Abstract
Human Dmc1 protein, a meiosis-specific homolog of Escherichia coli RecA protein, has previously been shown to promote DNA homologous pairing and strand-exchange reactions that are qualitatively similar to those of RecA protein and Rad51. Human and yeast Rad51 proteins each form a nucleoprotein filament that is very similar to the filament formed by RecA protein. However, recent studies failed to find a similar filament made by Dmc1 but showed instead that this protein forms octameric rings and stacks of rings. These observations stimulated further efforts to elucidate the mechanism by which Dmc1 promotes the recognition of homology. Dmc1, purified to a state in which nuclease and helicase activities were undetectable, promoted homologous pairing and strand exchange as measured by fluorescence resonance energy transfer (FRET). Observations on the intermediates and products, which can be distinguished by FRET assays, provided direct evidence of a three-stranded synaptic intermediate. The effects of helix stability and mismatched base pairs on the recognition of homology revealed further that human Dmc1, like human Rad51, requires the preferential breathing of A⋅T base pairs for recognition of homology. We conclude that Dmc1, like human Rad51 and E. coli RecA protein, promotes homologous pairing and strand exchange by a “synaptic pathway” involving a three-stranded nucleoprotein intermediate, rather than by a “helicase pathway” involving the separation and reannealing of DNA strands.
Two kinds of macromolecular protein structures play prominent roles in homologous genetic recombination: toroids and filaments. Among the toroidal structures, the best understanding of the relation of function to structure exists for Escherichia coli RuvB protein, which forms a hexameric ATPase/helicase. Two such hexameric rings, interacting with RuvA protein, drive the migration of Holliday structures, the 4-fold branched DNA intermediate formed after initial synapsis and processing have occurred. The consequence of this driven migration is the reciprocal exchange of a pair of like strands between two duplex molecules (1). Rings are also formed by the exonuclease of phage λ (2), the Erf protein of phage P22 (3), and human Rad52 protein (4, 5). On the basis of the crystal structure of toroidal λ exonuclease, Kovall and Matthews suggested a mechanistic explanation of the high processivity of this enzyme (2).
Some recombination proteins form both rings and filaments: the β protein of phage λ forms large rings and left-handed helical filaments (6). β protein promotes annealing of complementary single strands (7, 8) and migration of a single-stranded branch (9). This protein, which does not hydrolyze ATP, binds more strongly to the product of its annealing activity, which may help to explain how it drives branch migration (10), but there are no correlations of structure with function that help us to understand the role of either the rings or the left-handed filaments. RecT protein, the product of a cryptic prophage in E. coli and a functional homolog of β protein, also forms rings and filaments. The precise nature of the RecT filament is less clear than that of β but has been judged to be helical (11). In this case as well, few clues link structure and function.
Among the filamentous structures, the best understanding of the relation of function to structure exists for E. coli RecA protein and human Rad51 protein. Although these proteins form rings as well as right-handed helical filaments (12–14), genetic (15), electronmicroscopic (16), and biochemical evidence (17) indicates strongly that the filament promotes homologous pairing and strand exchange. The role or significance of the rings is unknown. RecA protein was the first-discovered member of a ubiquitous family of proteins found in prokarya, eukarya, and archea. The much-studied filament that is formed by RecA protein on single-stranded DNA promotes a search for homology in duplex DNA and incorporates the homologous duplex. Incorporation of the duplex produces a synaptic nucleoprotein structure containing three homologously aligned strands of DNA in a right-handed helix. In the case of RecA protein, this synaptic structure leads to extensive strand exchange that dissociates one strand of the parental duplex to replace it with the single strand that nucleates formation of the filament (for review, see ref. 18). Solution of the structure of the single strand has shown that the bases, whose axial spacing is 50% greater than in B-form DNA, are stacked alternately with the deoxyribose moieties of the phosphodiester backbone, a configuration that links the angular displacement of bases to a change in the pucker of the sugar (19). Recent studies of human Rad51 indicate that the filament formed on single-stranded DNA provides a catalytic surface, which, on collision with duplex DNA, promotes the opening of base pairs to check for homology at the point of contact anywhere along the filament (20).
Dmc1 protein, a true homolog of RecA protein that shares extensive sequence homology with it, is expressed specifically in meiotic cells (21). Discovered in baker's yeast, it has been found also in mammals (22). Homozygous knockout of Dmc1 in the mouse leads to asynapsis and sterility (23, 24), whereas knockout of Rad51 leads to embryonic lethality (25, 26). Both Dmc1 and Rad51 are essential for meiosis in yeast.
