Abstract
Heteroduplex joints are general intermediates of homologous genetic recombination in DNA genomes. A heteroduplex joint is formed between a single-stranded region (or tail), derived from a cleaved parental double-stranded DNA, and homologous regions in another parental double-stranded DNA, in a reaction mediated by the RecA/Rad51-family of proteins. In this reaction, a RecA/Rad51-family protein first forms a filamentous complex with the single-stranded DNA, and then interacts with the double-stranded DNA in a search for homology. Studies of the three-dimensional structures of single-stranded DNA bound either to Escherichia coli RecA or Saccharomyces cerevisiae Rad51 have revealed a novel extended DNA structure. This structure contains a hydrophobic interaction between the 2′ methylene moiety of each deoxyribose and the aromatic ring of the following base, which allows bases to rotate horizontally through the interconversion of sugar puckers. This base rotation explains the mechanism of the homology search and base-pair switch between double-stranded and single-stranded DNA during the formation of heteroduplex joints. The pivotal role of the 2′ methylene-base interaction in the heteroduplex joint formation is supported by comparing the recombination of RNA genomes with that of DNA genomes. Some simple organisms with DNA genomes induce homologous recombination when they encounter conditions that are unfavorable for their survival. The extended DNA structure confers a dynamic property on the otherwise chemically and genetically stable double-stranded DNA, enabling gene segment rearrangements without disturbing the coding frame (i.e., protein-segment shuffling). These properties may give an extensive evolutionary advantage to DNA.
Keywords: base-pair switch, homologous pairing, strand exchange, NMR, three-dimensional molecular structure
DNA as a General Molecular Carrier of Genetic Information
Genomic information generally is carried by double-stranded DNA. The double-stranded DNA structure discovered by Watson and Crick clearly explains the mechanisms of heredity, which include both the encoding of genetic information and its duplication as chemical properties of the molecule (1). In addition, the double-stranded structure enables cellular systems to recognize, as structural irregularities, erroneously incorporated nucleotides or lesions in bases, sugars, or the backbone strand, and to correct these errors by using the partner strand as a template. The genetic stability and the chemical inactivity of double-stranded DNA have been regarded as favorable molecular properties for its role as the carrier of genomic information. Evolution, which is a general attribute of the genome as well, has resulted in a variety of organisms, whose diversity arose not only as a result of changes in the genomic information, but also as a result of increased content and complexity. The faithful duplication and repair exhibited by the double-stranded DNA structure would seem to be incompatible with the process of evolution. Thus, evolution has been explained by the occurrence of “errors” during DNA replication and repair, which were subsequently stabilized as mutations and selected for by the process of natural selection (e.g., ref. 2).
If mutations played a key function in evolution, organisms with RNA genomes, which show a higher mutation frequency than DNA genomes, would have evolved into higher organisms much faster than those with DNA genomes, but this is not the case. One explanation why organisms with RNA genomes did not evolve beyond the level of viruses is that their high rate of spontaneous mutation prevents the maintenance of a genome of the required complexity. The low level of successful mutations in the DNA genome is unlikely to be caused by its chemical stability, but rather by correction systems acquired during evolution, such as proofreading and repair systems for mismatches and lesions in DNA (3, 4). The double-stranded structure required for repair or correction is also not a specific property of DNA, because the genomic RNA of some viruses is also double-stranded. Moreover, it is generally believed that primordial creatures consisted of RNA, and that RNA as a molecular carrier of genomic information eventually was supplanted by DNA. However, the specific molecular properties that give a critical evolutionary advantage to DNA are still unknown.
Homologous Recombination
Homologous recombination, which occurs between homologous chromosomes or sister chromatids, is another general attribute of a DNA genome. Homologous recombination is a type of genetic rearrangement that occurs through the breakage and rejoining of DNA molecules within a stretch (some hundreds to thousands of base pairs) of identical or very similar (i.e., homologous) sequences. Homologous recombination maintains the integrity of the genome through the accurate repair of various types of DNA damage, especially double-stranded breaks. For the repair of double-stranded breaks, an intramolecular intact strand is not available as a template. At the same time, homologous recombination is a mechanism that can confer genetic diversity on the genome of a species by the rearrangement of alleles, variations of which were acquired by mutations. This is believed to contribute to evolution.
Sexual reproduction occurs in a variety of organisms, especially eukaryotes. It has a tight connection with homologous recombination, in that, at an early stage of meiosis, cells induce homologous recombination by which each chromosome recombines with its homologue. Mutations causing deficiencies in meiotic recombination generally result in the nondisjunction (or random sorting) of homologous chromosomes during meiosis (see refs. 5–7 for review). Thus, sexual reproduction is regarded as a system that ensures that all chromosomes undergo recombination in each sexual generation.
Homologous recombination occurs through a general intermediate, the Holliday intermediate, in which a pair of parental DNA molecules are connected by heteroduplex joints. These are duplexes of complementary strands each of which is derived from each parental DNA molecule (8–10). The heteroduplex has been shown to extend to a size large enough to cover an entire gene, as revealed by the size of co-conversion tracts. The heteroduplex joint ensures the exact alignments of multiple genes at a homologous sequence, allowing them to be recombined without disturbing their coding frames.
Various genes involved in homologous recombination are well conserved from viruses and bacteria to higher eukaryotes, including human beings. In particular, heteroduplex joints generally are formed by a reaction between homologous double-stranded DNA and single-stranded DNA, mediated in vivo as well as in vitro by the RecA/Rad51-family of proteins (11–15). Proteins in this family include UvsX of a bacterial virus (Escherichia coli phage T4: ref. 16), RecA of various bacteria, RadA of archaea, and the Rad51-family proteins found in various eukaryotes from yeast to human beings (refs. 17 and 18, and see ref. 19). Some of the RecA/Rad51-family proteins were demonstrated by electron microscopy to form conserved, right-handed, spiral filamentous structures either by themselves or around DNA that is either single-stranded or double-stranded (20, 21), and to some extent to have conserved amino acid sequences within the core region. It is remarkable that UvsX is only slightly conserved in amino acid sequence (22), but is well conserved in its biochemical functions and three-dimensional structure (16, 23, 24). The three-dimensional structure of DNA-free RecA, as solved by x-ray crystallography, revealed a right-handed helical filament (25), which is consistent with that obtained by electron microscopy as described above.
