Abstract
Orchidaceae, one of the largest angiosperm families, has significant commercial value. Isolation of genes involved in orchid floral development and morphogenesis, scent production, and colouration will advance knowledge of orchid flower formation and facilitate breeding new varieties to increase the commercial value. With high-throughput virus-induced gene silencing (VIGS), this study identified five transcription factors involved in various aspects of flower morphogenesis in the orchid Phalaenopsis equestris. These genes are PeMADS1, PeMADS7, PeHB, PebHLH, and PeZIP. Silencing PeMADS1 and PebHLH resulted in reduced flower size together with a pelaloid column containing petal-like epidermal cells and alterations of epidermal cell arrangement in lip lateral lobes, respectively. Silencing PeMADS7, PeHB, and PeZIP alone resulted in abortion of the first three fully developed flower buds of an inflorescence, which indicates the roles of the genes in late flower development. Furthermore, double silencing PeMADS1 and PeMADS6, C- and B-class MADS-box genes, respectively, produced a combinatorial phenotype with two genes cloned in separate vectors. Both PeMADS1 and PeMADS6 are required to ensure the normal development of the lip and column as well as the cuticle formation on the floral epidermal cell surface. Thus, VIGS allows for unravelling the interaction between two classes of MADS transcription factors for dictating orchid floral morphogenesis.
Key words: Cymbidium mosaic virus, PebHLH, PeMADS1, PeMADS6, Phalaenopsis, virus-induced gene silencing.
Introduction
The Orchidaceae, well known for fascinating flowers, is one of the largest and most diverse families of flowering plants. The family contains more than 25000 species that occupy wide ranges of ecological habitats and exhibit highly specialized morphological, structural, and physiological characteristics (Dressler, 2005). The orchid flowers of the species usually have two lateral petals with a third larger petal modified into a lip to attract and in some cases trap potential pollinators. The column contains the male stamens attached to the female pistil in the centre. Because of their unique flowers, orchid reproductive biology is of special interest in terms of flower colour, flower morphological features, size and number of flowers, and floral fragrance (Hsiao et al., 2011).
The orchid industry has become an international business, comprising approximately 8% of the world’s floriculture trade (Chugh et al., 2009). In particular, Phalaenopsis and Oncidium are the cash crops. The identification and analysis of orchid gene functions could help improve orchids in terms of flower shape, size, scent, and colouration. Such investigation helps understand orchid biology and enhances the breeding of new varieties to increase the commercial value. These traits need to be studied to have a better understanding of the genetic factors and how morphological traits are determined in the orchid flower. However, the challenges are the lack of information on the genetic background of various cultivars, large genome size, long life cycle, and the inefficient transformation system of orchids. The study of functional genomics of orchid genes is difficult and inefficient.
Virus-induced gene silencing (VIGS) is a reverse genetics approach used for functional analysis of genes of plants, especially those with long life cycles and few genetic resources such as orchids. The technique has many advantages over other techniques used for functional analysis, such as cost-effectiveness with the use of expressed sequence tags (ESTs), good for orchid research because whole-genome sequences are not yet available. The analyses of floral bud ESTs in the OrchidBase (http://orchidbase.itps.ncku.edu.tw/est/home2012.aspx) have been initiated for Phalaenopsis aphrodite subsp. formosana, Phalaenopsis bellina, and Phalaenopsis equestris, which are usually used as parents for breeding (Hsiao et al., 2006; Tsai et al., 2006, 2013; Fu et al., 2011). A Cymbidium mosaic virus (CymMV)-based VIGS vector has been established (Lu et al., 2007) and was used to determine the effects of PeUFGT3 on anthocyanin biosynthesis (Chen et al., 2011), PeMADS5 and PeMADS6 on floral morphogenesis (Hsieh et al., 2013), and PhaTF15 on disease resistance in Phalaenopsis (Lu et al., 2012).
Several functional studies of floral transcription factors (TFs) have revealed their critical roles in a number of developmental processes in orchid plants. The MADS-box gene family encodes highly conserved TFs. The floral MADS-box genes of Arabidopsis have been divided into several major classes (A, B, C, D, and E) by phylogenetic analysis (Becker and Theissen, 2003); the function of ABCDE genes is partially conserved between Arabidopsis and orchid (Xu et al., 2006; Tsai et al., 2010; Xu et al., 2010). From the ABCDE model in Phalaenopsis, PeMADS1 is a C-class AGAMOUS-like MADS-box gene involved in the development of reproductive organs (Song et al., 2006; Chen et al., 2012), and PeMADS6 is a B-class GLOBOSA/PISTILLATA-like MADS-box gene ubiquitously expressed in the sepals, petals, lip, and column (Tsai et al., 2005). Silencing PeMADS6 confers leaf-like characteristics with greenish and discoloured areas in sepals, petals, and lip (Lu et al., 2007; Hsieh et al., 2013). PeMADS7 is a D-class gene that plays important roles in column and ovule development in Phalaenopsis (Chen et al., 2012). Except for MADS-box genes, little is known about the functions of floral TFs in orchids as compared with those involved in floral development in Arabidopsis.
To understand individual TFs that affect orchid floral morphogenesis and development, this study used high-throughput VIGS to silence the expression of 126 floral ESTs that encode TFs and examined their functions during floral morphogenesis in Phalaenopsis. The high-throughput VIGS process involves cloning libraries of short gene fragments, such as ESTs, directly into a VIGS vector, inoculating plants, and then identifying phenotypes of interest (Bernacki et al., 2010; Lu et al., 2012). Moreover, this work studied the cooperative action of different classes of MADS-box TFs by co-silencing with separate VIGS vectors. This process helped to unravel the functions of TFs involved in orchid floral morphogenesis.
Materials and methods
Plant materials
The Phalaenopsis plants purchased from Oxen Biotechnology (Tainan, Taiwan) were Doritaenopsis I-Hsin Sunrise Cinderella ‘OX1357’, Dtps. OX Red Shoe ‘OX1407’, and Phalaenopsis Sogo Yukidian ‘V3’. All plants were kept in a greenhouse with a controlled temperature of 27/22 °C (day/night). Plants were determined to be virus-free by reverse-transcription (RT)-PCR with two primer pairs, CymMV-CPF/CymMV-CPR and ORSV-CPF/ORSV-CPR (Supplementary Table S1, available at JXB online), for detecting the two prevalent orchid viruses, CymMV and ORSV, respectively, before VIGS experiments.
Construction of pCymMV-Gateway plasmids
The putative TFs of 152 ESTs were identified from 5593 orchid floral ESTs in the Arabidopsis thaliana TF database at the Arabidopsis Gene Regulatory Information Server (AGRIS) (http://arabidopsis.med.ohio-state.edu) (Tsai et al., 2006). Each EST was then cloned into pBluescript SK+ plasmid. Clones underwent PCR amplification with the primers for AttB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCTC TAGAACTAGTGGATCCCCCG-3′) and AttB2 (5′-GGGGACC ACTTTGTACAAGAAAGCTGGGTGCGAATTGG GTACCGGGCCCCCCC-3′). The length of EST sequences ranged from 0.5 to 3.3kb (Supplementary Fig. S1). The amplified PCR products were purified with use of polyethylene glycol/MgCl2 to remove primers. Purified PCR products were recombined into the pCymMV-Gateway vector by use of BP recombinase (Invitrogen, Carlsbad, CA, USA). As a result, only 126 ESTs were successfully cloned into the pCymMV-Gateway vector. After recombination into the pCymMV-Gateway vector, each construct was confirmed by PCR with the primer pairs for AttB1 and CymMV-5351 (5′-CTTCTGTACCATACACATAG-3′).
Agro-infiltration of plants
Agrobacterium tumefaciens containing the pCymMV-Gateway-EST was cultured overnight at 28 °C. After centrifugation, bacterial cell pellets were resuspended by adding 300 μl Murishige and Skoog medium containing 100 μM acetosyringone to OD600 = 1 and allowed to stand at room temperature for 0.5h without shaking before agro-infiltration. Then plants were infected following the standard protocol established previously (Hsieh et al., 2013). The suspensions were injected into the leaf right above the inflorescence emerging site by use of a 1-ml syringe with a needle. Plants with inflorescences containing eight nodes with one visible floral bud were inoculated (Supplementary Fig. S2D).
This work silenced 126 TF ESTs and one control, PeMADS6 (GenBank AY678299.1). For quantification of agro-infiltration to determine efficacy, the positive control plants were infiltrated with a pCymMV-Gateway vector inserted with a fragment of PeMADS6 cDNA, which showed morphological changes with greenish and discoloured areas in the sepal, petal and lip (Hsieh et al., 2013). Mock-treated plants were injected with an empty pCymMV-Gateway vector as a negative control to confirm that floral morphogenesis was not due to the viral infection. Each experiment involved five plants, and the silencing experiments for TFs with distinctive phenotypes on silencing were repeated at least twice in different cultivars. The TFs included PeMADS1 (GenBank AF234617.1), PeMADS7 (GenBank JN983500.1), PebHLH (OrchidBase 2.0 lcl|EFCPTF-001B04-02B1), PebZIP (OrchidBase 2.0 lcl|Unigene59946_Pe_fb), and PeHB (OrchidBase 2.0 lcl|AECP-22H12 AECP-22H12) (Fu et al., 2011; Tsai et al., 2013). The total number of inoculated plants and replicates from each treatment group are in Supplementary Tables S2 and S3.
Real-time quantitative RT-PCR
RNA was extracted from dissected floral organs and floral buds (8–12mm) by use of the RNeasy Plant Mini Kit (Qiagen) and treated with RNase-free DNase (Invitrogen) to remove residual DNA. RNA was used as a template for cDNA synthesis with reverse transcriptase and the SuperScript II kit (Invitrogen). For quantitative PCR, the cDNA template was mixed with 2X SYBR Green PCR master mix in an ABI Prism 7000 sequence detection system (Applied Biosystems). The primers used for quantification are given in Supplementary Table S1. Each pair of primers for the TFs PeMADS1, PeMADS6, and PebHLH was developed from specific regions of their nucleotide sequences for real time RT-PCR. For gene quantification, the first blooming flower was collected from three silenced plants that showed VIGS phenotypes. For the real-time RT-PCR reaction, each sample was analysed in triplicate. Reactions involved incubation at 50 °C for 2min, then 95 °C for 10min, and thermal cycling for 40 cycles at 95 °C for 15 s and 60 °C for 1min. After amplification, melting curve analysis was used to verify amplicon specificity and primer dimer formation. Every amplicon in this study yielded similar single peak melting curves, so only the specific products were amplified. Relative quantification followed the manufacturer’s instructions (Applied Biosystems). For controlling the integrity of RNA and normalizing target RNA copy numbers in mock-treated and gene-silenced flowers, the housekeeping gene PeActin (PACT4, AY134752) was amplified by real-time RT-PCR to generate a standard curve of Actin mRNA levels (Chen et al., 2005).
