Abstract
Proceedings of the National Academy of Sciences Colloquium on the roles of homologous recombination in DNA replication are summarized. Current findings in experimental systems ranging from bacteriophages to mammalian cell lines substantiate the idea that homologous recombination is a system supporting DNA replication when either the template DNA is damaged or the replication machinery malfunctions. There are several lines of supporting evidence: (i) DNA replication aggravates preexisting DNA damage, which then blocks subsequent replication; (ii) replication forks abandoned by malfunctioning replisomes become prone to breakage; (iii) mutants with malfunctioning replisomes or with elevated levels of DNA damage depend on homologous recombination; and (iv) homologous recombination primes DNA replication in vivo and can restore replication fork structures in vitro. The mechanisms of recombinational repair in bacteriophage T4, Escherichia coli, and Saccharomyces cerevisiae are compared. In vitro properties of the eukaryotic recombinases suggest a bigger role for single-strand annealing in the eukaryotic recombinational repair.
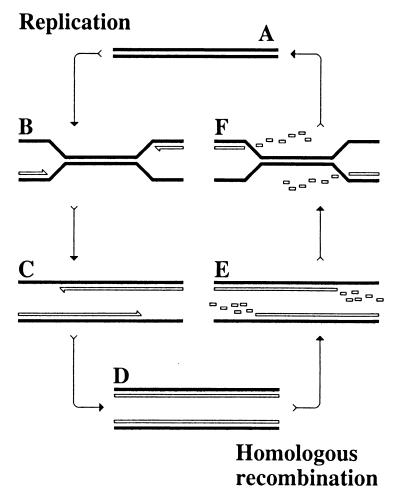
Replication makes identical copies of chromosomes, whereas genetic exchange, working in the opposite direction, scrambles homologous chromosomes to create new combinations of independently arisen alleles. Enzymatic mechanisms are opposite, too (Fig. 1): whereas replication separates the two strands of DNA duplex to synthesize their complements, homologous recombination [in this case, by single-strand annealing (1)] removes the complements to reestablish the original pairing. And, yet, there is a hidden unity in the apparent divergence, according to the participants of the National Academy of Sciences Colloquium entitled “Links Between Recombination and Replication: Vital Roles of Recombination,” organized by Charles Radding (chair), Nicholas Cozzarelli, Michael Cox, Kenneth Marians, and James Haber and held at the Beckman Center of the Academy in Irvine, California, on November 10–12, 2000. The recent surge in works on interdependence of DNA replication and homologous recombination, conducted in experimental systems ranging from bacteriophages to mammalian cell lines, highlighted faltering replication forks as the connecting points between the two seemingly opposite domains of DNA metabolism. A replication fork falters when it encounters an unrepaired DNA lesion or when its progress is blocked by a DNA-bound protein. As it turns out, the main mechanism of repair of faltering replication forks in all domains of life operates via homologous recombination.
Figure 1.
DNA replication vs. homologous recombination. Chromosomes are shown as double lines. Parental strands are filled; daughter strands are open. (A) A chromosome. (B) Chromosome replication has been initiated. (C) Chromosome replication is nearing completion. (D) Chromosome replication is complete. (E) Strand degradation in preparation for homologous recombination has started. (F) Strand degradation is nearing completion, whereas annealing of the complementary strands is going on.
The ideas, that replication forks can falter whereas homologous recombination can repair faltering replication forks, are not new. In 1966, Hanawalt proposed a scheme of replication fork collapse at single-strand interruptions in template DNA (2). In 1972, Strauss independently suggested replication fork collapse at nicks and proposed breakage of stalled replication forks (3). In 1974, Skalka suggested that the cell uses homologous recombination to repair collapsed replication forks (4). In 1976, Higgins elaborated the mechanism of stalled replication fork resetting to its present form (5). Now the time has come to appreciate these ideas.
Current hypotheses represent replication fork faltering and repair as follows:
(i) When a replication fork encounters a single-strand interruption in template DNA, it collapses, generating a double-strand end (Fig. 2 A → B → C). The idea of replication fork collapse is based on observations that single-strand interruptions in replicating chromosomes cause chromosome fragmentation (6, 7).
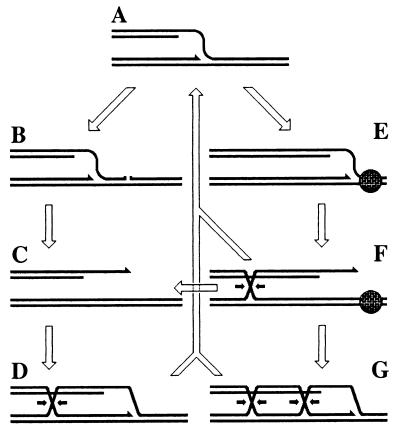
Figure 2.
The pathways of replication fork stalling/disintegration with subsequent resetting/repair. DNA duplexes are shown as double lines; a protein tightly bound to DNA is shown as a bricked circle. For all Holliday junctions, one of the two possible resolution directions is indicated by the small arrows. (A) A replication fork. (B) The replication fork approaching a single-strand interruption in template DNA. (C) The replication fork has collapsed at the interruption. (D) Double-strand end invasion to restore the replication fork structure. (E) A stalled replication fork. (F) Regression of the stalled replication fork forms a double-strand end and a Holliday junction. (G) Double-strand end invasion to restore the replication fork structure. Resolution of the Holliday junction in F leads to replication fork breakage (C). Resolution of the Holliday junctions in D or G, or exonucleolytic degradation of the linear tail in F leads to restoration of the replication fork structure (A).
(ii) When a replication fork is stalled because of a block in template DNA, it regresses, forming a Holliday junction and extruding the newly synthesized DNA in a duplex (Fig. 2 E → F). The replication fork structure can be restored by exonucleolytic degradation of the extruded duplex (ref. 8; Fig. 2 F → A), or the extrusion can be reversed by branch migration; in both cases, the replication fork is reset and given a chance to overcome the block. This idea is based on the observations that exonucleases prevent chromosomal fragmentation when replication forks are inhibited (9).
(iii) Alternatively, resolution of the Holliday junction by DNA junction-processing enzymes breaks the replication fork (Fig. 2 F → C). The idea of replication fork breakage is based on observations that inhibition of replication fork progress causes chromosome fragmentation (7, 10, 11), which in Escherichia coli can be prevented by inactivation of Holliday junction-processing enzymes (12).
(iv) Collapsed or broken replication forks are repaired via homology-guided invasion of the double-strand end into the intact sister duplex (Fig. 2 C → D), with subsequent resolution of the Holliday junction behind the reassembled fork structure (Fig. 2 D → A). There could be a similar way to repair regressed replication forks (Fig. 2 F → G → A). This idea is based on the recombination dependence of cells that experience replication-induced chromosomal fragmentation and on the apparent ability of homologous recombination to generate new replication forks (6, 7, 10, 11).