Heteroduplex Joint Formation Is Promoted by the RecA/Rad51-Family Proteins
Single-stranded DNA tails are created by the processing of double-stranded breaks for the initiation of homologous recombination during meiosis in vivo (26, 27). It has been demonstrated in vitro that heteroduplex joints are formed by a reaction between homologous double-stranded DNA and single-stranded DNA mediated by the RecA/Rad51-family of proteins (11–16). E. coli RecA is the prototypical protein that promotes heteroduplex joint formation (11, 12). Various combinations of DNA molecules have been shown to be substrates for heteroduplex joint formation by RecA. The formation is especially efficient when one of a pair of DNA molecules has a single-stranded region within a sequence homologous to its partner double-stranded DNA (28–30). RecA forms a Holliday intermediate in vitro when the substrates are a pair of linear double-stranded DNA molecules, one of which has at least one single-stranded tail (31). Natural closed-circle double-stranded DNA is negatively supercoiled, which stimulates the RecA-mediated heteroduplex joint formation. When double-stranded DNA is negatively supercoiled, RecA by itself can pair more than 50% of substrate double-stranded DNA molecules (ca. 10 kbp) with homologous single-stranded DNA molecules to form heteroduplex joints (D loops) in vitro within 2–3 min at 37°C in Mg2+- and ATP-dependent reactions (12, 30). Heteroduplex joint formation by Rad51 from yeast or human is much slower and less efficient compared with RecA, and efficient heteroduplex joint formation by Rad51 requires coactivator proteins such as a single-stranded binding protein (RPA) and Rad54 (13–15, 32).
The molecular mechanisms of heteroduplex joint formation have been extensively studied by using RecA-mediated reactions as a model system. The heteroduplex joint formation promoted by RecA has been experimentally separated into two phases: homologous pairing and strand exchange (33, 34). Homologous pairing is the formation of the core of the heteroduplex, which then is stabilized by strand exchange. Strand exchange is associated with the unidirectional extension of the joint (unidirectional branch migration) by several kilobase pairs (34, 35) and can integrate mismatches (36) and heterologous sequences (37) into a heteroduplex. RecA has DNA-dependent ATPase activity (38), and ATP is required for heteroduplex joint formation (11, 12, 35). ATP hydrolysis is not required for homologous pairing but is required for unidirectional branch migration to bypass heterologous sequences (35, 39, 40), and ATP hydrolysis stimulates the detachment of the protein from the DNA for the recycling of RecA (41–43).
The reaction steps have been identified in detail in the case of homologous pairing and strand exchange by RecA. In homologous pairing, the RecA polypeptide first polymerizes on single-stranded DNA (29, 44–46) to form a spiral filamentous structure around the DNA (47–49). The secondary structure of the single-stranded DNA is removed during this step (48), which is stimulated by SSB (single-strand binding protein; refs. 50 and 51). Then double-stranded DNA binds to the RecA–single-stranded DNA filament without any need for sequence homology (44). The search for sequence homology between the single-stranded and double-stranded DNA occurs within this complex, and upon the recognition of homology, a core heteroduplex is formed (44, 52–54). For the stabilization of the core heteroduplex, strand exchange follows as described above.
Homologous Pairing vs. Annealing
In the formation of double-stranded DNA from complementary single-stranded DNA molecules (annealing), once the secondary structure of the DNA has been unfolded, all of the bases of the single-stranded DNA are available to form a stretch of Watson–Crick base pairs. The mechanism of annealing thus can be explained by simple collisions of DNA molecules. On the other hand, in homologous pairing, all of the bases in the double-stranded DNA are already involved in Watson–Crick base pairs and do not appear to be available to form base pairs with the single-stranded DNA. Thus, how sequence homology (identity or complementarity) is recognized between double-stranded and single-stranded DNA, and how the base pairs of the parental double-stranded DNA switch to new base pairs during heteroduplex formation (base pair switch), are still unanswered questions in homologous recombination.
Two different mechanisms have been considered. In the first mechanism, RecA disrupts the base pairs of double-stranded DNA and promotes an annealing reaction between single-stranded DNA and a strand of double-stranded DNA. In the other mechanism, the recognition of sequence homology occurs through non-Watson–Crick interactions without the need for the disruption of the base pairs of the double-stranded DNA, such as interactions that form a triplex. Various and extensive attempts, such as chemical or enzymatic probing experiments, have been carried out to elucidate the molecular mechanism of heteroduplex joint formation by RecA (see refs. 55–58 for review), but these studies have not succeeded in giving a unified view of the mechanism involved. For example, whereas many studies have supported the idea that an incoming single-stranded DNA interacts with double-stranded DNA in its minor groove for the recognition of sequence homology by RecA (59–63), results contradicting this were published recently (64).
The Three-Dimensional Molecular Structure of Single-Stranded Oligo-DNA Bound to RecA
In an attempt to understand the mechanisms of homologous pairing and strand exchange, we analyzed the three-dimensional molecular structure of single-stranded oligo-DNA bound to RecA in the presence of an ATP analogue, ATPγS, by use of the transferred nuclear Overhauser effect (NOE) method, a technique of NMR spectroscopy. This NMR technique is frequently used for the structural analysis of small ligands bound to proteins (65, 66). We used single-stranded oligo-DNA with 3–6 bases and some variations in sequence. The NOEs obtained are clearly different from those of either B-form or A-form DNA (ref. 67 and T.N., unpublished observations; Table 1). The structure calculations applying a standard simulated annealing protocol gave a unique well-defined extended DNA structure for each single-stranded oligo-DNA (67). When RecA was replaced by Saccharomyces cerevisiae Rad51, the spectra were very similar to those obtained with RecA, indicating that RecA and Rad51 induce a common extended structure in single-stranded DNA upon binding (68).
Table 1.
Comparison of NOEs derived from various forms of DNA
| Extended form by RecAp | B form | A form | |
|---|---|---|---|
| Intra-residue | |||
| di (6, 8; 2′) | Strong | Strong | Medium |
| di (6, 8; 3′) | Medium | Medium | Strong |
| di (6, 8; 1′) | Medium | Medium | Medium |
| Inter-residue | |||
| ds (2"; 6, 8) | Medium | Strong | Medium |
| ds (2′; 6, 8) | Medium | Medium | Strong |
| ds (3′; 6, 8) | Medium | Weak | Medium |
| ds (1′; 6, 8) | None or weak | Strong | Medium |
| ds (1′; 5") | Medium | Strong | Medium |
| ds (2"; 5") | Medium | Medium | Strong |
| ds (2"; 2′) | None | Medium | None |
| ds (2"; 4′) | Weak | None | Medium |
| ds (1′; 4′) | Weak | Medium | None |
| ds (2′; 3′) | None | None | Medium |
| ds (2′; 4′) | None | None | Medium |
Strong: distance <3.0 Å; medium: <4.5 Å; weak: >4.5 Å. References: Extended form by RecA, (ref. 67 and unpublished observation); B form and A form: T. E. Ferrin and N. Pattabiraman, gennuc in University of California, San Francisco MidasPlus.
