Abstract
Cyclophilins constitute a subgroup of large family of proteins called immunophilins, which also include FKBPs and Parvulins. They are remarkably conserved in all genera, highlighting their pivotal role in important cellular processes. Most cyclophilins display PPIase enzymatic activity, multiplicity, diverse cellular locations and active role in protein folding which render them to be included in the class of diverse set of proteins called molecular chaperones. Due to their distinct PPIase function, besides protein disulfide isomerases and protein foldases, cyclophilins have been deemed necessary for in vivo chaperoning activity. Unlike other cellular chaperones, these proteins are specific in their respective targets. Not all cyclophilin proteins possess PPIase activity, indicating a loss of their PPIase activity during the course of evolution and gain of function independent of their PPIase activity. The PPIase function of cyclophilins is also compensated by their functional homologs, like FKBPs. Multiple cyclophilin members in plants like Arabidopsis and rice have been reported to be associated with diverse functions and regulatory pathways through their foldase, scaffolding, chaperoning or other unknown activities. Although many functions of plant cyclophilins were reported or suggested, the physiological relevance and molecular basis of stress-responsive expression of plant cyclophilins is still largely unknown. However, their wide distribution and ubiquitous nature signifies their fundamental importance in plant survival. Several of these members have also been directly linked to multiple stresses. This review attempts to deal with plant cyclophilins with respect to their role in stress response.
Keywords: cyclophilin, stress, protein folding
Introduction
Cyclophilins, one of the three protein subfamilies of immunophilin superfamily, (besides FK506 binding proteins and Parvulins) consist of highly conserved proteins, many but not all of which possess peptidyl prolyl cis-trans isomerase (PPIase) or rotamase activity.1 They are categorized as immunophilins owing to their ability to bind specific immunosuppressant molecules of fungal origin. The cyclophilins (Cyps) bind cyclosporin A (CsA), a cyclic undecapeptide, the FK506-binding proteins (FKBPs) bind macrolides such as FK506 (tacrolimus) and rapamycin that are structurally unrelated to CsA,2 while the third group of proteins, Parvulins bind Juglone, 5-hydroxy-1,4-naphthoquinone which irreversibly inhibits the enzymatic activity of several parvulins.3 Cyclophilins are the first subfamily of immunophilins discovered in Bovine thymocytes.4 They were initially discovered as ligands for immunosuppresent drugs and later recognized for their PPIase activity.5 Cyclophilins and FK506-binding proteins (FKBPs) have structurally distinct PPIase domains that are unrelated in their amino acid sequence. The drugs bind to the catalytic pocket of the PPIase domain and inhibit the PPIase activity. Additionally, the drug-immunophilin complexes, Cyp-CsA and FKBP-FK506, dock onto protein phosphatase 2B (PP2B) (also known as calcineurin), thereby inhibiting the phosphatase activity of PP2B.6 It results in the elevated phosphorylation of a number of PP2B substrates; as a consequence, immunologically important genes remain silent, suppressing the immune reaction. The drug-dependent functions, which gave rise to the name “immunophilin,” have clinical but no physiological relevance.7 Biochemical and sequence analyses have led to the identification of a novel family of chimeric immunophilins that contain both Cyp and FKBP domains. Due to their dual nature, these latter enzymes are names as FCBPs (FK506 and CsA binding proteins).8 Despite the apparent difference in their sequence and three-dimensional structure, the three families encode similar enzymatic and biological functions. Recent studies have revealed that many immunophilins possess a chaperone function independent of PPIase activity.7
Cyclophilins (PPIases) are Ubiquitous Proteins and Perform Diverse Functions
The term “Peptidyl-prolyl cis-trans isomerases” (PPIases) is used interchangeably for cyclophilins, FKBPs as well as Parvulins. The identification of the first protein that showed PPIase activity over 27 y ago5 was reported from bovine thymocytes as an intracellular protein with a high affinity for the immunosuppressive drug cyclosporin A (CsA).4 Five years later, 18 kDa protein with PPIase activity and CypA were found to be one and the same.9 All cyclophilins share a conserved domain, the cyclophilin-like domain (CLD). Although cyclophilins have been implicated in apoptotic genome degradation through their nuclease activity which in turn requires direct binding to DNA,10 this cannot be attributed to all cyclophilins unless they are explored for their nuclease activity. Cyclophilins possessing nuclease activity have two different active sites in spite of their relatively small molecular size. One site is responsible for their peptidyl-prolyl cis-trans isomerase activity, and the other active site is capable of catalytic degradation of DNA in a calcium/magnesium-dependent manner.10 The larger members of each family are modular in nature, having multiple PPIase and/or protein-protein interaction domains or domains unique to each member of the family that are associated with subcellular compartmentalization and functional specialization.11
Depending upon the functional modules present, cyclophilins have been broadly divided into two classes viz. single domain cyclophilins and multidomain cyclophilins. Single domain cyclophilins are characterized by the presence of a single catalytic CLD exhibiting PPIase activity. Multidomain cyclophilins, in addition to a conserved CLD, possess other functional domains like WD40, TPR, RRM, Zinc Finger etc.