RT-PCR
RNA samples were treated with RQ1 DNase (Promega) to remove remnant DNA. Synthesizing the first-strand cDNA involved use of the Superscript II kit (Invitrogen). Gene-specific primer pairs were designed for PeMADS1, PeMADS6, PeMADS7, PebHLH, PebZIP, and PeHB. The PeActin gene of Phalaenopsis was used as an internal control (Supplementary Table S1). The PCR protocol was initial denaturation at 94 °C for 3min, then 30 cycles of amplification at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1min, and 72 °C for 3min. The amplified products were separated on 1% agarose gel, visualized and photographed.
Scanning electron microscopy
For traditional scanning electron microscopy (SEM), samples were fixed in formalin/acetic acid/alcohol. After being dehydrated in an alcohol/acetone series, samples were critical-point dried, sputter-coated with platinum, and observed under a scanning electron microscope (S-4200, Hitachi, Tokyo, Japan) with an accelerating voltage of 15kV. Photographs were taken with use of a Verichrome pan film (Kodak, Rochester, NY, USA).
For cryo-SEM, samples were dissected and loaded on stubs, frozen with liquid nitrogen slush, then transferred to a sample preparation chamber at –160 °C. After 5min, when the temperature rose to –130 °C, the samples were fractured. They were etched for 10min at –85 °C. After being coated at –130 °C, samples were transferred to the SEM chamber and observed at –160 °C with cryo-SEM (Quanta 200 SEM/Quorum Cryo System PP2000TR, FEI, Hillsboro, OR, USA) at 20kV (Ku et al., 2008).
Characterization of epidermal cell types
The types of superimposed micro-structuring on cell surfaces can be classified into wax film, epicuticular wax crystals, and cuticular folds (Prüm et al., 2012). The shape of the epidermis can be divided into seven types depending on the outline of the epidermis cells and their ratio of width to height. The aspect ratio (β = width/height) of cell types and their terminology is as follows: convex (β ≥ 3/1), hemispherical (β ≧ 2/1), cupola (β < 3/2), conical (β > 3/2), papilla (β < 3/2 and > 1/2), hair papilla (β < 1/3 and > 1/6), and hair (β < 1/7) (Koch et al., 2008).
Measurement of floral organ morphological changes in PeMADS1-, PeMADS6-, and double-silenced flowers
To quantify the phenotypic changes of PeMADS1-, PeMADS6-, and double-silenced plants in various floral organs of the sequence of flowers in an inflorescence, this work tested five plants of two different cultivars (Dtps. I-Hsin Sunrise Cinderella ‘OX 1357’ and Dtps. OX Red Shoe ‘OX1407’). The proportion of greenish and discoloured areas in floral organs was calculated by use of ImageJ (http://imagej.en.softonic.com/).
The morphological changes of sepals were graded by estimating the proportion of greenish area on the abaxial surface (Supplementary Fig. S3A1), and of petals by estimating the proportion of greenish and discoloured areas on the adaxial surface (Supplementary Fig. S3B1). Lip morphological changes were characterized as three different types and were graded by calculating: (1) the proportion of greenish areas on the abaxial surface of the midlobe and lateral lobes (Supplementary Fig. S3C1); (2) the proportion of flowers that did not develop lateral lobes but contained cirrus (Supplementary Fig. S3C2); and (3) the proportion of flowers that did not develop cirrus and lateral lobes (Supplementary Fig. S3C3). Column morphological changes were characterized as three types and were graded by calculating: (1) the percentage change in length of the column (Supplementary Fig. S3D1); (2) the proportion of flowers showing an abnormal stigmatic cavity (Supplementary Fig. S3D2); and (3) the proportion of flowers showing an abnormal stigmatic cavity and an extra petal-like organ (Supplementary Fig. S3D3).
Results
High-throughput silencing of 126 Phalaenopsis ESTs
To identify genes involved in orchid flower morphogenesis, 5593 ESTs had been previously obtained from P. equestris flower buds; 152 were predicted as putative TF-related genes (Tsai et al., 2006). This work investigated by high-throughput VIGS whether these TFs play important roles in flower morphogenesis.
Nucleotide sequences from 500 to 3300bp for the 152 floral TF ESTs were cloned into the CymMV-Gateway vector. The cloning efficiency for inserts <1500bp was 100% and decreased with inserts >1500bp; the cloning efficiency was 0 for inserts >3000bp (Supplementary Fig. S1). Of the 152 TF ESTs studied, 126 were classified into 9 TF families and accounted for 56% of the TF ESTs. The silencing of PeMADS1, PeMADS7, PebHLH, PeHB, and PeZIP affected flower morphological features and thus flower development (Figs. 1–3 and Supplementary Fig. S4).
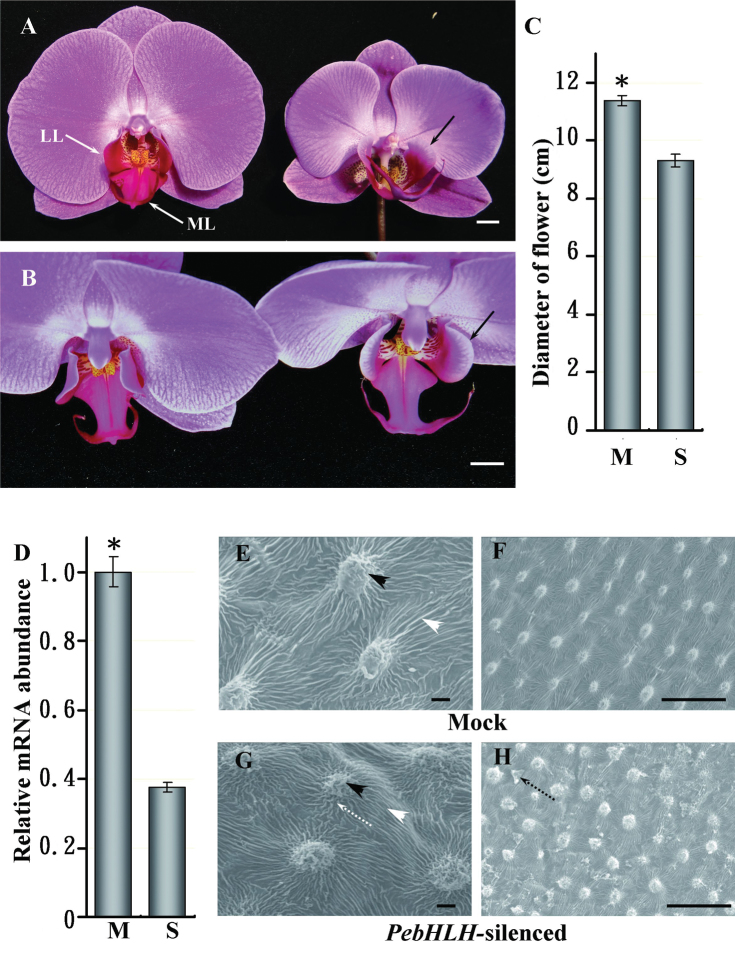
Fig. 1.
VIGS phenotypes of PebHLH-silenced flowers. (A) Front view of mock-treated (left) and PebHLH-silenced flowers (right) in Doritaenopsis I-Shin Sunrise Cinderella ‘OX1357’; solid white arrows indicate the lateral lobe (LL) and midlobe (ML); solid black arrows indicate the lateral lobes of the lip bowed outward. (B) Top-down view of the lateral lobes of the mock-treated (left) and PebHLH-silenced flowers (right); solid black arrows indicate the lateral lobes of the lip bowed outward; white bars, 1cm. (C) Diameter of flowers in mock-treated and PebHLH-silenced plants; a total of four flowers (the 3rd flower at blooming stage) from four plants were examined for each treatment; data are mean ± SD (n = 4); significance was accepted (one-tailed t-test) if P < 0.01 (*). (D) Relative level of PebHLH in mock-treated and PebHLH-silenced flowers. mRNA was extracted from the 3rd floral bud at 15 d post inoculation; data are mean ± SD (n = 4); significance was accepted (one-tailed t-test) if P < 0.01 (*). (E–H) SEM of the adaxial surface cells of lateral lobes in lips from mock-treated (E and F) and PebHLH-silenced flowers (G and H) at the flower-blooming stage; black arrowheads indicate node-like cuticular folds in the central fields of cells; white arrowheads indicate parallel cuticular folds in the anticline fields of cells; dotted black arrows indicate irregular cuticular folds in the central fields of cells; dotted black arrows indicate epicuticular wax crystals in the anticline field. Bars, 30 μm (this figure is available in colour at JXB online).