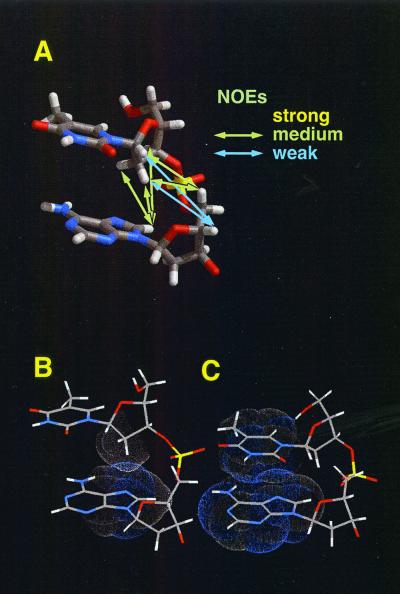
The results from the analysis (Fig. 1; Protein Data Bank ID 3REC; refs. 67 and 68) revealed that (i) the extended single-stranded DNA structure contains hydrophobic deoxyribose-base stacking in which the 2′ methylene moiety of each deoxyribose is placed above the base of the following residue, instead of the base-base stacking found in B-form DNA; (ii) the distance between neighboring bases is expanded to about 5 Å from the 3.4 Å of B-form DNA; (iii) the structure is specific to DNA, because the 2′ methylene-base interaction (the interaction of the 2′ methylene moiety with the aromatic ring of the next base) is likely to be a CH/π interaction, a weak attractive molecular force occurring between the CH groups and π-systems observed in various biomacromolecules, which would contribute to the stabilization of the extended structure; and (iv) the extended structure requires RecA or Rad51 and (at least in the case of RecA) an ATP analogue (69). To understand the roles of the proteins that are required for the induction and maintenance of the extended DNA structure, structural information about the DNA binding sites on the proteins is required, but was not obtained in this study, because of a technical limit of transferred NOE method.
Figure 1.
A model for the extended single-stranded DNA structure induced by binding of RecA/Rad51. (A) The model structure and summary of NOE intensities. (B and C) Comparison of the model structure of the extended single-stranded DNA (B) with a part of B-form DNA (C). The extended single-stranded DNA structure, in which the distance between bases is ca. 5 Å, contains hydrophobic 2′ methylene-base interactions, instead of the base-base stacking found in B-form DNA (the distance between bases: 3.4 Å). [Reproduced with permission from ref. 67 (Copyright 1997, National Academy of Sciences).]
The distance between bases by itself explains the well-known 1.5-fold longer length of single-stranded DNA as compared with B-form double-stranded DNA with the same sequence (48), because the bases have been shown to be almost perpendicular to the helical axis (70–72). When the DNA was replaced by RNA with the same sequence, the NOE signals were much smaller (67). This result is consistent with the observation that RNA showed lower affinity for RecA than DNA (73) and served as a poor cofactor for the ATPase activity of RecA (38, 74). This lower affinity and poor cofactor-activity are explained by RNA's structure: a bulky and hydrophilic OH group at the 2′ position of the pentose ring of RNA prevents its folding into the same extended structure.
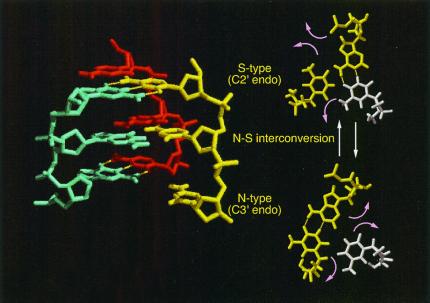
In B-form or A-form DNA, base rotation is sterically hindered. It is apparent that the extended structure is favorable for the free rotation of bases that is required for the base-pair switch. The sugar rings of DNA are generally not flat but have sugar puckers. In B-form and A-form DNA, sugar puckers are of the S type (C2′-endo) and N type (C3′-endo), respectively. In the calculated extended structure, sugar puckers of the deoxyribose ring in RecA-bound DNA are neither in the S type nor the N type, but are in a form in between (67). The structural calculation includes an assumption that there is only one major conformation for each oligo-DNA corresponding to the RecA-bound state. It is generally difficult to exclude the possibility that two or more conformations exist in the bound state. In addition, exchange systems (for this case, between bound and free states) have a limitation in that NMR-derived structural information lacks the spin-spin coupling constants, which are used for the determination of sugar puckers of nucleotides. Thus, the calculated structure could mean that the sugar puckers are stable in the intermediate form, or that the DNA is in a dynamic state in which sugar puckers convert between the two forms. During the model building described in the following section, we found that the interconversion of sugar puckers between the N type and the S type rotates bases horizontally without any steric hindrance, while maintaining the deoxyribose-base stacking interaction (68).