Decades of genetic and biochemical studies identified recombination-deficient mutants in bacteriophage T4, E. coli, and Saccharomyces cerevisiae and characterized corresponding enzymes (Table 1), revealing a striking cross-kingdom uniformity of DNA metabolism. And yet, the only enzymes with a clear cross-kingdom homology are recombinases: UvsX of bacteriophage T4, RecA of E. coli, and Rad51 of budding yeast. Remarkably, these recombinases were found to catalyze single-stranded DNA (ssDNA) invasion into a homologous duplex DNA (Fig. 2 C → D and F → G), rather than single-strand annealing (Fig. 1), which turned out to be mostly restricted to concatemeric genomes of bacteriophages (7, 13). The results reported at the Colloquium strengthen the emerging recombinational repair paradigm and, surprisingly, suggest a bigger role for single-strand annealing in eukaryotic recombinational repair.
Table 1.
T4—E. coli—S. cerevisiae dictionary of DNA replication and homologous recombination
| Activity | T4 | E. coli | Budding yeast |
|---|---|---|---|
| Replicative DNA polymerase | gp43 | DNA pol III | DNA polα, δ/ɛ |
| Sliding clamp | gp45 | DnaN (β) | PCNA |
| Clamp loader | gp44/62 | γ complex | RFC |
| Replicative helicase | gp41 | DnaB | MCM |
| Replicative primase | gp61 | DnaG | DNA polα |
| ssDNA-binding protein | gp32 | SSB | RPA |
| Producing 3′-ssDNA tails | gp46/47 | RecBCD | Rad50/58/60 |
| Anti-SSB activity | UvsY | RecBCD or RecFOR | Rad52/59 |
| Recombinase | UvsX | RecA | Rad51 |
| Recombinase regulators | ? | ? | Rad55/57 |
| Auxiliary helicases | UvsW | RecG | Rad54/Tid1 |
| DNA junction resolution | gp49 | RuvABC | ? |
| Primosome-assembly factor | gp59 | PriABC + DnaT | ? |
?, not known.
The scope of this review encompasses the mechanisms of homologous recombination, replication-dependent DNA damage formation, and recombinational repair but falls short of all-inclusive coverage, a daunting task for a broad meeting. Such fascinating phenomena of DNA metabolism as instability of repetitive DNA, adaptive mutagenesis, trans-lesion DNA synthesis, and yeast meiotic recombination, which had to be left out because of size limitations, are covered in recent comprehensive or specialized reviews (7, 13–16). Other papers in this Colloquium issue offer various perspectives.
Homologous Recombination in Phage T4
Gisela Mosig reviewed what is known about the intimate connection between replication and homologous recombination in the life cycle of T4 and outlined why this bacteriophage continues to be the system of choice to study the interdependence of these two branches of DNA metabolism (17). Whereas the early DNA replication in T4 is initiated from replication origins, the late DNA replication of this phage is primed exclusively from recombination intermediates.
Mechanisms of Recombination-Dependent DNA Replication.
In 1980, Mosig hypothesized that the late T4 replication is primed by homologous recombination (18), the idea being later confirmed in several ways (19–21). Kenneth Kreuzer described a particular T4-driven system in which the replication substrate is a circular plasmid that stops origin-dependent replication during the T4 infection, but resumes replication if T4 and the plasmid share homology (20, 22). Plasmid replication depends on the T4 recombination functions, implicating priming via T4 DNA invasion; the replication products are long plasmid concatemers, suggesting rolling-circle replication. The beauty of the T4 system is that any DNA replicated by the T4 replisomes contains hydroxymethyl cytosines in place of cytosines, which makes it resistant to many restriction enzymes and thus distinguishes it from unreplicated DNA. Kreuzer now reports an inquiry into the mechanisms of recombination-dependent replication, by using double-strand break-stimulated events between partially homologous plasmids during T4 infection (23). In addition to priming unidirectional replication from recombination intermediates, T4 is capable of using homology-dependent end-invasion to prime bidirectional chromosomal replication, as elaborated by Mosig in this issue (24).
Enzymology of DNA Replication Priming by Recombination.
When single-strand DNA cannot be replicated, it triggers homologous recombination. The first protein to bind ssDNA in the cell is always the ssDNA-binding protein (SSB; gp32 in T4, SSB in E. coli, and replication protein A (RPA) in yeast; see Table 1), thanks to its high affinity to ssDNA multiplied by high cooperativity of ssDNA binding. SSB is displaced from ssDNA by DNA replication and degradation enzymes but generally prevents binding of recombinases and helicases. Therefore, to start homologous recombination, an activity is required to help a recombinase to replace SSB on ssDNA. This activity should also be a sensor that targets the recombinase only to the ssDNA regions that have difficulty replicating. The role of the recombination-mediator protein UvsY in phage T4 is to sense replication problems and to help UvsX recombinase to replace gp32 on ssDNA (25).
Scott Morrical described how UvsY promotes gp32-to-UvsX replacement on ssDNA: wrapping of gp32-complexed DNA around UvsY hexamers is hypothesized to disrupt gp32 cooperativity, thus lowering the overall affinity of gp32 to ssDNA and allowing UvsX to displace gp32 (25). UvsX then promotes invasion of the ssDNA into a homologous duplex, whereas the displaced strand is complexed by gp32. Morrical now reports that, during this loading, UvsY stabilizes UvsX bound to the invading strand, preventing primosome assembly in the incorrect position (26). This simple sequence of enzymatic events leading to replication fork introduction is thought to be repeated, in greater complexity, both in E. coli and in yeast (25).
The Evolving E. coli Paradigm
E. coli continues to be the prime experimental organism to study mechanisms of DNA replication and homologous recombination and a testing ground for new ideas. Homologous recombination in E. coli was elucidated by using the conjugational system, which asks a cell to integrate a linear homologous DNA into the chromosome—in other words, to repair a double-strand DNA break (27, 28). In wild-type cells, recombination after conjugation depends on recA and recBC genes, whereas in recBC mutants this recombination depends on recA and recFOR genes. Conjugational recombination is only moderately affected by ruvABC or recG mutations, but it is completely blocked in ruv recG double mutants. Therefore, it is thought that E. coli's recombination and double-strand break repair are controlled by the recA gene and in the early phase are channeled along either the major RecBC or the minor RecFOR pathways, whereas in the late phase they are completed by either the RuvABC or the RecG pathways (28, 29).
Recombinational mutants show varying degree of DNA damage sensitivity (7), suggesting recruitment of homologous recombination to repair certain DNA lesions (4, 30). Michael Cox reviewed the current state of the idea that E. coli depends on homologous recombination to repair problems during DNA replication. Cox argues that homologous recombination, rather than being a special system to increase genetic diversity, is a DNA repair system that sometimes generates diversity as a byproduct (31). The elaborate pathways of homologous recombination nicely fit into the requirements for the pathways of recombinational repair. In particular, the current mechanism of double-strand end repair in E. coli includes: (i) the double-strand end processing by RecBCD to generate an ssDNA overhang, which is then complexed by SSB; (ii) RecBCD-catalyzed replacement of SSB with RecA on the ssDNA tail; and (iii) RecA-promoted invasion of the double-strand end into the intact sister duplex.
