] [Google Scholar]
- 64.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21. 1. Science. 1996;271(5247):350–3. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 65.Miyaki M, Iijima T, Konishi M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18(20):3098–103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 66.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 67.Schutte M, Hruban RH, Hedrick L, et al. DPC4 gene in various tumor types. Cancer Res. 1996;56(11):2527–30. [PubMed] [Google Scholar]
- 68.Nagatake M, Takagi Y, Osada H, et al. Somatic in vivo alterations of the DPC4 gene at 18q21 in human lung cancers. Cancer Res. 1996;56(12):2718–20. [PubMed] [Google Scholar]
- 69.Thiagalingam S, Lengauer C, Leach FS, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13(3):343–6. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 70.Hahn SA, Bartsch D, Schroers A, et al. Mutations of the DPC4/Smad4 gene in biliary tract carcinoma. Cancer Res. 1998;58(6):1124–6. [PubMed] [Google Scholar]
- 71.Howe JR, Roth S, Ringold JC, et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280(5366):1086–8. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 72.Gomis RR, Alarcon C, He W, et al. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci USA. 2006;103(34):12747–52. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pardali K, Kurisaki A, Moren A, et al. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J Biol Chem. 2000;275(38):29244–56. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- 74.Seoane J, Le HV, Shen L, et al. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117(2):211–23. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 75.Staller P, Peukert K, Kiermaier A, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3(4):392–9. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 76.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419(6908):729–34. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 77.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–21. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 78.Jang CW, Chen CH, Chen CC, et al. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat Cell Biol. 2002;4(1):51–8. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 79.Boulanger CA, Wagner KU, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-beta1 expression. Oncogene. 2005;24(4):552–60. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- 80.Glick AB, Weinberg WC, Wu IH, et al. Transforming growth factor beta 1 suppresses genomic instability independent of a G1 arrest, p53, and Rb. Cancer Res. 1996;56(16):3645–50. [PubMed] [Google Scholar]
- 81.Kirshner J, Jobling MF, Pajares MJ, et al. Inhibition of transforming growth factor-beta1 signaling attenuates ataxia telangiectasia mutated activity in response to genotoxic stress. Cancer Res. 2006;66(22):10861–9. doi: 10.1158/0008-5472.CAN-06-2565. [DOI] [PubMed] [Google Scholar]
- 82.Bornstein S, White R, Malkoski S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119(11):3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joseph H, Gorska AE, Sohn P, et al. Overexpression of a kinase-deficient transforming growth factor-beta type II receptor in mouse mammary stroma results in increased epithelial branching. Mol Biol Cell. 1999;10(4):1221–34. doi: 10.1091/mbc.10.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bhowmick NA, Chytil A, Plieth D, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303(5659):848–51. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 85.Li MO, Wan YY, Sanjabi S, et al. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 86.Monteleone G, Kumberova A, Croft NM, et al. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108(4):601–9. doi: 10.1172/JCI12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110(1):19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 88.Latres E, Malumbres M, Sotillo R, et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19(13):3496–506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen CR, Kang Y, Massague J. Defective repression of c-myc in breast cancer cells: A loss at the core of the transforming growth factor beta growth arrest program. Proc Natl Acad Sci USA. 2001;98(3):992–9. doi: 10.1073/pnas.98.3.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gomis RR, Alarcon C, Nadal C, et al. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10(3):203–14. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 91.Jen J, Harper JW, Bigner SH, et al. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994;54(24):6353–8. [PubMed] [Google Scholar]