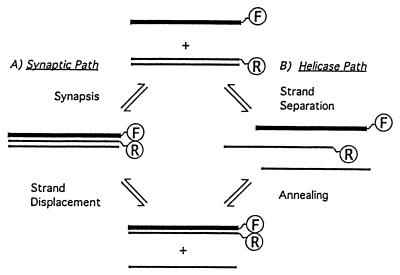
In vitro, Dmc1 protein promotes homologous pairing and limited strand exchange (27, 28). On the basis of sequence homology and preliminary biochemical characterization, it seemed likely that Dmc1 protein would catalyze homologous recognition by the same mechanism as that of E. coli RecA and eukaryotic Rad51 proteins. Prominent aspects of this mechanism include the formation of helical filaments on single-stranded DNA and the subsequent search for homology in the duplex DNA, which results in formation of a synaptic intermediate consisting of three strands of DNA within a protein filament. Surprisingly, however, recent studies from several laboratories failed to detect helical filaments made by Dmc1 (28, 29). These studies found instead that Dmc1 forms octameric rings and stacks of rings. A stack of rings with DNA running down the interior seems an unlikely structure for catalyzing homologous pairing. However, rings themselves, in particular rings that hydrolyze ATP, suggested the possibility of a helicase activity that might promote strand exchange in a very different way than helical filaments (Fig. 1). That notion led us to test more exhaustively for helicase activity of Dmc1, to look for positive evidence of synaptic complexes, and to determine directly whether Dmc1 uses the same mechanism as Rad51 to recognize homology.
Figure 1.
Alternative paths for homologous pairing and strand exchange promoted by human Dmc1 in vitro. (A) The synaptic path (Left) is believed to mediate homologous recognition and strand exchange catalyzed by RecA protein and the human and yeast Rad51 homologs. (B) The octameric ring structure of human Dmc1 (28, 29) suggests the need to evaluate an alternative helicase pathway to account for the same outcome. In this case, the duplex DNA might be opened up first to separate single strands, followed by a simple reannealing. The letters F and R contained within circles represent the fluorescent dyes fluorescein and rhodamine, positioned to detect homologous pairing by either pathway (see also Fig. 2).
Materials and Methods
Oligonucleotides.
The single-stranded 83-mer oligonucleotides used to form the filaments are designated as (−) strand and the respective complementary strands as (+) strand. The sequences of G16(−), G26(−), and G37(−) have been described earlier (20). The duplex oligonucleotides used in this study were made by thermal annealing of two complementary strands. The double-stranded DNA was radiolabeled by use of polynucleotide kinase and was subjected to PAGE to detect the presence of any excess of unannealed single-stranded DNA. The duplex substrates used in this study were free from any detectable single-stranded contaminant. Inosine was substituted in the place of guanine in G37(−) strand to form G13.I24(−). The numbers following the letters G and I indicate the percent of GC and IC content, respectively. To study the effect of mismatches, A⋅T transversions were made in the G16 and G26 duplex substrates. The position of mismatches is presented in Table 1. All concentrations of single-stranded oligonucleotides refer to mols of nucleotide residues, whereas concentrations of duplex oligonucleotides refer to mols of base pairs. The (−) and (+) strands contained a six-carbon primary amine linker at the 3′ and the 5′ ends, respectively. The labeling of these oligonucleotides with fluorescein and rhodamine was done as described before (30).
Table 1.
Location of A⋅T transversions made in G16 and G37 duplexes
| Number of transversions | Location of transversions (from 5′ end of minus strands) |
|---|---|
| In G16 | |
| 2 | 25, 56 |
| 4 | 15, 33, 51, 67 |
| 6 | 12, 24, 37, 48, 59, 72 |
| In G26 | |
| 2 | 25, 57 |
| 4 | 16, 33, 50, 67 |
| 6 | 12, 24, 36, 47, 59, 72 |
Purification of Dmc1.
The human Dmc1 coding sequence was amplified by PCR from a human testis cDNA library followed by the insertion into pQE-30 plasmid (Qiagen, Chatsworth, CA), as described before (27). The resulting construct, pEG 8–4, encoding HsDmc1 protein fused at its N terminus with six histidine residues, was transformed in E. coli strain EG1271, a derivative of E. coli F810(ΔrecA) (14), carrying the constitutively expressed lacIq gene in plasmid pREP4 (Qiagen). The strain thus obtained was grown in LB medium. Expression of Dmc1 was induced by the addition of isopropyl β-d-thiogalactoside, and the cells were harvested as described (27).
Batches of 40 grams of cells, wet weight, were lysed by the addition of three volumes of lysis buffer (20 mM potassium phosphate, pH 7.4/0.5 mM DTT/10% sucrose/0.5 M KCl/1 mM phenylmethylsulfonyl fluoride/1 mM EDTA/1 mg of lysozyme per milliliter), followed by sonication. The precipitate produced by ammonium sulfate at 40% saturation was redissolved in buffer A (20 mM potassium phosphate, pH 7.4/0.5 mM DTT/10% glycerol/50 mM KCl/1 mM phenylmethylsulfonyl fluoride/0.2 mM EDTA) and passed through an SP-Sepharose (Amersham Pharmacia) column (60 ml) preequilibrated with buffer A. The column was washed, and bound proteins were fractionated by gradient elution with 50–750 mM KCl in buffer A (400 ml). Fractions containing Dmc1 were pooled, diluted with three volumes of buffer B (20 mM potassium phosphate, pH 7.4/5 mM β-mercaptoethanol/10% glycerol/50 mM KCl/0.2 mM EDTA), and loaded on a 4-ml Ni-NTA (Stratagene) column equilibrated with 40 ml of buffer C (buffer B containing 200 mM KCl). The column was washed successively with 40 ml of buffer C containing 5 mM, 20 mM, and 50 mM imidazole. This was followed by gradient elution with buffer C containing 50–500 mM imidazole (40 ml), which eluted Dmc1 at about 150 mM imidazole. The fractions were pooled, dialyzed against buffer A containing 250 mM KCl, concentrated, and stored at −80°C.