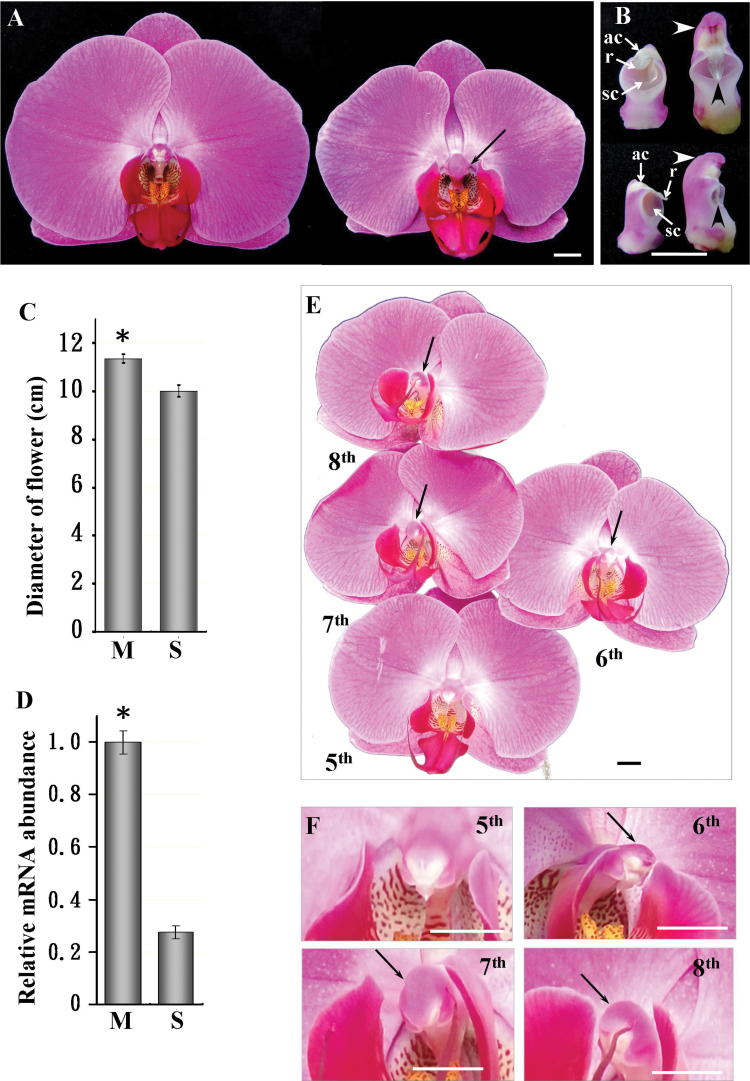
Fig. 3.
VIGS phenotypes of PeMADS1-silenced flowers. (A) Front view of mock-treated (left) and PeMADS1-silenced (right) flowers; solid black arrow indicates a longer and wider column. (B) Front and back view of columns in mock-treated (left) and PeMADS1-silenced flowers (right); solid white arrows indicate the anther cap (ac), rostellum (r), and stigmatic cavity (sc); white arrowheads indicate that the opposite site of stigmatic cavity in the column became longer and curved; black arrowheads indicate that the rostellum became longer. (C) Diameter of flowers in mock-treated (M) and PeMADS1-silenced plants (S); a total of six flowers (the 7th flower at blooming stage) from six plants were examined for each treatment; data are mean ± SD (n = 6); significance was accepted (one-tailed t-test) if P < 0.01 (*). (D) Relative mRNA level of PeMADS1 in M and S flowers; mRNA was extracted from the 7th floral bud on 35 d post inoculation; data are mean ± SD (n = 6); significance was accepted (one-tailed t-test) if P < 0.01 (*). (E) The 5th to 8th blooming flowers in a PeMADS1-silenced inflorescence and (F) columns of the 5th to 8th flowers; solid black arrows indicate that the column became longer and wider. Bars, 1cm.
Silencing PeMADS7, PeHB, and PebZIP resulted in floral abortion
Silencing PeMADS7, PeHB, or PebZIP caused abortion of the first three floral buds from the bottom of the raceme inflorescence within 2 weeks after agro-infiltration. These aborted flower buds had fully formed sepals, petals, lips, and columns, and their morphological features were unaffected (data not shown). When their expression was restored as the effect of VIGS gradually decreased, the 4th and subsequent floral buds could bloom normally (Supplementary Table S2). The expression of these three genes was detected in the various stages of visible floral buds (Supplementary Fig. S2C). Therefore, knockdown of PeMADS7, PeHB, or PebZIP expression allowed the flower to develop normally initially but affected further flower development and growth.
This work performed a BLAST search of the nucleotide sequences of PeMADS7, PeHB, and PebZIP in OrchidBase 2.0 and found no sequences with high similarity to PeMADS7 or PebZIP. Four nucleotide sequences showed high similarity to PeHB, with three having more than 21 nt that were 100% homologous to each other (Supplementary Fig. S5). Therefore, the VIGS effects could have resulted from the silencing of PeHB and three other HB family genes rather than gene-specific effects.
VIGS of PebHLH affected regulation of floral size and cell arrangement
Flower size was significantly reduced in PebHLH-silenced plants (Fig. 1A and Supplementary Fig. S4A). In addition, the lateral lobes of the lip bowed outward and their angle to the column was larger than in mock-treated plants (Fig. 1A and B). Adaxial epidermal cells of lip lateral lobes in PebHLH-silenced plants lost their organized arrangement (Fig. 1F and H), but they retained their convex shape (Fig. 1E and G) and accumulated epicuticular wax crystals in the anticline field of cells (Fig. 1F and H). The diameter was smaller, by 17.9%, for PebHLH-silenced than mock-treated flowers (Fig. 1A and C). The relative mRNA level of PebHLH in the floral organs was significantly reduced, by 62.3±4.7% (Fig. 1D).
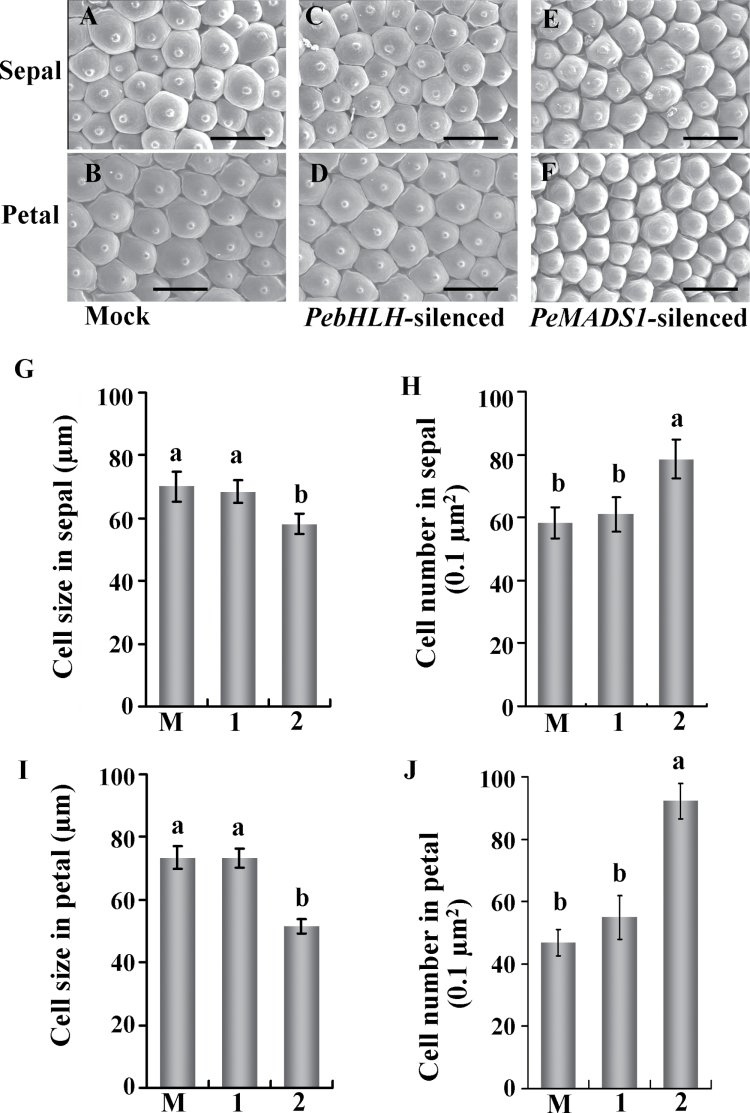
To analyse whether the reduced floral organ size in the PebHLH-silenced flowers was due to a defect in cell expansion, this work examined cell size and cell number per 0.1 μm2 surface area of the adaxial epidermis of sepals and petals by SEM. The epidermal cell shape in PebHLH-silenced flowers remained papilla shaped, as for mock-treated flowers (Fig. 2A–D). In addition, the adaxial epidermal cell size and cell number per 0.1 μm2 surface area was similar for PebHLH-silenced and mock-treated sepals and petals (Fig. 2G–J). Thus, silencing PebHLH did not affect cell morphological features, cell size, or cell number per unit area. The reduced flower size could be due to premature termination of proliferation or reduction of final cell number in sepals and petals.
Fig. 2.
Silencing of PebHLH and PeMADS1 affects cell size and number in the floral organ. (A–F) SEM of the sepal and petal adaxial surface cells at the flower-blooming stage in mock-treated (A, B), PebHLH-silenced (C, D), and PeMADS1-silenced flowers (E, F). (G–J) Cell size and cell number in the distal adaxial surface of sepals (G, I) and petals (H, J) in mock-treated (M), PebHLH-silenced (1), and PeMADS1-silenced flowers (2); data are mean ± SD (n = 6); the same letters above the bars indicate that there is no statistical difference by Duncan’s multiple range test (P < 0.05) Bar, 30 μm. Materials were Dtps. I-Hsin Sunrise Cinderella ‘OX 1357’.
The phylogeny of PebHLH was determined with all existing 158 (26 subfamilies) and 173 basic helix–loop–helix (bHLH) (27 subfamilies) gene sequences from Arabidopsis and rice, respectively (Pires and Dolan, 2010). PebHLH was in the same clade as AtbHLH149 set apart from the 27 subfamilies, and thus could be classified as an ‘orphan’ (Supplementary Fig. S6). PebHLH was expressed in roots, leaves, and all developmental stages of floral organs in Phalaenoopsis (Supplementary Fig. S2C and F).
VIGS of PeMADS1 resulted in a homeotic conversion in column and reduced cell size
PeMADS1 expression was detected in the floral meristem and all developmental stages of floral buds (Supplementary Fig. S2C). In PeMADS1-silenced flowers, the 6th to 8th flowers in a raceme inflorescence had an enlarged and wide column, with the most severe phenotypic changes on the 7th flower (Fig. 3A, B and E, F). The diameters of flowers were reduced by 11.8% in PeMADS1-silenced plants (Fig. 3A and C), and the mRNA level of PeMADS1 was significantly decreased by 72.4% in silenced plants (Fig. 3D).