DNA Replication: the Role of Sliding Clamps and Clamp Loaders.
One of the most surprising recent realizations of the DNA replication field is the similarity in the designs of the replication machinery from bacteriophages to eukaryotes. Mike O'Donnell described a uniform composition of replicative DNA polymerases, which always comprise three basic units (each could be a multiprotein complex on its own): a polymerization unit, a sliding clamp, and a clamp loader (Table 1). Using radiolabeled sliding clamps of E. coli, it was shown that (i) clamps are loaded (inefficiently) on 10-nt-long primers on ssDNA; (ii) with longer primers, loading happens at the 3′ end; (iii) the loading is inhibited if there is a steric hindrance on the primer (a bound protein or a hairpin) 14 nts or closer to the 3′ end (32). The exquisite choreography of the sliding clamp loading is studied with physical techniques in vitro, both with the complex from T4 phage, as has been discussed by Steven Benkovic (33) and with the E. coli complex (34). Besides the polymerization unit, the sliding clamp of E. coli is known to interact with two auxiliary DNA polymerases, pol II and pol V; O'Donnell now reports that the Okazaki fragment maturation enzymes, DNA pol I and DNA ligase, both interact with the sliding clamp in vitro (35). Therefore, similar to polA or lig mutations (see below), certain mutations in the sliding clamp gene (dnaN) or in the clamp loader complex genes are expected to show synthetic lethality with recombinational repair genes.
In Vivo Studies of Replication Fork Disintegration and Repair.
When a replication fork approaches a single-strand interruption in template DNA, will the fork stop short of the nick, inhibited by the lack of negative supercoiling in the downstream DNA, or will the fork run into the nick and collapse? Processing of Okazaki fragments is slow in conditional polA or lig mutants in E. coli; as a result, replicating chromosomes accumulate single-strand interruptions. E. coli mutants with increased levels of single-strand interruptions in DNA, like polA or lig, are dependent on recA and recBC genes for viability, suggesting replication fork collapse with subsequent recombinational repair (4, 6, 7). Andrei Kuzminov now reports that double-strand breaks form in vivo in replicating chromosomes at the locations of preexisting single-strand interruptions, suggesting that replication fork collapse does happen in the cell (36). The idea of recombinational repair of collapsed replication forks further requires that RecBC-promoted recombination would prime DNA synthesis, which was indirectly confirmed in vivo (37).
What happens with a replication fork if its progress is inhibited? In null rep and in certain dnaB mutants, in which replication forks are expected to malfunction, inactivation of the RecBCD enzyme leads to inviability (38, 39). This inviability is associated with chromosome fragmentation (9), which can be partially prevented by inactivation of the RuvABC resolvasome (12). Bénédicte Michel interpreted these observations as signifying reversal of inhibited replication forks with their subsequent breakage because of the RuvABC-promoted resolution (ref. 11; Fig. 2 E → F → C). Michel now reports that holD mutants, defective in a component of the γ complex clamp loader (Table 1), which is a part of the DNA pol III holoenzyme, are hyper-rec and inviable if the RecBCD enzyme is inactivated (40). RecBCD enzyme inactivation in holD cells leads to accumulation of fragmented chromosomal DNA; this accumulation is reduced about 50% if RuvABC resolvasome is inactivated (40).
Why and How Are the Stalled Replication Forks Reversed?
Robert Lloyd described how RecG helicase, working on a replication fork structure in vitro, catalyzes regression of the fork to form a Holliday junction (ref. 41; Fig. 2 E → F). Nicholas Cozzarelli reported that replication fork regression is also favored if the DNA template ahead of the fork is positively supercoiled (42). Replication forks stalled in vitro because of accumulation of positive supercoils become substrates for breakage by Holliday junction resolvases (43). Even though the overall E. coli DNA is kept negatively supercoiled in vivo, positive supercoiling in front of the fork may be generated if there is a converging replication fork or a protein tightly bound to both DNA and some macromolecular structures. In fact, RecG promotes replication fork regression in vitro even when the replication intermediates are negatively supercoiled (43).
Yet another possible mechanism for the replication fork regression is suggested by the partial recA-dependence of chromosomal fragmentation in dnaBts recB mutants (44). In a stalled replication fork, a portion of the template for the lagging strand DNA synthesis is left single-stranded (Fig. 2E). Cox now reports that, in vitro, the single-strand region of a replication fork structure attracts RecA, which then catalyzes strand exchange with the fully duplex sister strand, leading to replication fork regression (45). The findings that both RecA (44) and RuvABC (12) influence replication fork breakage in vivo makes the phenomenon complicated, because both functions are also required for the subsequent repair of broken replication forks.
There are also enzymes that should be able to reset stalled replication forks, decreasing the chances of replication fork breakage. One possible candidate is the Rep helicase; as discussed above, replication forks are prone to regression in rep mutants. Another helicase, PriA, is implicated in loading replisomes onto replication fork structures at places other than replication origins (46) [at the origins, replisomes are loaded by origin-recognition proteins like DnaA in E. coli (47, 48)]. Hiroshi Nakai describes how PriA attracts and loads the primosome and the replisome on replication fork structures that are intermediates in the transposition of bacteriophage Mu (49). The ability to attract replisomes to replication fork structures makes PriA indispensable for the completion of any replication fork repair.
Because replication fork repair in E. coli is absolutely dependent on RecA protein, one could expect the viability of priA mutants to be as low as, but not significantly worse than, the viability of recA mutants, which is about 50% (50). Paradoxically, priA mutants are very sick: they cannot grow in rich media and they post only 10% viability in minimal media (51, 52). Steven Sandler now reports that priA recG or priA ruv double mutants are inviable (53), which is surprising, because all other tested pairwise combinations of recombination mutants are viable. All this suggests that PriA has functions other than completion of recombinational repair of disintegrated replication forks. Kenneth Marians discussed the two in vitro PriA activities: (i) primosome assembly and (ii) DNA helicase, which is not required for primosome assembly (48). At replication forks, the helicase activity of PriA counteracts RecG (54), suggesting that, in addition to the primosome assembly role of PriA in replication fork restart, PriA helicase may act to prevent replication fork breakage by resetting regressed replication forks.
New Approaches To Study Recombinases.