A Model for Homologous Pairing and Strand Exchange Through Base-Pair Switching Meditated by the Interconversion of Sugar Puckers
Considering the above structural properties of the extended single-stranded DNA bound to RecA or Rad51, we constructed a model for homologous pairing and strand exchange by a computer-assisted two-step procedure (ref. 68; see this reference about the assumptions and the software used). First, we tried to construct a model of double-stranded DNA in an extended form that included deoxyribose-base stacking, assuming that all sugar puckers were in the S type (C2′-endo; like the case of B-form DNA), but it was not possible to construct a DNA model having structural parameters (95 Å of helical pitch and 18.6 bp/turn) identical to those obtained by electron microscopic studies of double-stranded DNA in an active RecA-filament. Instead, we obtained an extended double-stranded DNA model (64 Å of helical pitch and 12.5 bp/turn; Protein Data Bank ID 1I1U) that fits the inactive RecA filament. However, when we assumed that all sugar puckers were in the N type (C3′ endo), we were able to obtain an extended double-stranded DNA model (Protein Data Bank ID 1I1T) fitting the data of DNA in the active RecA filament. In the second step, a single-stranded DNA was placed in the minor groove of an extended double-stranded DNA molecule with N-type sugar puckers, within a distance that permitted intermolecular base-pairing upon the interconversion of the sugar puckers to the S type (which causes rotation of the bases toward the minor groove), provided that the base of the single-stranded DNA was complementary (Fig. 2; Protein Data Bank ID 1I1V). Thus, in this model structure, trials for the base pair switch induced by the interconversion of sugar puckers well explain the mechanism of homology search and heteroduplex joint formation, i.e., homologous pairing and strand exchange (Fig. 2 and step 3 in Fig. 3). If the single-stranded DNA is placed in the major groove of an extended double-stranded form with all of the sugar puckers in the S form, this model is still valid; i.e., the interconversion of sugar puckers to the N form will induce the rotation of the bases toward the major groove. Because the kinetic barrier of the interconversion of sugar puckers and the opening of a Watson–Crick base pair can be overcome by thermal molecular motion, a small region or core of heteroduplex will be formed through the above mechanism without any need for a supply of chemical energy by ATP-hydrolysis (see Fig. 3). In experiments, homologous pairing to form a core heteroduplex was shown to be independent of ATP hydrolysis as described in a previous section.
Figure 2.
Model of heteroduplex joint formation induced by the base rotation associated with sugar pucker interconversion. (Left) The side view. (Right) The bottom view. Single-stranded DNA approaches from the minor groove of the extended double-stranded DNA with the N-type (C3′ endo) sugar pucker (Lower Left) or from the major groove of the double-stranded DNA with the S-type (C2′ endo) sugar pucker (Upper Left). The interconversion of sugar puckers induces the horizontal rotation of bases, which tests whether a base in the single-stranded DNA is complementary to a base engaged in a base-pair interaction in the double-stranded DNA. Complementarity would result in a base-pair switch. Thermal molecular motions are predicted to be sufficient to induce the base rotation, because the kinetic barrier for the interconversion of sugar puckers and that for the disruption of each base pair are sufficiently low. [Reproduced with permission from ref. 68 (Copyright 1998, National Academy of Sciences).]
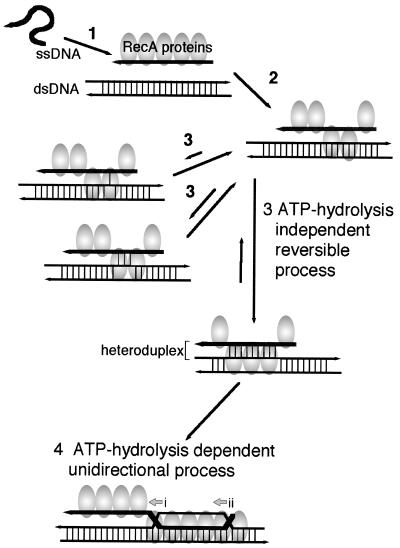
Figure 3.
Proposed ATP hydrolysis-independent and -dependent steps in heteroduplex joint formation and dissociation by RecA. Step 1: In the presence of ATP, RecA binds to single-stranded DNA (ssDNA) to form nucleoprotein filaments, resulting in the unfolding of secondary structures. Step 2: The nucleoprotein filament interacts with double-stranded DNA (dsDNA) without any need for sequence homology. These steps were well established by previous studies (see text). Step 3: A proposed ATP hydrolysis-independent reversible step. Repeated trials of base-pair switching induced by base rotation in the reaction lead to the formation of a core heteroduplex joint. The stability of the heteroduplex joint depends on the size of the base-paired region within the heteroduplex. Step 4: ATP hydrolysis-dependent unidirectional branch migration. Once a core heteroduplex is formed, the heteroduplex is extended in the 5′ to 3′ direction of parental single-stranded DNA (junction and direction are indicated by arrow i). When double-stranded DNA is under topological constraint (e.g., it is a closed circle or is anchored to the chromatin scaffold at points), the heteroduplex is extended until a supercoil is removed. Then, as the leading junction (indicated by arrow i) moves, the trailing junction (indicated by arrow ii) follows keeping the distance between the two junctions constant, and resulting in the migration of the heteroduplex. This step was characterized by various studies (see text). Step 4′: Proposed model of ATP hydrolysis-dependent dissociation at the end of the homologous sequence. If the heteroduplex joint is formed with a homologous sequence of a limited length, the heteroduplex joint is dissociated (41). This probably occurs by RecA-promoted migration of the trailing junction beyond the region of homology (41).
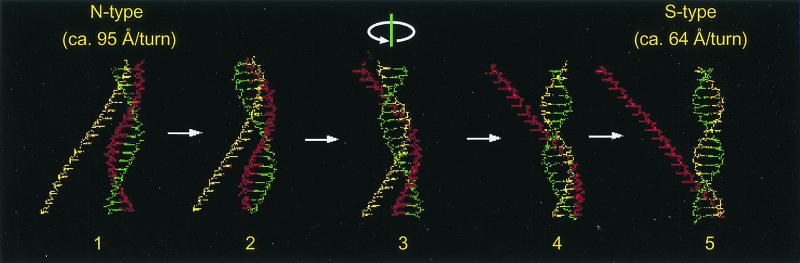
Once a homologous sequence is found that can form the core of the heteroduplex, the heteroduplex is extended by ATP hydrolysis-dependent unidirectional branch migration (5′ to 3′ of incoming single-stranded DNA; step 4 in Fig. 3). If the single-stranded DNA approaches the double-stranded DNA at the minor groove, the successive interconversion of the sugar puckers from the N type to the S type (from 18.6 bp/turn to 12.5 bp/turn) will create a rotational motion that helps the strand exchange, and thus, branch migration (ref. 68; Fig. 4). Because the duplex DNA extended by the 2′ methylene-base interaction with the S-type sugar puckers does not fit the description of the active RecA filament, but does fit with that of the inactive RecA filament, the hydrolysis of ATP bound to RecA will stimulate rotation of the double-stranded DNA, and thus will help strand exchange (Fig. 4).
Figure 4.