Assays of Homologous Pairing and Strand Exchange by Fluorescence Resonance Energy Transfer (FRET).
For the pairing assay, Dmc1 (1.2 μM) was preincubated at 37°C for 5 min with 83-mer minus strand (3 μM), labeled at its 3′ end with fluorescein, in a reaction mixture (150 μl) containing 1 mM MgCl2, 25 mM Hepes (pH 7.4), 1 mM DTT, 2 mM ATP, and 100 μg of BSA per ml. The concentration of MgCl2 was increased to 10 mM, duplex 83-mer was added at 3 μM, and incubation was continued at 37°C. The duplex was labeled at the 5′ end of the plus strand with rhodamine. To measure strand exchange, the filament was formed as described above, but on unlabeled minus strand, while the duplex contained fluorescein at the 3′ end of the minus strand and rhodamine at the 5′ end of the plus strand (Fig. 2).
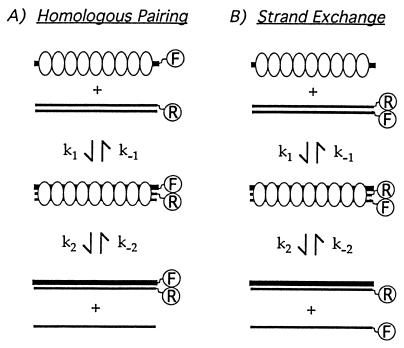
Figure 2.
Assays of homologous pairing and strand displacement on the basis of FRET. The fluorescein and rhodamine fluorophores were present on the 3′ and 5′ ends of the complementary strands, respectively. The assays are based on the nonradiative transfer of energy from fluorescein to rhodamine when the fluorescein is excited at its absorption maximum and the two dyes are in close proximity. (A) The quenching in fluorescein intensity or the subsequent increase in rhodamine emission, the so-called sensitized emission, is measured to detect homologous pairing. (B) The increase in fluorescein intensity or the decrease in the sensitized emission from rhodamine is measured to detect strand exchange.
Fluorescence emission spectra were taken from 502–620 nm on excitation at 493 nm on an SLM 8000C spectrofluorometer (SLM–Aminco, Urbana, IL). To determine the emission because of FRET on pairing, we subtracted from the spectrum of the complete reaction mixture at steady state, the individual spectra of reaction mixtures lacking either the rhodamine- or fluorescein-labeled DNA. The difference spectrum obtained is the so-called “sensitized emission,” i.e., the signal caused by FRET. The term “nonsensitized emission,” which is the background or noise, was used for the spectrum of the excited reaction mixture that lacked fluorescein-labeled DNA but contained everything else, including most particularly the rhodamine-labeled DNA.
The determination of FRET associated with strand exchange is a bit more complicated, because the duplex substrate at the beginning of the reaction is labeled with both dyes, which generates an initial FRET signal that is diminished by strand exchange. The reaction mixtures were constituted to determine the sensitized emission at the outset and the degree of its reduction when the reaction reached steady state. The reaction lacking Dmc1 was used to calculate FRET before the reaction, whereas the complete reaction in the presence of Dmc1 at equilibrium or steady state (1,800 sec) was used to determine FRET after the reaction. In both the cases, we subtracted from the spectra obtained with reactions containing both fluorophores the spectra from two mixtures, each containing either fluorescein- or rhodamine-labeled DNA. The difference in the sensitized emission or FRET before and after the reaction is the signal caused by strand exchange.
For both pairing and strand-displacement assays, corrections for small variations in the concentration of the two fluorophores between different samples were made by normalizing the experimental spectra on the basis of the reference measurements of emission at 520 and 585 nm, respectively, of standard reaction mixtures containing DNA labeled only with fluorescein or rhodamine.
For the measurement of time courses, the fluorescence emission was recorded at 520 nm on excitation at 493 nm, which measure the change in the emission from fluorescein. Homologous pairing is attended by quenching in fluorescein intensity as energy is transferred from fluorescein to rhodamine; strand displacement is attended by enhancement of emission from fluorescein as it ceases to transfer energy to rhodamine (see Fig. 2). In control reaction mixtures lacking Dmc1 protein, the change in fluorescence emission from fluorescein, which was less than 5%, was subtracted from the signal seen in complete reactions.
Assays for Helicase and Nuclease Activities.
For helicase activity, the substrates were 5′ radiolabeled 83-mer duplex G16 or partial duplexes made by annealing 83-mer G16(−) strand to complementary 43-mer to produce 5′ or 3′ tailed duplex. Substrates, at 3 μM, were incubated in 20 μl of the reaction mixture described in the previous section, except that MgCl2 was added at a final concentration of 10 mM. After the addition of 1–2 μM Dmc1, the mixture was incubated at 37°C for 1 h, and the reaction was terminated by the addition of 0.5% SDS and 25 mM EDTA. The reaction product was then subjected to 10% native PAGE. To assay nuclease activity, we used radiolabeled G16(−) single-stranded 83-mer or G16 duplex incubated in the same reaction mixture as used for the helicase assays, but the products were analyzed by denaturing PAGE. In both cases, reaction products were quantitated by use of a PhosphorImager (Molecular Dynamics) and imagequant software.