Takehiko Shibata discussed NMR studies revealing the basis for the extended structure of ssDNA within the filaments of RecA or its yeast homolog Rad51 (55). Charles Radding reviewed the current understanding of the most critical and still mysterious step of recombinational repair, the RecA-catalyzed homology search with subsequent strand exchange. Fluorescence energy transfer between strands of a DNA duplex reveals that the RecA-catalyzed reaction is subdivided into two, possibly three phases: (i) a rapid, second order step, reflecting initial interactions of the ssDNA with the complementary strand of the duplex; (ii) a rapid (and cryptic) first order step, perhaps reflecting subsequent proximity of the ssDNA to the homologous strand of the duplex; and (iii) a slower first order strand exchange step (56). All of the steps are reversible, suggesting that the observed high specificity of the RecA-promoted homology search is the product of several steps of moderate specificity. Rate constants of the reaction between DNAs with mismatches are consistent with this idea, because it is reported that mismatches (i) reduce the rate of the pairing step; (ii) increase the off rate during the pairing step; and (iii) reduce the rate of subsequent strand exchange (57). Recent studies employing oligonucleotides with varying GC-content as recombination substrates revealed that both the E. coli RecA and its human homologs Rad51 and Dmc1 rely on A:T base pairs for recognition of homology (58–60).
Stephen Kowalczykowski reviewed what is known about the RecBCD enzyme, a highly processive DNA helicase/exonuclease that modifies duplex DNA ends, so that they become substrates for RecA polymerization, and then promotes RecA polymerization on these ends in the presence of SSB (61). The mechanism of DNA degradation/modification by RecBCD has been studied in vitro for many years. Kowalczykowski demonstrated the latest approach—a direct visualization in real time of a DNA molecule being unwound by a single RecBCD molecule (62). Direct visualization has also been used by others to inquire about the fate of the RecD subunit of the enzyme during the postulated switch from DNA degradation to recombination (63).
Site-Specific Chromosome Monomerization.
Circular chromosomes are sensitive to odd numbers of interchromosomal exchanges, because the resulting dimeric chromosome cannot segregate into daughter cells. In E. coli, the crossover vs. non-crossover decision during recombinational repair appears to be regulated: in certain setups, RuvABC resolvasome favors non-crossovers (64, 65), whereas a novel protein RarA appears to favor the crossover resolution, leading to chromosome dimerization (66).
David Sherratt spoke about the chromosome of E. coli, where the dimers are resolved to monomers by a two-enzyme site-specific recombination system, acting on the dif site in the replication terminus (7, 67). XerC recombinase makes a half-crossover (a Holliday junction) between two dif sites on the sister chromatids, whereas XerD recombinase, acting on this Holliday junction, splits the “figure-eight” chromosome into two monomers. XerC appears to catalyze its half of the reaction independently of whether the chromosome is a dimer or a pair of monomers, whereas XerD seems to act only when the chromosome is a dimer—otherwise XerC simply reverses its part of the reaction (68).
E. coli plasmids employ the same pair of resolution enzymes, XerC and XerD, to monomerize; accessory proteins determine the directionality of the reaction in the plasmid systems (67). No such directionality-determining proteins are known for the chromosomal dimer resolution, suggesting that the chromosomal system is guided by a different kind of signal. Sherratt and others now report that FtsK protein, which is a part of the cell constriction machinery, is required for the XerD-catalyzed reaction (69, 70). Yet, FtsK is likely to be a trigger of the final resolution rather than a signal determining the direction of the resolution. The finding of a XerCD-dependent DNA topo IV cleavage site right at dif (71), a proposed functional polarization of the terminus region around dif (72), and the restricted access of the dif region to the FtsK at the septum (66) could provide clues to understanding the nature of the elusive directionality signal.
Yeast
Budding yeast, although a rather atypical eukaryote, is a gateway to the complicated world of eukaryotic recombinational repair. Most of the yeast homologous recombination genes were isolated for their role in resistance to ionizing radiation, hence the name “RAD” (73). Ionizing radiation, among other DNA lesions, generates double-strand breaks, and recombinational genes are responsible for double-strand break repair. All types of homologous recombination in yeast, whether the underlying mechanism is single-strand annealing or double-strand invasion, are virtually eliminated in rad52 mutants. Further, recombination between homologous chromosomes in diploid yeast is severely affected in rad51, rad54, rad55, and rad57 mutants (13). At the same time, there is another group of radiation-sensitive mutants—rad50, mre11 (rad58), rad59, and xrs2 (rad60)—posting slightly elevated levels of interhomolog recombination. Yet, when intersister recombination is measured, all RAD genes show comparable defects. Thus, the overall perception is that there are two pathways in the yeast homologous recombination, both controlled by the RAD52 gene: the mostly interhomolog RAD51/54/55/57 pathway and the intersister RAD50/58/59/60 pathway (13) [for the sake of uniformity, I will use the recently suggested “RAD” nomenclature for MRE11 and XRS2 (73)].
Replication-Dependent Chromosomal Fragmentation in Yeast.
The elaborate checkpoint machinery of eukaryotes halts the cell cycle in response to significant DNA damage, preventing the bulk of replication fork faltering events. Moreover, the sexuality of the model eukaryotic organisms implies that recombination in these organisms is for evolutionary purposes, to generate genetic diversity. Yet, phenomena indicative of replication fork disintegration are well-known in yeast, and there are multiple reports that recombination primes extensive DNA synthesis in this organism, pointing to its primarily repair role.
rad27 (flap endonuclease), pol30 (DNA clamp), rfc1 (DNA clamp loader), and cdc9 (DNA ligase) mutants of budding yeast accumulate single-strand interruptions in replicating DNA (74, 75), because of defective maturation of Okazaki fragments. Like the mutants in Okazaki fragment maturation in E. coli (see above), these yeast mutants are synthetically lethal if combined with mutations inactivating double-strand break repair (75–77), suggesting that single-strand interruptions in the yeast chromosomal DNA are somehow converted into double-strand breaks. Alain Nicolas now reports that rad27Δ strains require all genes of double-strand break repair, as well as certain auxiliary genes of the DNA metabolism (78).
RPA and Pol δ are recent additions to the list of S. cerevisiae proteins, whose inactivation makes cells dependent on recombination (79, 80). Michael Resnick and Dmitry Gordenin now report that a class of DNA polymerase δ mutants, deficient in the 3′ → 5′ exonuclease, is inviable in combination with rad27 mutations (81). Double mutants with partial defects in both Rad27 and 3′ → 5′ Exo of Pol δ are viable, but hyper-rec, and dependent on RAD50, RAD51, and RAD52. A defect in Okazaki fragment maturation is again suspected (81).
Mec1 is a key DNA damage checkpoint protein in S. cerevisiae; mec1-srf mutants depend on RAD52 and accumulate single-strand interruptions in the newly synthesized DNA, apparently because of their inability to lift the inhibition of ribonucleotide reductase (82). When budding yeast cells are treated with hydroxyurea to inhibit ribonucleotide reductase, they also accumulate single-strand interruptions in DNA and also become dependent on RAD52 (82). DNA of rad52 mutants, treated with hydroxyurea, is fragmented, suggesting conversion of single-strand breaks into double-strand breaks (82). The most economical interpretation of these observations is replication fork collapse (Fig. 2 B → C).