Cyclophilins are ubiquitous proteins12 found in mammals, plants, insects, fungi and bacteria; they are structurally conserved throughout evolution and most of them, if not all, have PPIase activity. There are total of 16 cyclophilin isoforms in human genome, 7 major cyclophilin isoforms in humans include hCypA (also called hCyp-18a, 18 denotes molecular mass of 18 kDa), hCypB (also called hCyp-22/p, 22 kDa), hCypC, hCypD, hCypE, hCyp40 (40 kDa) and hCypNK (first identified from human natural killer cells).1 Little is known about the genomic structure of human cyclophilin genes; they are generally not linked to each other in the genome. Drosophila has at least 9 cyclophilins,1 C. elegans has 11,13 whereas 8 cyclophilins, Cpr1-Cpr8, have been found in Saccharomyces cerevisiae.11
Cyclophilins are present in all cellular compartments,14 involved in processes including protein trafficking and maturation,15 receptor complex stabilization,16 apoptosis,17 receptor signaling,18 RNA processing,19 muscle differentiation, detoxification of reactive oxygen species (ROS),20 immune response,21 spliceosome assembly,22 miRNA activity23 and RISC assembly.24 Cyclophilins also have proposed functions in facilitating protein folding and trafficking.25 Some of the cyclophilin members may serve as scaffolding proteins for assembly of large supramolecular complexes.26 The diverse roles and still emerging novel functions suggest that the cyclophilins in plants encompass far more functions than already defined function of protein folding. However, the mechanisms of how cyclophilins contribute to these cellular events have been difficult to establish and are still largely unknown.
The 18-kDa archetypal cyclophilin CypA is cytosolic and found in all tissues, whereas other cyclophilins, whether they have a CLD alone or in combination with other domains, are found in the endoplasmic reticulum (ER), the mitochondria or the nucleus. The crystal structures of several cyclophilins have been determined.27 Human CypA has an eight-stranded antiparallel-barrel structure, with two helices enclosing the barrel from either side. Seven aromatic and other hydrophobic residues form a compact hydrophobic core within the barrel, usually in the area where CsA binds. A loop from Lys118 to His126 and four β-strands (β3-β6) make up the binding site for CsA.28 The overall structure of hCypB resembles that of hCypA, the main difference being in the two loop regions (residues 19–24 and 152–164) and at the amino and carboxyl termini.29 Murine CypC also has a structure similar to that of hCypA, differing mainly in the conformation of three surface loop regions.30 The large cyclophilin Cyp40 consists of a CLD with a structure similar to that of hCypA linked to tetratricopeptide repeats (TPRs), which are also found in proteins involved in stress responses.31
Cyclophilins (PPIases): Role in Protein Folding
Protein folding in vitro is mediated by an array of proteins that act as molecular chaperones, as foldases or both.32 For many proteins, in vitro folding is a spontaneous process that does not require energy or assisting factors.33 However, protein folding in vivo is a more complex process and may require ATP and is aided by several proteins. The foldases are catalysts accelerating slow steps in the folding by rearrangements of disulfide bonds brought in by disulfide isomerases or isomerisation of prolyl peptide bonds by PPIases.34 Protein disulfide isomerase activity has generally been found associated with the ER of lower eukaryotes and mammals while proteins with PPIase activity have wider distribution.35 In folded proteins, the peptide bonds occur in two conformations, cis or trans, and the dihedral angles for the rotation about the CN bond are tightly clustered around 0° (cis) and 180° (trans).36 Peptide bonds not preceding proline are almost always trans in folded proteins, but 5.7% of all Xaa-Pro peptide bonds show the cis conformation in the proteins with known 3D structure.37 There is good evidence now that during slow isomerisation steps in protein folding, cyclophilins stabilize the cis-trans transition state and accelerate isomerisation (Fig. 1), a process that is considered important not only in protein folding but also during the assembly of multidomain proteins.38 Protein folding requires the assistance of both foldases and molecular chaperones and, in some cases cyclophilins serve in both capacities in the maintenance or assembly of supermolecular complexes. Cyp40 is capable of maintaining the protein in a folding competent state with efficiency comparable to that of hsp90. Interestingly, the molecular chaperoning activity of Cyp40 was not suppressed by CsA, which indicates that it may be separate from its peptidyl-prolyl cis-trans isomerase activity.39 Studies in yeast and mammals have shown that the Cyp40 protein has both PPIase and protein chaperone activity and it binds to HSP90 via its TPR domain.40 AtCyp38 is involved in accumulation of PSII supercomplex.41 Cyp20–3 of chloroplast stroma has been shown to assist the folding or assembly of SAT1 enzyme to form the hetero-oligomeric complex between cysteine synthase and OASTL, the cysteine synthase complex.42 Experiments with isolated mitochondria from Saccharomyces cerevisiae and Neurospora crassa indicated that the PPIase activity accelerates the refolding of dihydrofolate reductase that was imported into the mitochondrial matrix.43 Additionally, in yeast, cyclophilin Cpr3 deletion mutants exhibited slower rates of refolding.44 Slower growth rates were also reported for yeast strains lacking FKBP12 and the cyclophilin 40 homolog, Cpr7.45 As cis-trans isomerisation of peptide bonds takes place spontaneously, and at a much slower pace, it is possible that cyps are not absolutely required under normal conditions. Rather, they might be generally required for selective prolyl peptide bond isomerisation in processes where fast changes in protein-protein interactions or protein phosphorylation/dephosphorylation take place.46
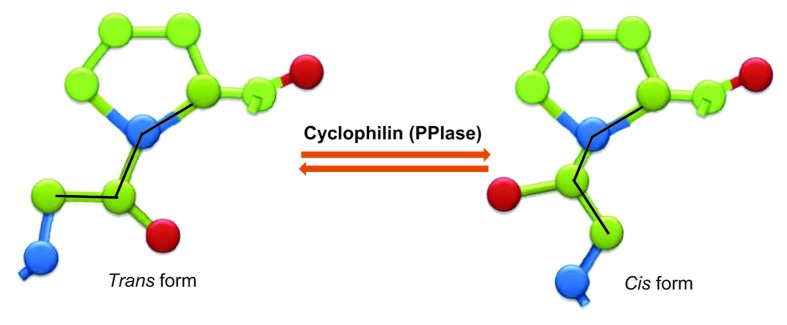
Figure 1. A schematic illustration of the trans and cis isomers of the peptidyl prolyl bond. The interconversion between the two forms is catalyzed by cyclophilins and other peptidyl-prolyl isomerases (PPIases).