- 92.Padua D, Zhang XH, Wang Q, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeCoteau JF, Knaus PI, Yankelev H, et al. Loss of functional cell surface transforming growth factor beta (TGF-beta) type 1 receptor correlates with insensitivity to TGF-beta in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 1997;94(11):5877–81. doi: 10.1073/pnas.94.11.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rooke HM, Vitas MR, Crosier PS, Crosier KE. The TGF-beta type II receptor in chronic myeloid leukemia: analysis of microsatellite regions and gene expression. Leukemia. 1999;13(4):535–41. doi: 10.1038/sj.leu.2401384. [DOI] [PubMed] [Google Scholar]
- 95.Capocasale RJ, Lamb RJ, Vonderheid EC, et al. Reduced surface expression of transforming growth factor beta receptor type II in mitogen-activated T cells from Sezary patients. Proc Natl Acad Sci USA. 1995;92(12):5501–5. doi: 10.1073/pnas.92.12.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernandez T, Amoroso S, Sharpe S, et al. Disruption of transforming growth factor beta signaling by a novel ligand-dependent mechanism. J Exp Med. 2002;195(10):1247–55. doi: 10.1084/jem.20011521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mori N, Morishita M, Tsukazaki T, et al. Human T-cell leukemia virus type I oncoprotein Tax represses Smad-dependent transforming growth factor beta signaling through interaction with CREB-binding protein/p300. Blood. 2001;97(7):2137–44. doi: 10.1182/blood.v97.7.2137. [DOI] [PubMed] [Google Scholar]
- 98.Kurokawa M, Mitani K, Irie K, et al. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 1998;394(6688):92–6. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 99.Jakubowiak A, Pouponnot C, Berguido F, et al. Inhibition of the transforming growth factor beta 1 signaling pathway by the AML1/ETO leukemia-associated fusion protein. J Biol Chem. 2000;275(51):40282–7. doi: 10.1074/jbc.C000485200. [DOI] [PubMed] [Google Scholar]
- 100.Inman GJ. Switching TGFbeta from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev. 2011;21(1):93–9. doi: 10.1016/j.gde.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 101.Adorno M, Cordenonsi M, Montagner M, et al. A Mutant-p53/ Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 102.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat Cell Biol. 2007;9(9):1000–4. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 103.Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12(1):22–9. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 104.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 105.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–72. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512–20. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Valcourt U, Kowanetz M, Niimi H, et al. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16(4):1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cheng N, Chytil A, Shyr Y, et al. Transforming growth factor-beta signaling-deficient fibroblasts enhance hepatocyte growth factor signaling in mammary carcinoma cells to promote scattering and invasion. Mol Cancer Res. 2008;6(10):1521–33. doi: 10.1158/1541-7786.MCR-07-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7(10):1118–22. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 111.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–80. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 112.Arteaga CL, Hurd SD, Winnier AR, et al. Anti-transforming growth factor (TGF)-beta antibodies inhibit breast cancer cell tumorigenicity and increase mouse spleen natural killer cell activity. Implications for a possible role of tumor cell/host TGF-beta interactions in human breast cancer progression. J Clin Investig. 1993;92(6):2569–76. doi: 10.1172/JCI116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee JC, Lee KM, Kim DW, Heo DS. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172(12):7335–40. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 114.Castriconi R, Cantoni C, Della Chiesa M, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci USA. 2003;100(7):4120–5. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10(8):554–67. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hagedorn HG, Bachmeier BE, Nerlich AG. Synthesis and degradation of basement membranes and extracellular matrix and their regulation by TGF-beta in invasive carcinomas (Review) Int J Oncol. 2001;18(4):669–81. doi: 10.3892/ijo.18.4.669. [DOI] [PubMed] [Google Scholar]
- 117.Porsch H, Bernert B, Mehić M, et al. Efficient TGFβ-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene. 2012 doi: 10.1038/onc.2012.475. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 119.Sanchez-Elsner T, Botella LM, Velasco B, et al. Synergistic cooperation between hypoxia and transforming growth factor-beta pathways on human vascular endothelial growth factor gene expression. J Biol Chem. 2001;276(42):38527–35. doi: 10.1074/jbc.M104536200. [DOI] [PubMed] [Google Scholar]