Dna2p is a DNA helicase/nuclease essential for DNA replication; S. cerevisiae dna2ts mutants complete DNA replication at nonpermissive temperatures, but their chromosomes contain multiple single-strand interruptions (83, 84). In budding yeast, Dna2p is thought to work in a complex with Rad27p to process Okazaki fragments (85); interestingly, dna2ts mutants depend on RAD52 at sublethal temperatures (86). Inactivation of dna2 in fission yeast results in chromosome fragmentation reminiscent of that observed in DNA ligase I mutants of this organism (87). Growth of S. pombe dna2ts mutants at high temperature is restored by increased production of the enzymes participating in maturation of Okazaki fragments (87). dna2+ was also isolated as a multicopy suppressor of a cdc24 mutant in S. pombe (88); cdc24 mutants arrest after completing chromosome replication with signs of chromosome fragmentation (88). In addition to dna2+, cdc24 mutants are also rescued by multiple copies of pcn1+ (DNA clamp) and rfc1+ (DNA clamp loader) genes; Cdc24p binds to Pcn1p and Rfc1p in vivo (89). A defect in Okazaki fragment maturation is suspected to lead to the phenotypes of cdc24 mutants. One may predict that cdc24 and dna2 mutants of S. pombe are dependent on homologous recombination. In summary, yeast mutants with increased levels of single-strand interruptions depend on double-strand break repair, suggesting replication fork collapse as the ultimate DNA lesion.
Mechanisms of Recombinational Repair in Yeast.
Edward Egelman compared the structure of RecA and Rad51 filaments (90). In response to DNA damage, Rad51 assembles in foci in the S-phase cells, suggesting that Rad51 functions in repair of replication-induced DNA damage (91). James Haber outlined the hypothetical sequence of events for the double-strand end repair in yeast, which is analogous to that of T4 and E. coli: (i) the double-strand end is processed by Rad50/Rad58/Rad60 to generate an ssDNA overhang, which is complexed by RPA; (ii) Rad52 catalyzes replacement of RPA with Rad51; (iii) Rad51, with the help of Rad54, Rad55, and Rad57, catalyzes invasion of the double-strand end into the intact sister duplex (13). In this mechanism, Rad51 plays the central role, both as a protein complex organizer and a catalyst.
However, genetic data discussed by Haber indicate that the central activity of the yeast double-strand end repair is Rad52 rather than Rad51. A chromosomal double-strand break is efficiently repaired in the wild-type diploid yeast; because the broken chromosome is lost in rad52 mutants, the repair must be via recombination with the homologous intact chromosome (92, 93). If only one end of the break is homologous to the intact chromosome, the chromosomal arm with the nonhomologous end is likewise lost, again implicating homologous recombination in repair. The product of the one-end repair is, nevertheless, a complete chromosome because the lost arm is replaced with an arm copied from the homologous chromosome (92, 94). Thus, there are two stages of the double-strand break repair in yeast: (i) invasion of one of the ends into the intact homolog forms a recombination intermediate resembling a replication fork; and (ii) recruitment of the other end prevents maturation of the recombination intermediate into a replication fork. If, because of a limited homology, the second end fails to join the recombination intermediate, the replication fork matures and proceeds till the end of the chromosome.
Remarkably, 35% of the double-strand breaks are repaired in rad51 mutant cells; however, even with homology on both sides of the break, the rad51-independent events are one-sided (93). The partial Rad51-independence of the double-strand break repair process implies existence of a repair mechanism distinct from Rad51-catalyzed strand invasion. In vitro enzymatic activities of Rad52 and Rad54 proteins suggest participation of single-strand annealing in the eukaryotic replication fork repair.
Enzymology of the Yeast Recombination: Rad52-Promoted Annealing.
Rodney Rothstein reviewed what is known about Rad52, the master activity of recombinational repair in budding yeast. Like Rad51, Rad52 relocalizes from a diffuse nuclear distribution to distinct foci in response to DNA damaging treatments, but again only during the S-phase, suggesting that it also participates in repair of replication-induced DNA lesions (95). However, RAD52 gene has no homology to recA and RAD51 recombinases, implying a different kind of control and/or mechanism of recombinational repair in yeast. Purified Rad52 proteins from yeast and humans form oligomeric rings that bind to single-stranded DNA (96, 97). Stephen West now reports that, in the case of the human protein, the rings are Rad52 heptamers (98); they specifically bind linear ssDNA at the end, apparently wrapping the terminal 36 nt of the DNA strand around the heptamer (99). Such a Rad52 heptamer bound at a ssDNA end could, on the one hand, mark the end for signal purposes, whereas, on the other hand, could provide a nucleation site for Rad51 filament assembly on the RPA-bound ssDNA. Rad52 also catalyzes strand annealing reactions (100), which are enhanced by RPA (96, 101), but Rad52 cannot catalyze strand invasion reactions, characteristic of prokaryotic RecA or eukaryotic Rad51 proteins. Because rad52 mutations completely block homologous recombination in budding yeast, the ability of Rad52 to catalyze only single-strand annealing implies that the central reaction of recombinational repair in yeast is not the Rad51-catalyzed double-strand end invasion.
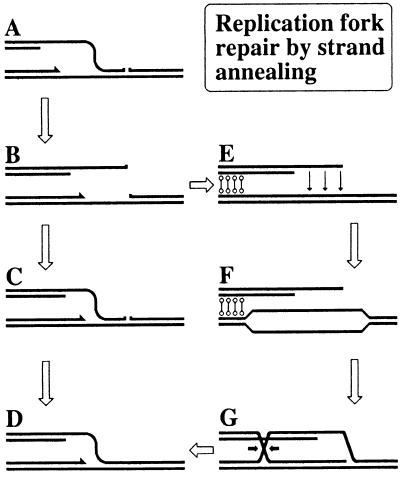
In fact, a replication fork could be repaired by single-strand annealing if (i) there were regions of single-stranded DNA at the replication forks (and there are always such regions); and (ii) the rate of DNA synthesis was slow (and it is relatively slow in eukaryotes). Therefore it is possible that, before the single-strand regions in a collapsed replication fork have been converted into duplexes, Rad52 would reattach the end to the chromosome by annealing the ssDNA tail to the complementary single strand gap (Fig. 3 B → C). Similarly, Rad52 should be able to attach a loner double-strand end to a homologous chromosome by annealing the ssDNA tail of the end with the complementary ssDNA regions exposed at replication forks. Such a SSA end-capture would explain the RAD52-dependent but RAD51-independent break-induced replication (93).
Figure 3.