Regardless of their origin, the structural conservation of cyclophilins throughout evolution and the PPIase activity of all members, underlines the importance of this enzymatic reaction. Unlike chaperones, PPIases are classical enzymes. They do not require energy (ATP) and in their enzymatic properties follow the simple Michaelis-Menten kinetics. Studies have shown that conformational folding steps and prolyl isomerisation are mutually interdependent.47 The close relationship between structure formation and prolyl isomerisation is a key feature of these slow folding steps and is of central importance for understanding the role of PPIases in these processes.48 Cyclophilins not only catalyze the rate-limiting steps in the folding pathway but also participate in the folding process as chaperone.49 Chaperone like activity has been associated with several PPIases.50 Some chloroplast immunophilins have also been postulated to comprise a target of the thioredoxin system: as cysteine-containing protein foldases, they may provide a link between protein tertiary structure formation and chloroplast redox state.
Cyclophilins (PPIases) in Plants
Plant cyclophilins were first identified in 1990 with the isolation of cyclophilin cDNA sequences from tomato (Lycopersicon esculentum), maize (Zea mays) and oilseed rape Brassica napus.33 Photosynthetic organisms harbour a significantly large number of cyclophilins e.g., 29 in Arabidopsis, 27 in rice and 26 in Chlamydomonas indicating toward some function specific to plants. The largest cyclophilin family identified in any organism to date has been reported from a higher plant, Arabidopsis. Usually, a large number of cyclophilins are targeted to thylakoid lumen e.g., 5 in Arabidopsis, largest immunophilin family in any cellular organelle. The first direct role for any immunophilin in a photosynthetic process was reported for TLP40, a multifunctional enzyme involved both in chloroplast biogenesis and intracellular signaling. TLP40 is also so far the best-characterized plant cyclophilin, a multidomain spinach 40 kD thylakoid lumenal protein that contains a poorly conserved C-terminal cyclophilin domain, a predicted N-terminal Leu zipper and a phosphatase-binding domain. TLP40 plays a key role in turnover of the D1 photosystem II protein by regulating its dephosphorylation.51 Several Arabidopsis cyclophilin proteins have been functionally characterized. AtCyp38, Arabidopsis homolog of the extensively characterized spinach TLP40, plays a critical role in the assembly and maintenance of photosystem II (PSII) supercomplexes (SCs) in Arabidopsis.41 Mutant plants with the AtCyp38 gene interrupted by T-DNA insertion showed stunted growth and were hypersensitive to high light. Leaf chlorophyll fluorescence analysis and thylakoid membrane composition indicated that cyp38 mutant plants had defects in PSII SCs.41 The Arabidopsis cyclophilin Cyp20–3 (also known as “ROC4”) is located in the stroma (soluble phase) of chloroplasts and it was found to be hypersensitive to oxidative stress conditions created by high light levels, rose bengal, high salt levels and osmotic shock. Cyp20–3 is shown to link light and stress to serine acetyl transferase (SAT1) activity and cysteine biosynthesis. Consequently, Cyp20–3 links photosynthetic electron transport and redox regulation to the folding of SAT1, thereby enabling the cysteine-based thiol biosynthesis pathway to adjust to light and stress conditions.52 Recent mass spectroscopy studies identified peptidyl-prolyl cis-trans isomerase (PPIases) as potential molecular chaperones functioning in the phloem translocation stream.53 Castor bean phloem cyclophilin, RcCyp1 that has high peptidyl-prolyl cis–trans isomerase activity and it plays a role in the refolding of non-cell-autonomous proteins after their entry into the phloem translocation stream. RcCyp1 belongs to the cytosolic cyclophilin family54 and it functions within the context of the phloem translocation stream.55 Recently, a gravitropic tomato mutant (diageotropica) with a defect in auxin signaling for plant growth, development and stress response was found to be mutated in a cytosolic cyclophilin, LeCyp1.56 Peptidyl prolyl cis-trans isomerase (PPIase) activity was also detected in the cytosol, mitochondria and chloroplast of pea plants.35 Wheat Cyp-45 was specifically induced during combined stress treatment, suggesting that cyclophilins may be the potential candidates to be exploited in making various strategies for engineering stress tolerance in plants.57 AtCyp59 and its orthologs from different organisms belong to a family of modular proteins consisting of a peptidyl-prolyl cis-trans isomerase (PPIase) domain, followed by an RNA recognition motif (RRM), and a C-terminal domain enriched in charged amino acids. AtCyp59 interacts with the C-terminal domain (CTD) of the largest subunit of RNA polymerase II. Ectopic expression of the tagged protein in Arabidopsis cell suspension resulted in highly reduced growth that is most probably due to reduced phosphorylation of the CTD. It suggests a possible function of AtCyp59 in activities connecting transcription and pre-mRNA processing.16 Cyp71 regulates plant morphogenesis and serves as a histone remodelling factor involved in chromatin based silencing. Thus, indicating that cyclophilin like foldases play a fundamental role in the chromatin regulating processes that influence the expression of regulatory genes.58 Proteomic, enzymatic and mutant analyses revealed that peptidyl-prolyl isomerase (PPIase) activity in the chloroplast thylakoid lumen of Arabidopsis is determined by two immunophilins: AtCYP20–2 and AtFKBP13. Profound redox-dependence of PPIase activity implies oxidative activation of protein folding catalysis under oxidative stress and photosynthetic oxygen production in the thylakoid lumen of plant chloroplasts.59 The data from different organisms leads to the conclusion that cyclophilins besides playing an essential role in protein folding may perform specific functions through interactions with unique sets of restricted partner proteins.