Possible stimulation of heteroduplex joint extension by continuous conversion of sugar puckers of the N type to the S type. Because the twist of extended double-stranded DNA is increased by sugar pucker interconversion from the N type to the S type, the sugar pucker type interconversion may create the rotational motion that stimulates branch migration. As extended double-stranded DNA with the N-type sugar puckers fits well with the active form (ATP form) of the RecA filament, and extended double-stranded DNA with S-type sugar puckers fits well with the inactive form (ADP form) of the RecA filament, ATP hydrolysis is predicted to stimulate rotation and, therefore, branch migration. It should be noted that this model is compatible with heteroduplex joint formation with single-stranded DNA approaching from the minor groove of double-stranded DNA with the N-type sugar pucker.
Further studies are required to test the validity of these models of heteroduplex joint formation.
Reversibility of the Homology Search Through Base Rotation and ATP Hydrolysis-Dependent Branch Migration
Because double-stranded DNA is helical, the recognition of sequence homology between double-stranded DNA and single-stranded DNA requires one of them to wrap around the other, and thus, to avoid topological problems, the primary recognition of sequence homology would involve only a small number of base pairs. The reported minimum DNA size required for the recognition of sequence homology is eight bases (75). On the other hand, genomic DNA contains numerous repeated sequences, whose repeating unit size varies from one to thousands of base pairs. Such repeated sequences prevent the alignment of DNA molecules at homologous sites in homologous chromosomes or sister chromatids and would cause chromosome aberrations. Because chromosome aberrations are very rare, cells must have an efficient system for maximizing the homologous alignment of parental DNA molecules. RecA appears to have a function to eliminate the misalignment of DNA molecules that would occur from the matching of short homologous sequences at nonhomologous sites. This might be carried out by at least two levels of reversibility in the heteroduplex joint formation by RecA. A heteroduplex joint, with a limited length formed through the primary contact, is likely to be reversible and would be dissociated unless the joint were stabilized by continuous base pairs of the required length. Because RecA-promoted branch migration is unidirectional (5′ to 3′ of incoming single-stranded DNA), the core heteroduplex joint is expanded unless the junction encounters the end of homologous sequences (Fig. 3). Thus, longer homologous sequences will form more stable heteroduplex joints. This view is consistent with the results of kinetic analyses of heteroduplex joint formation from oligo-DNA molecules (ca. 80 mers and 80 bp) by RecA; i.e., they revealed a rapid and reversible second-order pairing step followed by a slower and reversible first-order strand exchange step, and the discrimination of homology from heterology during both steps (76, 77).
Another level of reversibility is that observed in the D-loop cycle. After the formation of a D loop from closed-circle double-stranded DNA and a short, homologous, single-stranded DNA fragment (ca. 200 bases), the D loop is dissociated in the 5′ to 3′ direction by RecA in an ATP hydrolysis-dependent reaction (41, 78). Just after the dissociation, the double-stranded DNA is extensively unwound (or rather untwisted) and transiently does not interact with single-stranded DNA, but is used in the meantime in another round of D-loop formation and dissociation (41, 78). If the size of the homologous sequence is very large (e.g., if the homologous single-stranded DNA is circular), the heteroduplex joint is not dissociated (T.S., unpublished observation). This D-loop cycle explains the preference for heteroduplex joint formation between completely homologous DNA molecules over that between molecules that have only a short homologous sequence (41). The mechanism of the dissociation of the D loop is likely to be branch migration promoted by RecA. The D loop has two junctions of the parental strand and the homologous single-strand of the heteroduplex, and both junctions move unidirectionally (5′ to 3′ of the single-stranded DNA). The topological constraint of double stranded DNA maintains a constant distance between the two junctions. When the trailing junction moves beyond the end of the homologous single-stranded DNA, the D loop is dissociated (41).
Recently, a study suggested another class of ATP hydrolysis-dependent reversible alignment of DNA strands, through which RecA facilitates the realignment of DNA molecules to correct for misalignments at repeated sequences consisting of mono-, di-, and trinucleotides (79). Thus, RecA is likely to have a function that maximizes the alignment of homologous stretches in parental DNA molecules.
RecA is capable of efficiently carrying out the whole process of core heteroduplex formation and branch migration by itself. In contrast, homologous pairing in the phage T4 system is mediated by UvsX, a member of the RecA/Rad51 family, but branch migration is mediated by a DNA helicase (80).
The Conserved and Diverse Molecular Structures of the RecA/Rad51-Family Proteins
E. coli RecA and eukaryotic Rad51 share conserved regions of amino acid sequence in their core regions, which contain a presumed nucleotide-binding site (17, 18, 20, 81, 82). Electron microscopic studies have revealed extensive conservation in the three-dimensional organization of the filamentous structures that RecA/Rad51-family proteins form upon binding to single- or double-stranded DNA (20). Recently, x-ray crystallographic studies have revealed significant conservation between the core domain of E. coli RecA and DNA helicases. In particular, the structure of the helicase domain of T7 replicative primase-helicase closely resembles that of RecA (122 Cα atoms superimposed with an rms deviation of 1.6 Å; ref. 83). This conservation suggests that RecA/Rad51 and a group of DNA helicases evolved from a common ancestor.
On the other hand, structural analyses of an external DNA-binding site of RecA-DNA filament and that of the human Rad51 (HsRad51) protein-DNA filament obtained by NMR spectroscopy and site-directed mutagenesis suggest diversity in the domain structures of RecA/Rad51-family proteins, which probably was created by domain shuffling (84–86). A comparison of the structures of E. coli RecA and Rad51 homologues shows that RecA has a C-terminal domain that Rad51s does not have, and that Rad51s have a common N-terminal domain that RecA does not have (17, 18, 81, 82). Crystallographic studies and electron microscopic studies have shown that the C-terminal domain of RecA protrudes into a groove of RecA-helical filament in a lobe-like manner (25, 87). The determination of the solution structure and DNA titration by NMR techniques revealed that the C-terminal domain of RecA and the N-terminal domain of HsRad51 have sites for DNA binding, and site-directed mutagenesis has demonstrated that the binding of DNA to these sites is essential for the function of RecA (84) and Rad51 (86). In contrast to the functional similarity, the three-dimensional structures of the C-terminal domain of RecA and the N-terminal domain of HsRad51 are very different. An interesting point is that the three-dimensional structure of the N-terminal domain of HsRad51 resembles that of a part of the DNA binding site of another E. coli DNA repair endonuclease, endonuclease III (86). Thus, although the similarity in amino acid sequence is not significant, the N-terminal domain of HsRad51 and a part of the DNA binding site of endonuclease III might have a common ancestor. This could be an example of the shuffling of protein domains during evolution, which will be discussed later.