Results
Characterization of Purified Dmc1 and Lack of Helicase or Nuclease Activities.
Purified Dmc1 protein was checked for helicase activity as described in Materials and Methods. No helicase activity was detected even after 1 h of incubation by assays using blunt-ended 83-mer duplex or partial duplexes formed from an 83- and 43-mer with either a 5′ or 3′ single-stranded tail (data not shown). Bacterial RecQ helicase, used as a positive control in these experiments, dissociated more than half of a blunt-ended duplex or a duplex with a 3′ tail (data not shown). The lack of helicase activity was confirmed by fluorometric assay with the substrates labeled with fluorescein and rhodamine. Similarly, assays using radiolabeled single-stranded, double-stranded, and partially double-stranded DNA failed to detect any nuclease activity. The His-tagged Dmc1 protein hydrolyzed ATP in a DNA-dependent manner with a catalytic rate constant (kcat) of 0.7 mols/mol/min, similar to the kcat for nontagged protein, described before (27).
Formation of Synaptic Complexes by Dmc1.
In previous studies, we have used assays based on FRET to distinguish and kinetically characterize two phases of recombination reactions catalyzed by members of the RecA family of proteins (20, 30, 31). These phases are the recognition of homology and the exchange of strands. In the first phase, protein, a single strand, and a duplex molecule form a synaptic complex, in which homology is recognized; in the second phase, an exchange of strands converts the DNA within the complex into a heteroduplex molecule and a displaced single strand (Fig. 2). The formation of a synaptic complex is a key criterion that might help to distinguish a “synaptic pathway” from a “helicase pathway,” as illustrated in Fig. 1.
The FRET assays, as used to study pairing and strand exchange catalyzed by proteins like E. coli RecA protein, make use of oligonucleotide substrates labeled with fluorescein and rhodamine. FRET assays are based on the transfer of resonance energy from fluorescein to rhodamine, which occurs on excitation of fluorescein at its absorption maximum when the two fluors are in close proximity (Fig. 2). In the pairing assay, the formation of a synaptic intermediate generates a FRET signal (Fig. 2A). In the strand-exchange assay, the fluorescein and rhodamine fluors, which are juxtaposed in the substrate duplex, generate a FRET signal at the outset, and strand exchange diminishes that signal (Fig. 2B.) Reactions can be monitored by measuring either the change in FRET signal or the change in fluorescence emission from fluorescein itself. The fluorescein emission diminishes when energy is transferred from fluorescein to rhodamine. The change in fluorescein emission at 520 nm was used to monitor the time courses of these reactions.
Although FRET assays can distinguish the two phases of the reactions, as just discussed, they do not directly detect the synaptic complex because the FRET signal that is observed on pairing persists even after strand exchange occurs (Fig. 2A). However, we have observed previously that deproteinization diminishes part of the signal generated in the pairing assay but has no effect on the signal generated in the assay for strand exchange, which we have taken as evidence of dissociation of synaptic complexes (31).
With oligonucleotides containing 16% GC as substrates, we used the FRET assay to measure the steady-state levels of homologous pairing and strand exchange promoted by Dmc1 (Fig. 3A). Homologous pairing catalyzed by Dmc1 resulted in a FRET signal that was 60% above the background rhodamine emission. Fluorescence emission from controls lacking Dmc1 protein or containing heterologous DNA substrates, respectively, was indistinguishable from the background emission from rhodamine (see Materials and Methods). A strand-exchange reaction diminished the FRET signal about 40% below the background emission from rhodamine. This was qualitatively consistent with the results from a polyacrylamide gel assay of reactions of the same substrates (data not shown).
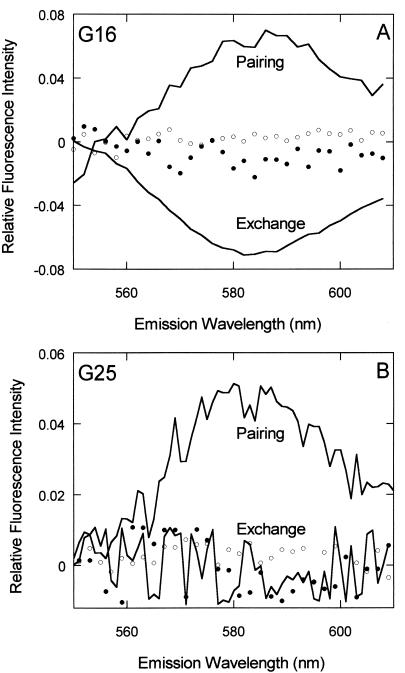
Figure 3.
Paranemic joints: direct evidence of synaptic complexes. This figure shows the FRET signal for homologous and heterologous reactions promoted by Dmc1. Homologous pairing and strand-exchange reactions are plotted as solid lines, whereas heterologous reactions are plotted as dotted lines. Open dots represent the heterologous controls for pairing, and filled dots represent the heterologous controls for strand exchange. (A) FRET signals for reactions of a duplex oligonucleotide substrate containing 16% GC (see Materials and Methods). For pairing, the spectrum represents the sensitized emission at steady state, whereas for strand exchange, the spectrum indicates the difference in sensitized emission before and after the reaction. The heterologous reactions used G26(−) instead of G16(−) to form the filament. The precise measurement of the sensitized and nonsensitized emission for pairing and strand exchange was done as described in Materials and Methods. (B) Identical measurements made with a duplex substrate containing 26% GC. Dmc1 can pair such substrates but cannot carry out strand exchange. G16(−) was used as the filament strand to calculate FRET in the heterologous reactions.