Genome Wide Analysis of Cyclophilin Genes in Higher Plants
Cyclophilins are present as large multigene families in plants. Few cyclophilins and FKBPs members have been found to be produced specifically in the green tissues and induced by light when etiolated seedlings were illuminated. These immunophilin proteins were shown to be chloroplast residents.32 Comparative genomic tools have established that the divergence of red and green algae led to a steep increase in the number and diversity of immunophilin isoforms in photosynthetic organisms.15 Remarkably, a total of 52 putative immunophilin isoforms have been reported in Chlamydomonas reinhardtii genome with a high number of distinct orthologs of Arabidopsis immunophilins among them.60 Genome wide analysis of cyclophilin gene members has been done in few plant species where complete genome sequence is available. It has been attempted in Arabidopsis61 and rice.62 In Arabidopsis, 29 different cyclophilin gene members have been reported which yield 39 protein products. However, rice Cyp gene family has 27 genes which encode 46 protein products suggesting a higher incidence of alternative splicing. In our laboratory, few new members from both Arabidopsis and rice genomes have been predicted using bioinformatic analysis (data unpublished). In silico analysis of cellular localization of these proteins indicates toward their distribution in diverse cellular organelles thereby suggesting their specific and diverse cellular functions. However, majority of the proteins encoded by these genes were found to be cytosolic in both Arabidopsis and rice. Though, several Arabidopsis cyclophilin members have been functionally characterized as discussed earlier, only one member has been functionally characterized in rice.63,64 Other immunophilin protein family, FKBPs have also been reported in Arabidopsis, wheat and rice. With the availability of complete sequences of more plant genera, concerted efforts need to be done to gain in-depth understanding of functional relevance of presence of such large cyclophilin gene families in plants.
“Loss of function” Studies for Immunophilin Genes
Although cyclophilin genes show high level of conservation across genera in both prokaryotes and eukaryotes, few are indispensible for growth and development under normal conditions. An octuplet yeast mutant lacking all 8 cyclophilins was viable, contrary to the expectation that cyclophilins are indispensable. However, in yeast, absence of cytosolic cyclophilin members has been found to be concomitant with enhanced heat sensitivity.65 Higher organisms are more seriously affected by cyclophilin gene deletion. “Loss of function” mutants have been studied widely in PPIase proteins particularly in FKBPs. FKBP12 mice mutants show early embryonic death due to cardiac defects. FKBP42 and FKBP70 show severe morphogenic defects in Arabidopsis. PAS1 (AtFKBP70) leads to tumor growth in plants indicating its role in cell division and cell differentiation.66 The Arabidopsis FKBP42 “loss of function” mutant twisted dwarf 1 (twd1) displays a drastic reduction of cell elongation combined with disoriented growth behavior of all plant organs.67 In contrast, only few mutants have been reported for plant cyclophilin genes. In Arabidopsis, AtCyp40 function has been demonstrated in vivo using a reverse genetics approach; its primary structure is characterized by three C-terminal tetratricopeptide repeats and a conserved cyclophilin domain.68 Loss of function mutants of AtCyp40, show defects in the transition from the juvenile to adult stages of vegetative development as well as in inflorescence morphology.23 The roc1 and roc1-D loss of function mutants in Arabidopsis show altered patterns of phosphorylation of the transcription factor BES1, a known point of control of sensitivity to brassinosteroids.69 Atcyp20–3 mutants show enhanced sensitivity to oxidative stressors.52
Cyclophilins: Stress Dependent Regulation and Genetic Manipulation
Cyclophilin expression has been shown to be induced by both biotic and abiotic stresses including HgCl2, viral infection, ethephon (an ethylene releaser), salicylic acid, salt stress, heat and cold shock,70 light,32,54 drought,71 wounding, fungal infection, abscisic acid and methyl jasmonate.70 AtCyp5 is localized in ER, expressed mainly in young stems especially in the apical region and weakly in leaves and roots and it is induced by cold stress and salt stress, but not by heat stress.72 Bean cyclophilin expression has been reported to be induced in response to salicylic acid, ethephon and might play a role in defense mechanisms.73 The expression of ThCyp1 is highly induced by salt, abscissic acid, H2O2 and heat shock.74 Arabidopsis cyclophilin gene, AtCyp20-3 expression is regulated by high intensity light.61 PCypB, a chloroplast localized cyclophilin from fava bean is induced in response to heat shock.32 Transcript of GjCyp-1 is induced by plant hormones such as gibberellic acid (GA3), indoleacetic acid (IAA) and zeatin (ZA). Constitutive overexpression of GjCyp-1 appeared to be beneficial to seed germination.75 Rice OsCyp2 gene has been shown to be induced by multiple abiotic stresses.63,64
Few studies have now established a direct co-relation of cyclophilin overexpression with that of stress protection. Table 1 presents some recent examples where overexpression of cyclophilin gene in diverse organisms has resulted in enhanced tolerance toward various stresses. Overexpression of CypD in Xenopus has been shown to provide tolerance toward oxidative stress. Point mutations in CyPD that eliminated PPIase activity did not have similar protective effects, clearly demonstrating that CyPD requires functional PPIase activity.17 Overexpression of ThCyp1, a single domain, nuclear localized cyclophilin isolated from Thullengiella halophila in BY2 tobacco cells and yeast provides multiple abiotic stress tolerance.74 GjCyp-1 from the red alga (Griffithsia japonica), was overexpressed under CaMV35S promoter in Nicotiana tabacum. The ratio of emergence of cotyledon from seeds overexpressing GjCyp-1 was almost three times higher than that of transgenic seeds carrying only the vector. In addition, it was found that seedlings overexpressing GjCyp-1 showed thermotolerance and transgenic plants were short statured with altered root system.75 Overexpression of rice cyclophilin gene, OsCyp2, has been shown to enhance multiple stress tolerance in E. coli and S. cerevisiae.63 Rice OsCyp2 gene has also been shown to impart salinity tolerance to rice plants.64 Overexpression studies with GhCyp1 performed in tobacco showed its role in salinity stress protection as well as enhanced tolerance to Psuedomonas syringae pv tabaci infection.76 Overexpression of CcCyp1, isolated from Cajanus cajan, in pigeon pea lead to a marked increase in its ability to withstand abiotic stresses.72 Yeast cyclophilin gene, Cpr1, provides stress tolerance to a range of stressors including cadmium, cobalt, copper, hydrogen peroxide, tert-butyl hydroperoxide (t-BOOH) and sodium dodecyl sulfate (SDS) when overexpressed in Saccharomyces cerevisiae.77
Table 1. A few reports where overexpression of cyclophilin genes isolated from diverse genera has been implicated in enhanced stress tolerance.