On SEM, adaxial epidermal cells in the sepal and petal in PeMADS1-silenced flowers were 17% and 30% smaller, respectively, than those of mock-treated flowers (Fig. 2G and I). The reduced cell size was accompanied by increased cell number in the same surface area in PeMADS1-silenced flowers (Fig. 2H and J). Therefore, small floral organ size in PeMADS1-silenced plants was primarily caused by reduced cell expansion.
The epidermal cell shape of sepals and petals was the same in PeMADS1-silenced and mock-treated flowers (Fig. 2E and F). However, the rostellum of the column was longer in PeMADS1-silenced than mock-treated flowers. The anther cap, which covers the end of the column, was located on the same side as the stigmatic cavity in PeMADS1-silenced flowers (Fig. 3B).
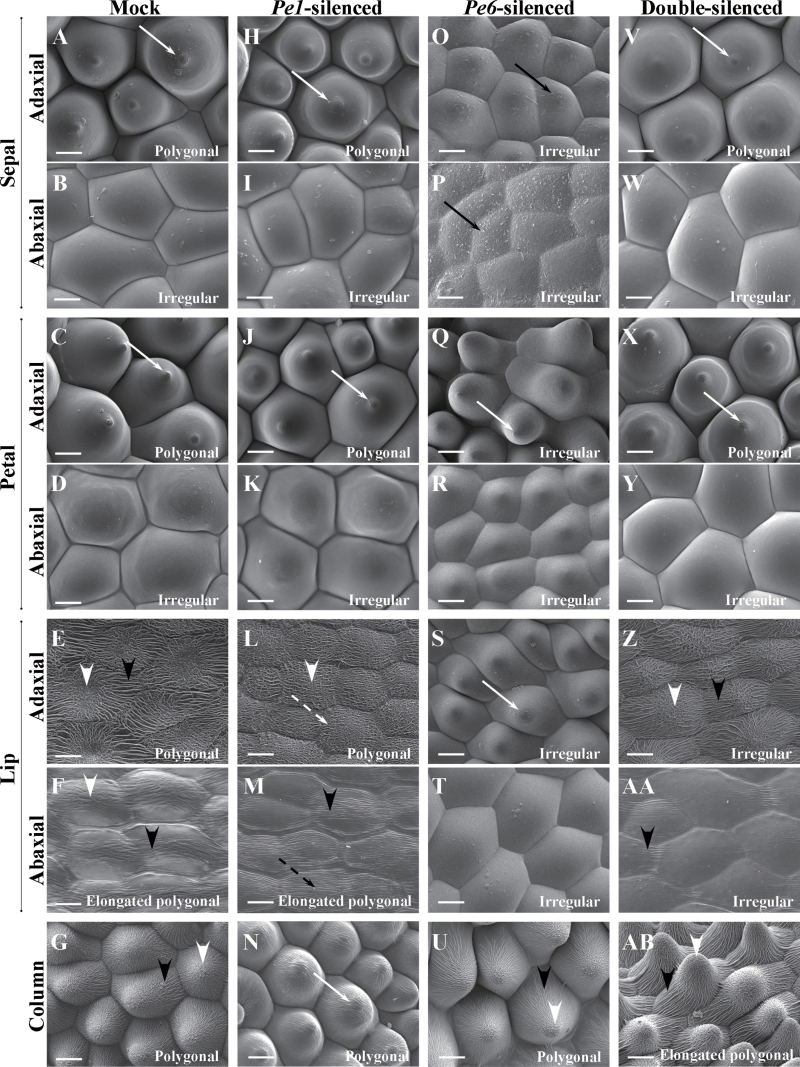
In addition, the epidermal cells of PeMADS1-silenced columns (Fig. 4N) were changed and similar to that of the adaxial surface of mock-treated petals (Fig. 4C). Epidermal cells of mock-treated columns showed cuticular folds in the central field (Fig. 4G). In contrast, PeMADS1-silenced columns showed a protuberance on the top of cells and no cuticular folds in the central field (Fig. 4N). Thus, the column of PeMADS1-silenced flowers exhibited a partial homeotic transformation to petal-like.
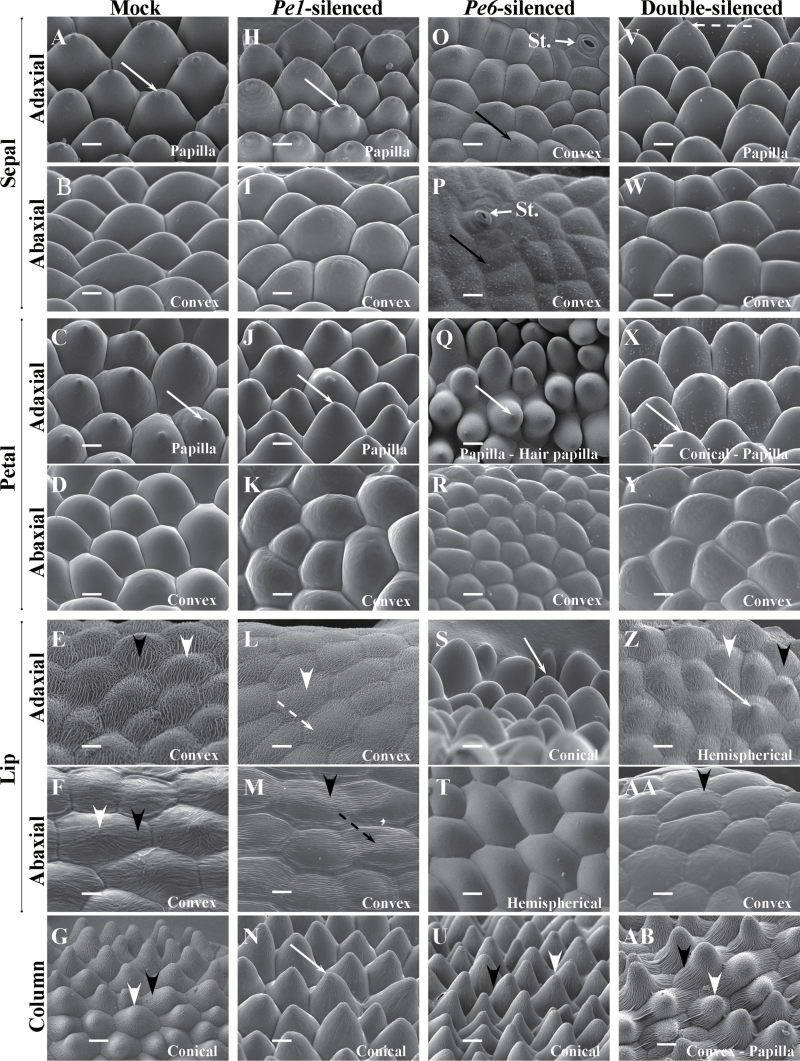
Fig. 4.
Cyro-SEM micrographs of top views of epidermal cells of floral organs of the 7th blooming flowers of mock-treated (A–G), PeMADS1-silenced (H–N), PeMADS6-silenced (O–U), and double-silenced floral organs (V–AB). (A, H, O, V) Sepal adaxial epidermal cells had polygonal, polygonal, irregular, and polygonal forms. (B, I, P, W) Sepal abaxial epidermal cells all had irregular forms. (C, J, Q, X) Petal adaxial epidermal cells had polygonal, polygonal, irregular, and polygonal forms. (D, K, R, Y) Petal abaxial epidermal cells all had irregular forms. (E, L, S, Z) Lip adaxial epidermal cells had polygonal, polygonal, irregular, and irregular forms. (F, M, T, AA) Lip abaxial epidermal cells had elongated polygonal, elongated polygonal, irregular, and irregular forms. (G, N, U, AB) Column epidermal cells had polygonal, polygonal, polygonal, and elongated polygonal forms. Solid white arrows indicate a protuberance on the top of the cell; solid black arrow indicates epicuticular wax crystals; white arrowheads indicate irregular cuticular folds in the central field; black arrowheads indicate parallel cuticular folds in the central field; dashed white arrows indicate irregular cuticular folds in the anticline field; dashed black arrows indicate parallel cuticular folds in the central field. Bars, 25 μm. Materials were Dtps. OX Red Shoe ‘OX1407’.
Co-silencing B- and C-class MADS-box genes caused a petal-like lip and column
To investigate the additive effect of B- and C-class MADS-box gene expression on orchid floral morphogenesis, PeMADS1 and PeMADS6 fragments constructed in separate vectors were co-infiltrated. These two genes were expressed in various stages of visible floral buds (Supplementary Fig. S2C). Previously, silencing PeMADS6 alone caused leaf-like sepals and a petal-like lip (Hsieh et al., 2013). In this study, silencing PeMADS1 alone caused a petal-like column. For double silencing, Phalaenopsis plants were inoculated with a mixture of Agrobacterium cultures containing pCAMBIA-CymMV-PeMADS1 and -PeMADS6 at a 1:1 ratio. In the positive control, the greenish and discoloured phenotype appeared in the sepals, petals, and lip of the first eight blooming flowers of PeMADS6-silenced plants, with the first three not able to bloom normally (Supplementary Fig. S7E). In contrast, with double-silencing PeMADS1 and PeMADS6, a similar effect was detected only from the 1st to the 6th blooming flowers; the phenotypic changes were most severe in the first flower and weakened with each consecutive flower (Supplementary Fig. S7F). In the first blooming flower, the margins of sepals and petals were curly and discoloured (Fig. 5E–H), and the lip was greenish and discoloured (Fig. 5G) as compared with mock-treated flowers (Fig. 5A–D).