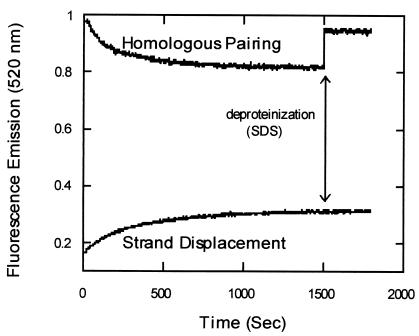
To detect the presence of synaptic complex, we monitored the change in fluorescein fluorescence, and when the reactions reached steady state, we deproteinized them (Fig. 4). In a reaction to measure pairing, the addition of SDS resulted in restoration of more than half of the signal, whereas in a reaction to measure strand exchange, SDS had no effect on the signal from fluorescein at steady state. These results suggest that Dmc1 produced a heteroduplex product at least in part by a reaction pathway involving the formation of a synaptic intermediate. To rule out the possibility that the effect is specific to the arrangement of fluors or to the DNA sequence, experiments were done in which fluorescein was present at the 5′ end of the G16(+) filament DNA, and rhodamine was present at the 3′ end of the G16(−) strand in the duplex. We observed amplitudes of reactions and effects of SDS (data not shown) similar to those shown above. Because SDS has an effect on the fluorescein intensity, the concentration and volume of SDS used in these reactions were standardized so that it had no net effect on the fluorescence signal from the reaction in the absence of Dmc1.
Figure 4.
Reversal of homologous pairing on deproteinization. The pairing and strand displacement reactions were performed with oligonucleotide G16 (16% GC) by fluorometric assay as described in Materials and Methods. The fluorescein emission was recorded every 2 sec at 520 nm on excitation at 493 nm. At 1,500 sec (arrow), the reaction was deproteinized by the addition of SDS (final concentration of 0.3%).
Paranemic Joints: Direct Evidence of Synaptic Complexes.
With circular single-stranded DNA and circular duplex DNA as substrates, RecA protein was shown to promote the recognition of homology and the formation of joints whose stability depends on the continued binding of the protein (32, 33). Such joints, in which there is no net change in linking number and no net strand exchange, were called paranemic joints. Paranemic joints, of course, are archetypal synaptic complexes.
The same name, paranemic joints, and the same concept have been applied to joints that are formed between a single-strand and linear duplex DNA in which heterologous ends of the linear molecule block complete strand displacement and confine the product of the reaction to an intermediate containing three strands. With such substrates, we showed that human Rad51 protein could form paranemic joints (31).
As described above, Dmc1 promoted pairing and strand exchange with an 83-mer oligonucleotide substrate containing 16% G⋅C base pairs (Fig. 3A), but when GC content was increased to 26%, no product was seen by the gel assay (data not shown). Because the gel assay measures the overall reaction efficiency and cannot differentiate between the two phases of the reaction, we resorted to the fluorometric assays of pairing and strand exchange. We found that Dmc1 recognized homology in duplex DNA, as evidenced by a significant pairing signal (≈44% above background rhodamine emission) but did not promote any detectable strand exchange (Fig. 3B). The pairing signal was completely reversed when the reaction was deproteinized (data not shown). Thus, Dmc1 could promote the formation of paranemic joints in which homology was recognized in the absence of strand exchange and whose stability depended on the continued presence of protein.
Mechanism of Recognition of Homology.
We previously showed that the recognition of homology in duplex DNA by the single strand in the Rad51 filament requires the breathing of base pairs, particularly A⋅T base pairs, in the duplex DNA (19). That mechanism of recognition plus the ability of Rad51 to form paranemic joints implies that the nucleoprotein filament provides an extended catalytic surface that can promote breathing and subsequent homologous recognition anywhere along the length of the DNA molecules involved. The breathing mechanism seems incompatible with a searching process mediated exclusively by rings or stack of rings, as formed by Dmc1 (29). For that reason in particular, we asked whether Dmc1 promotes recognition by the same breathing mechanism as Rad51
As just described, when the GC content of the substrates was 16%, Dmc1 promoted both recognition and strand exchange, whereas at 26% GC, Dmc1 still promoted pairing but not strand exchange. At higher GC content, no reaction can be detected (see Fig. 5). This pattern mirrors that seen for Rad51, suggesting that Dmc1 and Rad51 use the same mechanism to recognize homology. To examine that inference further, we used two tests that we used in the case of Rad51 (20).
Figure 5.
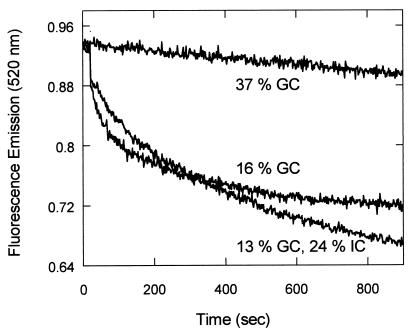
Effect of helix stability on the recognition of homology by Dmc1. Homologous pairing reactions promoted by Dmc1 were done with three different pairs of single-stranded and duplex oligonucleotides; G16 (16% GC), G37 (37% GC), and inosine-substituted G37 (13% GC, 24% IC). The inosine residues were substituted in G37 in the place of guanines. (The Tm values of G37 and its inosine-substituted derivative were 70.5 and 55.7°C, respectively) (20).