| Cyclophilin Gene | Source | Host for Overexpression | Tolerance Conferred | Reference |
|---|---|---|---|---|
|
Cyp D |
Rat |
Xenopus |
Oxidative stress |
17 |
|
OsCyp2 |
Oryza sativa |
E. coli, Yeast |
Multiple abiotic stresses |
63 |
|
OsCyp2 |
Oryza sativa |
Oryza sativa |
Salinity tolerance |
64 |
|
ThCyp1 |
Thellungiella halophila |
BY2 cells Tobacco, Yeast |
Multiple abiotic stresses |
74 |
|
GjCyp1 |
Griffithsia japonica |
Tobacco |
Heat stress |
75 |
| GhCyp1 |
Gossypium hirsutum |
Tobacco |
Salinity, Psuedomonas syringae pv tabaci |
76 |
|
CcCYP |
Cajanus cajan |
Arabidopsis |
Multiple abiotic stresses |
77 |
| Cpr1 | Saccharomyces cerevisiae | Saccharomyces cerevisiae | Cadmium, cobalt, copper, hydrogen peroxide, tert-butyl hydroperoxide and sodium dodecyl sulfate | 78 |
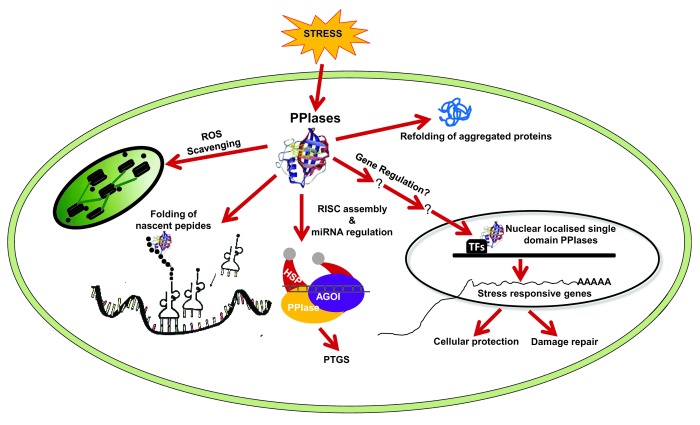
However, the mechanism by which cyclophilins participates in stress protection is not clear yet. Because cyclophilins possess the PPIase activity that catalyzes the rate limiting step in protein folding, one possible function of cyclophilins may be to act as chaperone-like proteins to facilitate the folding of stress related proteins or to protect these proteins from proteolytic degradation or aggregation under stress conditions as has been predicted for some other cyp member, e.g., AtCyp20–3.52 A hypothetical model proposing possible mechanisms through which cyclophilins exert their stress protective properties, based on studies conducted on cyclophilin genes from diverse genera, has been illustrated in Figure 2. To further understand the role of cyclophilins in stress responsive signaling, experiments need to be undertaken to study loss of function mutants of cyclophilins as well as their interaction with other protein components. Such studies will provide a greater insight into the mechanism of stress tolerance involving cyclophilins in plants.
Figure 2. A hypothetical model for cyclophilin mediated stress protective mechanism operative at cellular level. Cyclophilin proteins possibly employ diverse strategies to prevent stress-induced injury via its gene regulatory and repair pathways.
Conclusions
Cyclophilins, a subfamily of peptidyl-prolyl isomerases, highly conserved ubiquitous protein folding catalysts were discovered as ligands for immunosuppressive drugs. Presence of such proteins in plant system that too as large multigene families was intriguing as well as perplexing. Probably this apparent anomaly served as an impetus for several studies in plant cyclophilins worldwide. These studies established that cyclophilins entail a range of novel functions not yet fully understood. Since protein foldases have directly been linked to stress response, involvement of cyclophilins in stress response was investigated by several workers. These studies provided a basis for a positive correlation between this protein family and stress protection. But the underlying mechanism which is targeted by cyclophilin proteins to bring about stress protection has not been worked out yet. Besides stress protection, a number of these plant proteins have been found to play diverse but specific roles. This presents the most basic question about their role in stress response pertaining to their specificity. Do they control specific pathways or they perform a more generalized function in maintaining the correct conformation of newly formed proteins under stress condition? Additionally, does the presence of multiple members provide redundancy to compensate for loss of any particular member? Since many of the smaller stress induced proteins of this family have also been shown to be localized in the nucleus, there is a possibility that they might also act as regulatory proteins. However, studies on the precise role of these proteins are still in infancy stage and these issues can only be addressed through integrated functional genomics, proteomics and metabolomics approaches. The search for their interacting partners under stress conditions and large scale gene disruption studies can aid in understanding their physiological roles and potential function in stress alleviation.
Acknowledgments
Authors would like to thank Department of Biotechnology, Department of Science and Technology (Government of India) and Jawaharlal Nehru University (through Capacity building, CAS and PURSE) for supporting research work in the laboratory. Award of research fellowship from Council of Scientific and Industrial Research (CSIR) to SR is also thankfully acknowledged.