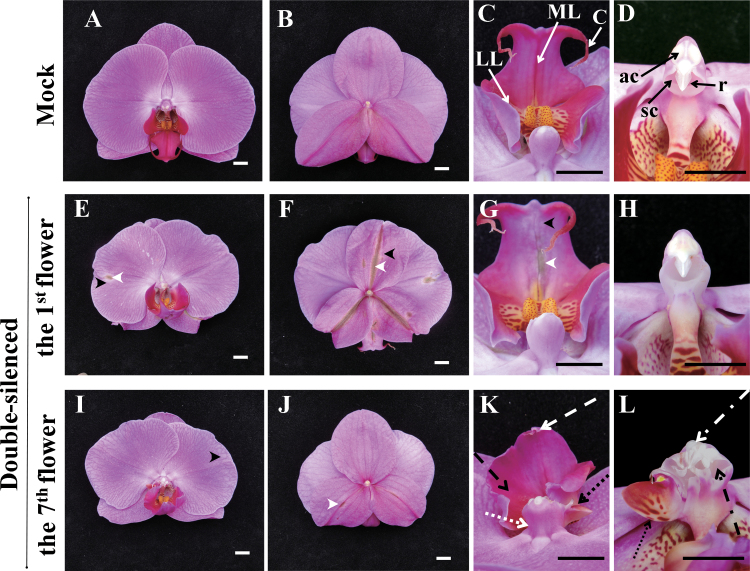
Fig. 5.
Concurrent silencing of both PeMADS1 and PeMADS6 with a pCymMV-Gateway vector. (A–D) Mock-treated plants: top view (A), back view (B) of the flower and front view of lip (C) and column (D); solid white arrows indicate the lateral lobe (LL) and midlobe (ML); solid black arrows indicate the anther cap (ac), rostellum (r), stigmatic cavity (sc), and cirrus (C). (E–H) 1st blooming flower of double-silenced plants: top view (E), back view (F), and front view of lip (G) and column (H); black arrowheads indicate discoloured characteristics of sepals, petals, and lips; white arrowheads indicate dark green colouration (including leaf-like characteristics) of sepals, petals, and lips. (I–L) 7th blooming flower of double-silenced plants: top view (I), back view (J), and front view of lip (K) and column (L); dashed white arrow indicates that no cirrus developed in the tip of the midlobe; dashed black arrow indicates that the lateral lobes and midlobe were fused into a petal-like lip; dotted white arrow indicates that the base of the column became wider; dotted black arrows indicate that the side of the column developed an extra petal-like organ; dash-dotted white arrow indicates that the anther cap became wider and the appearance of more than one rostellum-like organ; dash-dotted black arrow indicates that the stigmatic cavity did not develop normally. White bars and black bars, 1cm.
The combined silencing phenotype of lip and columns was detected in the 7th blooming flowers in the same inflorescence approximately 58 d after agro-infiltration (Fig. 5I–L). The lip lacked cirrus and the lateral lobes fused with the mid-lobe into a petal-like lip (Fig. 5K). The column did not develop a circle-shaped stigmatic cavity, and in some cases, extra anther cap-like and rostellum-like structures appeared at the end of the column (Fig. 5L). Therefore, both PeMADS1 and PeMADS6 play important roles to ensure the normal development of lip and column. Similar phenomena were observed with the double silencing of PeMASD1 and PeMADS6 in three different cultivars, Dtps. I-Hsin Sunrise Cinderella ‘OX1357’, Dtps. OX Red Shoe ‘OX1407’, and P. Sogo Yukidian ‘V3’ (Supplementary Fig. S7 and S8 and Supplementary Table S3).
Epidermal cellular changes with co-silencing B- and C-class MADS-box genes
In mock-treated flowers, the sepal adaxial epidermal cells had a polygonal and papilla shape with a protuberance on the top of cells (Figs. 4A and 6A), and the abaxial ones had an irregular and convex shape (Figs. 4B and 6B). The shapes of adaxial and abaxial epidermal cells of the petal were similar to those of the sepal (Figs. 4C, D and 6C, D). The lip adaxial and abaxial epidermal cells contained irregular cuticular folds in the central fields and parallel cuticular folds in the anticline fields (Figs. 4E, F and 6E, F). However, the lip adaxial epidermal cells had a polygonal and convex shape (Figs. 4E and 6E), and the abaxial ones had an elongated polygonal and convex shape (Figs. 4F and 6F). The column epidermal cells had a polygonal and conical shape with irregular cuticular folds in the central fields and parallel cuticular folds in the anticline fields (Figs. 4G and 6G).
Fig. 6.
Cyro-SEM micrograph of side views of epidermal cells of floral organs of the 7th blooming flowers of mock-treated (A–G), PeMADS1-silenced (H–N), PeMADS6-silenced (O–U), and double-silenced floral organs (V–AB). (A, H, O, V) Sepal adaxial epidermal cells had papilla, papilla, convex, and papilla shapes. (B, I, P, W) Sepal abaxial epidermal cells all had convex shape; St., stomata. (C, J, Q, X) Petal adaxial epidermal cells had papilla, papilla, papilla/hair papilla, and conical/papilla shapes. (D, K, R, Y) Petal abaxial epidermal cells all had convex shapes. (E, L, S, Z) Lip adaxial epidermal cells had convex, convex, conical, and hemispherical shapes. (F, M, T, AA) Lip abaxial epidermal cells had convex, convex, hemispherical, and convex shapes. (G, N, U, AB) Column epidermal cells had conical, conical, conical, and different shapes (including convex, hemispherical cupola, conical, and papilla). Solid white arrows indicate a protuberance on the top of the cell; solid black arrow indicates epicuticular wax crystals; white arrowheads indicate irregular cuticular folds in the central field; black arrowheads indicate parallel cuticular folds in the central field; dashed white arrows indicate irregular cuticular folds in the anticline field; dashed black arrows indicate parallel cuticular folds in the central field. Bars, 25 μm. Materials were Dtps. OX Red Shoe ‘OX1407’.
PeMADS1-silenced flowers showed significant phenotypic changes in the lip and column epidermal cells (Figs. 4L–N and 6L–N). In the adaxial epidermal cells of the lip, the parallel epicuticular folds were replaced with irregular ones in the anticline fields (Figs. 4L and 6L). The irregular epicuticular folds in the abaxial epidermal cells of the lip were replaced with parallel ones in the central fields (Figs. 4M and 6M). A protuberance appeared on the top of column epidermal cells (Figs. 4N and 6N).
PeMADS6-silenced flowers showed significant phenotypic changes in the sepal, petal and lip epidermal cells (Figs. 4O–U and 6O–U). The adaxial and abaxial epidermal cells of sepals in greenish areas (Supplementary Fig. S7E) were convex shaped, with epicuticular wax crystals (Figs. 4O, P and 6O, P). The adaxial epidermal cells of petals in greenish areas had papilla and hair papilla shapes (Figs. 4Q and 6Q) and the surface of epidermis was not flat (Supplementary Fig. S9K). The adaxial and abaxial epidermal cells of the lip in greenish areas lost the cuticular folds covering the cells (Figs. 4S, T and 6S, T). In addition, the adaxial cells had a conical shape (Fig. 4S and 6S) and an irregular arrangement in epidermis (Supplementary Fig. S9S).
The first blooming flowers of double-silenced plants showed significant phenotypic changes in the sepal (Supplementary Fig. S10Q, R and X, Y) and lip (Supplementary Fig. S10U, V and AB, AC). The adaxial and abaxial epidermal cells of sepals in greenish areas were all irregular and convex shaped, with cuticular wax crystals (Supplementary Fig. S10Q, R and X, Y), which was similar to those of leaves (Supplementary Fig. S10H and I). The adaxial epidermal cells of the lips with greenish areas were irregular and cupola shaped, with a protuberance on the top of cells (Supplementary Fig. S10U and AB).
Petal-like lip and column epidermal cells in the 7th blooming flowers of double-silenced plants showed significant phenotypic changes (Figs. 4Z–AB and 6Z–AB). The adaxial epidermal cells of the petal-like lip had an irregular and hemispherical shape with a protuberance on the top of cells and thin, dense cuticular folds (Figs. 4Z and 6Z), while the abaxial ones lost irregular cuticular folds in the central fields (Figs. 4AA and 6AA). The column epidermal cells had an elongated polygonal form (Fig. 4AB) and different shapes including convex, hemispherical, cupola, conical, and papilla (Fig. 6AB and S9AB).
Thus, PeMADS6-silenced plants showed a homeotic transformation in the epidermal cells in the greenish areas of the lip. The epidermal cells of PeMADS1-silenced lips showed slight changes in structure. Double-silenced plants went through a homeotic transformation from a lip to a petal-like lip in greenish areas. However, their epidermal cells showed only slight changes in shape and structure.
Double-silenced flowers showed more significant changes in the column and lip than single-silenced flowers
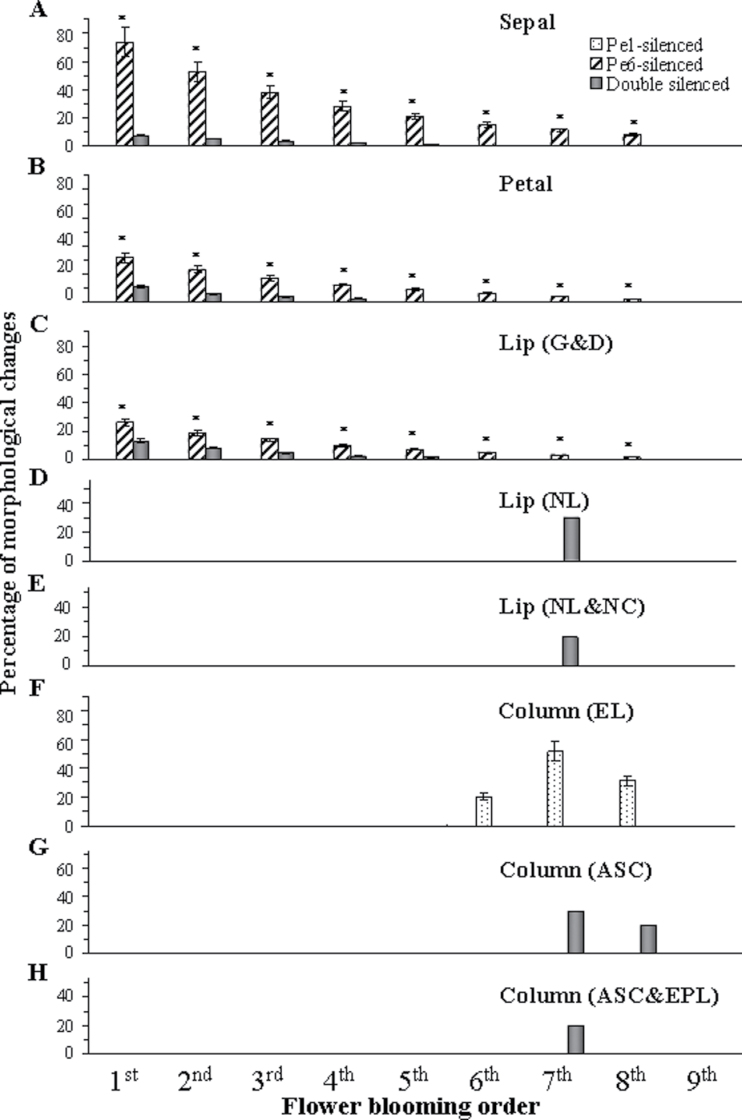
The severity of floral organ morphological changes was quantified for sequences of flowers from PeMADS1-, PeMADS6-, and double-silenced plants (Supplementary Fig. S3A–D). Greenish areas of the sepal, petal and lip appeared only in PeMADS6- and double-silenced plants but not PeMADS1-silenced plants (Fig. 7A–C). Approximately eight flowers of each inflorescence showed the VIGS phenotype in PeMADS6- and double-silenced plants, beginning with the bottom of the raceme (Fig. 7A–E and G–H). However, three PeMADS1-silenced flowers showed morphological changes in the column from the 6th to the 8th flowers (Fig. 7F). The most severe morphological change occurred in the 7th flower, followed by the 8th and then 6th flower (Fig. 7F). The VIGS phenotype lasted longer for PeMADS6- than PeMADS1-silenced plants (Fig. 7A–C and F), so the period of floral development affected by loss of function lasted longer for PeMADS6- than PeMADS1-silenced plants.