Two ways to repair a replication fork by single-strand annealing. DNA duplexes are shown as double lines; sister chromatid cohesion is indicated by thin dumbbells. (A) A replication fork approaching a single-strand interruption in template DNA. (B) The replication fork has collapsed. (C) Rad52-promoted reannealing of the detached end with the complementary single-strand gap on the full-length chromatid. (D) Repair of the single-strand interruption. (E) Filling-in the single-strand gap on the full-length chromatid. Sister chromatid alignment is shown. The thin arrows indicate a hypothetical signal from the Rad52-bound double-strand end to the intact sister chromatid. (F) Rad54-catalyzed unwinding of the intact chromatid in the vicinity of the double-strand end. (G) Rad52-catalyzed annealing of the double-strand end with the open sister duplex to generate a replication fork structure.
Enzymology of the Yeast Recombination: Rad54-Promoted Duplex Opening.
Patrick Sung spoke about the in vitro reactions catalyzed by Rad54, which is a DNA helicase of the SWI2/SNF2 family of chromatin-remodeling proteins. DNA in chromatin is less accessible to any reaction, including Rad51-catalyzed strand invasion—therefore, the need for nucleosome reorganization/removal in the region to be repaired. Rad54 enhances Rad51-catalyzed insertion of a single strand into a naked covalently closed circular (ccc) DNA (102, 103). Remarkably, the cccDNA does not have to be negatively supercoiled in the Rad54-supplemented reaction, although high negative supercoiling is generally required for the efficient D-loop formation in cccDNA molecules. Sung reports that Rad54 overcomes this requirement for high negative supercoiling by generating a highly underwound domain in the cccDNA substrate (103). Generation of unconstrained plectonemic supercoils in DNA by human Rad54 is also reported (104). If Rad54 unwinds the region of the duplex, homologous to the incoming single strand, the subsequent Rad51-promoted reaction is more single-strand annealing than end-invasion.
If Rad54 were to promote this DNA unwinding randomly, the activity would have been biologically irrelevant. However, Rad54 interacts with Rad51 in vivo (105, 106), and the Rad54 unwinding activity is targeted toward the DNA duplexes bound by the Rad51 filament (102, 103). Incorporation of the unwinding step should slow the homology search process significantly; on the other hand, because the sister chromatids in eukaryotes are held together in the aligned position along their entire length (107), the homology search should be simpler. In fact, the double-strand end attachment would not even require Rad51 if Rad52, bound at the end of a ssDNA tail, could attract Rad54 to unwind the aligned sister duplex (Fig. 3E). Rad54-promoted unwinding would generate a strand complementary to the ss-tailed end, whereas Rad52 could catalyze the annealing, reestablishing a replication fork structure. Rad51-independent but Rad52-dependent Holliday junctions were reported in the yeast ribosomal DNA (108).
One prediction of this Rad51-independent scheme is that it should not work for inter-homolog recombination, because homologous chromosomes are not aligned. Interestingly, overproduction of Rad54 compensates for the rad51 defect in DNA damage repair that relies on interactions between sisters, but not in repair that relies on inter-homolog recombination (105). Another prediction is that mutants defective in sister chromatid cohesion should be defective in replication fork repair. Interestingly, trf4 and mcd1 yeast mutants, defective in sister chromatid cohesion, are exquisitely sensitive to camptothecin and to methyl methanesulfonate [the agents that are thought to cause replication fork collapse (109, 110)] but are not sensitive to UV (111).
Homeologous End-Joining in Yeast: Rad58 (MRE11) Promotes Strand-Assimilation.
What to do with a double-strand break that, because of some reasons, cannot be repaired by homologous recombination with its intact sister or a homolog? Repair of such double-strand breaks is associated with misalignment at regions of microhomology and is severely affected by rad50, rad58(mre11), or rad60(xrs2) mutations (112). As discussed by Tomoko Ogawa, the properties of the Rad58p (Mre11p) make it a perfect match for the role: this multifunctional enzyme not only degrades one DNA strand, but can also unwind DNA and anneal complementary DNA strands (113). Martin Gellert reviewed the properties of the human Mre11/Rad50/Nbs1 complex, which are very similar to those of the yeast Rad50/58/60 complex (114, 115). Thus, Mre11 should be able to catalyze strand assimilation, the type of single-strand annealing recombination promoted by phage λ Exo + Beta proteins (116). The Rad50/Rad58/Rad60 system in yeast or the Rad50/Mre11/Nbs1 system of higher eukaryotes are thought to join the ends of a double-strand break after a limited hydrolysis of the 5′-ending strands by annealing the two ss-tails at regions of microhomology (13).
Recombinational Repair in Vertebrate Cells
Recombinational repair in vertebrate cells follows the yeast paradigm with an added complexity. Maria Jasin reviewed the data suggesting that, in mouse embryonic stem cells and in two hamster cell lines, the majority of double-strand breaks are repaired faithfully by homologous recombination (117–119). The mechanism of this repair is revealed by shrinking the size of homology on one side of the break: the resulting repair products carry varying duplications of the nonhomologous side of the break (118). Microhomologies are frequently found bracketing the duplicated regions. Apparently, the homologous end of the break invades the intact sister chromatid and starts a replication fork, which is disintegrated after proceeding for varying distances. The generated end is joined with the nonhomologous end at regions of microhomology, resulting in a duplication bracketed by short direct repeats (118). Haber now reports a similar finding in yeast (120).
Shunichi Takeda described his mutational studies with the chicken lymphoma cell line. The two major deviations of vertebrate cells from the yeast paradigm are: (i) rad51 mutant vertebrate cells die rapidly, developing gross chromosomal abnormalities before death (121, 122); and (ii) rad52 mutant vertebrate cells are not hypersensitive to DNA damaging agents (123, 124). Another noteworthy difference is the presence of multiple Rad51p paralogs (distant homologs) in the vertebrate cells. XRCC2 is one such paralog; Jasin reports that double-strand break-induced homologous recombination is decreased 100-fold in xrcc2-mutant hamster cells, whereas nonhomologous end joining is unaffected (125). XRCC3 is another paralog of Rad51p; in hamster cells, it is also required for double-strand break repair by homologous recombination (126).
Conclusion
Lesions in one DNA strand are efficiently removed by excision repair. However, once encountered by a replication fork, originally one-strand lesions spread to the second DNA strand and require a different type of repair. One can say that damage replicates with the DNA. The consequences of DNA damage replication are grave, because two-strand damage interferes with subsequent rounds of chromosomal replication. Because of the immediate danger to the integrity of the whole chromosome, two-strand damage can be called “chromosomal damage.”
Removal of the chromosomal damage requires chromosomal repair. The major chromosomal repair pathway operates via homologous recombination. Richard Kolodner describes that the search for mutants with dramatically increased frequency of gross chromosomal rearrangements in yeast yields defects in DNA metabolism (which contribute to one-strand DNA damage) as well as mutants in recombinational repair (127). Thus, an increased frequency of one-strand damage contributes to chromosomal damage, whereas unrepaired chromosomal damage in the best-case scenario leads to genome rearrangements.