To test whether the inhibitory effect of GC content is because of its effect on the stability of duplex DNA, which makes the breathing of base pairs more difficult, we destabilized duplex DNA by substituting inosine in the place of guanine. Because I⋅C base pairs contain only two hydrogen bonds, the inosine substitution in the place of guanine destabilizes the duplex and should stimulate strand exchange. Inosine was substituted in G37(−) strand, in the place of 20 guanine residues to construct G13.I24(−) strand. In this experiment, the G13⋅I24(−) strand was used to form the filament while the duplex contained G13⋅I24(−) strand and unsubstituted G37(+) strand. This experimental design guarantees that no energetic bias will be introduced to favor either the forward or reverse reaction. With 16% GC substrate (G16), a significant pairing signal was observed (Figs. 3 and 5), but when GC content was raised to 37%, the pairing reaction was completely abolished (Fig. 5). However, when inosine residues were substituted for guanine in the same oligonucleotide (G13⋅I24), the reaction was strongly stimulated, with an amplitude that was equivalent to that of G16. The strong stimulation by inosine substitution indicates that the stability of the helix plays an important role in recognition of homology.
By virtue of their lesser stability, A⋅T base pairs play a bigger role than G⋅C base pairs in homology recognition promoted by Rad51 (20). Mismatches opposite A⋅T base pairs in more G⋅C-rich DNA had a bigger effect than similar mismatches in more A⋅T-rich DNA. We applied the same test to homologous recognition promoted by Dmc1.
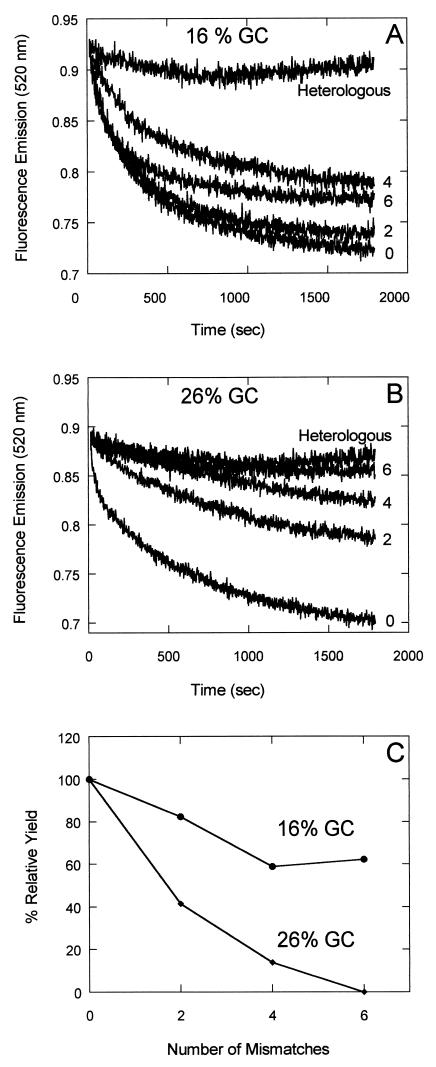
Two, four, or six transversions were made at A⋅T base pairs in G16 (16% GC) and G26 (26% GC) duplexes to create mismatched substrates (see Table 1). The mismatches were made in this way so that the initial duplex substrates remained fully paired, and the DNA that was used to form filament remained the same for each set of reactions with a varying number of mismatches. Using the fluorometric assay for pairing, we found that for any given number of mismatches, the pairing with G16 was more tolerant to mismatches than the pairing with G26. Six mismatches in the 26% GC substrate eliminated homologous pairing whereas six mismatches in the 16% GC substrate had relatively little effect (Fig. 6). This experiment reveals the special role of A⋅T base pairs in homologous recognition promoted by Dmc1.
Figure 6.
Differential effects on recognition of homology resulting from A⋅T transversion mismatches in “AT-rich” vs. “GC-rich” DNA. Two, four, or six mismatches at the sites of A⋅T base pairs were created by transversions in the duplex substrates, G16 (16% GC) and G37 (37% GC) (see Materials and Methods). A and B show the original data on the effects of increasing number of mismatches. In C, the yield of these reactions at 30 min was plotted against the number of mismatches. The yield at 30 min with no mismatched substrate was taken as 100%.
Discussion
Although eukaryotic Dmc1 protein was discovered some years ago, little is known about its biochemical properties. Human Dmc1 hydrolyzes ATP in a DNA-dependent manner and promotes the formation of D-loops and reactions leading to strand exchange (27, 28). The catalytic rate constant for ATPase activity is similar to that of Rad51 protein (≈0.7), which is considerably smaller than that of E. coli RecA protein (≈30). Human Dmc1 protein binds preferentially to single-stranded DNA with a stoichiometry of approximately one molecule of protein per three nucleotide residues. Dmc1 physically interacts with Rad51 and Tid1 proteins (28, 34). Almost nothing was known as to how Dmc1 promotes homologous pairing and strand exchange. The studies described here were further motivated by the failure thus far to find a Dmc1 helical nucleoprotein filament like those formed by E. coli RecA protein and human or yeast Rad51 (see Introduction).