Recombination in Organisms with DNA Genomes vs. Recombination of RNA Viruses
In addition to its function in the error-free repair of DNA damage, homologous recombination is induced and shut off as a programmed function during differentiation, development, and sexual reproduction.
Unfavorable conditions for survival, such as nutrient deprivation and high cell density, induce homologous recombination in some bacteria, such as Bacillus subtilis. B. subtilis cells take up exogenous DNA, and, when the DNA is homologous to their chromosomal DNA, integrate almost all of it into the chromosomal DNA by recA-dependent homologous recombination. In this organism, the ability to take up exogenous DNA (“competence”) and homologous recombination are coactivated with sporulation under such unfavorable conditions (see refs. 88 and 89). Competence for transformation also is induced in Streptococcus pneumoniae. During the competence induction, the expression of RecA and another protein called Exp10 (or colligrin) are coinduced and the two are translocated as a complex to the membrane, where clusters of RecA are formed in an Exp10-dependent step (90). A gene for the homologue of Exp10 has been found in B. subtilis, and the genetic organization of the locus that encodes Exp10 in B. subtilis is identical to that of S. pneumoniae (see ref. 90). Upon the maturation of a spore, DNA of the mother cell is released to the environment and is ready to be taken up by other cells.
In eukaryotes with a sexual life cycle, homologous recombination that depends on RecA/Rad51-family proteins is induced at an early stage of meiosis to a level several orders of magnitude higher than in mitotic cells; this is indispensable to meiosis. In higher eukaryotes, meiosis is programmed as a step in development, but in simple eukaryotes such as the budding yeast S. cerevisiae and the fission yeast Schizosaccharomyces pombe, meiosis is induced by nutrient deprivation (see ref. 91 for review). The induction of meiotic recombination depends on several genes and is regulated by a complex network of cellular signaling systems, as revealed by genetic studies in both yeasts (see refs. 5–7 and 91–93 for review). The ability to induce homologous recombination in response to unfavorable environmental changes would be adaptive for each species, as it would increase genetic diversity and would help to avoid species' extinction (e.g., ref. 94).
Homologous recombination would be more efficient for evolution than random mutagenesis or nonhomologous recombination. Although the latter two will mostly disrupt previously existing genes rather than creating new ones, homologous recombination can use previously existing genes as building blocks, thus enabling the creation of new proteins with more complex functions in a step-by-step manner. Homologous recombination occurs at much higher frequencies than nonhomologous recombination during the lifecycles of organisms with DNA genomes, probably because of the existing induction system for homologous recombination and the accuracy of homologous recombination that is ensured by heteroduplex joint formation.
The above descriptions are more than a hypothesis. The quick acquisition of a new function by homologous recombination between genes with slightly different sequences was demonstrated in a series of in vitro experiments and observed in vivo during B-cell development in avian species and rabbits. In the in vitro experimental system called DNA shuffling, a pool of DNA molecules carrying a gene with different mutations was randomly fragmented and reassembled by a self-priming polymerase reaction (95). This system is not exactly the same as the homologous recombination involving double-stranded DNA, but incorporates the formation of a heteroduplex by the annealing of complementary strands bearing different mutations. Thus, the consequence of the reactions is equivalent to homologous recombination plus random mutagenesis. The DNA shuffling was shown to be much more efficient than simple random mutagenesis for the directed evolution of a gene product with enhanced activity, altered substrate specificity, and so on, and when a mixture of DNA bearing a common gene(s) from related organisms was used as starting DNA, the efficiency of the directed evolution of the gene(s) was extensively enhanced (96). During B-cell development in chicken, for an example, a unique rearranged V gene is diversified through repeated homologous recombination (gene-conversion type) with a group of homologous pseudogenes serving as donors with various mutations (see refs. 97 and 98 for review).
The shuffling of protein parts by homologous recombination does not require introns, which might play an important role in exon shuffling for later stages of protein evolution (99), and would have played a significant role, especially at an early stage of evolution when genetic variability was much more limited than now.
The fact that most complex organisms have a DNA genome instead of the RNA genome that very primitive organisms have indicates that DNA has a critical evolutionary advantage over RNA as a molecular carrier of genomic information. Although homologous recombination through heteroduplex joint formation is a general and essential feature of organisms with DNA genomes, homologous recombination of RNA viruses (that replicate without DNA intermediates, thus excluding retroviruses) is generally very rare (see refs. 100 and 101 for review). Significant levels of homologous recombination have been detected only in retroviruses and in a limited group of RNA viruses.
We assume that the critical advantage of DNA is its double-stranded structure and capacity for homologous recombination. The double-stranded structure provides a template for the correction of erroneously incorporated bases during duplication and for the repair of base or strand damage. This is essential for maintaining the integrity of a genome whose size is sufficiently large to encode for all of the genomic information necessary for independent cellular life. On the other hand, homologous recombination is essential for the well-organized dynamic property of double-stranded DNA that is necessary for the evolution of genomic information as discussed above.
Unlike DNA genomes, homologous recombination of RNA viruses is carried out by a copy-choice (replicative template switch) mechanism. In the copy-choice mechanism, RNA replication is initiated on a template RNA by an RNA replicase, followed by a template switch (see refs. 101–103 for review), and thus, both parental RNA molecules have to be single-stranded. In addition, it is claimed that nonhomologous recombination and homologous recombination of RNA viruses occur at comparable frequencies (101, 104). The presence of the massive and hydrophilic hydroxyl group at the 2′ position of the sugar ring prevents RNA from taking on the extended structure that is induced in DNA upon the binding of RecA/Rad51. These facts suggest that the 2′ methylene-base interaction is essential for the efficient and accurate homologous recombination of double-stranded polynucleotides and gives a critical advantage to DNA over RNA for evolution.
Concluding Remarks
A transferred NOE study on the three-dimensional structure of RecA-bound oligo-DNA revealed a unique extended DNA structure containing a 2′ methylene-base interaction. This interaction plays a pivotal role in heteroduplex joint formation through homologous pairing and strand exchange by base-pair switch. The observed requirements of RecA/Rad51 to induce the extended DNA structure and the general requirement for RecA/Rad51 for homologous recombination in various organisms suggest that homologous recombination through heteroduplex joint formation is an intrinsic property of a DNA structure induced by RecA/Rad51-family proteins. This function confers on double-stranded DNA, which is otherwise chemically and genetically stable, a well-organized dynamic property that enables the rearrangements of gene segments to create new genes without disturbing their coding frame. We suggest that the two faces of DNA, its stability and its propensity for homologous recombination, might give DNA a critical advantage over RNA for evolution.