- 120.Larsson J, Goumans MJ, Sjostrand LJ, et al. Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J. 2001;20(7):1663–73. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oshima M, Oshima H, Taketo MM. TGF-beta receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev Biol. 1996;179(1):297–302. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- 122.Stearns ME, Garcia FU, Fudge K, et al. Role of interleukin 10 and transforming growth factor beta1 in the angiogenesis and metastasis of human prostate primary tumor lines from orthotopic implants in severe combined immunodeficiency mice. Clin Cancer Res. 1999;5(3):711–20. [PubMed] [Google Scholar]
- 123.Buck MB, Fritz P, Dippon J, et al. Prognostic significance of transforming growth factor beta receptor II in estrogen receptor-negative breast cancer patients. Clin Cancer Res. 2004;10(2):491–8. doi: 10.1158/1078-0432.ccr-0320-03. [DOI] [PubMed] [Google Scholar]
- 124.Dalal BI, Keown PA, Greenberg AH. Immunocytochemical localization of secreted transforming growth factor-beta 1 to the advancing edges of primary tumors and to lymph node metastases of human mammary carcinoma. Am J Pathol. 1993;143(2):381–9. [PMC free article] [PubMed] [Google Scholar]
- 125.Biswas S, Guix M, Rinehart C, et al. Inhibition of TGF-beta with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. J Clin Invest. 2007;117(5):1305–13. doi: 10.1172/JCI30740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giampieri S, Manning C, Hooper S, et al. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11(11):1287–96. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Siegel PM, Shu W, Cardiff RD, et al. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci USA. 2003;100(14):8430–5. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Forrester E, Chytil A, Bierie B, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 2005;65(6):2296–302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 129.Kingsley LA, Fournier PG, Chirgwin JM, Guise TA. Molecular biology of bone metastasis. Mol Cancer Ther. 2007;6(10):2609–17. doi: 10.1158/1535-7163.MCT-07-0234. [DOI] [PubMed] [Google Scholar]
- 130.Kang Y, He W, Tulley S, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci USA. 2005;102(39):13909–14. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saito H, Tsujitani S, Oka S, et al. An elevated serum level of transforming growth factor-beta 1 (TGF-beta 1) significantly correlated with lymph node metastasis and poor prognosis in patients with gastric carcinoma. Anticancer Res. 2000;20(6B):4489–93. [PubMed] [Google Scholar]
- 132.Tsai JF, Jeng JE, Chuang LY, et al. Elevated urinary transforming growth factor-beta1 level as a tumour marker and predictor of poor survival in cirrhotic hepatocellular carcinoma. Br J Cancer. 1997;76(2):244–50. doi: 10.1038/bjc.1997.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Papadopoulou E, Anagnostopoulos K, Tripsianis G, et al. Evaluation of predictive and prognostic significance of serum TGF-beta1 levels in breast cancer according to HER-2 codon 655 polymorphism. Neoplasma. 2008;55(3):229–38. [PubMed] [Google Scholar]
- 134.Saunier EF, Akhurst RJ. TGF beta inhibition for cancer therapy. Curr Cancer Drug Targets. 2006;6(7):565–78. doi: 10.2174/156800906778742460. [DOI] [PubMed] [Google Scholar]
- 135.Calone I, Souchelnytskyi S. Inhibition of TGFbeta signaling and its implications in anticancer treatments. Exp Oncol. 2012;34(1):9–16. [PubMed] [Google Scholar]
- 136.Hau P, Jachimczak P, Schlingensiepen R, et al. Inhibition of TGF-beta2 with AP 12009 in recurrent malignant gliomas: from preclinical to phase I/II studies. Oligonucleotides. 2007;17(2):201–12. doi: 10.1089/oli.2006.0053. [DOI] [PubMed] [Google Scholar]
- 137.Schlingensiepen KH, Jaschinski F, Lang SA, et al. Transforming growth factor-beta 2 gene silencing with trabedersen (AP 12009) in pancreatic cancer. Cancer Sci. 2011;102(6):1193–200. doi: 10.1111/j.1349-7006.2011.01917.x. [DOI] [PubMed] [Google Scholar]
- 138.Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, et al. TGF-beta signalling in colon carcinogenesis. Cancer Lett. 2012;314(1):1–7. doi: 10.1016/j.canlet.2011.09.041. [DOI] [PubMed] [Google Scholar]
- 139.Oettle H, Seufferlein T, Luger T, et al. Final results of a phase I/II study in patients with pancreatic cancer, malignant melanoma, and colorectal carcinoma with trabedersen. J Clin Oncol. 2012;30 [abstract: 2012 ASCO meeting] [Google Scholar]