Because Dmc1, in the absence of DNA forms abundant octameric rings (29), we were led to consider seriously the possibility that pairing and strand exchange in vitro might result from helicase activity either extrinsic or intrinsic to Dmc1 (see Fig. 1). However, no helicase activity could be detected in Dmc1 preparations by using DNA substrates with blunt-ended, 5′ tailed, or 3′ tailed duplexes.
To get positive evidence on the nature of the pathway catalyzed by Dmc1, we looked for the formation of synaptic complexes. Two kinds of experiment provided evidence of synaptic complexes. The first was one in which the reactions for pairing and strand exchange were deproteinized after the reactions reached steady state or equilibrium. Previous studies of Rad51, by using FRET assays, produced evidence that a three-stranded synaptic intermediate exists in which stable homologous recognition is accomplished by the partial switching of base pairs (20). As might be expected, deproteinization of this complex leads in some measure to the regeneration of substrates, whereas deproteinization of the duplex product of the reaction has no effect (31). Similarly, the failure of deproteinization to reverse strand exchange promoted by Dmc1 argues that reversal of the pairing assay is because of the presence of synaptic intermediate which that assay detects. We did a further test by repeating this experiment with an oligonucleotide of different sequence and with the fluorophores located at the other end of the substrates. Nucleases and helicases, for the most part, are specific to the 5′ or 3′ end of the DNA molecule to initiate the action. The action of such nucleases or helicases on duplex DNA, followed by annealing, could plausibly produce a heteroduplex product indistinguishable from that made by a synaptic pathway as already indicated (Fig. 1). The fluorescent dyes at the ends of DNA in our assays might be expected to interfere sterically with the directional specificity of a pathway driven by nuclease or helicase activity. The data argue against such pathways because there was no effect of altering the positions of the dyes.
The second kind of experiment used to test for synaptic complexes was one that looked for paranemic joints, the joints in which homologous pairing occurs in the absence of net strand exchange. In such joints, the three strands are held together by the proteins to form homology-specific synaptic complexes (32, 33). The formation of paranemic joints by Dmc1 therefore provides direct evidence of synaptic complexes. The similar yield of pairing reactions irrespective of the DNA end at which fluors are present, along with the formation of paranemic joints, indicates that homologous recognition driven by Dmc1 is independent of DNA ends. This further suggests that Dmc1 functions similarly to other RecA-like proteins.
Although synaptic joints appear diagnostic of a RecA-like pathway of pairing and strand exchange, one can imagine a helicase pathway that goes through some variety of nucleoprotein intermediate that has three strands of DNA in it. For that reason, our experiments on the mechanism by which Dmc1 promotes recognition of homology are of further importance in establishing the nature of the pathway. Previously, we found that recognition of homology and initiation of strand exchange as promoted by human Rad51 are integral aspects of a concerted mechanism that involves the preferential switching of A⋅T base pairs (20). In the present experiments on Dmc1, the substitution of inosine for guanine demonstrated a general effect of helix stability on recognition of homology, and A⋅T mismatches demonstrated a special role of A⋅T base pairs in recognition of homology. These findings, plus the inhibition of Dmc1 reactions by GC content, are identical in form to the observations on human Rad51. Thus the present findings support the model that posits that recognition of homology requires the breathing and switching of base pairs, accomplished preferentially by A⋅T base pairs. In the case of Rad51, as well as RecA, the helical filament formed on single-stranded DNA forms a long catalytic surface that is coextensive with the DNA sequence. Collision of that catalytic surface anywhere along its length with duplex DNA can promote the opening of the latter and test for homology by Watson–Crick complementarity. A similar scenario for Dmc1 based on a filament made by a stack of rings seems inconceivable, unless the DNA were wrapped around the outside of the protein filament, which also appears unlikely (29). Similarly, it is difficult to imagine that an octameric protein ring, migrating like a helicase, could provide the requisite opening of duplex DNA specifically at the sites of collision of the single-stranded and duplex substrates. The similarity of the mechanisms by which Dmc1 and Rad51 recognize homology, despite the apparent differences in the superstructures they form, is striking. Perhaps Dmc1 in vitro forms a helical filament that is either less stable or more dynamic and hence has eluded detection. Clearly, more studies are needed to understand the protein–protein and protein–DNA interactions of Dmc1. Taken together, however, the observations described here, including the lack of helicase activity, the formation of synaptic complexes, and the similarity of the pairing mechanism of Dmc1 to that of Rad51, argue that Dmc1, acting alone, follows the same reaction path as its homologs.