Fig. 7.
Severity of floral organ morphological changes in ordered blooming flowers in PeMADS6- (dotted bars), PeMADS1- (slash bars), and double-silenced plants (black bars). (A) Sepal morphological changes graded by estimating the percentage of greenish area on the abaxial surface. (B) Petal morphological changes graded by estimating the percentage of greenish and discoloured areas on the adaxial surface. (C–E) Lip morphological changes graded by estimating the percentage of greenish and discoloured areas on the abaxial surface of the midlobe and lateral lobes (C, G&D, greenish and discoloured), calculating the percentage of flowers that did not develop lateral lobes (D, NL, no lateral lobes), and he percentage of flowers that did not develop cirrus as well as the lateral lobes (E, NL&NC, no lateral lobes and no cirrus). (F–H) Column morphological changes were graded by calculating the percentage change in the length of the column (F, EL, elongated length of column; the formula is (length of VIGS column – length of mock column)/(length of mock column) ×100, calculating the percentage of flowers that developed an abnormal stigmatic cavity (G, ASC), and the percentage of flowers that developed an abnormal stigmatic cavity as well as an extra petal-like organ (H, ASC&EPL). Significance was accepted (one-tailed t-test) if P < 0.01 (*).
In PeMADS6-silenced plants, the greenish areas in the sepal were gradually reduced from 73.9% in the 1st flower to zero in the 9th flower (Fig. 7A), while the greenish and discoloured areas in the petal and lip were reduced from 32.3% and 25.8% to zero, respectively (Fig. 7B, C). In double-silenced plants, the greenish areas of the sepal were gradually reduced from 7.4% in the 1st flower to zero in the 9th flower (Fig. 7A), while the greenish and discoloured areas in the petal and lip were reduced from 10.7% and 13.1% to zero, respectively (Fig. 7B, C). Thus, VIGS phenotypic changes were less in flowers of double-silenced than PeMADS6-silenced plants.
In addition, in 30% of the double-silenced plants, the 7th flower did not develop lateral lobes (Fig. 7D), 20% did not develop cirrus and lateral lobes on the lip (Fig. 7E), 30% developed an abnormal stigmatic cavity (Fig. 7G), and 20% developed an abnormal stigmatic cavity as well as an extra petal-like organ on the column (Fig. 7H). Thus, both genes were silenced simultaneously within both the 7th and 8th blooming flowers.
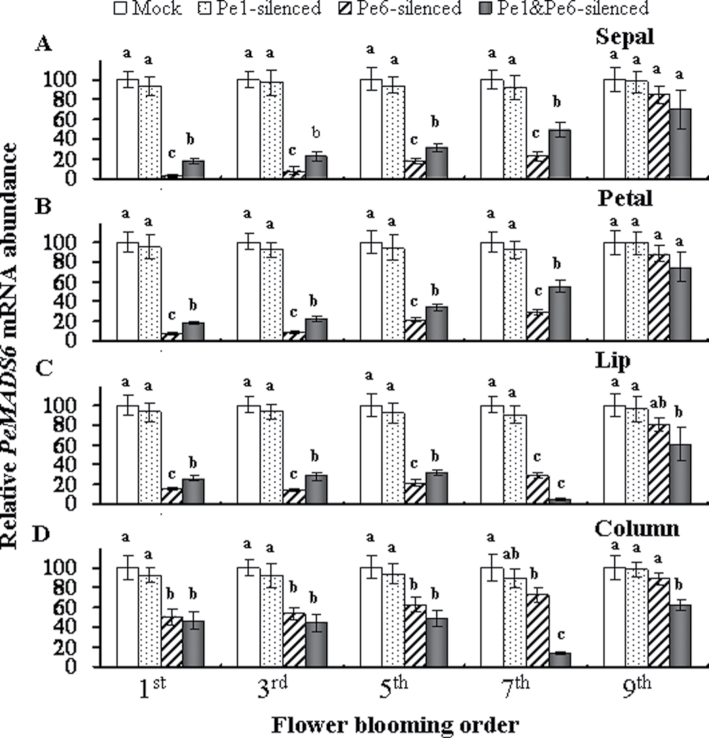
The silencing effect of PeMADS6 was assessed by comparing the mRNA levels of odd-numbered mock-treated and PeMADS1-, PeMADS6-, and double-silenced flowers (Fig. 8A-D). The PeMADS6 mRNA levels were lower in PeMADS6- and double-silenced sepals, petals, lips, and columns from the 1st to the 5th blooming flowers than in mock-treated and PeMADS1-silenced parts (Fig. 8A–D). Interestingly, the PeMADS6 mRNA levels were sharply reduced to 4.7% and 13.9% in the 7th blooming flower of double-silenced lip and column, respectively. As expected for very little phenotypic changes, the PeMADS6 mRNA levels increased again and reached just over 60% in the 9th blooming flower (Fig. 8C, D).
Fig. 8.
Expression of PeMADS6 in PeMADS1-, PeMADS6-, and double-silenced plants according to flower blooming order (1st, 3rd, 5th, 7th, and 9th) in inflorescences of mock-treated plants (Mock) and PeMADS1-, PeMADS6-, and double-silenced plants (Pe1-silenced, Pe6-silenced, and Pe1&Pe6-silenced) in the (A) sepal, (B) petal, (C) lip, and (D) column. mRNA was extracted at flower blooming date in Dtps. I-Hsin Sunrise Cinderella ‘OX 1357’. Data are mean ± SD from three plants for each experiment (n = 3); the same letters above the bars indicate that there is no statistical difference by Duncan’s multiple range test (P < 0.05).
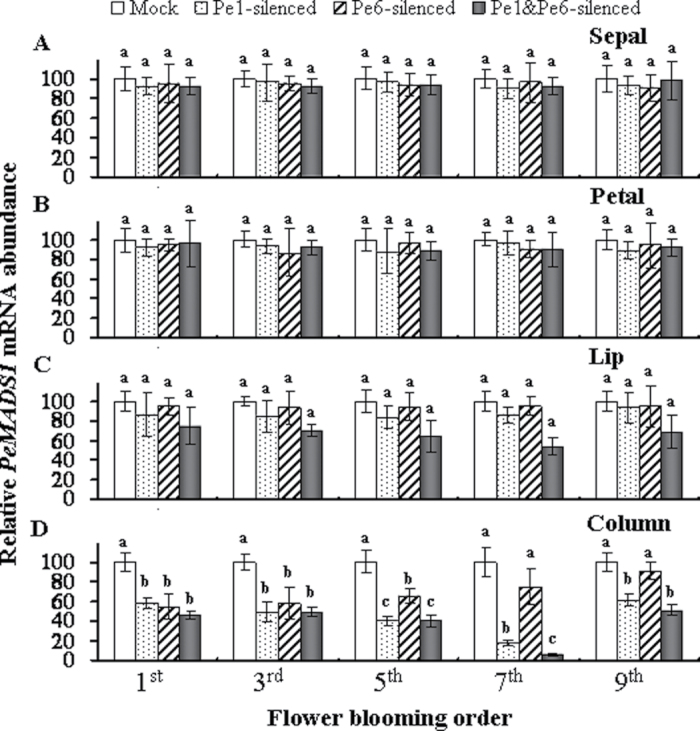
Real-time RT-PCR analysis showed that the levels of PeMADS1 expression in the sepal, petal, and lip were generally at least 2000–5000-fold lower than found in the column in mock treated flowers (data not shown). The PeMADS1 mRNA levels were significantly lower in PeMADS1-, PeMADS6-, and double-silenced columns from the 1st to the 5th blooming flowers in mock-treated columns (Fig. 9D). Interestingly, the PeMADS1 mRNA levels of PeMADS1-silenced and double-silenced flowers were further reduced to 17.7% and 6.2% in the 7th blooming flower (Fig. 9D). These results suggests that the PeMADS1 mRNA levels were mainly affected in the 7th blooming flowers of double-silenced plants, but the PeMADS6 mRNA levels were all affected from the 1st to the 7th blooming flowers.
Fig. 9.
Expression of PeMADS1 in PeMADS1-, PeMADS6-, and double-silenced plants according to flower blooming order (1st, 3rd, 5th, 7th, and 9th) in inflorescences of mock-treated plants (Mock) and PeMADS1-, PeMADS6-, and double-silenced plants (Pe1-silenced, Pe6-silenced, and Pe1&Pe6-silenced) in the (A) sepal, (B) petal, (C) lip, and (D) column. mRNA was extracted at flower blooming date in Dtps. I-Hsin Sunrise Cinderella ‘OX 1357’. Data are mean ± SD from three plants for each experiment (n = 3); the same letters above the bars indicate that there is no statistical difference by Duncan’s multiple range test (P < 0.05).