Glossary
Abbreviations:
- Cyp
cyclophilin
- FKBP
FK506 binding protein
- CsA
cyclosporin A
- FK506
Tacrolimus
- PPIase
Peptidyl prolyl cis-trans isomerase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22734
References
- 1.Galat A. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity--targets--functions. Curr Top Med Chem. 2003;3:1315–47. doi: 10.2174/1568026033451862. [DOI] [PubMed] [Google Scholar]
- 2.Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–60. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 3.Hennig L, Christner C, Kipping M, Schelbert B, Rücknagel KP, Grabley S, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37:5953–60. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 4.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–7. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 5.Fischer G, Bang H, Mech C. [Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides] Biomed Biochim Acta. 1984;43:1101–11. [PubMed] [Google Scholar]
- 6.McCaffrey PG, Perrino BA, Soderling TR, Rao A. NF-ATp, a T lymphocyte DNA-binding protein that is a target for calcineurin and immunosuppressive drugs. J Biol Chem. 1993;268:3747–52. [PubMed] [Google Scholar]
- 7.Barik S. Immunophilins: for the love of proteins. Cell Mol Life Sci. 2006;63:2889–900. doi: 10.1007/s00018-006-6215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams B, Musiyenko A, Kumar R, Barik S. A novel class of dual-family immunophilins. J Biol Chem. 2005;280:24308–14. doi: 10.1074/jbc.M500990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi N, Hayano T, Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–5. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 10.Montague JW, Gaido ML, Frye C, Cidlowski JA. A calcium-dependent nuclease from apoptotic rat thymocytes is homologous with cyclophilin. Recombinant cyclophilins A, B, and C have nuclease activity. J Biol Chem. 1994;269:18877–80. [PubMed] [Google Scholar]
- 11.Arevalo-Rodriguez M, Wu X, Hanes SD, Heitman J. Prolyl isomerases in yeast. Front Biosci. 2004;9:2420–46. doi: 10.2741/1405. [DOI] [PubMed] [Google Scholar]
- 12.Galat A. Variations of sequences and amino acid compositions of proteins that sustain their biological functions: An analysis of the cyclophilin family of proteins. Arch Biochem Biophys. 1999;371:149–62. doi: 10.1006/abbi.1999.1434. [DOI] [PubMed] [Google Scholar]
- 13.Page AP, MacNiven K, Hengartner MO. Cloning and biochemical characterization of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans. Biochem J. 1996;317:179–85. doi: 10.1042/bj3170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gullerova M, Barta A, Lorkovic ZJ. AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA. 2006;12:631–43. doi: 10.1261/rna.2226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira PA, Nakayama TA, Pak WL, Travis GH. Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature. 1996;383:637–40. doi: 10.1038/383637a0. [DOI] [PubMed] [Google Scholar]
- 16.Leverson JD, Ness SA. Point mutations in v-Myb disrupt a cyclophilin-catalyzed negative regulatory mechanism. Mol Cell. 1998;1:203–11. doi: 10.1016/S1097-2765(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 17.Lin DT, Lechleiter JD. Mitochondrial targeted cyclophilin D protects cells from cell death by peptidyl prolyl isomerization. J Biol Chem. 2002;277:31134–41. doi: 10.1074/jbc.M112035200. [DOI] [PubMed] [Google Scholar]
- 18.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA. 2002;99:1899–904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzywicka A, Beisson J, Keller AM, Cohen J, Jerka-Dziadosz M, Klotz C. KIN241: a gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol Microbiol. 2001;42:257–67. doi: 10.1046/j.1365-2958.2001.02634.x. [DOI] [PubMed] [Google Scholar]
- 20.Hong F, Lee J, Song JW, Lee SJ, Ahn H, Cho JJ, et al. Cyclosporin A blocks muscle differentiation by inducing oxidative stress and inhibiting the peptidyl-prolyl-cis-trans isomerase activity of cyclophilin A: cyclophilin A protects myoblasts from cyclosporin A-induced cytotoxicity. FASEB J. 2002;16:1633–5. doi: 10.1096/fj.02-0060fje. [DOI] [PubMed] [Google Scholar]
- 21.Wiederrecht G, Hung S, Chan HK, Marcy A, Martin M, Calaycay J, et al. Characterization of high molecular weight FK-506 binding activities reveals a novel FK-506-binding protein as well as a protein complex. J Biol Chem. 1992;267:21753–60. [PubMed] [Google Scholar]
- 22.Horowitz DS, Lee EJ, Mabon SA, Misteli T. A cyclophilin functions in pre-mRNA splicing. EMBO J. 2002;21:470–80. doi: 10.1093/emboj/21.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MR, Willmann MR, Wu G, Berardini TZ, Möller B, Weijers D, et al. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:5424–9. doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lki T, Yoshikawa M, Meshi T, Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2012;31:267–78. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bose S, Weikl T, Bügl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–7. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 26.Goel M, Garcia R, Estacion M, Schilling WP. Regulation of Drosophila TRPL channels by immunophilin FKBP59. J Biol Chem. 2001;276:38762–73. doi: 10.1074/jbc.M104125200. [DOI] [PubMed] [Google Scholar]
- 27.Dornan J, Taylor P, Walkinshaw MD. Structures of immunophilins and their ligand complexes. Curr Top Med Chem. 2003;3:1392–409. doi: 10.2174/1568026033451899. [DOI] [PubMed] [Google Scholar]
- 28.Kallen J, Spitzfaden C, Zurini MGM, Wider G, Widmer H, Wüthrich K, et al. Structure of human cyclophilin and its binding site for cyclosporin A determined by X-ray crystallography and NMR spectroscopy. Nature. 1991;353:276–9. doi: 10.1038/353276a0. [DOI] [PubMed] [Google Scholar]