The results described here and previously (20, 31, 35) provide insight into functional differences between E. coli RecA protein and several of its eukaryotic homologs. It was immediately apparent in various studies that eukaryotic homologs of RecA hydrolyze ATP at rates that are between one and two orders of magnitude slower (14, 27, 35, 36). In the absence of protein cofactors, the yields of reactions promoted by eukaryotic homologs are notably lower, and at least in the cases of human Rad51 and human Dmc1 proteins, the extent of strand exchange is limited (27, 28, 31, 35, 37). It is now clear that the yields of these reactions are sensitive to the GC content of the substrates. In our studies, the degree of sensitivity to GC content is in the order Dmc1 > Rad51 > RecA. The limited strand exchange by Dmc1 and Rad51 correlates with this sensitivity. At 26% GC, Dmc1 still catalyzes recognition of homology but is unable to initiate strand exchange (this study), whereas, under the same conditions, 40% GC content has the same effect on human Rad51 (31). We know from studies in vitro that there are several proteins that play important roles in enhancing the activities of Rad51 (38–42) and from genetic studies, especially in Saccharomyces cerevisiae, that multiple proteins govern homologous recombination (43–45). An understanding of the possible biological significance of the effect of base composition as observed in vitro must await studies of a reconstituted complete set of recombination proteins. For the moment, the principal significance of the effects of base composition lies in what it has told us about the mechanism of recognition of homology, which appears to be a mechanism shared by homologs of RecA (20).
Acknowledgments
We thank our colleague Ewa Folta-Stogniew for helpful discussions and Jan Zulkeski for data processing. This research was sponsored by Grant R37-GM33504 from the National Institute of General Medical Sciences.
Abbreviation
- FRET
fluorescence resonance energy transfer
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 2.Kovall R, Matthews B W. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 3.Poteete A R, Sauer R T, Hendrix R W. J Mol Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- 4.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 5.Van Dyck E, Hajibagheri N M A, Stasiak A, West S C. J Mol Biol. 1998;284:1027–1038. doi: 10.1006/jmbi.1998.2203. [DOI] [PubMed] [Google Scholar]
- 6.Passy S I, Yu X, Li Z, Radding C M, Egelman E H. Proc Natl Acad Sci USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kmiec E, Holloman W K. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 8.Muniyappa K, Radding C M. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- 9.Li Z, Karakousis G, Chiu S K, Radding C M. J Mol Biol. 1998;276:733–744. doi: 10.1006/jmbi.1997.1572. [DOI] [PubMed] [Google Scholar]
- 10.Karakousis G, Ye N, Li Z, Chiu S K, Reddy G, Radding C M. J Mol Biol. 1998;276:721–731. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 11.Thresher R J, Makhov A M, Hall S D, Kolodner R, Griffith J D. J Mol Biol. 1995;254:364–371. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- 12.Stasiak A, Di Capua E. Nature (London) 1982;299:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 13.Heuser J, Griffith J. J Mol Biol. 1989;210:473–484. doi: 10.1016/0022-2836(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 14.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutreix M, Burnett B, Bailone A, Radding C M, Devoret R. Mol Gen Genet. 1992;232:489–497. doi: 10.1007/BF00266254. [DOI] [PubMed] [Google Scholar]
- 16.Stasiak A Z, Rosselli W, Stasiak A. Biochimie. 1991;73:199–208. doi: 10.1016/0300-9084(91)90203-d. [DOI] [PubMed] [Google Scholar]
- 17.Tsang S S, Muniyappa K, Azhderian A, Gonda D K, Radding C M, Flory J, Chase J W. J Mol Biol. 1985;185:295–310. doi: 10.1016/0022-2836(85)90405-x. [DOI] [PubMed] [Google Scholar]
- 18.Roca A I, Cox M M. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 19.Shibata T, Nishinaka T, Mikawa T, Aihara H, Kurumizaka H, Yokoyama S, Ito Y. Proc Natl Acad Sci USA. 2001;98:8425–8432. doi: 10.1073/pnas.111005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta R C, Folta-Stogniew E, O'Malley S O, Takahashi M, Radding C M. Mol Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- 21.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 22.Habu T, Taki T, West A, Nishimune Y, Morita T. Nucleic Acids Res. 1996;24:470–477. doi: 10.1093/nar/24.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pittman D L, Cobb J, Schimenti K J, Wilson L A, Cooper D M, Brignull E, Handel M A, Schimenti J C. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 25.Lim D, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Golub E I, Gupta R, Radding C M. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masson J, Davies A A, Hajibagheri N, Benson F, Stasiak A Z, Stasiak A, West S. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passy S I, Yu X, Li Z, Radding C M, Masson J-Y, West S C, Egelman E E. Proc Natl Acad Sci USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bazemore L R, Takahashi M, Radding C M. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 31.Gupta R C, Folta-Stogniew E, Radding C M. J Biol Chem. 1999;274:1248–1256. doi: 10.1074/jbc.274.3.1248. [DOI] [PubMed] [Google Scholar]
- 32.Bianchi M, DasGupta C, Radding C M. Cell. 1983;34:931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- 33.Riddles P W, Lehman I R. J Biol Chem. 1985;260:165–169. [PubMed] [Google Scholar]
- 34.Dresser M E, Ewing D J, Conrad M N, Dominguez A M, Barstead R, Jiang H, Kodadek T. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta R, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 37.Baumann P, Benson F, West S. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 38.Baumann P, West S C. J Mol Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 39.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 40.Song B, Sung P. J Biol Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- 41.Petukhova G, Stratton S, Sung P. Nature (London) 1998;393:81–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 42.Petukhova G, Sung P, Klein H. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Broach J R, Jones E W, Pringle J R, editors. I. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 407–520. [Google Scholar]
- 44.Game J C. Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 45.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]