- 140.Nam JS, Terabe M, Mamura M, et al. An anti-transforming growth factor beta antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 2008;68(10):3835–43. doi: 10.1158/0008-5472.CAN-08-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ganapathy V, Ge R, Grazioli A, et al. Targeting the transforming growth factor-beta pathway inhibits human basal-like breast cancer metastasis. Mol Cancer. 2010;9:122. doi: 10.1186/1476-4598-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morris JC, Shapiro GI, Tan AR, et al. A human anti-transforming growth factor-beta (TGFbeta) monoclonal antibody (mAB) in patients with advanced malignant melanoma (MM) or renal cell carcinoma (RCC) J Clin Oncol. 2008;26:9028. doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang YA, Dukhanina O, Tang B, et al. Lifetime exposure to a soluble TGF-beta antagonist protects mice against metastasis without adverse side effects. J Clin Invest. 2002;109(12):1607–15. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Muraoka RS, Dumont N, Ritter CA, et al. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002;109(12):1551–9. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rowland-Goldsmith MA, Maruyama H, Matsuda K, et al. Soluble type II transforming growth factor-beta receptor attenuates expression of metastasis-associated genes and suppresses pancreatic cancer cell metastasis. Mol Cancer Ther. 2002;1(3):161–7. [PubMed] [Google Scholar]
- 146.Bandyopadhyay A, Zhu Y, Malik SN, et al. Extracellular domain of TGFbeta type III receptor inhibits angiogenesis and tumor growth in human cancer cells. Oncogene. 2002;21(22):3541–51. doi: 10.1038/sj.onc.1205439. [DOI] [PubMed] [Google Scholar]
- 147.van Meeteren LA, Thorikay M, Bergqvist S, et al. Anti-human activin receptor-like kinase 1 (ALK1) antibody attenuates bone morphogenetic protein 9 (BMP9)-induced ALK1 signaling and interferes with endothelial cell sprouting. J Biol Chem. 2012;287(22):18551–61. doi: 10.1074/jbc.M111.338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Goff LW, De Braud FG, Cohen RB, et al. Phase I study of PF-03446962, a fully human mAb against ALK 1, a TGFbeta receptor involved in tumor angiogenesis. J Clin Oncol. 2012;30:e13606. [Google Scholar]
- 149.Hjelmeland MD, Hjelmeland AB, Sathornsumetee S, et al. SB-431542, a small molecule transforming growth factor-beta-receptor antagonist, inhibits human glioma cell line proliferation and motility. Mol Cancer Ther. 2004;3(6):737–45. [PubMed] [Google Scholar]
- 150.Matsuyama S, Iwadate M, Kondo M, et al. SB-431542 and Gleevec inhibit transforming growth factor-beta-induced proliferation of human osteosarcoma cells. Cancer Res. 2003;63(22):7791–8. [PubMed] [Google Scholar]
- 151.Tanaka H, Shinto O, Yashiro M, et al. Transforming growth factor beta signaling inhibitor, SB-431542, induces maturation of dendritic cells and enhances anti-tumor activity. Oncol Rep. 2010;24(6):1637–43. doi: 10.3892/or_00001028. [DOI] [PubMed] [Google Scholar]
- 152.Takeuchi K, Abe M, Hiasa M, et al. Tgf-Beta inhibition restores terminal osteoblast differentiation to suppress myeloma growth. PLoS One. 2010;5(3):e9870. doi: 10.1371/journal.pone.0009870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65(3):744–52. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- 154.Ehata S, Hanyu A, Fujime M, et al. Ki26894, a novel transforming growth factor-beta type I receptor kinase inhibitor, inhibits in vitro invasion and in vivo bone metastasis of a human breast cancer cell line. Cancer Sci. 2007;98(1):127–33. doi: 10.1111/j.1349-7006.2006.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shinto O, Yashiro M, Kawajiri H, et al. Inhibitory effect of a TGFbeta receptor type-I inhibitor, Ki26894, on invasiveness of scirrhous gastric cancer cells. Br J Cancer. 2010;102(5):844–51. doi: 10.1038/sj.bjc.6605561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bandyopadhyay A, Agyin JK, Wang L, et al. Inhibition of pulmonary and skeletal metastasis by a transforming growth factor-beta type I receptor kinase inhibitor. Cancer Res. 2006;66(13):6714–21. doi: 10.1158/0008-5472.CAN-05-3565. [DOI] [PubMed] [Google Scholar]
- 157.Uhl M, Aulwurm S, Wischhusen J, et al. SD-208, a novel transforming growth factor beta receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res. 2004;64(21):7954–61. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]
- 158.Mohammad KS, Javelaud D, Fournier PG, et al. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011;71(1):175–84. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Melisi D, Ishiyama S, Sclabas GM, et al. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol Cancer Ther. 2008;7(4):829–40. doi: 10.1158/1535-7163.MCT-07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Korpal M, Yan J, Lu X, et al. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nat Med. 2009;15(8):960–6. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 161.Zhang B, Halder SK, Zhang S, Datta PK. Targeting transforming growth factor-beta signaling in liver metastasis of colon cancer. Cancer Lett. 2009;277(1):114–20. doi: 10.1016/j.canlet.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Connolly EC, Saunier EF, Quigley D, et al. Outgrowth of drug-resistant carcinomas expressing markers of tumor aggression after long-term TbetaRI/II kinase inhibition with LY2109761. Cancer Res. 2011;71(6):2339–49. doi: 10.1158/0008-5472.CAN-10-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yakymovych I, Engstrom U, Grimsby S, et al. Inhibition of transforming growth factor-beta signaling by low molecular weight compounds interfering with ATP-or substrate-binding sites of the TGF beta type I receptor kinase. Biochemistry. 2002;41(36):11000–7. doi: 10.1021/bi025936u. [DOI] [PubMed] [Google Scholar]