- 29.Mikol V, Kallen J, Walkinshaw MD. The X-ray structure of (MeBm2t)1-cyclosporin complexed with cyclophilin A provides an explanation for its anomalously high immunosuppressive activity. Protein Eng. 1994;7:597–603. doi: 10.1093/protein/7.5.597. [DOI] [PubMed] [Google Scholar]
- 30.Ke H, Mayrose D, Cao W. Crystal structure of cyclophilin A complexed with substrate Ala-Pro suggests a solvent-assisted mechanism of cis-trans isomerization. Proc Natl Acad Sci USA. 1993;90:3324–8. doi: 10.1073/pnas.90.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure. 2001;9:431–8. doi: 10.1016/S0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- 32.Luan S, Albers MW, Schreiber SL. Light-regulated, tissue-specific immunophilins in a higher plant. Proc Natl Acad Sci USA. 1994;91:984–8. doi: 10.1073/pnas.91.3.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasser CS, Gunning DA, Budelier KA, Brown SM. Structure and expression of cytosolic cyclophilin/peptidyl-prolyl cis-trans isomerase of higher plants and production of active tomato cyclophilin in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:9519–23. doi: 10.1073/pnas.87.24.9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker KW, Gilbert HF. Protein disulfide isomerase. In AL Fink, Y Goto, eds, Molecular Chaperones in the Life Cycle of Proteins: Structure, Function and Mode of Action. Marcel Dekker, New York 1998; pp 331-60. [Google Scholar]
- 35.Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK. Plant organelles contain distinct peptidylprolyl cis,trans-isomerases. J Biol Chem. 1992;267:21293–6. [PubMed] [Google Scholar]
- 36.Stewart DE, Sarkar A, Wampler JE. Occurrence and role of cis peptide bonds in protein structures. J Mol Biol. 1990;214:253–60. doi: 10.1016/0022-2836(90)90159-J. [DOI] [PubMed] [Google Scholar]
- 37.MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-H. [DOI] [PubMed] [Google Scholar]
- 38.Göthel SF, Herrler M, Marahiel MA. Peptidyl-prolyl cis-trans isomerase of Bacillus subtilis: identification of residues involved in cyclosporin A affinity and catalytic efficiency. Biochemistry. 1996;35:3636–40. doi: 10.1021/bi9520803. [DOI] [PubMed] [Google Scholar]
- 39.Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–20. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann K, Handschumacher RE. Cyclophilin-40: evidence for a dimeric complex with hsp90. Biochem J. 1995;307:5–8. doi: 10.1042/bj3070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu A, He Z, Lima A, Buchanan BB, Luan S. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15947–52. doi: 10.1073/pnas.0707851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leustek T, Martin MN, Bick JA, Davies JP. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:141–65. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- 43.Matouschek A, Rospert S, Schmid K, Glick BS, Schatz G. Cyclophilin catalyzes protein folding in yeast mitochondria. Proc Natl Acad Sci USA. 1995;92:6319–23. doi: 10.1073/pnas.92.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rassow J, Mohrs K, Koidl S, Barthelmess IB, Pfanner N, Tropschug M. Cyclophilin 20 is involved in mitochondrial protein folding in cooperation with molecular chaperones Hsp70 and Hsp60. Mol Cell Biol. 1995;15:2654–62. doi: 10.1128/mcb.15.5.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13093–8. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gullerova M, Barta A, Lorkovic ZJ. Rct1, a nuclear RNA recognition motif-containing cyclophilin, regulates phosphorylation of the RNA polymerase II C-terminal domain. Mol Cell Biol. 2007;27:3601–11. doi: 10.1128/MCB.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayr LM, Odefey C, Schutkowski M, Schmid FX. Kinetic analysis of the unfolding and refolding of ribonuclease T1 by a stopped-flow double-mixing technique. Biochemistry. 1996;35:5550–61. doi: 10.1021/bi953035y. [DOI] [PubMed] [Google Scholar]
- 48.Kurek I, Aviezer K, Erel N, Herman E, Breiman A. The wheat peptidyl prolyl cis-trans-isomerase FKBP77 is heat induced and developmentally regulated. Plant Physiol. 1999;119:693–704. doi: 10.1104/pp.119.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freskgård PO, Bergenhem N, Jonsson BH, Svensson M, Carlsson U. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science. 1992;258:466–8. doi: 10.1126/science.1357751. [DOI] [PubMed] [Google Scholar]
- 50.Kruse M, Brunke M, Escher A, Szalay AA, Tropschug M, Zimmermann R. Enzyme assembly after de novo synthesis in rabbit reticulocyte lysate involves molecular chaperones and immunophilins. J Biol Chem. 1995;270:2588–94. doi: 10.1074/jbc.270.6.2588. [DOI] [PubMed] [Google Scholar]
- 51.Fulgosi H, Vener AV, Altschmied L, Herrmann RG, Andersson B. A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 1998;17:1577–87. doi: 10.1093/emboj/17.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominguez-Solis JR, He Z, Lima A, Ting J, Buchanan BB, Luan S. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc Natl Acad Sci USA. 2008;105:16386–91. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giavalisco P, Kapitza K, Kolasa A, Buhtz A, Kehr J. Towards the proteome of Brassica napus phloem sap. Proteomics. 2006;6:896–909. doi: 10.1002/pmic.200500155. [DOI] [PubMed] [Google Scholar]
- 54.Chou IT, Gasser CS. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol Biol. 1997;35:873–92. doi: 10.1023/A:1005930024796. [DOI] [PubMed] [Google Scholar]
- 55.Gottschalk M, Dolgener E, Xoconostle-Cázares B, Lucas WJ, Komor E, Schobert C. Ricinus communis cyclophilin: functional characterisation of a sieve tube protein involved in protein folding. Planta. 2008;228:687–700. doi: 10.1007/s00425-008-0771-8. [DOI] [PubMed] [Google Scholar]