In prokaryotes, the main activity of recombinational repair is the strand invasion protein RecA. In budding yeast, the main recombinational activity is the strand annealing protein Rad52, implying that the mechanisms of the eukaryotic recombinational repair are different from prokaryotic ones. It is suggested that eukaryotes, because of their slow DNA replication and because of the sister chromatid alignment, rely more on single-strand annealing than on strand invasion in their recombinational repair. The prediction is that mutants with defective sister-chromatid cohesion should be also defective in the RAD50/58/59/60-dependent DNA repair, but have elevated RAD51/54/55/57-dependent interhomolog recombination.
Acknowledgments
I thank Charles Radding, Mike Cox, and Bénédicte Michel for helpful comments on the manuscript. Work in the author's laboratory is supported by National Science Foundation Award MCB-01 96020.
Abbreviations
- ssDNA
single-stranded DNA
- SSB
ssDNA-binding protein
- cccDNA
covalently closed circular DNA
- RPA
replication protein A
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Szybalski W. Abh Dtsch Akad Wiss Berlin, Kl Med. 1964;4:1–19. [Google Scholar]
- 2.Hanawalt P C. Photochem Photobiol. 1966;5:1–12. [PubMed] [Google Scholar]
- 3.Strauss B S. In: DNA-Repair Mechanisms. Altmann H, editor. Stuttgart: F.K. Schattauer; 1972. pp. 151–171. [Google Scholar]
- 4.Skalka A. In: Mechanisms in Recombination. Grell R F, editor. New York: Plenum; 1974. pp. 421–432. [Google Scholar]
- 5.Higgins N P, Kato K, Strauss B. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 6.Kuzminov A. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 7.Kuzminov A. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louarn J-M, Louarn J, François V, Patte J. J Bacteriol. 1991;173:5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzminov A. BioEssays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 11.Michel B. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 12.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 13.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster P L. Annu Rev Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman M F. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- 16.Gordenin D A, Resnick M A. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 17.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 18.Mosig G, Benedict S, Ghosal D, Luder A, Dannenberg R, Bock S. In: Mechanistic Studies of DNA Replication and Genetic Recombination. Alberts B, editor. Vol. 19. New York: Academic; 1980. pp. 527–543. [Google Scholar]
- 19.Dannenberg R, Mosig G. J Virol. 1983;45:813–831. doi: 10.1128/jvi.45.2.813-831.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreuzer K N, Yap W Y, Menkens A E, Engman H W. J Biol Chem. 1988;263:11366–11373. [PubMed] [Google Scholar]
- 21.Luder A, Mosig G. Proc Natl Acad Sci USA. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W E. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George J W, Stohr B A, Tomso D J, Kreuzer K N. Proc Natl Acad Sci USA. 2001;98:8290–8297. doi: 10.1073/pnas.131007598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosig G, Gewin J, Luder A, Colowick N, Vo D. Proc Natl Acad Sci USA. 2001;98:8306–8311. doi: 10.1073/pnas.131007398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beernink H T H, Morrical S W. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 26.Bleuit J S, Xu H, Ma Y, Wang T, Liu J, Morrical S W. Proc Natl Acad Sci USA. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark A J, Low K B. In: The Recombination of Genetic Material. Low K B, editor. San Diego: Academic; 1988. pp. 155–215. [Google Scholar]
- 28.Clark A J, Sandler S J. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 29.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard-Flanders P. Brit Med Bull. 1973;29:226–235. doi: 10.1093/oxfordjournals.bmb.a071012. [DOI] [PubMed] [Google Scholar]
- 31.Cox M M. Proc Natl Acad Sci USA. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao N, Leu F P, Anjelkovic J, Turner J, O'Donnell M. J Biol Chem. 2000;275:11440–11450. doi: 10.1074/jbc.275.15.11440. [DOI] [PubMed] [Google Scholar]
- 33.Trakselis M A, Alley S C, Abel-Santos E, Benkovic S J. Proc Natl Acad Sci USA. 2001;98:8368–8375. doi: 10.1073/pnas.111006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertram J G, Bloom L B, Hingorani M M, Beechem J M, O'Donnell M, Goodman M F. J Biol Chem. 2000;275:28413–28420. doi: 10.1074/jbc.M910441199. [DOI] [PubMed] [Google Scholar]
- 35.López de Saro F J, O'Donnell M. Proc Natl Acad Sci USA. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzminov A. Proc Natl Acad Sci USA. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuzminov A, Stahl F W. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saveson C J, Lovett S T. Genetics. 1999;152:5–13. doi: 10.1093/genetics/152.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uzest M, Ehrlich S D, Michel B. Mol Microbiol. 1995;17:1177–1188. doi: 10.1111/j.1365-2958.1995.mmi_17061177.x. [DOI] [PubMed] [Google Scholar]
- 40.Flores M-J, Bierne H, Ehrlich S D, Michel B. EMBO J. 2001;20:619–629. doi: 10.1093/emboj/20.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 42.Postow L, Ullsperger C, Keller R W, Bustamante C, Vologodskii A V, Cozzarelli N R. J Biol Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 43.McGlynn P, Lloyd R G, Marians K J. Proc Natl Acad Sci USA. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seigneur M, Ehrlich S D, Michel B. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- 45.Robu M E, Inman R B, Cox M M. Proc Natl Acad Sci USA. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seufert W, Messer W. EMBO J. 1986;5:3401–3406. doi: 10.1002/j.1460-2075.1986.tb04656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 48.Marians K J. Trends in Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 49.Nakai H, Doseeva V, Jones J M. Proc Natl Acad Sci USA. 2001;98:8247–8254. doi: 10.1073/pnas.111007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capaldo-Kimball F, Barbour S D. J Bacteriol. 1971;106:204–212. doi: 10.1128/jb.106.1.204-212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee E H, Kornberg A. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nurse P, Zavitz K H, Marians K J. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCool J D, Sandler S J. Proc Natl Acad Sci USA. 2001;98:8203–8210. doi: 10.1073/pnas.121007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Deib A A, Mahdi A A, Lloyd R G. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibata T, Nishinaka T, Mikawa T, Aihara H, Kurumizaka H, Yokoyama S, Ito Y. Proc Natl Acad Sci USA. 2001;98:8425–8432. doi: 10.1073/pnas.111005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bazemore L R, Takahashi M, Radding C M. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 57.Bazemore L R, Folta-Stogniew E, Takahashi M, Radding C M. Proc Natl Acad Sci USA. 1997;94:11863–11868. doi: 10.1073/pnas.94.22.11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta R C, Folta-Stogniew E, O'Malley S, Takahashi M, Radding C M. Mol Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- 59.Gupta R C, Folta-Stogniew E, Radding C M. J Biol Chem. 1999;274:1248–1256. doi: 10.1074/jbc.274.3.1248. [DOI] [PubMed] [Google Scholar]