Acknowledgments
This study was supported in part by a grant from the Biodesign Research Program of RIKEN (The Institute of Physical and Chemical Research) and by a grant for CREST from JST (Japan Science and Technology Corporation) to T.S.
Abbreviation
- NOE
nuclear Overhauser effect
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Watson J D, Crick F H C. Nature (London) 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Kimura M. Genome. 1989;31:24–31. doi: 10.1139/g89-009. [DOI] [PubMed] [Google Scholar]
- 3.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 4.Setlow R B. Prog Nucleic Acid Res Mol Biol. 1968;8:257–295. doi: 10.1016/s0079-6603(08)60548-6. [DOI] [PubMed] [Google Scholar]
- 5.Smith K N, Nicolas A. Curr Opin Genet Dev. 1998;8:200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- 6.Haber J E. Cell. 1997;89:163–166. doi: 10.1016/s0092-8674(00)80194-4. [DOI] [PubMed] [Google Scholar]
- 7.Roeder G S. Proc Natl Acad Sci USA. 1995;92:10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 9.Potter H, Dressler D. Proc Natl Acad Sci USA. 1976;73:3000–3004. doi: 10.1073/pnas.73.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell L R, Byers B. Cold Spring Harbor Symp Quant Biol. 1983;47:829–840. doi: 10.1101/sqb.1983.047.01.095. [DOI] [PubMed] [Google Scholar]
- 11.McEntee K, Weinstock G M, Lehman I R. Proc Natl Acad Sci USA. 1979;76:2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibata T, DasGupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 14.Baumann P, Benson F E, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R C, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonesaki T, Minagawa T. EMBO J. 1985;4:3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 19.Thacker J. Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 21.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujisawa H, Yonesaki T, Minagawa T. Nucleic Acids Res. 1985;13:7473–7481. doi: 10.1093/nar/13.20.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith J, Formosa T. J Biol Chem. 1985;260:4484–4491. [PubMed] [Google Scholar]
- 24.Yu X, Egelman E H. J Mol Biol. 1993;232:1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- 25.Story R M, Weber I, Steitz T A. Nature (London) 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 27.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 28.Cassuto E, West S C, Mursalim J, Conlon S, Howard-Flanders P. Proc Natl Acad Sci USA. 1980;77:3962–3966. doi: 10.1073/pnas.77.7.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West S C, Cassuto E, Mursalim J, Howard-Flanders P. Proc Natl Acad Sci USA. 1980;77:2569–2573. doi: 10.1073/pnas.77.5.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata T, DasGupta C, Cunningham R P, Williams J G K, Osber L, Radding C M. J Biol Chem. 1981;256:7565–7572. [PubMed] [Google Scholar]
- 31.DasGupta C, Wu A M, Kahn R, Cunningham R P, Radding C M. Cell. 1981;25:507–516. doi: 10.1016/0092-8674(81)90069-6. [DOI] [PubMed] [Google Scholar]
- 32.Petukhova G, Stratton S, Sung P. Nature (London) 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 33.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahn R, Cunningham R P, DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1982;79:762–766. doi: 10.1073/pnas.79.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bianchi M E, Radding C M. Cell. 1983;35:511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa T, Wabiko H, Tsurimoto T, Horii T, Masukata H, Ogawa H. Cold Spring Harbor Symp Quant Biol. 1979;43:909–915. doi: 10.1101/sqb.1979.043.01.099. [DOI] [PubMed] [Google Scholar]
- 39.Rosselli W, Stasiak A. EMBO J. 1991;10:4391–4396. doi: 10.1002/j.1460-2075.1991.tb05017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J I, Cox M M, Inman R B. J Biol Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 41.Shibata T, Ohtani T, Iwabuchi M, Ando T. J Biol Chem. 1982;257:13981–13986. [PubMed] [Google Scholar]
- 42.Cox M M, Soltis D A, Lehman I R, DeBrosse C, Benkovic S J. J Biol Chem. 1983;258:2586–2592. [PubMed] [Google Scholar]
- 43.Cazenave C, Toulme J-J, Helene C. EMBO J. 1983;2:2247–2251. doi: 10.1002/j.1460-2075.1983.tb01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata T, Cunningham R P, DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radding C M, Flory J, Wu A, Kahn R, DasGupta C, Gonda D, Bianchi M, Tsang S S. Cold Spring Harbor Symp Quant Biol. 1983;47:821–828. doi: 10.1101/sqb.1983.047.01.094. [DOI] [PubMed] [Google Scholar]
- 46.Kahn R, Radding C M. J Biol Chem. 1984;259:7495–7503. [PubMed] [Google Scholar]
- 47.Howard-Flanders P, West S C, Stasiak A. Nature (London) 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 48.Flory J, Tsang S S, Muniyappa K. Proc Natl Acad Sci USA. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang S S, Muniyappa K, Azhderian E, Gonda D K, Radding C M, Flory J, Chase J W. J Mol Biol. 1985;185:295–309. doi: 10.1016/0022-2836(85)90405-x. [DOI] [PubMed] [Google Scholar]
- 50.McEntee K, Weinstock G M, Lehman I R. Proc Natl Acad Sci USA. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata T, DasGupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1980;77:2606–2610. doi: 10.1073/pnas.77.5.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonda D K, Shibata T, Radding C M. Biochemistry. 1985;24:413–420. doi: 10.1021/bi00323a026. [DOI] [PubMed] [Google Scholar]
- 53.Tsang S S, Chow S A, Radding C M. Biochemistry. 1985;24:3226–3232. doi: 10.1021/bi00334a023. [DOI] [PubMed] [Google Scholar]
- 54.Chow S A, Radding C M. Proc Natl Acad Sci USA. 1985;82:5646–5650. doi: 10.1073/pnas.82.17.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radding C M. J Biol Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 56.Dunderdale H J, West S C. Curr Opin Genet Dev. 1994;4:221–228. doi: 10.1016/s0959-437x(05)80048-6. [DOI] [PubMed] [Google Scholar]