- 56.Oh KC, Ivanchenko MG, White TJ, Lomax TL. The diageotropica gene of tomato encodes a cyclophilin: a novel player in auxin signaling. Planta. 2006;224:133–44. doi: 10.1007/s00425-005-0202-z. [DOI] [PubMed] [Google Scholar]
- 57.Sharma AD, Kaur P. Combined effect of drought stress and heat shock on cyclophilin protein expression in Triticum aestivum. Gen Appl Plant Physiol. 2009;35:88–92. [Google Scholar]
- 58.Li H, He Z, Lu G, Lee SC, Alonso J, Ecker JR, et al. A WD40 domain cyclophilin interacts with histone H3 and functions in gene repression and organogenesis in Arabidopsis. Plant Cell. 2007;19:2403–16. doi: 10.1105/tpc.107.053579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shapiguzov A, Edvardsson A, Vener AV. Profound redox sensitivity of peptidyl-prolyl isomerase activity in Arabidopsis thylakoid lumen. FEBS Lett. 2006;580:3671–6. doi: 10.1016/j.febslet.2006.05.054. [DOI] [PubMed] [Google Scholar]
- 60.Vallon O. Chlamydomonas immunophilins and parvulins: survey and critical assessment of gene models. Eukaryot Cell. 2005;4:230–41. doi: 10.1128/EC.4.2.230-241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romano PGN, Edvardsson A, Ruban AV, Andersson B, Vener AV, Gray JE, et al. Arabidopsis AtCYP20-2 is a light-regulated cyclophilin-type peptidyl-prolyl cis-trans isomerase associated with the photosynthetic membranes. Plant Physiol. 2004;134:1244–7. doi: 10.1104/pp.104.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn JC, Kim DW, You YN, Seok MS, Park JM, Hwang H, et al. Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress. BMC Plant Biol. 2010;10:253. doi: 10.1186/1471-2229-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumari S, Singh P, Singla-Pareek SL, Pareek A. Heterologous expression of a salinity and developmentally regulated rice cyclophilin gene (OsCyp2) in E. coli and S. cerevisiae confers tolerance towards multiple abiotic stresses. Mol Biotechnol. 2009;42:195–204. doi: 10.1007/s12033-009-9153-0. [DOI] [PubMed] [Google Scholar]
- 64.Ruan SL, Ma HS, Wang SH, Fu YP, Xin Y, Liu WZ, et al. Proteomic identification of OsCYP2, a rice cyclophilin that confers salt tolerance in rice (Oryza sativa L.) seedlings when overexpressed. BMC Plant Biol. 2011;11:34. doi: 10.1186/1471-2229-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sykes K, Gething MJ, Sambrook J. Proline isomerases function during heat shock. Proc Natl Acad Sci USA. 1993;90:5853–7. doi: 10.1073/pnas.90.12.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faure JD, Vittorioso P, Santoni V, Fraisier V, Prinsen E, Barlier I, et al. The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development. 1998;125:909–18. doi: 10.1242/dev.125.5.909. [DOI] [PubMed] [Google Scholar]
- 67.Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell. 2003;14:4238–49. doi: 10.1091/mbc.E02-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berardini TZ, Bollman K, Sun H, Poethig RS. Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science. 2001;291:2405–7. doi: 10.1126/science.1057144. [DOI] [PubMed] [Google Scholar]
- 69.Trupkin SA, Mora-García S, Casal JJ. The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 2012;71:712–23. doi: 10.1111/j.1365-313X.2012.05013.x. [DOI] [PubMed] [Google Scholar]
- 70.Godoy AV, Lazzaro AS, Casalongue CA, Segundo BS. Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Sci. 2000;152:123–34. doi: 10.1016/S0168-9452(99)00211-3. [DOI] [Google Scholar]
- 71.Sharma AD, Singh P. Effect of water stress on expression of a 20 kD cyclophilin-like protein in drought susceptible and tolerant cultivars of Sorghum. Journal of Plant Biotechnology. 2003;12:77–80. [Google Scholar]
- 72.Saito T, Niwa Y, Ashida H, Tanaka K, Kawamukai M, Matsuda H, et al. Expression of a gene for cyclophilin which contains an amino-terminal endoplasmic reticulum-targeting signal. Plant Cell Physiol. 1999;40:77–87. doi: 10.1093/oxfordjournals.pcp.a029477. [DOI] [PubMed] [Google Scholar]
- 73.Marivet J, Frendo P, Burkard G. Effects of abiotic stresses on cyclophilin gene-expression in maize and bean and sequence analysis of cyclophilin cDNA. Plant Sci. 1992;84:171–8. doi: 10.1016/0168-9452(92)90131-5. [DOI] [Google Scholar]
- 74.Chen AP, Wang GL, Qu ZL, Lu CX, Liu N, Wang F, et al. Ectopic expression of ThCYP1, a stress-responsive cyclophilin gene from Thellungiella halophila, confers salt tolerance in fission yeast and tobacco cells. Plant Cell Rep. 2007;26:237–45. doi: 10.1007/s00299-006-0238-y. [DOI] [PubMed] [Google Scholar]
- 75.Cho EK, Kim M. A red algal cyclophilin has an effect on development and growth in Nicotiana tabacum. Plant Physiol Biochem. 2008;46:868–74. doi: 10.1016/j.plaphy.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 76.Zhu C, Wang Y, Li Y, Bhatti KH, Tian Y, Wu J. Overexpression of a cotton cyclophilin gene (GhCyp1) in transgenic tobacco plants confers dual tolerance to salt stress and Pseudomonas syringae pv. tabaci infection. Plant Physiol Biochem. 2011;49:1264–71. doi: 10.1016/j.plaphy.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Sekhar K, Priyanka B, Reddy VD, Rao KV. Isolation and characterization of a pigeonpea cyclophilin (CcCYP) gene, and its over-expression in Arabidopsis confers multiple abiotic stress tolerance. Plant Cell Environ. 2010;33:1324–38. doi: 10.1111/j.1365-3040.2010.02151.x. [DOI] [PubMed] [Google Scholar]
- 78.Kim IS, Kim HY, Shin SY, Kim YS, Lee DH, Park KM, et al. A cyclophilin A CPR1 overexpression enhances stress acquisition in Saccharomyces cerevisiae. Mol Cells. 2010;29:567–74. doi: 10.1007/s10059-010-0071-6. [DOI] [PubMed] [Google Scholar]