Discussion
Among angiosperms, orchids are unique in their floral patterning, particularly in the lip and column structures. To uncover the role of specific TFs in floral development, this work used high-throughput VIGS with the CymMV-Gateway vector and shows the genetic interactions between B- and C-class MADS-box genes, namely PeMADS6 and PeMADS1, in determining proper patterning of floral organs by transiently downregulating both genes in a non-model plant, the orchid.
In high-throughput VIGS for floral genomics, PeMADS1, a C-class AGAMOUS-like MADS-box gene (Tsai et al., 2008), was identified as being involved in the normal development of the column and affecting floral size. Previous studies showed that loss of function of C-class MADS-box genes caused homeotic transformations of reproductive organs into perianth organs in petunia (Kapoor et al., 2002) and tomato (Pnueli et al., 1994) and reduced floral size in Antirrhinum (Bradley et al., 1993), Arabidopsis (Yanofsky et al., 1990), wheat (Aida et al., 2003), and Chinese narcissus (Deng et al., 2011). In Phalaenopsis, PeMADS1 may also regulate floral organ size primarily by affecting cell size.
In PeMADS1/PeMADS6-silenced plants, this study present a schema of period of development that is most sensitive to loss of function with agro-infiltration in an inflorescence containing eight internodes and one visible floral bud (Supplementary Fig. S11). In observing the anatomy of a Phalaenopsis inflorescence tip, the development of the inflorescence containing one visible floral bud can be divided into four major stages: stage 1, the formation of floral primordia; stage 2, the formation of early floral organ primordia with sepals, petals, lip, and column; stage 3, the formation of late floral organ primordia with pollina and callus developing on the column and the lip, respectively; and stage 4, the development and enlargement of floral buds (Pan et al., 2011). The floral buds in an inflorescence could be sensitive to loss of function from stage 2 to 4 with PeMADS6 silencing and sensitive to loss of function from stage 2 to 3 with PeMADS1 silencing (Supplementary Fig. S11).
The flowers of double mutants of B- and C-class genes, such as pi ag and ap3 ag, consist entirely of sepal-like organs, and their flowers are indeterminate with flowers within flowers in Arabidopsis (Bowman et al., 1989). Similarly, in rice flowers, both B- and C-class gene mutants (spw1-1 osmads3-4 and spw1-1 osmads58) show carpelloid structures and glume-like organs but appear to have morphological features similar to that of the marginal region of the wild-type palea (Yun et al., 2013). The current work observed extra petal-like organs at the position of column and petal-like epidermal cells in the lip in PeMADS6 and PeMADS1 double-silenced flowers. In addition, the lip was transformed into a petal-like organ. Although the flowers showed partial homeotic conversions of floral organs into petals, they were unlike the bc mutants of Arabidopsis (ap3 ag and the pi ag) or rice (spw1-1 osmads3-4 and spw1-1 osmads58), that have indeterminate flowers composed only of sepal-like organs or carpelloid flowers, respectively (Bowman et al., 1991; Coen and Meyerowitz, 1991; Meyerowitz et al., 1991; Yun et al., 2013). PeMADS6-silenced plants show petal-like lip and leaf-like sepal epidermal cells (Hsieh et al., 2013). In PeMADS1-silenced plants, the column became longer and wider. However, PeMADS6 and PeMADS1 double-silenced flowers showed severe phenotypic changes in the column as compared with PeMADS1-silenced flowers because PeMADS6 and PeMADS1 are both expressed only in the column (Tsai et al., 2005; Song et al., 2006). In addition, phenotypic changes were more severe in double-silenced than PeMADS6-silenced lips, which further suggests that the interaction of B- and C-class genes may dictate the lip morphogenesis.
The epidermal cell surfaces of sterile whorl organs in Phalaenopsis flowers are distinctive (Bercu et al., 2011). The varieties of cell shape and superimposed micro-structuring are important factors that affect how flowers interact with the abiotic environment (influencing light capture and reflectance, evaporation, temperature, and wetness) and the biotic environment (providing visual, tactile, and olfactory cues to pollinating animals) (Whitney et al., 2011). The changes in epidermal features alone can be considered a homeotic transformation in floral morphogenesis (Vandenbussche et al., 2004). In addition, epidermal cell shape is frequently used to determine floral organ identity in floral organs showing a homeotic transformation (Chang et al., 2010). In the PhGLO1 mutant plants of Petunia hybrida, flowers showed a conversion of the typical conical petal epidermal cells into sepal-like epidermal cells with the presence of stomata as well as the development of trichomes, which suggests a shift of the petal toward a sepal identity in these regions (Vandenbussche et al., 2004). Double NbGLO1-NbGLO2-VIGS (B-class MADS gene-silenced) Nicotiana benthamiana flowers showed a strong conversion of petals into sepals with the presence of stomata, numerous trichomes as well as puzzle-shaped epidermal cells. The petal-to-sepal transformation was also evident in the abaxial and adaxial epidermal cells of the petal, which were morphologically similar to those of wild-type sepals (Geuten and Irish, 2010). In rice flowers, the shape and the arrangement of epidermal cells of the carpel-like organs of C-class gene mutants (osmads58-s1) were almost identical to those of the wild-type carpel wall. Furthermore, the surface morphological aspects of the carpel-like organs differed from those of wild-type palea and lemma (Yamaguchi et al., 2006). In both spw1-1 osmads3-4 and spw1-1 osmads58 (B- and C-class gene mutants) flowers, the epidermal cells of both abaxial and adaxial surfaces of glume-like organs showed a smooth morphological aspect and no trichomes, which was similar to cells of the marginal region of wild-type paleas.
In Phalaenopsis, the shape of PeMADS6-silenced epidermal cells from the greenish or discoloured areas of the lip was almost identical to that of mock-treated petals. In addition, sepal epidermal cells covered by a cuticle film were similar to those of mock-treated leaves. This finding could be due to B-class gene functions possibly acting in primordia to establish identity and also late in development to achieve the proper final morphological features of conical cells in the petal epidermis, which occurs in Antirrhinum (Zachgo et al. 1995). The conical epidermal cell may be a hallmark for petaloid organs, possibly aiding pollinator orientation on flowers (Kramer and Hodges, 2010; Whitney et al., 2011).
In cotton, an AGAMOUS (AG)-like gene, GbAGL2, specifies the identity of floral organs and potentially plays a key role in fibre development (Liu et al., 2009). In Phalaenopsis amabilis, the sepal and petal structures are similar, with papillose cells in the adaxial epidermis and a few stomata in the abaxial surface. Lip epidermal cells appear elliptical in form, not papillose shaped, and are covered by a striate cuticle (Bercu et al., 2011). In double-silenced Phalaenopsis, the conical and convex shapes of the lip cells of the 1st blooming flowers were similar to the epidermal cells of mock-treated petals and PeMADS6-silenced lips. In the 7th blooming flowers, the abaxial epidermal cells of the petal-like lip lacked cuticular folds in the central fields, and the adaxial ones had fewer and thinner cuticular folds in the anticline fields than in the mock-treated lip. PeMADS6 and PeMADS1 genes might specify the identity of floral organs and potentially play a key role in cuticle development.
This study identified PebHLH involved in the expression of floral development by high-throughput VIGS. The functions of most bHLH genes are conserved during angiosperm evolution (Li et al., 2006). SPATULA (SPT), a bHLH transcription factor, controls the development of the carpel margins in Arabidopsis (Heisler et al., 2001). Silencing bHLH transcription factors, such as ANTHOCYANIN1, BIGPETAL, and SPT, can cause several changes depending on the plant. In Arabidopsis, silencing BIGPETAL increased petal size because of increased cell size and number (Szecsi et al., 2006; Varaud et al., 2011). Loss of function of SPT increased leaf size and total cell number, but cell size was not affected (Ichihashi et al., 2010). PebHLH is in the same clade as AtbHLH149 (AIF4) of Arabidopsis. AIF4 is a bHLH DNA-binding superfamily protein, which negatively regulates cell elongation (Wang et al., 2009; Ikeda et al., 2013). PebHLH-silenced plants showed curved floral organs, for a change in flower size. Therefore, PebHLH might affect total cell number for the whole floral organ and have a wide variety of functions in Phalaenopsis.
In conclusion, in a large-scale screening of gene functions related to floral morphogenesis in Phalaenopsis, this study found that knocking down the expression of individual TFs, including PebHLH and PeMADS1, affects floral morphological features. Floral growth during later developmental stages may be stopped by silencing PeMADS7, PeHB, and PeZIP. In addition, co-silencing B- and C-class MADS-box genes revealed the importance of studying lip morphogenesis in orchid flowers, which cannot be addressed using model plants such as rice and Arabidopsis.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Primers for semi-quantitative RT-PCR and real-time RT-PCR amplification.
Supplementary Table S2. Ratios of features of plant organs affected by transcription-factor silencing.
Supplementary Table S3. Ratios of features of plant organs affected by transcription-factor silencing in three cultivars.
Supplementary Fig. S1. Effect of gene fragment length on successful insertion into the pCymMV-Gateway vector.
Supplementary Fig. S2. Analysis of PeMADS7, PebZIP, PeHB, PeMADS1, PeMADS6, PebHLH expression during flower development and in different tissues in Phalaenopsis.
Supplementary Fig. S3. Severity of floral organ morphological changes in PeMADS1-, PeMADS6-, and double-silenced flowers.
Supplementary Fig. S4. VIGS phenotypes of PeMADS1- and PebHLH-silenced plants in Doritaenopsis OX Red Shoe ‘OX1407’.
Supplementary Fig. S5. The multiple sequence alignment of homeobox (HB) transcription factor nucleotide sequences from OrchidBase 2.0.
Supplementary Fig. S6. Phylogenetic analysis of PebHLH in bHLH gene families of Arabidopsis and rice.
Supplementary Fig. S7. Concurrent silencing of both PeMADS1 and PeMADS6 with a pCymMV-Gateway vector in Doritaenopsis OX Red Shoe ‘OX1407’.