- 60.Gupta R C, Golub E, Bi B, Radding C M. Proc Natl Acad Sci USA. 2001;98:8433–8439. doi: 10.1073/pnas.121005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 62.Bianco P R, Brewer L R, Corzett M, Balhorn R, Yeh Y, Kowalczykowski S C, Baskin R J. Nature (London) 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 63.Dohoney K M, Gelles J. Nature (London) 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 64.Cromie G A, Leach D R F. Mol Cell. 2000;6:815–826. doi: 10.1016/s1097-2765(05)00095-x. [DOI] [PubMed] [Google Scholar]
- 65.Michel B, Recchia G D, Penel-Colin M, Ehrlich S D, Sherratt D J. Mol Microbiol. 2000;37:180–191. doi: 10.1046/j.1365-2958.2000.01989.x. [DOI] [PubMed] [Google Scholar]
- 66.Barre F-X, Søballe B, Michel B, Aroyo M, Robertson M, Sherratt D J. Proc Natl Acad Sci USA. 2001;98:8189–8195. doi: 10.1073/pnas.111008998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sherratt D J, Arciszewska L K, Blakely G, Colloms S, Grant K, Leslie N, McCulloch R. Philos Trans R Soc London. 1995;347:37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- 68.Barre F-X, Aroyo M, Colloms S D, Helfrich A, Cornet F, Sherratt D J. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Recchia G D, Aroyo M, Wolf D, Blakely G, Sherratt D J. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steiner W, Liu G, Donachie W D, Kuempel P. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 71.Hojgaard A, Szerlong H, Tabor C, Kuempel P. Mol Microbiol. 1999;33:1027–1036. doi: 10.1046/j.1365-2958.1999.01545.x. [DOI] [PubMed] [Google Scholar]
- 72.Pérals K, Cornet F, Merlet Y, Delon I, Louarn J-M. Mol Microbiol. 2000;36:33–43. doi: 10.1046/j.1365-2958.2000.01847.x. [DOI] [PubMed] [Google Scholar]
- 73.Game J C. Mutat Res. 2000;451:277–293. doi: 10.1016/s0027-5107(00)00055-5. [DOI] [PubMed] [Google Scholar]
- 74.Johnston L H, Nasmyth K A. Nature (London) 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 75.Merrill B J, Holm C. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Montelone B A, Prakash S, Prakash L. J Bacteriol. 1981;147:517–525. doi: 10.1128/jb.147.2.517-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 78.Debrauwère H, Loeillet S, Lin W, Lopes J, Nicolas A. Proc Natl Acad Sci USA. 2001;98:8263–8269. doi: 10.1073/pnas.121075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C, Umezu K, Kolodner R D. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 80.Giot L, Chanet R, Simon M, Facca C, Faye G. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin Y H, Obert R, Burgers P M J, Kunkel T A, Resnick M A, Gordenin D A. Proc Natl Acad Sci USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. . (First Published April 17, 2001; 10.1073/pnas.091095198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Merrill B J, Holm C. Genetics. 1999;153:595–605. doi: 10.1093/genetics/153.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Budd M E, Campbell J L. Proc Natl Acad Sci USA. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiorentino D F, Crabtree G R. Mol Biol Cell. 1997;8:2519–2537. doi: 10.1091/mbc.8.12.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Budd M E, Campbell J L. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Budd M E, Campbell J L. Mutat Res. 2000;459:173–186. doi: 10.1016/s0921-8777(99)00072-5. [DOI] [PubMed] [Google Scholar]
- 87.Kang H-Y, Choi E, Bae S-H, Lee K-H, Gim B-S, Kim H-D, Park C, MacNeill S A, Seo Y-S. Genetics. 2000;155:1055–1067. doi: 10.1093/genetics/155.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gould K L, Burns C G, Feoktistova A, Hu C-P, Pasion S G, Forsburg S L. Genetics. 1998;149:1221–1233. doi: 10.1093/genetics/149.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanaka H, Tanaka K, Murakami H, Okayama H. Mol Cell Biol. 1999;19:1038–1048. doi: 10.1128/mcb.19.2.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu X, Jacobs S A, West S C, Ogawa T, Egelman E H. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gasior S L, Olivares H, Ear U, Hari D M, Weichselbaum R, Bishop D K. Proc Natl Acad Sci USA. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bosco G, Haber J E. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Malkova A, Ivanov E L, Haber J E. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morrow D M, Connelly C, Hieter P. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lisby M, Rothstein R, Mortensen U H. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 97.Van Dyck E, Stasiak A Z, Stasiak A, West S C. Nature (London) 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 98.Stasiak A Z, Larquet E, Stasiak A, Müller S, Engel A, Van Dyck E, West S C, Egelman E H. Curr Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- 99.Parsons C A, Baumann P, Van Dyck E, West S C. EMBO J. 2000;19:4175–4181. doi: 10.1093/emboj/19.15.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugiyama T, New J H, Kowalczykowski S C. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mazin A V, Bornarth C J, Solinger J A, Heyer W-D, Kowalczykowski S C. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 103.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 104.Ristic D, Wyman C, Paulusma C, Kanaar R. Proc Natl Acad Sci USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clever B, Interhal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer W-D. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 107.Carson D R, Christman M F. Proc Natl Acad Sci USA. 2001;98:8270–8275. doi: 10.1073/pnas.131022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zou H, Rothstein R. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 109.D'Arpa P, Liu L F. Biochim Biophys Acta. 1989;989:163–177. doi: 10.1016/0304-419x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- 110.Saffhill R, Ockey C H. Chromosoma. 1985;92:218–224. doi: 10.1007/BF00348697. [DOI] [PubMed] [Google Scholar]
- 111.Walowsky C, Fitzhugh D J, Castaño I, Ju J Y, Levin N A, Christman M F. J Biol Chem. 1999;274:7302–7308. doi: 10.1074/jbc.274.11.7302. [DOI] [PubMed] [Google Scholar]
- 112.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 114.Paull T T, Gellert M. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 115.Paull T T, Gellert M. Proc Natl Acad Sci USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. . (First Published May 23, 2000; 10.1073/pnas.110144297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cassuto E, Radding C M. Nat New Biol. 1971;229:13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- 117.Johnson R D, Jasin M. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Richardson C, Jasin M. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richardson C, Moynahan M E, Jasin M. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kraus E, Leung W-Y, Haber J E. Proc Natl Acad Sci USA. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lim D-S, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sonoda E, Sasaki M S, Buerstedde J-M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rijkers T, Ouweland V D, Morolli B, Rolink A G, Baarends W M, Van Sloun P P H, Lohman P H M, Pastink A. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yamaguchi-Iwai Y, Sonoda E, Buerstedde J-M, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson R D, Liu N, Jasin M. Nature (London) 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 126.Pierce A J, Johnson R D, Thompson L H, Jasin M. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Flores-Rozas H, Kolodner R D. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]