- 164.Bueno L, de Alwis DP, Pitou C, et al. Semi-mechanistic modelling of the tumour growth inhibitory effects of LY2157299, a new type I receptor TGF-beta kinase antagonist, in mice. Eur J Cancer. 2008;44(1):142–50. doi: 10.1016/j.ejca.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 165.Rodon Ahnert J, Baselga J, Calvo E, et al. First human dose (FHD) study of the oral transforming growth factor-beta receptor I kinase inhibitor LY2157299 in patients with treatment-refractory malignant glioma. J Clin Oncol. 2011;29:abstract 3011. [Google Scholar]
- 166.Zhao BM, Hoffmann FM. Inhibition of transforming growth factor-beta1-induced signaling and epithelial-to-mesenchymal transition by the Smad-binding peptide aptamer Trx-SARA. Mol Biol Cell. 2006;17(9):3819–31. doi: 10.1091/mbc.E05-10-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5(12):997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 168.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.White LK, Wright WE, Shay JW. Telomerase inhibitors. Trends Biotechnol. 2001;19(3):114–20. doi: 10.1016/s0167-7799(00)01541-9. [DOI] [PubMed] [Google Scholar]
- 170.Gupta S, Jain MM, Maru A, et al. A phase I study of selective cyclin dependent kinase inhibitor P1446A-05 administered on an intermittent schedule in patients with advanced refractory tumors. J Clin. 2012;30:abstract 3011. [Google Scholar]
- 171.Shah S, Islam MN, Dakshanamurthy S, et al. The molecular basis of vitamin D receptor and beta-catenin crossregulation. Mol Cell. 2006;21(6):799–809. doi: 10.1016/j.molcel.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 172.Larriba MJ, Valle N, Palmer HG, et al. The inhibition of Wnt/beta-catenin signalling by 1alpha,25-dihydroxyvitamin D3 is abrogated by Snail1 in human colon cancer cells. Endocr Relat Cancer. 2007;14(1):141–51. doi: 10.1677/ERC-06-0028. [DOI] [PubMed] [Google Scholar]
- 173.Walia B, Wang L, Merlin D, Sitaraman SV. TGF-beta down-regulates IL-6 signaling in intestinal epithelial cells: critical role of SMAD-2. Faseb J. 2003;17(14):2130–2. doi: 10.1096/fj.02-1211fje. [DOI] [PubMed] [Google Scholar]
- 174.Tang Y, Kitisin K, Jogunoori W, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA. 2008;105(7):2445–50. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin L, Amin R, Gallicano GI, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28(7):961–72. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gold P, Freedman SO. Specific carcinoembryonic antigens of the human digestive system. J Exp Med. 1965;122(3):467–81. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thomas P, Forse RA, Bajenova O. Carcinoembryonic antigen (CEA) and its receptor hnRNP M are mediators of metastasis and the inflammatory response in the liver. Clin Exp Metastasis. 2011;28(8):923–32. doi: 10.1007/s10585-011-9419-3. [DOI] [PubMed] [Google Scholar]
- 178.Bajenova OV, Zimmer R, Stolper E, et al. Heterogeneous RNA-binding protein M4 is a receptor for carcinoembryonic antigen in Kupffer cells. J Biol Chem. 2001;276(33):31067–73. doi: 10.1074/jbc.M104093200. [DOI] [PubMed] [Google Scholar]
- 179.Li Y, Cao H, Jiao Z, et al. Carcinoembryonic antigen interacts with TGF-{beta} receptor and inhibits TGF-{beta} signaling in colorectal cancers. Cancer Res. 2010;70(20):8159–68. doi: 10.1158/0008-5472.CAN-10-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Roberts AB, Lamb LC, Newton DL, et al. Transforming growth factors: isolation of polypeptides from virally and chemically transformed cells by acid/ ethanol extraction. Proc Natl Acad Sci USA. 1980;77(6):3494–8. doi: 10.1073/pnas.77.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Moses HL, Branum EL, Proper JA, Robinson RA. Transforming growth factor production by chemically transformed cells. Cancer Res. 1981;41(7):2842–8. [PubMed] [Google Scholar]
- 182.Deharvengt S, Marmarelis M, Korc M. Concomitant targeting of EGF receptor, TGF-beta and SRC points to a novel therapeutic approach in pancreatic cancer. PloS One. 2012;7(6):e39684. doi: 10.1371/journal.pone.0039684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]