Supplementary Fig. S8. Concurrent silencing of both PeMADS1 and PeMADS6 with a pCymMV-Gateway vector in Phalaenopsis Sogo Yukidian ‘V3’.
Supplementary Fig. S9. Cyro-SEM micrograph of epidermal cell arrangements of floral organs.
Supplementary Fig. S10. Cyro-SEM micrograph top and side view of adaxial and abaxial epidermal cells of PeMADS1 and PeMADS6 double-silenced floral organs from the first blooming flowers.
Supplementary Fig. S11. Estimation of the period of floral development is most sensitive to loss of function by PeMADS1 and PeMADS6 silencing in orchid.
Acknowledgements
The authors are grateful to Dr Wen-Chieh Tsai [Institute of Tropical Plant Science, National Cheng Kung University (ITPS, NCKU)] and Dr Chi-Kuang Wen (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for helpful discussion of this work. The authors thank Dr Wann-Neng Jane (Plant Cell Biology Core Facility, Institute of Plant and Microbial Biology, Academia Sinica) for assisting with cryo-SEM of floral micro-structures. The authors thank Ms Jui-Ling Hsu (ITPS, NCKU) for assistance in agro-infiltration and real-time quantitative RT-PCR. The authors also thank Oxen Biotechnology (Tainan, Taiwan) for assistance in choosing plant materials.
References
- Aida R, Murai K, Kishimoto S, Ohmiya A. 2003. Introduction of WAG, a wheat AGAMOUS homolog, reduces corolla size in torenia. Bulletin of the National Institute of Floricultural Science 3, 21–27. [Google Scholar]
- Becker A, Theissen G. 2003. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution 29, 464–489. [DOI] [PubMed] [Google Scholar]
- Bercu R, Bavaru A, Broasca L. 2011. Anatomical aspects of Phalaenopsis amabilis (L.) Blume. Annals of the Romanian Society for Cell Biology 16, 102–109. [Google Scholar]
- Bernacki S, Karimi M, Hilson P, Robertson N. 2010. Virus-induced gene silencing as a reverse genetics tool to study gene function. Methods in Molecular Biology 655, 27–45. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1989. Genes directing flower development in Arabidopsis . The Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1991. Genetic interactions among floral homeotic genes of Arabidopsis . Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. 1993. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum . Cell 72, 85–95. [DOI] [PubMed] [Google Scholar]
- Chang YY, Kao NH, Li JY, Hsu WH, Liang YL, Wu JW, Yang CH. 2010. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiology 152, 837–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Hsu CY, Cheng HY, Chang H, Chen HH, Ger MJ. 2011. Downregulation of putative UDP-glucose: flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis . Plant Cell Reports 30, 1007–1017. [DOI] [PubMed] [Google Scholar]
- Chen YH, Tsai YJ, Huang JZ, Chen FC. 2005. Transcription analysis of peloric mutants of Phalaenopsis orchids derived from tissue culture. Cell Research 15, 639–657. [DOI] [PubMed] [Google Scholar]
- Chen YY, Lee PF, Hsiao YY, Wu WL, Pan ZJ, Lee YI, Liu KW, Chen LJ, Liu ZJ, Tsai WC. 2012. C- and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant and Cell Physiology 53, 1053–1067. [DOI] [PubMed] [Google Scholar]
- Chugh S, Guha S, Rao IU. 2009. Micropropagation of orchids: A review on the potential of different explants. Scientia Horticulturae 122, 507–520. [Google Scholar]
- Coen ES, Meyerowitz EM. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Deng X, Xiong L, Wang Y, Li X. 2011. Ectopic expression of an AGAMOUS homolog NTAG1 from Chinese narcissus accelerated earlier flowering and senescence in Arabidopsis . Molecular Plant Breeding 2, 14–21. [Google Scholar]
- Dressler RL. 2005. How many orchid species? Selbyana 26, 155–158. [Google Scholar]
- Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, Huang YM, Tsai WC, Chen HH. 2011. OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant and Cell Physiology 52, 238–243. [DOI] [PubMed] [Google Scholar]
- Geuten K, Irish V. 2010. Hidden variability of floral homeotic B genes in Solanaceae provides a molecular basis for the evolution of novel functions. The Plant Cell 22, 2562–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MGB, Atkinson A, Bylstra YH, Walsh R, Smyth DR. 2001. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Hsiao YY, Pan ZJ, Hsu CC, Yang YP, Hsu YC, Chuang YC, Shih HH, Chen WH, Tsai WC, Chen HH. 2011. Research on orchid biology and biotechnology. Plant and Cell Physiology 52, 1467–1486. [DOI] [PubMed] [Google Scholar]
- Hsiao YY, Tsai WC, Kuoh CS, Huang TH, Wang HC, Wu TS, Leu YL, Chen WH, Chen HH. 2006. Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biology 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Lu HC, Pan ZJ, Yeh HH, Wang SS, Chen WH, Chen HH. 2013. Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Science 201, 25–41. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Horiguchi G, Gleissberg S, Tsukaya H. 2010. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana . Plant and Cell Physiology 51, 252–261. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. 2013. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signaling and Behavior 8, e23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Tsuda S, Tanaka Y, Mayama T, Okuyama Y, Tsuchimoto S, Takatsuji H. 2002. Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. The Plant Journal 32, 115–127. [DOI] [PubMed] [Google Scholar]
- Koch K, Bhushan B, Barthlott W. 2008. Diversity of structure, morphology and wetting of plant surfaces. Royal Society of Chemistry 10, 1943–1963. [Google Scholar]
- Kramer EM, Hodges SA. 2010. Aquilegia as a model system for the evolution and ecology of petals. Philosophical Transactions of the Royal Society B 365, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SM, Kono Y, Liu Y. 2008. Begonia pengii (sect. Coelocentrum, Begoniaceae), a new species from limestone areas in Guangxi, China. Botanical Studies 49, 167–175. [Google Scholar]
- Li X, Duan X, Jiang H, et al. 2006. Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and Arabidopsis . Plant Physiology 141, 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zuo K, Zhang F, Li Y, Xu J, Zhang L, Sun X, Tang K. 2009. Identification and expression profile of GbAGL2, a C-class gene from Gossypium barbadense . Journal of Biosciences 34, 941–951. [DOI] [PubMed] [Google Scholar]
- Lu HC, Chen HH, Tsai WC, Chen WH, Su HJ, Chang DC, Yeh HH. 2007. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiology 143, 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HC, Hsieh MH, Chen CE, Chen HH, Wang HI, Yeh HH. 2012. A high-throughput virus-induced gene-silencing vector for screening transcription factors in virus-induced plant defense response in orchid. Molecular Plant–Microbe Interactions 25, 738–746. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM, Bowman JL, Brockman LL, Drews GN, Jack T, Sieburth LE, Weigel D. 1991. A genetic and molecular model for flower development in Arabidopsis thaliana . Development Supplement 1, 157–167. [PubMed] [Google Scholar]
- Pan ZJ, Cheng CC, Tsai WC, Chung MC, Chen WH, Hu JM, Chen HH. 2011. The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant and Cell Physiology 52, 1515–1531. [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. 2010. Origin and diversification of basic-helix–loop–helix proteins in plants. Molecular Biology and Evolution 27, 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF. 1994. lsolation of the tomato AGAMOUS gene TAG7 and analysis of its homeotic role in transgenic plants. American Society of Plant Physiologists 6, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüm B, Seide l.R, Bohn HF, Speck T. 2012. Impact of cell shape in hierarchically structured plant surfaces on the attachment of male Colorado potato beetles (Leptinotarsa decemlineata). Beilstein Journal of Nanotechnology 3, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song IJ, Nakamura T, Fukuda T, Yokoyama J, Ito T, Ichikawa H, Horikawa Y, Kameya T, Kanno A. 2006. Spatiotemporal expression of duplicate AGAMOUS orthologues during floral development in Phalaenopsis . Development Genes and Evolution 216, 301–313. [DOI] [PubMed] [Google Scholar]
- Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. 2006. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. The EMBO Journal 25, 3912–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Fu CH, Hsiao YY, Huang YM, Chen LJ, Wang M, Liu ZJ, Chen HH. 2013. OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant and Cell Physiology 54, e7. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Hsiao YY, Lee SH, Tung CW, Wang DP, Wang HC, Chen WH, Chen HH. 2006. Expression analysis of the ESTs derived from the flower buds of Phalaenopsis equestris . Plant Science 170, 426–432. [Google Scholar]
- Tsai WC, Hsiao YY, Pan ZJ, Chen HH. 2010. Molecular mechanisms underlying orchid floral morphogenesis. Acta Horticulturae 878, 115–123. [Google Scholar]
- Tsai WC, Lee PF, Chen HI, Hsiao YY, Wei WJ, Pan ZJ, Chuang MH, Kuoh CS, Chen WH, Chen HH. 2005. PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant and Cell Physiology 46, 1125–1139. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Pan ZJ, Hsiao YY, Jeng MF, Wu TF, Chen WH, Chen HH. 2008. Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant and Cell Physiology 49, 814–824. [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. 2004. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. The Plant Cell 16, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szecsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp . The Plant Cell 23, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J. 2009. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix–loop–helix proteins. The Plant Cell 21, 3781–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney HM, Bennett KM, Dorling M, Sandbach L, Prince D, Chittka L, Glover BJ. 2011. Why do so many petals have conical epidermal cells? Annals of Botany 108, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Teo LL, Zhou J, Kumar PP, Yu H. 2006. Floral organ identity genes in the orchid Dendrobium crumenatum . The Plant Journal 46, 54–68. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yu H, Kumar PP. 2010. Characterization of floral organ identity genes of the orchid Dendrobium crumenatum . Asia-Pacific Journal of Molecular Biology and Biotechnology 18, 185–187. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. 2006. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa . The Plant Cell 18, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. 1990. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yun D, Liang W, Dreni L, Yin C, Zhou Z, Kater MM, Zhang D. 2013. OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Molecular Plant 6, 743–756. [DOI] [PubMed] [Google Scholar]
- Zachgo S, de Andrade Silva E, Motte P, Tröbner W, Saedler H, Schwarz-Sommer Z. 1995. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121, 2861–2875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.