- 57.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roca A I, Cox M M. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 59.Adzuma K. Genes Dev. 1992;6:1679–1694. doi: 10.1101/gad.6.9.1679. [DOI] [PubMed] [Google Scholar]
- 60.Kumar K A, Muniyappa K. J Biol Chem. 1992;267:24824–24832. [PubMed] [Google Scholar]
- 61.Baliga R, Singleton J W, Dervan P B. Proc Natl Acad Sci USA. 1995;92:10393–10397. doi: 10.1073/pnas.92.22.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podyminogin M A, Meyer R B, Gamper H B. Biochemistry. 1995;34:13098–13108. doi: 10.1021/bi00040a022. [DOI] [PubMed] [Google Scholar]
- 63.Zhou X H, Adzuma K. Biochemistry. 1997;36:4650–4661. doi: 10.1021/bi9630063. [DOI] [PubMed] [Google Scholar]
- 64.Malkov V A, Panyutin I G, Neumann R D, Zhurkin V B, Camerini-Otero R D. J Mol Biol. 2000;299:629–640. doi: 10.1006/jmbi.2000.3770. [DOI] [PubMed] [Google Scholar]
- 65.Clore G M, Gronenborn A M, Greipel J, Maass G. J Mol Biol. 1986;187:119–124. doi: 10.1016/0022-2836(86)90411-0. [DOI] [PubMed] [Google Scholar]
- 66.Ni F, Scheraga H A. Acc Chem Res. 1994;27:257–264. [Google Scholar]
- 67.Nishinaka T, Ito Y, Yokoyama S, Shibata T. Proc Natl Acad Sci USA. 1997;94:6623–6628. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishinaka T, Shinohara A, Ito Y, Yokoyama S, Shibata T. Proc Natl Acad Sci USA. 1998;95:11071–11076. doi: 10.1073/pnas.95.19.11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishio M, Umezawa Y, Hirota M, Takeuchi Y. Tetrahedron. 1995;51:8665–8701. [Google Scholar]
- 70.Takahashi M, Kubista M, Norden B. J Biol Chem. 1987;262:8109–8111. [PubMed] [Google Scholar]
- 71.Norden B, Elvingson C, Kubista M, Sjoberg B, Ryberg H, Ryberg M, Mortensen K, Takahashi M. J Mol Biol. 1992;226:1175–1191. doi: 10.1016/0022-2836(92)91060-3. [DOI] [PubMed] [Google Scholar]
- 72.Norden B, Elvingson C, Eriksson T, Kubista M, Sjoberg B, Takahashi M, Mortensen K. J Mol Biol. 1990;216:223–228. doi: 10.1016/S0022-2836(05)80311-0. [DOI] [PubMed] [Google Scholar]
- 73.McEntee K, Weinstock G M, Lehman I R. J Biol Chem. 1981;256:8835–8844. [PubMed] [Google Scholar]
- 74.Weinstock G M, McEntee K, Lehman I R. J Biol Chem. 1981;256:8829–8834. [PubMed] [Google Scholar]
- 75.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bazemore L R, Takahashi M, Radding C M. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 77.Bazemore L R, Folta-Stogniew E, Takahashi M, Radding C M. Proc Natl Acad Sci USA. 1997;94:11863–11868. doi: 10.1073/pnas.94.22.11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu A M, Kahn R, DasGupta C, Radding C M. Cell. 1982;30:37–44. doi: 10.1016/0092-8674(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 79.Sen S, Karthikeyan G, Rao B J. Biochemistry. 2000;39:10196–206. doi: 10.1021/bi000753y. [DOI] [PubMed] [Google Scholar]
- 80.Salinas F, Kodadek T. Cell. 1995;82:111–119. doi: 10.1016/0092-8674(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 81.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Basile G, Aker M, Mortimer R K. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawaya M R, Guo S, Tabor S, Richardson C C, Ellenberger T. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 84.Kurumizaka H, Aihara H, Ikawa S, Kashima T, Bazemore L R, Kawasaki K, Sarai A, Radding C M, Shibata T. J Biol Chem. 1996;271:33515–33524. doi: 10.1074/jbc.271.52.33515. [DOI] [PubMed] [Google Scholar]
- 85.Aihara H, Ito Y, Kurumizaka H, Terada T, Yokoyama S, Shibata T. J Mol Biol. 1997;274:213–221. doi: 10.1006/jmbi.1997.1403. [DOI] [PubMed] [Google Scholar]
- 86.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. J Mol Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 87.Yu X, Shibata T, Egelman E H. J Mol Biol. 1998;283:985–992. doi: 10.1006/jmbi.1998.2141. [DOI] [PubMed] [Google Scholar]
- 88.Lovett C M J, Love P E, Yasbin R E. J Bacteriol. 1989;171:2318–2322. doi: 10.1128/jb.171.5.2318-2322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dubnau D. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Masure H R, Pearce B J, Shio H, Spellerberg B. Mol Microbiol. 1998;27:845–852. doi: 10.1046/j.1365-2958.1998.00732.x. [DOI] [PubMed] [Google Scholar]
- 91.Esposito R E, Klapholz S. In: Meiosis and Ascospore Development. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 211–287. [Google Scholar]
- 92.Kleckner N. Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Nature (London) 1998;392:491–494. [Google Scholar]
- 95.Stemmer W P C. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 96.Crameri A, Raillard S A, Bermudez E, Stemmer W P. Nature (London) 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 97.Weill J C, Reynaud C A. Science. 1987;238:1094–1098. doi: 10.1126/science.3317827. [DOI] [PubMed] [Google Scholar]
- 98.Thompson C B. Trends Genet. 1992;8:416–422. doi: 10.1016/0168-9525(92)90324-w. [DOI] [PubMed] [Google Scholar]
- 99.Patthy L. Matrix Biol. 1996;15:301–310. doi: 10.1016/s0945-053x(96)90131-6. ; discussion 311–312. [DOI] [PubMed] [Google Scholar]
- 100.Lai M M C. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chetverin A B. FEBS Lett. 1999;460:1–5. doi: 10.1016/S0014-5793(99)01282-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nagy P D, Simon A E. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 103.Aaziz R, Tepfer M. J Gen Virol. 1999;80:1339–1346. doi: 10.1099/0022-1317-80-6-1339. [DOI] [PubMed] [Google Scholar]
- 104.Chetverin A B, Chetverina H V, Demidenko A A, Ugarov V I. Cell. 1997;88:503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]